Abstract
Dynamic Nuclear Polarization (DNP) enhanced solid state NMR increases experimental sensitivity, potentially enabling detection of biomolecules at their physiological concentrations. The sensitivity of DNP experiments is due to the transfer of polarization from electron spins of free radicals to the nuclear spins of interest. Here, we investigate the reduction of AMUPol in both lysed and intact HEK293 cells. We find that nitroxide radicals are reduced with first order reduction kinetics by cell lysates at a rate of ~12% of the added nitroxide radical concentration per hour. We also found that electroporation delivered a consistent amount of AMUPol to intact cells and that nitroxide radicals are reduced just slightly more rapidly (~15% per hour) by intact cells than by cell lysates. The two nitroxide radicals of AMUPol are reduced independently and this leads to considerable accumulation of the DNP-silent mono-radical form of AMUPol, particularly in preparations of intact cells where nearly half of the AMUPol is already reduced to the DNP silent mono-radical form at the earliest experimental time points. This confirms that the loss of DNP-active bi-radical form of AMUPol is faster than the nitroxide reduction rate. Finally, we investigate the effect of adding N-ethyl maleimide, a well-known inhibitor of thiol (-SH) group-based reduction of nitroxide bi-radicals in cells, on AMUPol reduction, cellular viability, and DNP performance. Although pre-treatment of cells with NEM effectively inhibited the reduction of AMUPol, exposure to NEM compromised cellular viability and, surprisingly, did not improve DNP performance. Collectively, these results indicate that currently, the most effective strategy to obtain high DNP enhancements for DNP-assisted in-cell NMR is to minimize room temperature contact times with cellular constituents and suggest that the development of bio-resistant polarization agents for DNP could considerably increase the sensitivity of DNP-assisted in-cell NMR experiments.
Keywords: DNP NMR, Polarizing agent, AMUPol, DNP NMR, In-cell NMR, In-cell EPR
Introduction
Dynamic nuclear polarization (DNP) based solid state NMR can enhance the sensitivity of NMR signals by several orders of magnitude. This enables the detection of signals from biomolecules at their endogenous concentrations in experimentally tractable acquisition times. Recent studies have capitalized on this technique to detect and study proteins and nucleic acids at low concentrations in complex biological environments that range from cellular lysates to intact cells 1-16. These studies provide proof of concept for the application of DNP NMR to study biomolecular structures in situ. Moreover, in some cases, these studies highlight the importance of doing so since the structure of the target molecules in their native environments differed from that of the target molecule in purified settings 1,3.
The sensitivity enhancements conferred by DNP result from the transfer of polarization from electron spins of free radicals to nuclear spins of the atoms of interest 17-19. Free radicals are typically introduced by doping samples with millimolar concentrations of polarization agents 1-4,19-21. Some of the most efficient polarization agents for DNP are nitroxide bi-radicals that rely upon the cross-effect for polarization transfer, a mechanism that couples two electrons to one nucleus 17-19,22,23. The commercially-available nitroxide biradical, AMUPol, is an efficient polarization agent for biological molecules in hydrated solutions 1,3,4,21. Moreover, it is non-toxic to mammalian cells; cells can regrow after exposure to DNP-relevant concentrations21. AMUPol is highly effective at enhancing the signal of all the mammalian biomass components21.
However, nitroxide radicals are prone to reduction in cellular environments 24-28. The reduction of nitroxides in cellular environments is thought to be largely driven by sulfhydryl groups on proteins and glutathione although other factors like ascorbic acid and radical oxygen species could also contribute15,26,28-33. This has been well-documented by EPR where nitroxide radicals are often used to assess protein conformations in situ24-27. In the context of DNP, the reduction of the radicals translates into a decrease in experimental sensitivity21,34. The effect of reduction is probably more pronounced for DNP polarization agents that rely upon the cross-effect since reduction of just one of the two nitroxide moieties inactivates the polarization agent and loss of experimental sensitivity is likely more rapid than the reduction rate.21,35 Despite growing interest in determining protein conformations in mammalian cells, the reduction of the bi-radical containing polarization agents used for DNP NMR by mammalian cells has not yet been systematically investigated.
