Abstract
The discovery that half of the mammalian protein-coding genome is regulated by the circadian clock has clear implications for medicine. Recent studies demonstrated that the circadian clock influences therapeutic outcomes in human heart disease and cancer. However, biological time is rarely given clinical consideration. A key barrier is the absence of information on tissue-specific molecular rhythms in the human body. We have applied the cyclic ordering by periodic structure (CYCLOPS) algorithm, designed to reconstruct sample temporal order in the absence of time-of-day information, to the gene expression collection of 13 tissues from 632 human donors. We identified rhythms in gene expression across the body; nearly half of protein-coding genes were shown to be cycling in at least 1 of the 13 tissues analyzed. One thousand of these cycling genes encode proteins that either transport or metabolize drugs or are themselves drug targets. These results provide a useful resource for studying the role of circadian rhythms in medicine and support the idea that biological time might play a role in determining drug response.
INTRODUCTION
Metabolic, endocrine, and behavioral functions show daily variation in humans (1). This variability is orchestrated by endogenous circadian clocks that drive tissue-specific rhythms in gene expression (2). The discovery that ~50% of mammalian genes are expressed with 24-hour rhythms (3, 4) has broad physiological and pharmacological implications. Circadian medicine aims to incorporate this information to develop safer and more efficacious therapeutics. Recent studies show that time of day is critical to outcomes in heart surgery (5) and cancer treatment (6, 7). Despite this, biological time is often overlooked in clinical practice and during drug development. One of the reasons is the lack of understanding of the mechanisms by which the circadian clock governs rhythms in physiology and pathophysiology. Human time-series data have not been feasible at the spatial and temporal resolution of animal models (8–10). To fill this gap, we used a state-of-the-art algorithm called cyclic ordering by periodic structure (CYCLOPS), able to reconstruct sample order in the absence of time-of-day information (11), to build a population-level human circadian atlas. Here, we have applied CYCLOPS to the Genotype-Tissue Expression (GTEx) collection (12) of 4292 post-mortem RNA sequencing (RNA-seq) samples from 632 donors across 13 distinct tissue types. We found extensive population-scale rhythms in gene expression, with ~50% of protein-coding genes cycling in at least 1 of the 13 tissues. We identified a set of oscillating genes across the majority of tissues. These included well-established “core clock” and many other genes.
Although the core clock network is shared across organ systems, the clear majority of rhythmic expression was tissue-specific. Circadian medicine hinges on reliable prediction of drug-target spatiotemporal dynamics. Focusing on the cardiovascular system, we connected more than 100 drugs to 136 genes oscillating robustly in at least one of atrial chamber, aorta, coronary artery, or tibial artery. Overall, our results identified specific opportunities for timed administration of drugs in pursuit of circadian medicine in cardiovascular disease and many other therapeutic areas.
RESULTS
Population-level rhythms in gene expression across human anatomy
From the full GTEx collection of RNA-sequenced human donor samples, we ordered 13 of 51 tissues with CYCLOPS (fig. S1). These 13 tissues were chosen because (i) they had at least 160 samples, (ii) the molecular clock network was preserved (13), and (iii) the orderings reproduced evolutionarily conserved phase relationships between core clock genes. In total, our analyses considered 632 donors characterized by a mix of ages, sex, and health status (Table 1).
Table 1.
GTEx donor demographics.
| Sex | Male | Female | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| 66% | 34% | |||||
| Age (years) | 20–29 | 30–39 | 40–49 | 50–59 | 60–69 | 70–79 |
| 8% | 7% | 17% | 34% | 32% | 3% | |
| Death | Fast | Intermediate | Slow | Sudden-Natural | Ventilator | NA |
| 4% | 5% | 10% | 26% | 53% | 1% | |
NA, not available.
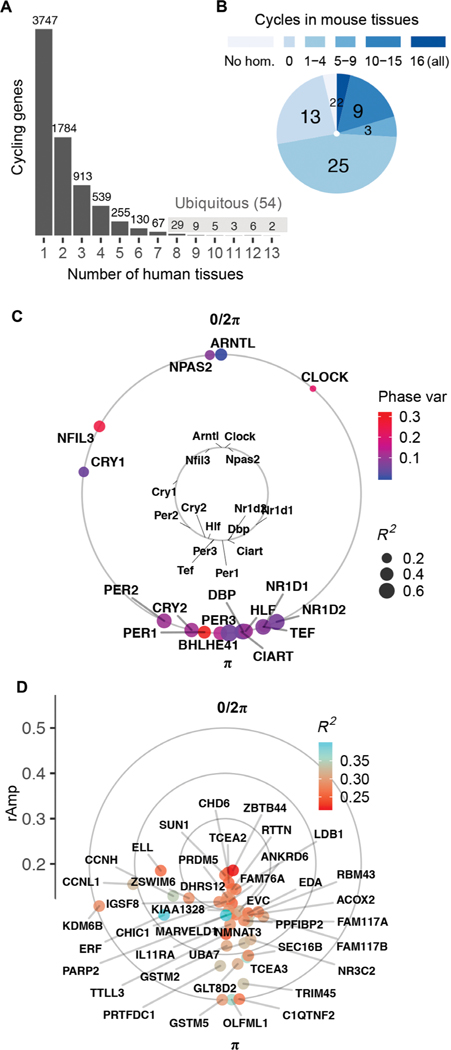
Cosinor analysis on ordered samples from each tissue identified thousands of genes whose expression met criteria for rhythmicity [false discovery rate (FDR) (14) < 0.05; relative amplitude (rAmp) ≥ 0.1; coefficient of determination (R2) ≥ 0.1; fig. S2, A to C, and data file S1]. We found that 7486 distinct genes cycle in at least 1 of the 13 tissues (Fig. 1A). This represents 44% of total distinct genes considered for analysis (16,906), which were limited to the top 15,000 expressed from each tissue. Although the vast majority of cycling genes were protein-coding, noncoding elements (including antisense RNAs, microRNAs, and long intergenic noncoding RNAs) were a small share in each tissue (fig. S2D and data file S1).
