Abstract
Testing of susceptibility to 13 antibiotics was performed with 90 isolates of Lactobacillus, Leuconostoc, and Pediococcus. MICs at which 90% of the isolates tested were inhibited by HMR3647, erythromycin, and ciprofloxacin were 0.015, 0.125 and 32 μg/ml, respectively. The penicillin MIC was ≥16 μg/ml against 26.2% of the studied Lactobacillus sp. isolates and 50% of Lactobacillus plantarum. HMR3647 showed excellent activity against these genera.
Lactobacillus, Leuconostoc, and Pediococcus spp. are commonly found as natural microflora in the mucous membranes of humans and animals, in dairy products, and on some plant surfaces (2, 21). Nevertheless, they are increasingly recognized as opportunistic pathogens involved in human infections (1, 3, 5, 8, 10, 11, 14, 17, 22). Intrinsic resistance to glycopeptides is well documented in these genera of lactic acid bacteria (LAB) (13, 26, 31); however, there are few reports on susceptibilities of LAB to other antibiotics. The purpose of this work was to determine the susceptibility pattern to 13 antibiotics in 90 LAB isolates: 60 Lactobacillus isolates (L. plantarum, 28; L. paracasei, 17; L. brevis, 8; L. rhamnosus, 3; L. casei, 1; L. fermentum, 1; L. curvatus, 1; L. pentosus, 1); 18 Leuconostoc isolates (L. mesenteroides, 10; L. pseudomesenteroides, 1; L. oenos, 1; Leuconostoc spp., 6); and 12 Pediococcus isolates (P. pentosaceus, 10; P. acidilactici, 2). They were identified as previously recommended (6, 15, 15a) and by using the API50 CH system (Biomérieux). MICs were determined by the National Committee for Clinical Laboratory Standards agar dilution method (19) using brain heart infusion agar (BHIA) and an atmosphere of 5% CO2. Results in BHIA were compared with those obtained in Mueller-Hinton agar supplemented with 5% sheep blood (MHA-B). Strains grew better in BHIA than in MHA-B, and therefore, the former agar medium was used for MIC determinations. Control strains used were Staphylococcus aureus ATCC 29213 and Enterococcus faecalis ATCC 29212. Tested antibiotics were the following: ketolide HMR3647, erythromycin A, clarithromycin, roxithromycin, azithromycin, levofloxacin, and teicoplanin (all supplied by Hoechst Marion Roussel); spiramycin and penicillin (Sigma); pristinamycin I (Rhône-Poulenc Rorer); vancomycin (Elli Lilly); and ciprofloxacin (Bayer).
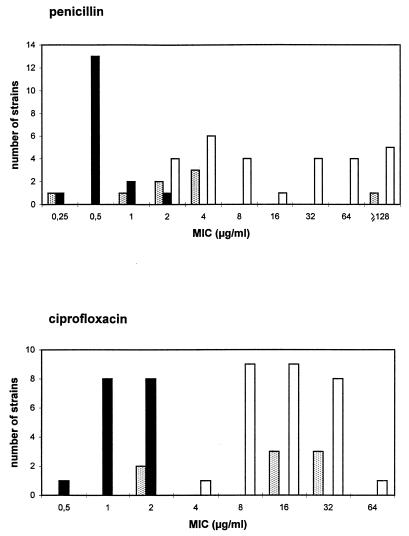
The susceptibility testing results are shown in Table 1 and 2. The MICs at which 90% of isolates were inhibited (MIC90s) of the macrolides tested for 60 Lactobacillus isolates were in the range of 0.03 to 0.5 μg/ml (Table 1). Similar data have been obtained by others (23, 26). The MIC90 of HMR3647 for these isolates was 0.015 μg/ml, 3 dilutions lower than that of erythromycin. One of the studied Lactobacillus isolates, L. paracasei J19, was resistant to all macrolides (MICs in the range of 16 to 256 μg/ml). The MIC of HMR3647 for this isolate was 0.5 μg/ml, lower than those of macrolides and higher than those found for susceptible isolates. Erythromycin resistance mediated by plasmids has been previously reported for Lactobacillus (7, 9, 20, 28), and different genes encoding methylases, such as ermGT (28), ermBC (7), and ermAM (20), have been reported. Only one Lactobacillus isolate was susceptible to vancomycin, and five were susceptible to teicoplanin. MICs of penicillin for 16 of 60 Lactobacillus isolates (26.6%) were ≥16 μg/ml, but no β-lactamase activity was detected. MICs of penicillin or ampicillin for most Lactobacillus isolates referred to in the literature were ≤2 μg/ml (12, 26). Ciprofloxacin had a poor activity against our Lactobacillus isolates (MIC ≥ 4 μg/ml in 60% of them). L. plantarum showed the highest level of resistance to all tested antibiotics except macrolides (Table 2). The respective MIC90s of penicillin and ciprofloxacin were ≥128 and 32 μg/ml for L. plantarum and were 1 and 2 μg/ml for L. paracasei. The modal MICs of penicillin and ciprofloxacin for L. brevis were 4 μg/ml and 16 to 32 μg/ml, respectively (Fig. 1).