In the handful of published examples, reduction of polarization agents by mammalian cells is slow relative to the time scale of sample preparation. In most of these studies, however, it is unclear how much of the added polarization agent was in contact with the cellular constituents during the measurement time since the plasma membrane is only semi-permeable to AMUPol. In experiments where all the DNP polarization agent in the sample was in contact with cellular constituents, the DNP enhancements conferred by AMUPol decrease with increasing room temperature incubation times 21. DNP enhancements are highest in samples of lysed mammalian cells that are flash frozen immediately upon addition of AMUPol. DNP enhancements decrease by half after incubation with lysed mammalian cells for an hour at room temperature before freezing and DNP enhancements decrease by half after incubation inside intact cells for just 45 minutes at room temperature before freezing21. The DNP enhancement in cellular lysates reaches its maximum of nearly 100 at 5 mM AMUPol and has a sharp dependence on AMUPol concentration. The DNP enhancement for intact cells where AMUPol is delivered by electroporation and then removed from the exterior of the cells before measurement has a lower maximum DNP enhancement of 60 and the dependence on AMUPol concentration is more difficult to be characterized due to uncertainty about the quantity of AMUPol delivered to the cells by this method. Moreover, because the intact cells are prepared differently, there is a significant difference in sample preparation time for lysed and intact cells21,36. Interestingly, however, even after controlling for the difference in sample preparation time, the DNP enhancements for intact mammalian cells decrease more rapidly than for lysed mammalian cells21. This set of experiments indicate that while it is possible to attain very high DNP enhancements in complicated biological mixtures, the current state of the art results in losses of about a third of the polarizing capacity of the polarization agent AMUPol inside intact cells before data collection. However, in this set of experiments, only the DNP performance was assessed. Neither the amount of AMUPol that was delivered to cells in the samples nor the rate at which the AMUPol was reduced were directly determined. Quantification of the amount of AMUPol that is delivered and the timescale of nitroxide biradical reduction in mammalian cells will help to identify strategies to combat polarization-agent reduction and enable detection of low concentration molecules in experimentally tractable acquisition times.
Indeed, the pioneering work that characterized the reduction of a related binitroxide radical, TOTAPOL, by both lysed and intact bacterial cells suggested that introduction of alkylating reagents like N-ethyl maleimide (NEM) prevented radical reduction 35. Interestingly, in that work, the stabilization of the radical did not translate to an improvement in the DNP performance, an observation that was tentatively attributed to poor delivery of the polarization agent to the cell 35. Because delivery of the polarization agent to mammalian cells via electroporation is efficient, the mammalian cell system is well suited to explore the strategy of stabilizing the radical in biological environment by incubation of lysates or intact mammalian cells with NEM prior to delivery of the polarization agent. Therefore, in an effort to increase the sensitivity of DNP-assisted in-cell NMR spectroscopy, here we quantify the amount of AMUPol delivered to intact cells by electroporation, determine the AMUPol reduction rate in intact and lysed HEK293 cells and test the efficacy of incubation with N-ethylmaleimide to prolong the lifetime and preserve the DNP properties of radicals in both lysed and intact mammalian cells.
Results
The reduction of AMUPol in cellular lysates is relatively slow
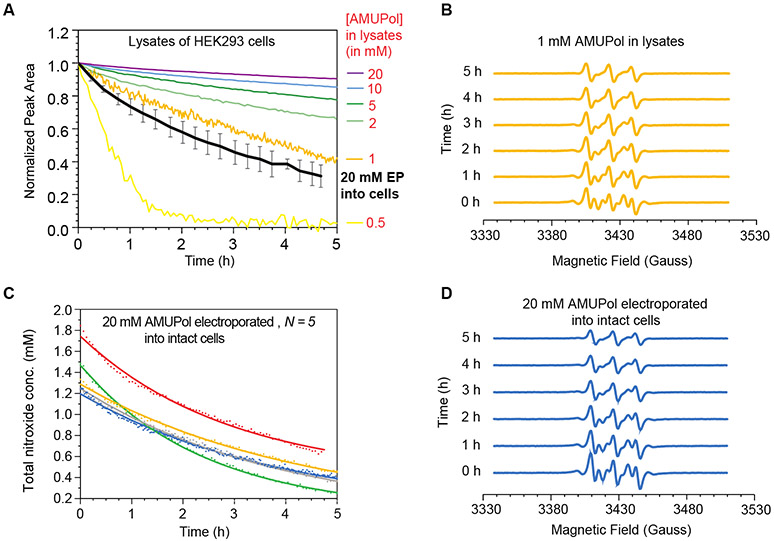