Fig. 1. Population-level rhythms in gene expression across human anatomy.
(A) Number of genes that met criteria for rhythmicity (FDR < 0.05; rAmp ≥ 0.1; R2 ≥ 0.1) in at least 1 of the 13 tissues. Periodic analysis was performed by cosinor regression on expression values for 160 CYCLOPS-ordered samples per tissue, as discussed in fig. S1 and Materials and Methods. See fig. S2 for numbers of rhythmic genes discovered at varying FDR thresholds. (B) Set of 54 ubiquitous genes cycling in at least 8 of 13 human tissues, grouped by prior circadian context as reported in CircaDB (darkest blue, homolog cycles in 16 of 16 mouse tissues; lightest blue, not cycling in any of the 16 mouse tissues). (C) Average acrophases of core clock genes (external circle) across all human tissues compared to mouse [internal circle, average across 12 tissues reported by Zhang et al. (3)]. Average coefficient of determination (R2, a measure of cycling robustness) is indicated by point size, and phase variability (Phase var) is indicated by color. Peak phase of ARNTL (human) or Arntl (mouse) was set as 0 for comparison. (D) Average acrophases of all other (noncore clock) human ubiquitous cycling genes. Genes located more distant from the center have larger average amplitudes of oscillation (rAmp) across tissues where they cycle.
We reported a set of 54 genes that cycle in 8 or more of the 13 tissues (Fig. 1, A and B), referred to hereafter as “ubiquitous.” Among these were well-established clock and clock-controlled genes, which showed phase correlations matching those observed in mouse—target transcriptional activators BMAL1, NPAS2, and CLOCK peak, on average, 12 hours out of phase with their E-box targets NR1D1, NR1D2, and PER3 (Fig. 1C). In addition to known clock factors, the ubiquitous set included genes that do not cycle robustly in mice (Fig. 1, B and D, and data file S2) (15).
Robust oscillator and divergent output programs
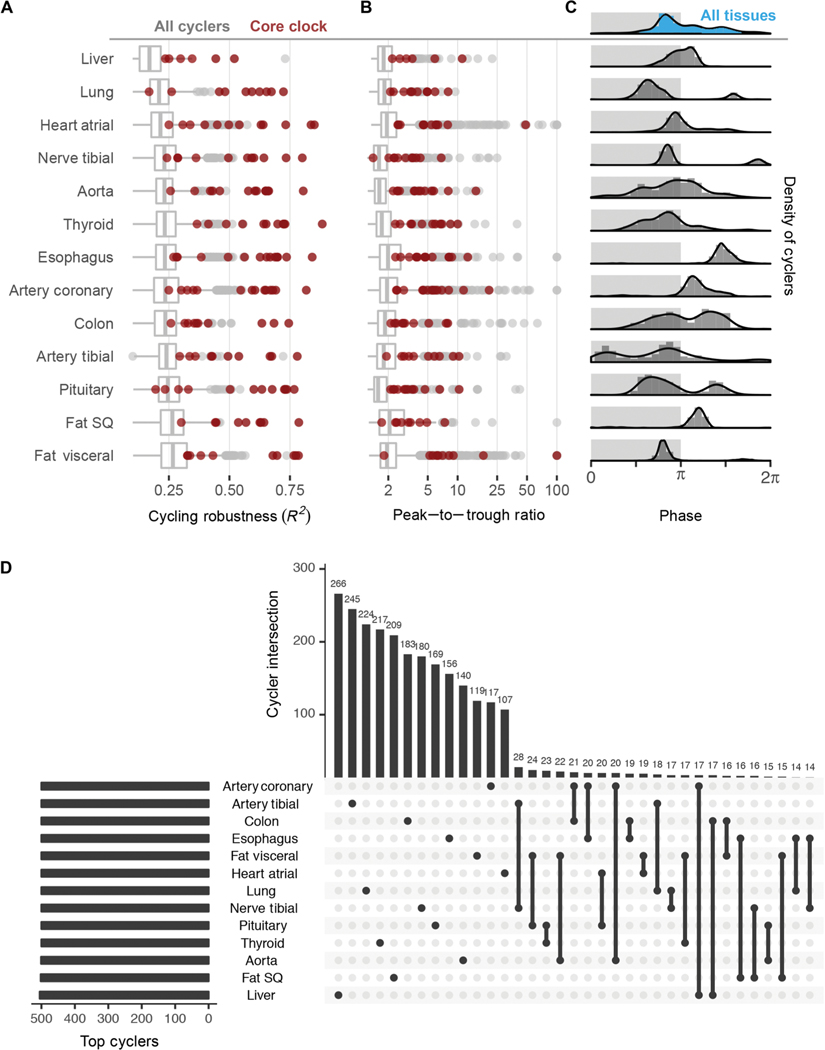
Core clock genes ranked among the most robustly cycling genes across most of the tissues analyzed (Fig. 2, A and B). This aligns with observations from time-series studies—from invertebrates to nonhuman primates (3, 4, 16). As the first large-scale population analysis, we demonstrate that clock gene oscillations persist despite wide interindividual variability.