TABLE 1.
In vitro activities of different antibiotics against LAB isolates from various generaa
| Microorganisms (no. of isolates) | Antimicrobial agent | MIC (μg/ml)b
|
||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| Lactobacillus spp. (60) | Erythromycin A | 0.03–256 | 0.06 | 0.125 |
| Clarithromycin | 0.0015–128 | 0.015 | 0.03 | |
| Roxithromycin | 0.007–128 | 0.06 | 0.125 | |
| Azithromycin | 0.0035–128 | 0.06 | 0.125 | |
| HMR3647 | ≤0.0007–0.5 | 0.007 | 0.015 | |
| Spiramycin | 0.06–16 | 0.25 | 0.5 | |
| Pristinamycin I | ≤0.25–16 | 2 | 8 | |
| Vancomycin | 4–≥4,096 | ≥4,096 | ≥4,096 | |
| Teicoplanin | 2–≥4,096 | 2,048 | ≥4,096 | |
| Penicillin | 0.25–≥128 | 2 | ≥128 | |
| Ciprofloxacin | 0.5–64 | 8 | 32 | |
| Levofloxacin | 0.125–16 | 4 | 8 | |
| Leuconostoc spp. (18) | Erythromycin A | 0.03–0.125 | 0.06 | 0.125 |
| Clarithromycin | 0.007–0.03 | 0.03 | 0.03 | |
| Roxithromycin | 0.03–0.5 | 0.125 | 0.5 | |
| Azithromycin | 0.03–0.125 | 0.125 | 0.125 | |
| HMR3647 | ≤0.0007–0.015 | 0.007 | 0.015 | |
| Spiramycin | 0.25–0.5 | 0.25 | 0.5 | |
| Pristinamycin I | ≤0.25–8 | 2 | 8 | |
| Vancomycin | 512–≥4,096 | 1,024 | ≥4,096 | |
| Teicoplanin | 512–≥4,096 | 1,024 | ≥4,096 | |
| Penicillin | 0.25–2 | 0.5 | 2 | |
| Ciprofloxacin | 1–32 | 4 | 16 | |
| Levofloxacin | 1–8 | 4 | 8 | |
| Pediococcus spp. (12) | Erythromycin A | 0.06–≥1,024 | 0.125 | 0.25 |
| Clarithromycin | 0.007–128 | 0.06 | 0.06 | |
| Roxithromycin | 0.007–1,024 | 0.06 | 0.5 | |
| Azithromycin | 0.06–128 | 0.125 | 0.25 | |
| HMR3647 | ≤0.0007–4 | 0.007 | 0.015 | |
| Spiramycin | 0.25–≥64 | 0.5 | 1 | |
| Pristinamycin I | 1–128 | 4 | 8 | |
| Vancomycin | 512–2,048 | 1,024 | 1,024 | |
| Teicoplanin | 512–≥4,096 | 2,048 | ≥4,096 | |
| Penicillin | 0.5–2 | 1 | 2 | |
| Ciprofloxacin | 32–64 | 32 | 64 | |
| Levofloxacin | 4–16 | 8 | 16 | |
| Total LAB (90) | Erythromycin A | 0.03–≥1,024 | 0.06 | 0.125 |
| Clarithromycin | 0.0015–128 | 0.015 | 0.06 | |
| Roxithromycin | 0.007–≥1,024 | 0.06 | 0.25 | |
| Azithromycin | 0.0035–128 | 0.06 | 0.125 | |
| HMR3647 | ≤0.0007–4 | 0.007 | 0.015 | |
| Spiramycin | 0.06–≥64 | 0.5 | 0.5 | |
| Pristinamycin I | ≤0.25–128 | 4 | 8 | |
| Vancomycin | 4–≥4,096 | 2,048 | ≥4,096 | |
| Teicoplanin | 2–≥4,096 | 2,048 | ≥4,096 | |
| Penicillin | 0.25–≥128 | 1 | 64 | |
| Ciprofloxacin | 0.5–64 | 8 | 32 | |
| Levofloxacin | 0.125–16 | 4 | 8 | |
Agar dilution in BHIA and 5% CO2.