To determine the rate of reduction of the nitroxide biradical AMUPol in biological environments, we added AMUPol at concentrations relevant for DNP to lysed HEK293 cells and measured the total nitroxide signal intensity by EPR over time. The integrated EPR signal reports on total nitroxide concentration, which, in the absence of reduction, is double the AMUPol concentration. Because the two nitroxide moieties of AMUPol can be reduced independently, for accuracy, we report values as the total nitroxide concentration. AMUPol is a stable nitroxide bi-radical-containing compound (Figure 1). When AMUPol is suspended in buffer, the total nitroxide concentration of AMUPol did not change with time (data not shown). However, when AMUPol was added to mammalian cells, which were then lysed, the total nitroxide concentration of AMUPol decreased in a time-dependent manner. For all samples, the experimental starting radical concentration matched the target concentration and then decreased with increasing incubation times at room temperature. After 12 hours of room temperature incubation, no radical remained in the lysate samples with starting concentrations of 0.5 or 1 mM AMUPol while for samples with starting concentrations of AMUPol of 2 mM and higher, more than half of the total nitroxide radical remained (Figure 2A). Thus, the nitroxide radical reduction curves for the lysate samples were fit to a mono-exponential expression with an offset. These reduction curves were generally well-described by this expression (average R2 = 0.99, Table S1). For the samples with starting concentrations of AMUPol of 2 mM and greater, the total nitroxide reduction rate was directly proportional to the starting concentration; the total nitroxide concentration decreased at a rate of 0.18 ± 0.01% of the starting concentration per minute. However, for these samples, during the initial 30 minutes of the reduction reaction, the total nitroxide concentration is systematically underestimated by a few percent (Figure S1). Lysates consistently reduced more total nitroxide over this period than was predicted by the fit to the mono-exponential equation by an amount equivalent to ~0.1 mM, regardless of the starting nitroxide radical concentration. The small systemic deviation from a mono-exponential fit during the first 30 minutes accounted for the same quantity of radical, independent of starting concentration. The offsets, which represent the final concentration of nitroxide radical in the sample after the reduction reaction is complete, were concentration dependent and ranged from close to 0 at the two lowest concentrations, where all the radical was reduced by the lysates, to 31.8 mM, or 79% of the total nitroxide starting concentration of 40 mM nitroxide (e.g. 20 mM of AMUPol). The dependence of the offset on the initial AMUPol concentrations fit well to a saturation curve and indicated that the maximal amount of radical that lysates of HEK293 cells at room temperature can reduce is 9.6 mM (Figure S2). For the samples with starting concentrations of 0.5 mM and 1 mM AMUPol, the total nitroxide reduction rates were well described by a mono-exponential decay even during the first 30 minutes. The reduction rates (2.4% and 0.3% of the starting total nitroxide concentration per minute, respectively) were faster than the nitroxide reduction rate for reactions with higher concentrations of AMUPol, probably because the reduction rates for the quantity that is not captured by the mono-exponential equation and the quantity that is captured cannot be distinguished. Overall, apart from a small amount of AMUPol that is rapidly reduced in the first 30 minutes, the radical is reduced at a rate of 0.2% of the starting concentration per minute; increasing the concentration caused a proportional change in reduction rate, indicating first-order reduction kinetics with respect to AMUPol. The reduction of AMUPol by mammalian cell lysates is slow relative to the time scale of cellular sample preparation for DNP NMR.
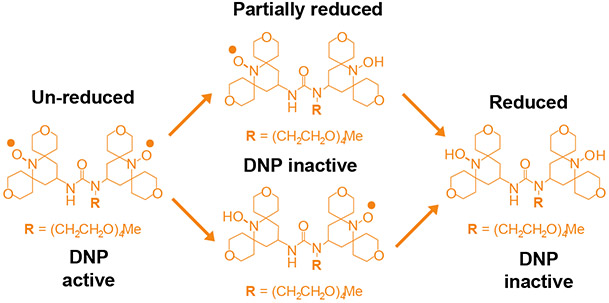
Figure 1:
Chemical structure of the nitroxide bi-radical, AMUPol. Upon exposure to reducing environments of cells, the bi-radical is reduced to mono-radical form or completely reduced form. Both the mono-radical and completely reduced forms are DNP-inactive.