Fig. 2. Robust tissue-specific rhythms.
(A) Distribution of robustness (R2, goodness of data fit to 24-hour sine wave) and (B) peak-to-trough ratio for all cyclers (gray) in each tissue; most clock genes (red) reside in the upper quartile in most tissues. (C) Distribution of acrophases for cyclers differs between tissue types. Light and dark shading represents inferred active and inactive phase based on Arntl expression, which is known to peak in anticipation of the inactive period in mammals. (D) UpSetR (18) to rank and visualize the intersection between multiple sets (tissues). Restricting input to the top 500 cyclers by FDR from each tissue, the largest intersections describe genes whose expression cycles only in a single tissue.
Although the core clock network was shared across tissues, overall transcriptional programs were not. The cycling transcriptomes of the 13 tissues differed in both phase distribution and genes (Fig. 2C). On the whole, most tissues displayed a prominent peak of rhythmic transcription at or near the end of the inferred inactivity phase, evident in single-tissue and composite (blue shaded) histograms (Fig. 2C and fig. S2E). There were exceptions to this. Peak phases in esophagus, coronary artery, and subcutaneous fat occurred later during the inferred activity phase. Similar to observations from mouse time-series studies (3), we also detected strong bimodal patterns, depending on tissue type. Although the physiological significance of this remains unknown, studies of heart rhythms, for example, suggest that these patterns reflect organ function (17, 18).
We applied UpSetR (19), a technique to visualize set intersections in a matrix-style layout, to the sets of cycling genes in each tissue (Fig. 2D). When restricting input to the top 500 cyclers (ranked by FDR) in each tissue, we found that the largest two-tissue intersection, between tibial artery and tibial nerve, was composed of just 28 genes (5%), near the percentage due to chance alone (Fisher’s exact test, P = 0.1). Most cycling genes belonged to nonintersecting sets. For example, 266 of the top 500 liver cyclers (53%) were exclusive to liver. Anatomic divergence in clock output is a well-characterized feature in model organisms, likely reflecting the nature of sampling across distinct organ systems. Although precise mechanisms governing tissue- specific circadian gene regulation require elaboration, its extent underscores the distinct functions of the human organs profiled.
Pathway analyses (20) identified evolutionarily conserved temporal regulation of numerous biological processes (fig. S3). Top enriched gene networks were tissue-specific, many with therapeutic relevance (for example, ion transport in the heart and aorta), prompting us to look at drug targets.
Dynamic drug targets
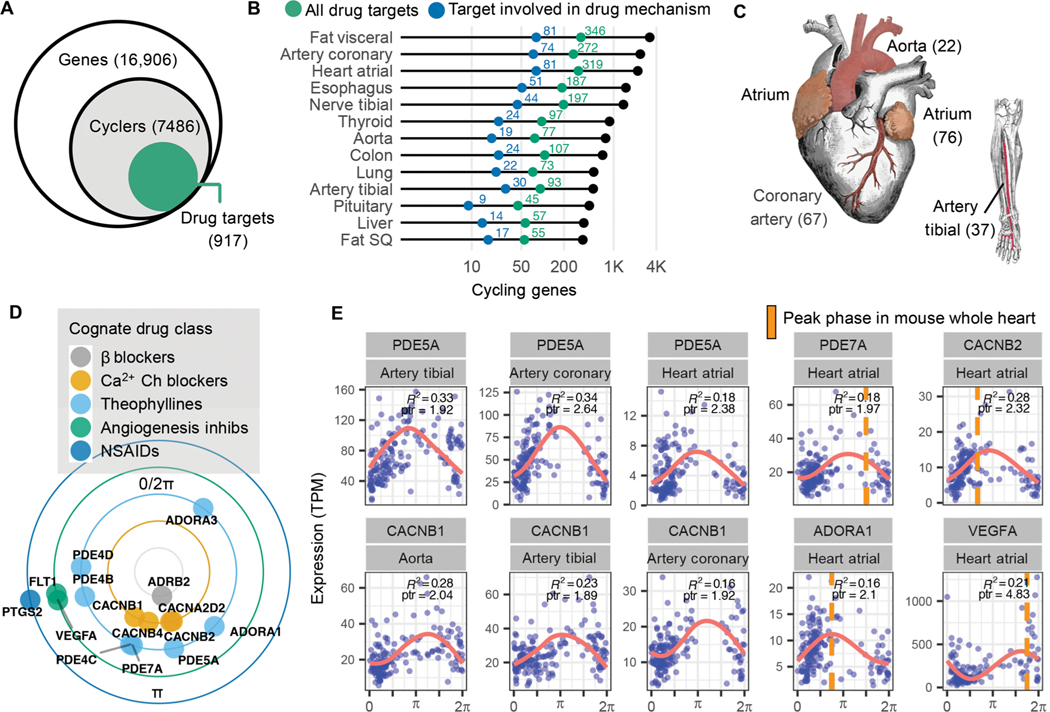
Previous analyses linked circadian genes in mouse, and recently baboon, to specific drug entities (3, 4). Here, we sought to identify pharmacological connections to rhythms in the human population. Of the thousands of genes that cycle in at least 1 of the 13 tissues, 917 (12%) of them encode proteins that either transport or metabolize drugs or are themselves drug targets (Fig. 3A) (21). Overall, this connects thousands of different drugs—both approved and experimental—to nearly 1000 cycling genes. Figure 3B depicts genes that mapped to drugs with a defined mechanism for each tissue (transport, metabolism, or drug action). Subsequent analyses focused on this set.