50% and 90%, MIC50 and MIC90, respectively.
TABLE 2.
In vitro activities of different antibiotics against Lactobacillus spp.a
| Microorganisms (no. of isolates) | Antimicrobial agent | MIC (μg/ml)b
|
||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| L. plantarum (28) | Erythromycin A | 0.03–0.125 | 0.06 | 0.125 |
| Clarithromycin | 0.0007–0.06 | 0.007 | 0.06 | |
| Roxithromycin | 0.03–0.5 | 0.06 | 0.25 | |
| Azithromycin | 0.0035–0.125 | 0.06 | 0.125 | |
| HMR3647 | ≤0.0007–0.015 | 0.007 | 0.015 | |
| Spiramycin | 0.25–1 | 0.25 | 0.5 | |
| Pristinamycin I | 2–16 | 4 | 16 | |
| Vancomycin | 1,024–≥4,096 | ≥4,096 | ≥4,096 | |
| Teicoplanin | 1,024–≥4,096 | ≥4,096 | ≥4,096 | |
| Penicillin | 2–≥128 | 8 | ≥128 | |
| Ciprofloxacin | 4–64 | 16 | 32 | |
| Levofloxacin | 0.125–16 | 4 | 8 | |
| L. paracasei (17) | Erythromycin A | 0.03–256 | 0.06 | 0.125 |
| Clarithromycin | 0.0035–128 | 0.03 | 0.03 | |
| Roxithromycin | 0.015–128 | 0.125 | 0.125 | |
| Azithromycin | 0.03–128 | 0.06 | 0.06 | |
| HMR3647 | ≤0.0007–0.5 | 0.007 | 0.015 | |
| Spiramycin | 0.25–16 | 0.5 | 0.5 | |
| Pristinamycin I | 0.25–2 | 0.5 | 2 | |
| Vancomycin | 4–≥4,096 | 1,024 | ≥4,096 | |
| Teicoplanin | 4–≥4,096 | 512 | 1,024 | |
| Penicillin | 0.25–2 | 0.5 | 1 | |
| Ciprofloxacin | 0.5–2 | 1 | 2 | |
| Levofloxacin | 0.5–2 | 2 | 2 | |
| L. brevis (8) | Erythromycin A | 0.03–0.125 | —c | — |
| Clarithromycin | ≤0.0007–0.06 | — | — | |
| Roxithromycin | 0.007–0.25 | — | — | |
| Azithromycin | 0.0035–0.125 | — | — | |
| HMR3647 | ≤0.0007–0.015 | — | — | |
| Spiramycin | 0.125–0.5 | — | — | |
| Pristinamycin I | 1–16 | — | — | |
| Vancomycin | 128–≥4,096 | — | — | |
| Teicoplanin | 8–≥4,096 | — | — | |
| Penicillin | 0.25–128 | — | — | |
| Ciprofloxacin | 2–32 | — | — | |
| Levofloxacin | 2–8 | — | — | |
| Lactobacillus spp. (7) | Erythromycin A | 0.03–0.06 | — | — |
| Clarithromycin | 0.015–0.06 | — | — | |
| Roxithromycin | 0.06–0.125 | — | — | |
| Azithromycin | 0.03–0.25 | — | — | |
| HMR3647 | ≤0.0007–0.007 | — | — | |
| Spiramycin | 0.06–0.5 | — | — | |
| Pristinamycin I | 0.25–8 | — | — | |
| Vancomycin | 128–≥4,096 | — | — | |
| Teicoplanin | 2–512 | — | — | |
| Penicillin | 0.25–≥128 | — | — | |
| Ciprofloxacin | 0.5–4 | — | — | |
| Levofloxacin | 0.25–4 | — | — | |
Agar dilution in BHIA and 5% CO2.