Figure 2:
Deactivation kinetics of AMUPol in HEK293 cells. (A). Reduction kinetics of AMUPol in cellular lysates. The average AMUPol reduction rate for intact cells (black) is plotted for comparison. Error bars are the standard deviation of 5 replicates from part C. (B) EPR spectral changes over the time course of the reduction reaction for cell lysates with 1 mM AMUPol. (C) Reduction kinetics of AMUPol inside intact HEK293 cells. AMUPol was introduced by electroporation of cells in the presence of 20 mM AMUPol, followed by a 10-minute room temperature recovery period. After recovery, extracellular AMUPol was removed and EPR measurement began. Total nitroxide concentration determined by double integration of the EPR spectra is indicated by dots and lines indicate best fit to an exponential decay. Each biological replicate is plotted in a different color. The concentration AMUPol delivered to cells was about an order of magnitude lower than the concentration present in the buffer at the time of electroporation as expected. (D) EPR spectral changes over the time course of the AMUPol reduction reaction in intact cells. Displayed are the individual spectra from the time course depicted in blue in C.
The reduction of AMUPol in intact cells is faster than in lysates
To determine the rate of reduction of AMUPol inside intact cells, we electroporated AMUPol into HEK293 cells and then removed extracellular AMUPol before monitoring the decrease in the total nitroxide signal intensity over time using EPR21,37-39. Delivery of AMUPol to cells by electroporation in the presence of 20 mM AMUPol followed by a 10-minute recovery period and then removal of extracellular AMUPol resulted in a starting concentration of 1.9 ± 0.3 mM total nitroxide in the volume of the EPR sample (Figure 2C). Thus, as expected electroporation resulted in delivery of consistent quantities of AMUPol to cells21. Because the extracellular AMUPol was removed before measurements, and AMUPol-free buffer accounts for about half of the volume of pelleted cells, the total cellular nitroxide concentration at the earliest time point was ~10% of the concentration present in the buffer during electroporation. The electroporated cells were incubated at room temperature inside the EPR spectrometer and the nitroxide radical signal was monitored. After 12 hours of room temperature incubation, the radical signal was no longer detectable. The rate of radical reduction of AMUPol inside intact cells was described by a mono-exponential decay with a reduction rate of 0.24 ± 0.06% per minute (or 0.005 ± 0.001 mM/min) (Table S2). The reduction in intact cells is well described by a single exponential decay rate (R2 = 0.99) and the systematic small underestimation of AMUPol concentration observed during the initial 30 minutes of the reduction reaction for lysates is not observed for intact cells (Figure S1). The reduction rate for AMUPol in intact cells is faster than the reduction rate for AMUPol in cellular lysates (Figure 1A) and the reductive capacity of intact cells is greater than that of cellular lysates for samples with similar starting concentrations of AMUPol. This indicates that AMUPol reduction in intact cells is both faster and more efficient than in lysates.
The contribution of the DNP-silent monoradical form of AMUPol is more prominent in intact cells than in lysates
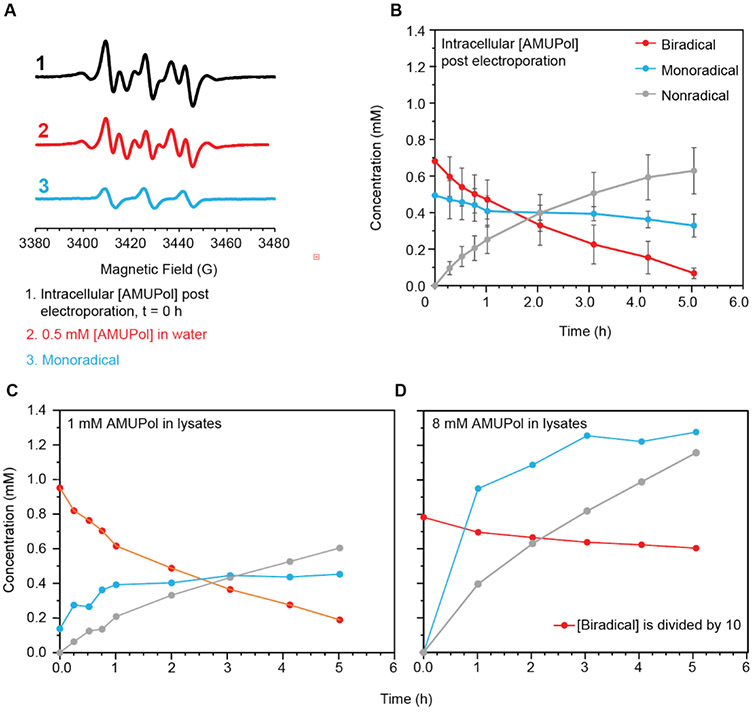
The EPR signal intensity reports on the radical concentration in the sample while the EPR line shape reports on interactions between the electron and the environment. The decrease in radical concentration for both intact and lysed cells was accompanied by changes in the EPR line shape. An isolated nitroxide radical has an EPR spectrum with three peaks. Interactions with other radicals in the sample can broaden or split these peaks. DNP-active AMUPol has two nitroxide radicals that are 11.6 angstroms (avg.) apart20,40. The EPR spectra at the initial time points for both intact and lysed cells had the characteristic splitting pattern of two nitroxide radicals in close proximity (Figure 2B, 2D) 41,42. At longer incubation times, not only did the EPR signal intensity decrease, but the EPR line shape for an isolated