Fig. 3. Pharmacological links to molecular rhythms in the human population.
(A) Of 7486 genes found to cycle in at least 1 of 13 human tissues sampled, 917 (12%) encode at least one drug target, transporter, or metabolizing enzyme (collectively referred to as “targets”). This represents a total of 2764 drug entities, both approved and experimental as logged in DrugBank (20), with targets that oscillate somewhere in the body. (B) Numbers of total (colored green) and pharmacologically active (colored blue) cycling drug targets by tissue type. (C) Cardiovascular-related tissues from ordered GTEx data. To enrich for high-amplitude cardio-cyclers, we limited to genes in the upper 50th percentile for rAmp within each tissue and FDR ≤0.1. Numbers of cardio-cyclers that meet these cycling criteria and are also drug targets are indicated in parentheses. (D) Cardio-cyclers targeted by select drug classes relevant to heart and vessel physiology. Plotted phase represents average peak phase across all cardiac tissues where that gene was found to cycle. (E) Expression values plotted as a function of sample phase for select cardio-cyclers. Coefficient of determination (R2 ) and peak-to-trough ratio (ptr) are indicated. Right: Genes that are also rhythmically expressed in whole mouse heart, with peak phases indicated by orange bars. ARNTL (human) or Arntl (mouse) was set to 0 for comparison.
Genes found to cycle in human cardiovascular tissue (atrial chamber, aorta, coronary artery, or tibial artery) are targeted by many of these drugs. The cardiovascular system demonstrates 24-hour rhythms in blood pressure (BP) and contractility (22). Mechanisms underlying daily variation in heart and peripheral vasculature, however, remain poorly understood. To reveal the strongest oscillating drug targets in cardiovascular tissue, we narrowed focus to the subset of “cardio-cyclers” in the upper 50th percentile for rAmp within each tissue and FDR ≤ 0.1. The more permissive FDR threshold was chosen to detect potentially noisier but very high amplitude oscillations. This identified 136 drug targets that were rhythmically expressed in at least one of the four tissues (data file S3, Fig. 3C, and fig. S4). A large number of these have established relevance to cardiovascular physiology— many being key targets of standard-of-care agents in heart disease (Fig. 3, D and E). For example, the family of calcium channel blockers, which inhibit Ca2+ influx into cells of the heart and blood vessels producing smooth muscle relaxation, target several rhythmically expressed Ca2+ channel subunits (CACNA2D2, CACNB1, CACNB2, and CACNB4). Peak-to-trough ratio represents the fold difference between the highest and lowest levels of expression in periodic data. These four Ca2+ channel subunits had an average peak-to-trough ratio of 2.2 in cardiovascular tissue, placing them in the upper 70th percentile of all cycling genes detected in this study. Many of the drugs that target these gene products have short half-lives (<6 hours) and may be influenced by time of administration.
We identified a separate group of drug classes that, although not necessarily indicated for cardiovascular pathology, actively target this system (Fig. 3, D and E). For example, theophyllines—bronchodilators prescribed for asthma and chronic obstructive pulmonary disease— function by inhibiting adenosine receptors (ADORA1/3) and multiple phosphodiesterases in the lung. These genes are also critical to normal heart function (23, 24), and their pharmacologic inhibition can be cardiotoxic (25). Additional examples include nonsteroidal anti-inflammatory drugs (NSAIDs) prescribed for pain, and anti-angiogenic agents commonly used in cancer, both associated with cardiovascular risk (25, 26) and both with cardio-cycling targets. Many of these targets also cycle in mouse heart or aorta with similar phase relationships [whole heart and/or aorta, Jonckheere-Terpstra- Kendall or JTK_CYCLE Q value (JTK Q) ≤ 0.1; Fig. 3E and data file S3] (3, 27, 28), indicating evolutionary conservation of these targets and significance for application in circadian medicine.