50% and 90%, MIC50 and MIC90, respectively.
—, too few isolates for MIC determinations.
FIG. 1.
Graph of penicillin and ciprofloxacin MICs for 53 Lactobacillus isolates. White bars, L. plantarum; black bars, L. paracasei; stippled bars, L. brevis.
All studied Leuconostoc isolates were susceptible to macrolides, with MIC90s in the range of 0.03 to 0.5 μg/ml (Table 1). The MIC90 of HMR3647 was 0.015 μg/ml, 3 dilutions lower than that of erythromycin. Similar results have previously been reported (18, 26). Penicillin MICs were always ≤2 μg/ml. Ciprofloxacin and levofloxacin were poorly active against Leuconostoc, showing MICs of ≥4 μg/ml for 72.2 and 61.1% of the isolates, respectively.
For all but one of the studied Pediococcus isolates erythromycin MICs were ≤0.25 μg/ml. The MIC90 of HMR3647 for these isolates was 0.015 μg/ml. Marie-Bigot et al. (18) have reported similar results. P. acidilactici AR63 was the only macrolide-resistant Pediococcus isolate found in our study (MICs of ≥64 μg/ml). The MIC of HMR3647 for this isolate was 4 μg/ml. The mechanism of resistance involved was studied by PCR, and a positive result was obtained when degenerate erm primers were used (4). Nevertheless, negative results were obtained when specific primers for amplification of ermA, ermB, ermC (25), or ermTR (16), as well as msrA (30) and mefA/E (24), were assayed. A high-molecular-weight plasmid was observed in P. acidilactici AR63, and its putative relationship with macrolide resistance was studied by filter conjugation using erythromycin-sensitive E. faecalis JH2-2 and E. faecium GE-1 as recipients. Results were negative in all cases. Erythromycin resistance has previously been reported for Pediococcus strains (26, 31). One P. acidilactici strain with macrolide resistance related to a determinant homologous to the ermAM gene located on a nontransferable 46 kb plasmid has previously been reported (27).
For all Leuconostoc and Pediococcus isolates penicillin MICs were ≤2 μg/ml. MIC ranges of ciprofloxacin for Pediococcus and Leuconostoc were 32 to 64 and 1 to 32 μg/ml, respectively. All isolates of both genera showed high-level glycopeptide resistance (MIC ≥ 512 μg/ml). This group of organisms is sometimes mistaken for enterococci, but the latter genus is usually susceptible to vancomycin.
MICs were determined in MHA-B medium and were compared with those obtained in BHIA for 78 LAB isolates in this study. MICs were similar in both media, although they tended to be 1 dilution higher in BHIA (data not shown).
In summary, with few exceptions, the LAB tested in this work were susceptible to all macrolides and the ketolide HMR3647. For all tested Pediococcus and Leuconostoc isolates penicillin MICs were ≤2 μg/ml, whereas for 26% of Lactobacillus isolates MICs were ≥16 μg/ml. Ciprofloxacin MICs for all Pediococcus isolates, 60% of Lactobacillus isolates, and 72% of Leuconostoc isolates were ≥4 μg/ml. Ecological balance modifications of the natural species in the human gut flora by antibiotic action are a matter of growing concern. This paper shows that differences in susceptibility to widely used groups of antibiotics occur among different genera, as well as among species within a given genus. Therefore, population replacement derived from antibiotic pressure would not come unexpectedly. Thus, for instance, L. plantarum (resistant to penicillin and ciprofloxacin) might replace L. paracasei (susceptible to both types of drugs). The clinical or evolutionary implications of these changes are currently unknown. More attention should be paid to the selection of naturally (or acquired) resistant microorganisms that are members of the normal human microbiota.