nitroxide radical became prominent. (e.g. Figure 2B and 2D, t = 5 hrs). The spectral changes are similar in both lysed and intact cells at similar time points when the starting AMUPol concentrations are similar. Because the monoradical form of AMUPol contributes to the EPR signal and will affect the NMR relaxation properties of nearby molecules, but is DNP-silent, we determined the individual contributions of the mono and bi radical forms of AMUPol to the EPR spectra. To do that, we scaled the spectrum of a pure sample of the biradical form of AMUPol to match the intensity of the most down-field and up-field features of the experimental EPR spectra and then subtracted the spectrum of the biradical form of AMUPol from the experimental spectrum. After subtraction of the biradical spectrum, the EPR spectrum had the characteristic features of an isolated nitroxide radical (Figure 3A). We determined the concentrations of the different forms of AMUPol individually by double integration. For lysates with 10 mM AMUPol, the biradical form of AMUPol was the major contributor to the EPR spectra throughout the reaction. The concentration of the biradical form of AMUPol decreased at a rate of 0.01 mM/min over the first hour. The monoradical form increased to a concentration of about 1 mM, but never became the dominant species (Figure 3D). For lysates with 1 mM AMUPol, the biradical form of AMUPol was the major contributor to the EPR spectra at the earliest time points (Figure 3C). The concentration of the biradical form of AMUPol decreased at a rate of 0.005 ± 0.001 mM/min within the first hour. The monoradical form of AMUPol was a minor contributor to the EPR spectra of AMUPol in lysates at the earliest time points but increased from 0.1 mM to 0.4 mM over the first hour of incubation and then remained relatively constant. In contrast, even at the earliest time point, the spectra of intact cells that had been electroporated with AMUPol had significant contributions from both the biradical and monoradical forms of AMUPol; more than half the AMUPol in the sample at the initial time point was in the mono-radical (DNP-silent) form (Figure 3B). The concentration of the biradical form of AMUPol decreased, while the concentration of the monoradical form of AMUPol remained at around 0.45 mM. While the starting monoradical concentration in the lysate sample mixed with 1 mM AMUPol was less than the starting monoradical concentration in the electroporated samples, the monoradical concentration in the lysate sample increased to the same steady-state concentration as in the intact cell sample. The difference in starting monoradical concentration between the two samples could be a result of a temporal difference in preparation times. Measurement of the lysate samples began approximately 5 minutes after the first exposure of AMUPol to cellular components. Lysates were prepared by mixing the cells with the desired concentration of AMUPol then performing 4 freeze-thaw cycles (1 min each) before measurement of the EPR signal. Measurement of the intact cell samples began approximately 15 minutes after first exposure of AMUPol to cellular components. After electroporation, intact cells were incubated for 10 minutes at room temperature to allow them to recover and then washed with PBS twice (2 min each time) before loading into the EPR spectrometer. However, taking this difference in preparation time into account does not explain the magnitude of the difference in the starting mono-radical concentration between the lysed and intact cells. Thus, the difference in starting mono-radical concentration between the lysed and intact cells must result from the reduced reductive capacity of lysates versus that of intact cells. The mono-radical form of AMUPol, which is not DNP-active, accounts for half of the AMUPol present inside intact cells at the earliest time points. This means that electroporation delivers closer to 10% of the amount of AMUPol present in the electroporation mixtures, in line with the existing literature of the delivery efficiency by electroporation. It also implies that the enhancements of in cell DNP NMR could double if AMUPol was protected from inactivation by the cellular constituents.