DISCUSSION
Efforts toward precision medicine have largely centered around linking genetic variants to therapeutic response. Time-series profiles from mice (3), and recently a nonhuman primate (4), showed extensive rhythms in transcript abundance for gene products targeted by many drugs. For drugs with rapid pharmacokinetics (PK), this suggests that circadian time could influence efficacy, toxicity, or the ratio between the therapeutic index. However, although animal models were instrumental to our understanding of circadian biology, results from their study have had limited translational impact. Typically, animal model experiments use small numbers of animals of similar sex, age, and environmental conditions. These are critical variables in all clinical translation, including circadian medicine.
To address these limitations, we are moving to population-based analyses. We applied CYCLOPS to thousands of unordered human samples taken from a heterogeneous population and found that nearly half of all analyzed genes are rhythmically expressed somewhere in the body. This approximates findings in mice, where the number of FDR-controlled cyclers discovered plateaus at ~50% of the protein-coding genome (3). Nearly 1000 of the human cyclers influence drug action. This far exceeds the number of human genes with allelic associations to a specific drug response (21). A few drugs targeting several of these mechanisms are already taken with time-of-day dependence (steroids and short-acting statins). Nevertheless, this leaves many opportunities to explore time of dosage to improve the therapeutic index. Further, the circadian effect sizes we see are often much larger than those seen in genetic studies. It stands to reason, then, that for high-amplitude cycling genes, genetic influences may have been masked by circadian variation. Whereas gene by environment (GxE) interactions are widely appreciated by the genetics community (29, 30), gene by circadian (GxC) interactions are not. Developing algorithms that account for this circadian variability may reveal previously unknown genetic associations.
Many drug-metabolizing enzymes, transporters, and targets were found cycling in the cardiovascular system. Circadian clock–dependent variation in cardiac ion channels has been shown in animal models (31–33) and is proposed to underlie time-dependent arrhythmias and sudden cardiac death in humans (34, 35). We found several L-type Ca2+ channel subunits—critical for cardiac polarization and targets of commonly prescribed antihypertensives—cycling in the human heart and vasculature at population scale. Ca2+ channel blockers, however, are not routinely administered with regard to time of day. We propose that harmonizing dosage time with the window of peak target abundance for fast-PK Ca2+ channel blockers such as verapamil, nifedipine, and others could improve their efficacy. In support of this notion, several papers report improved efficacy of nifedipine by timing dosage before bedtime (36).
The circadian system could also influence adverse events. Cardiotoxicity is the leading cause of drug discontinuation (37). Drugs designed to target other organ systems can provoke adverse cardiac problems. For example, theophylline, commonly prescribed for pulmonary disease, can elicit heart palpitations that limit or preclude its use in some patients (25). We note that NSAIDs, typically prescribed for pain and inflammation, and anti-angiogenic agents, typically prescribed for cancer, can cause cardiotoxic response (26, 38). Here, we show that their targets cycle in the cardiovascular system. For these agents, dosage time could be leveraged to avoid cardiotoxicity and perhaps retain efficacy.
Translational potential hinges on a correspondence between transcript and protein. Although previous work shows that an ~80% of rhythmic proteins in liver have corresponding mRNA rhythms (39), the amplitudes and phases of target proteins will need to be determined empirically. An additional limitation of our findings is that inferred relationships between estimated transcript phase and external clock time (the clock on the wall) relied upon the assumption that clocks in different tissues share a similar phase. Although based on time-series studies of clock gene expression from multiple tissues in mouse (BMAL1 peaks in anticipation of the inactive period), this assumption may not hold true at human population scale. To this point, Hughey and Butte showed phase differences between central (brain) and peripheral (blood and skin) clocks (40). The degree to which clocks in different organ systems are differentially phased remains an outstanding question in circadian biology but does not prevent application of circadian medicine.
Prospective trials designed to test the impact of time-of-day drug administration are required for clinical implementation. Wearable devices with continuous monitoring are being used to study a wide range of physiologic measures with daily rhythms (1). Ambulatory BP monitoring, the gold standard for hypertension diagnosis, or a recently developed cuffless BP monitoring system (41) might be used to test the effect of circadian dosage time on efficacy. In parallel, the development of robust circadian biomarkers might help clinicians administer treatment at optimal times. Recent work from our laboratory identified a biomarker set capable of reporting circadian phase to within 3 hours from a single sample (42). We hope that, collectively, these works foster prospective model organism and clinical studies and catalyze the field of circadian medicine.
MATERIALS AND METHODS
Study design
GTEx human donor criteria were as follows: (i) 21 ≤ age (years) ≤ 70; (ii) 18.5 < body mass index < 35; (iii) time between death and tissue collection less than 24 hours; (iv) no whole-blood transfusion within 48 hours before death; (v) no history of metastatic cancer; (vi) no chemotherapy or radiation therapy within the 2 years before death; and (vii) unselected for the presence or absence of diseases or disorders, except for potentially communicable diseases that disqualify someone to donate organs or tissues, was also disqualifying for GTEx (12). For each donor, multiple tissues were RNA-sequenced using Illumina TruSeq RNA protocol, averaging ~50 million aligned reads per sample.