(Part of this work was presented at the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Canada, 1997 [29].)
Acknowledgments
This work was supported in part by a grant of Hoescht Marion Rousell and a grant from the Fondo de Investigaciones Sanitarias of Spain (FIS 98/282).
REFERENCES
- 1.Aguirre M, Collins M D. Lactic acid bacteria and human clinical infections. J Appl Bacteriol. 1993;75:95–107. doi: 10.1111/j.1365-2672.1993.tb02753.x. [DOI] [PubMed] [Google Scholar]
- 2.Ahrné S, Novaek S, Jeppsson B, Adlerberth Y, Wold A E, Molin G. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J Appl Microbiol. 1998;85:88–94. doi: 10.1046/j.1365-2672.1998.00480.x. [DOI] [PubMed] [Google Scholar]
- 3.Antony S, Dummer S, Stratton C. Lactobacillus bacteremia and endocarditis. Clin Infect Dis. 1998;26:1483–1484. doi: 10.1086/517657. [DOI] [PubMed] [Google Scholar]
- 4.Arthur M, Molinas C, Mabilat C. PCR detection of erm erythromycin resistance genes by using degenerate oligonucleotide primers. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology. Washington, D.C.: American Society for Microbiology; 1993. pp. 534–538. [Google Scholar]
- 5.Atkins J T, Tillman J, Tan T Q, Demmler G J. Pediococcus pentosaceus catheter associated infection in an infant with gastroschisis. Pediatr Infect Dis J. 1994;13:75–76. [PubMed] [Google Scholar]
- 6.Axelsson L T. Lactic acid bacteria: classification and physiology. In: Salminen S, von Wright A, editors. Lactic acid bacteria. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 1–63. [Google Scholar]
- 7.Axelsson L T, Ahrné S E, Andersson M C, Stahl S R. Identification and cloning of a plasmid-encoded erythromycin resistance determinant from Lactobacillus reuteri. Plasmid. 1988;20:171–174. doi: 10.1016/0147-619x(88)90023-6. [DOI] [PubMed] [Google Scholar]
- 8.Coovadia Y M, Solwa Z, Ende J V D. Meningitis caused by vancomycin-resistant Leuconostoc sp. J Clin Microbiol. 1987;25:1784–1785. doi: 10.1128/jcm.25.9.1784-1785.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fons M, Hégé T, Ladiré M, Raibaud P, Ducluzeau R, Maguin E. Isolation and characterization of a plasmid from Lactobacillus fermentum conferring erythromycin resistance. Plasmid. 1997;37:199–203. doi: 10.1006/plas.1997.1290. [DOI] [PubMed] [Google Scholar]
- 10.Friedland I R, Snipelisky M, Khoosal M. Meningitis in a neonate caused by Leuconostoc sp. J Clin Microbiol. 1990;28:2125–2126. doi: 10.1128/jcm.28.9.2125-2126.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golledge C L, Stingemore N, Aravena M, Joske D. Septicemia caused by vancomycin-resistant Pediococcus acidilactici. J Clin Microbiol. 1990;28:1678–1679. doi: 10.1128/jcm.28.7.1678-1679.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green M, Barbadora K, Michaels M. Recovery of vancomycin-resistant gram-positive cocci from pediatric liver transplant recipients. J Clin Microbiol. 1991;29:2503–2506. doi: 10.1128/jcm.29.11.2503-2506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths J K, Daly J S, Dodge R A. Two cases of endocarditis due to Lactobacillus species: antimicrobial susceptibility, review, and discussion of therapy. Clin Infect Dis. 1992;15:250–255. doi: 10.1093/clinids/15.2.250. [DOI] [PubMed] [Google Scholar]
- 14.Handwerger S, Horowitz H, Coburn K, Kolokathis A, Wormser G P. Infection due to Leuconostoc species: six cases and review. Rev Infect Dis. 1990;12:602–610. doi: 10.1093/clinids/12.4.602. [DOI] [PubMed] [Google Scholar]
- 15.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T, editors. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. Group 17. Gram-positive cocci; pp. 527–533. [Google Scholar]
- 15a.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T, editors. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. Group 19. Regular, nonsporing Gram-positive rods; pp. 565–567. [Google Scholar]
- 16.Kataja J, Seppälä H, Skurnik M, Sarkkinen H, Huovinen P. Different erythromycin resistance mechanisms in group C and group G streptococci. Antimicrob Agents Chemother. 1998;42:1493–1494. doi: 10.1128/aac.42.6.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein G, Zill E, Schindler R, Louwers J. Peritonitis associated with vancomycin-resistant Lactobacillus rhamnosus in a continuous ambulatory peritoneal dialysis patient: organism identification, antibiotic therapy, and case report. J Clin Microbiol. 1998;36:1781–1783. doi: 10.1128/jcm.36.6.1781-1783.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marie-Bigot C, Decre D, Muller C, Salgado E, Bergogne-Berezin E. Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1997. In vitro activity of a novel ketolide antimicrobial agent HMR3647 compared to erythromycin (ery), roxithromycin (rox), azithromycin (azi) and clarithromycin (cla), abstr. F-117; p. 166. [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. 1997. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 20.Rinckel L A, Savage D C. Characterization of plasmids and plasmid-borne macrolide resistance from Lactobacillus sp. strain 100-33. Plasmid. 1990;23:119–125. doi: 10.1016/0147-619x(90)90030-g. [DOI] [PubMed] [Google Scholar]
- 21.Salminen S, Deighton M, Gorbach S. Lactic acid bacteria in health and disease. In: Salminen S, Wright A V, editors. Lactic acid bacteria. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 199–225. [Google Scholar]
- 22.Sarma P S, Mohanty S. Pediococcus acidilactici pneumonitis and bacteriemia in a pregnant woman. J Clin Microbiol. 1998;36:2392–2393. doi: 10.1128/jcm.36.8.2392-2393.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schülin T, Wennersten C B, Moellering R C, Jr, Eliopoulos G M. In vitro activity of RU 64004, a new ketolide antibiotic, against gram-positive bacteria. Antimicrob Agents Chemother. 1997;41:1196–1202. doi: 10.1128/aac.41.5.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swenson J M, Facklam R R, Thornsberry C. Antimicrobial susceptibility of vancomycin-resistant Leuconostoc, Pediococcus, and Lactobacillus species. Antimicrob Agents Chemother. 1990;34:543–549. doi: 10.1128/aac.34.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tankovic J, Leclercq R, Duval J. Antimicrobial susceptibility of Pediococcus spp. and genetic basis of macrolide resistance in Pediococcus acidilactici HM3020. Antimicrob Agents Chemother. 1993;37:789–792. doi: 10.1128/aac.37.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tannock G W, Luchansky J B, Miller L, Connell H, Thode-Andersen S, Mercer A A, Klaenhammer T R. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid. 1994;31:60–71. doi: 10.1006/plas.1994.1007. [DOI] [PubMed] [Google Scholar]
- 29.Torres C, Zarazaga M, Tenorio C, Sáenz Y, Portillo A, Baquero F. Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1997. Susceptibility of Lactobacillus, Leuconostoc, and Pediococcus to ketolide HMR 3647 and other antibiotics, abstr. F-247; p. 188. [Google Scholar]
- 30.Wondrack L, Massa M, Yang B V, Sutcliffe J. Clinical strain of Staphylococcus aureus inactivates and causes efflux of macrolides. Antimicrob Agents Chemother. 1996;40:992–998. doi: 10.1128/aac.40.4.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamane N, Jones R N. In vitro activity of 43 antimicrobial agents tested against ampicillin-resistant enterococci and gram-positive species resistant to vancomycin. Diagn Microbiol Infect Dis. 1991;14:337–345. doi: 10.1016/0732-8893(91)90025-b. [DOI] [PubMed] [Google Scholar]