The objective of this study was to identify tissue-specific molecular rhythms in the human body. We used CYCLOPS to order GTEx donor samples for 13 different tissues. To identify rhythmically expressed genes, we performed cosinor analysis on transcripts per million (TPM) values as a function of ordered sample phases for each of the top 15,000 expressed genes in each tissue.
Preprocessing for CYCLOPS
For each tissue, gene-level TPM data were filtered to exclude any gene with a read count of zero (TPM = 0) in any sample. Following this, only the top 15,000 expressed genes by median TPM were considered for each tissue.
CYCLOPS run parameters
We used circadian seed gene inputs to sharpen CYCLOPS ordering, as reported by Anafi et al., with the following deviations: Oscope (43) was used to select the eigengene clusters for ordering for each tissue. Each eigengene cluster yielded a separate sample ordering. For the final ordering, we selected the eigengene cluster that identified rhythmic expression of clock genes and phase relationships best matching those observed in mouse. This modified CYCLOPS approach was validated in human skin and reported by Wu et al. (42).
Phase set enrichment analysis
Phase set enrichment analysis was run on cycling genes in human (FDR < 0.05; rAmp ≥ 0.1; R2 ≥ 0.1) and mouse-matched (JTK Q ≤ 0.05) tissues to identify phase-enriched pathways from among 4653 GO:biological process gene sets from the Molecular Signatures Database.
Drug target analyses
We used DrugBank (21) version 5.0.11, released on 20 December 2017, to identify genes whose protein products are targets, transporters, carriers, or enzymes linked to specific drug entities. DrugBank contains a total of 10,991 drug entries linked to 4901 target protein sequences.
Statistical analysis
CYCLOPS and cosinor regression were run in Julia version 0.3.12. Cosinor regression was run as implemented in the Anafi et al.’s CYCLOPS package (11). R2 and rAmp (amplitude normalized to baseline gene expression) were computed for each gene. P values were adjusted for multiple hypothesis testing using Benjamini and Hochberg FDR. Criteria for rhythmic expressed genes were set to the following: Entrez ID, FDR < 0.05, rAmp ≥ 0.1, R2 ≥ 0.1. All other analyses were run in R version 3.3.1 (44), with packages tidyverse (45), CircStats (46), and UpSetR (18) and helper functions including drugbankR (47) and homologene (48).
Supplementary Material
Fig. S1. Pipeline for sample selection and CYCLOPS ordering for 13 tissues.
Fig. S2. Numbers of cycling genes by different statistical thresholds and noncoding gene type.
Fig. S3. Phase set enrichment in human and mouse counterpart tissues.
Fig. S4. Gene expression traces for top cardio-cycling drug targets.
Data file S1. Cosinor regression output for all genes and all tissues.
Data file S2. Set of 54 genes that cycle in 8 or more of the 13 tissues.
Data file S3. Set of 136 cardio-cycler drug targets.
Acknowledgments:
We thank J. Hughey, R. Irizarry, T. de Andrade, and G. Rogers for thoughtful discussions.
Funding: This work was supported by Cincinnati Children’s Hospital Medical Center, the National Institute of Neurological Disorders and Stroke (NINDS; 2R01NS054794 to J.B.H. and A. Liu), and the National Human Genome Research Institute (NHGRI; 2R01HG005220 to R. Irizarry and J.B.H.). The raw data used for the analyses described in this article were obtained from the GTEx Portal on 1 December 2017. The GTEx Project was supported by the Common Fund of the Office of the Director of the NIH and by National Cancer Institute, NHGRI, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and NINDS.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials and have been incorporated into CircaDB http://circadb.hogeneschlab.org/.
REFERENCES AND NOTES
- 1.Skarke C, Lahens NF, Rhoades SD, Campbell A, Bittinger K, Bailey A, Hoffmann C, Olson RS, Chen L, Yang G, Price TS, Moore JH, Bushman FD, Greene CS, Grant GR, Weljie AM, FitzGerald GA, A pilot characterization of the human chronobiome. Sci. Rep. 7, 17141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver DR, Introduction to circadian rhythms and mechanisms of circadian oscillations, in Circadian Clocks: Role in Health and Disease, Gumz ML, Ed. (Springer; New York, 2016), pp. 1–55. [Google Scholar]
- 3.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB, A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 111, 16219–16224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, Ngotho M, Kariuki T, Dkhissi-Benyahya O, Cooper HM, Panda S, Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359, eaao0318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montaigne D, Marechal X, Modine T, Coisne A, Mouton S, Fayad G, Ninni S, Klein C, Ortmans S, Seunes C, Potelle C, Berthier A, Gheeraert C, Piveteau C, Deprez R, Eeckhoute J, Duez H, Lacroix D, Deprez B, Jegou B, Koussa M, Edme J-L, Lefebvre P, Staels B, Daytime variation of perioperative myocardial injury in cardiac surgery and its prevention by Rev-Erbα antagonism: A single-centre propensity-matched cohort study and a randomised study. Lancet 39, 59–69 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Kobuchi S, Yazaki Y, Ito Y, Sakaeda T, Circadian variations in the pharmacokinetics of capecitabine and its metabolites in rats. Eur. J. Pharm. Sci. 112, 152–158 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Squire T, Buchanan G, Rangiah D, Davis I, Yip D, Chua YJ, Rich T, Elsaleh H, Does chronomodulated radiotherapy improve pathological response in locally advanced rectal cancer? Chronobiol. Int 34, 492–503 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Mauvoisin D, Wang J, Jouffe C, Martin E, Atger F, Waridel P, Quadroni M, Gachon F, Naef F, Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc. Natl. Acad. Sci. U.S.A. 111, 167–172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng D, Lazar MA, Clocks, metabolism, and the epigenome. Mol. Cell 47, 158–167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, Venkataraman A, Olarerin-George AO, Francey LJ, Mukherjee S, Girish S, Selby CP, Cal S, ER U, Sianati B, Sengupta A, Anafi RC, Kavakli IH, Sancar A, Baur JA, Dang CV, Hogenesch JB, Weljie AM, Clock regulation of metabolites reveals coupling between transcription and metabolism. Cell Metab. 25, 961–974.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anafi RC, Francey LJ, Hogenesch JB, Kim J, CYCLOPS reveals human transcriptional rhythms in health and disease. Proc. Natl. Acad. Sci. U.S.A. 114, 5312–5317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GTEx Consortium, The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shilts J, Chen G, Hughey JJ, Evidence for widespread dysregulation of circadian clock progression in human cancer. PeerJ 6, e4327 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benjamini Y, Hochberg Y, Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57, 289–300 (1995). [Google Scholar]
- 15.Pizarro A, Hayer K, Lahens NF, Hogenesch JB, CircaDB: A database of mammalian circadian gene expression profiles. Nucleic Acids Res. 41, D1009–D1013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald MJ, Rosbash M, Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107, 567–578 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Young ME, Temporal partitioning of cardiac metabolism by the cardiomyocyte circadian clock. Exp. Physiol. 101, 1035–1039 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Prosdocimo DA, Bai X, Fu C, Zhang R, Campbell F, Liao X, Coller J, Jain MK, KLF15 establishes the landscape of diurnal expression in the heart. Cell Rep. 13, 2368–2375 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Conway JR, Lex A, Gehlenborg N, UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang R, Podtelezhnikov AA, Hogenesch JB, Anafi RC, Discovering biology in periodic data through phase set enrichment analysis (PSEA). J. Biol. Rhythms 31, 244–257 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M, DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Degaute JP, van de Borne P, Linkowski P, Van Cauter E, Quantitative analysis of the 24-hour blood pressure and heart rate patterns in young men. Hypertension 18, 199–210 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Bobin P, Belacel-Ouari M, Bedioune I, Zhang L, Leroy J, Leblais V, Fischmeister R, Vandecasteele G, Cyclic nucleotide phosphodiesterases in heart and vessels: A therapeutic perspective. Arch. Cardiovasc. Dis. 109, 431–443 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Shryock JC, Belardinelli L, Adenosine and adenosine receptors in the cardiovascular system: Biochemistry, physiology, and pharmacology. Am. J. Cardiol. 79, 2–10 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Mladěnka P, Applová L, Patočka J, Costa VM, Remiao F, Pourová J, Mladěnka A, Karlíčková J, Jahodář L, Vopršalová M, Varner KJ, Štěrba M; TOX-OER and CARDIOTOX Hradec Králové Researchers and Collaborators, Comprehensive review of cardiovascular toxicity of drugs and related agents. Med. Res. Rev 38, 1332–1403 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sondergaard KB, Weeke P, Wissenberg M, Olsen A-MS, Fosbol EL, Lippert FK, Torp-Pedersen C, Gislason GH, Folke F, Non-steroidal anti-inflammatory drug use is associated with increased risk of out-of-hospital cardiac arrest: A nationwide case–time–control study. Eur. Heart J. Cardiovasc. Pharmacother. 3, 100–107 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Rudic RD, McNamara P, Reilly D, Grosser T, Curtis A-M, Price TS, Panda S, Hogenesch JB, FitzGerald GA, Bioinformatic analysis of circadian gene oscillation in mouse aorta. Circulation 112, 2716–2724 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Tsimakouridze EV, Straume M, Podobed PS, Chin H, LaMarre J, Johnson R, Antenos M, Kirby GM, Mackay A, Huether P, Simpson JA, Sole M, Gadal G, Martino TA, Chronomics of pressure overload–induced cardiac hypertrophy in mice reveals altered day/night gene expression and biomarkers of heart disease. Chronobiol. Int. 29, 810–821 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Caspi A, Hariri AR, Holmes A, Uher R, Moffitt TE, Genetic sensitivity to the environment: The case of the serotonin transporter gene and its implications for studying complex diseases and traits. Am. J. Psychiatry 167, 509–527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamza TH, Chen H, Hill-Burns EM, Rhodes SL, Montimurro J, Kay DM, Tenesa A, Kusel VI, Sheehan P, Eaaswarkhanth M, Yearout D, Samii A, Roberts JW, Agarwal P, Bordelon Y, Park Y, Wang L, Gao J, Vance JM, Kendler KS, Bacanu S-A, Scott WK, Ritz B, Nutt J, Factor SA, Zabetian CP, Payami H, Genome-wide gene-environment study identifies glutamate receptor gene GRIN2A as a Parkinson’s disease modifier gene via interaction with coffee. PLOS Genet. 7, e1002237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins HE, Rodrigo GC, Inotropic response of cardiac ventricular myocytes to β-adrenergic stimulation with isoproterenol exhibits diurnal variation: Involvement of nitric oxide. Circ. Res. 106, 1244–1252 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Zhu D, Yuan J, Han Z, Wang Y, Qian Z, Hou X, Wu T, Zou J, CLOCK-BMAL1 regulate the cardiac L-type calcium channel subunit CACNA1C through PI3K-Akt signaling pathway. Can. J. Physiol. Pharmacol. 94, 1023–1032 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Ko ML, Shi L, Grushin K, Nigussie F, Ko GY-P, Circadian profiles in the embryonic chick heart: L-type voltage-gated calcium channels and signaling pathways. Chronobiol. Int. 27, 1673–1696 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, Hu K, Lu Y, Eapen BL, Sharma N, Ficker E, Cutler MJ, Gulick J, Sanbe A, Robbins J, Demolombe S, Kondratov RV, Shea SA, Albrecht U, Wehrens XHT, Rosenbaum DS, Jain MK, Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature 483, 96–99 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willich SN, Levy D, Rocco MB, Tofler GH, Stone PH, Muller JE, Circadian variation in the incidence of sudden cardiac death in the Framingham Heart Study population. Am. J. Cardiol. 60, 801–806 (1987). [DOI] [PubMed] [Google Scholar]
- 36.Hermida RC, Ayala DE, Mojón A, Fernández JR, Chronotherapy with nifedipine GITS in hypertensive patients: Improved efficacy and safety with bedtime dosing. Am. J. Hypertens. 21, 948–954 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Laverty HG, Benson C, Cartwright EJ, Cross MJ, Garland C, Hammond T, Holloway C, McMahon N, Milligan J, Park BK, Pirmohamed M, Pollard C, Radford J, Roome N, Sager P, Singh S, Suter T, Suter W, Trafford A, Volders PGA, Wallis R, Weaver R, York M, Valentin JP, How can we improve our understanding of cardiovascular safety liabilities to develop safer medicines? Br. J. Pharmacol. 163, 675–693 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Girardi F, Franceschi E, Brandes AA, Cardiovascular safety of VEGF-targeting therapies: Current evidence and handling strategies. Oncologist 15, 683–694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robles MS, Cox J, Mann M, In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLOS Genet. 10, e1004047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughey JJ, Butte AJ, Differential phasing between circadian clocks in the brain and peripheral organs in humans. J. Biol. Rhythms 31, 588–597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandrasekhar A, Kim C-S, Naji M, Natarajan K, Hahn J-O, Mukkamala R, Smartphone-based blood pressure monitoring via the oscillometric finger-pressing method. Sci. Transl. Med. 10, eaap8674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu G, Ruben M, Schmidt R, Francey L, Smith D, Anaf R, Hughey J, Tassef R, Sherrill J, Oblong J, Mills K, Hogenesch J, Population level rhythms in human skin: Implications for circadian medicine. bioRxiv 301820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leng N, Chu L-F, Barry C, Li Y, Choi J, Li X, Jiang P, Stewart RM, Thomson JA, Kendziorski C, Oscope identifies oscillatory genes in unsynchronized single-cell RNA-seq experiments. Nat. Methods 12, 947–950 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R Core Team, R: A Language and Environment for Statistical Computing (2016); www.R-project.org/.
- 45.Wickham H, tidyverse: Easily Install and Load “Tidyverse” Packages (2017); https://CRAN.R-project.org/package=tidyverse. [Google Scholar]
- 46.S-plus original by Ulric Lund, R port by Claudio Agostinelli, CircStats: Circular Statistics, from “Topics in Circular Statistics” (2001) (2012); https://CRAN.R-project.org/ package=CircStats.
- 47.Duan Y, drugbankR: R package for querying drugbank database (2017); https://github.com/yduan004/drugbankR). [Google Scholar]
- 48.Mancarci O, homologene: Quick Access to Homology Information (2018); https://CRAN.R-project.org/package=homologene. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Pipeline for sample selection and CYCLOPS ordering for 13 tissues.
Fig. S2. Numbers of cycling genes by different statistical thresholds and noncoding gene type.
Fig. S3. Phase set enrichment in human and mouse counterpart tissues.
Fig. S4. Gene expression traces for top cardio-cycling drug targets.
Data file S1. Cosinor regression output for all genes and all tissues.
Data file S2. Set of 54 genes that cycle in 8 or more of the 13 tissues.
Data file S3. Set of 136 cardio-cycler drug targets.