Figure 3:
Determination of the contributions of the biradical and mono-radical populations of AMUPol to the EPR spectra. A) The EPR spectra at the earliest time points for cells electroporated with AMUPol had a significant contribution from a mono-radical form of AMUPol. The subtraction of the EPR spectrum of a completely unreduced sample of AMUPol in the biradical form (red) from the EPR spectrum of cells electroporated with AMUPol (black) results in a spectrum characteristic of an isolated nitroxide radical (blue). (B) The concentration (in millimolar) of the bi-radical (red), mono-radical (blue) and completely reduced non-radical species (gray) changes with time for cells electroporated with AMUPol. Error bars indicate the standard deviation of six independent measurements. The concentration of the bi-radical (red), mono-radical (blue) and completely reduced non-radical species (gray) for lysates after addition of (C) 1 mM AMUPol and of (D) 8 mM AMUPol over time. In D, note that the concentration of the biradical species is divided by 10 to plot the concentration data on the same scale.
NEM pre-treatment prevents intracellular AMUPol reduction
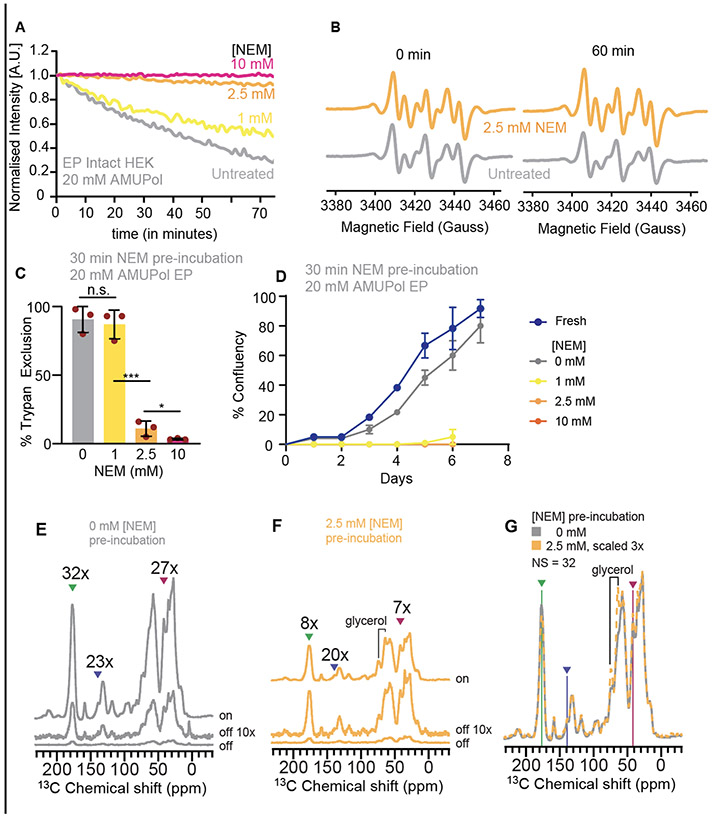
Pre-treatment of cells with oxidizing agent N-ethylmaleimide (NEM) neutralizes some of the reductive capacity of the cellular constituents. Pre-treatment of cellular lysates of bacteria slows or eliminates reduction of the nitroxide biradical, TOTAPOL 35. To determine if treatment of mammalian cells with NEM could slow or stop reduction of AMUPol in intact cells, we incubated intact mammalian cells with NEM for 30 minutes, washed the cells to remove the NEM, and then introduced AMUPol into the cells by electroporation and monitored the EPR signal. The total nitroxide concentration decreased with time for cells that were pre-incubated with 1 mM NEM while pretreatment of cells with 2.5 mM and 10 mM NEM largely prevented reduction of AMUPol (Fig 4 A and B). We determined the concentrations of the different forms of AMUPol individually by double integration. As before, for cells that were not pre-treated with NEM, about half of the AMUPol was in the monoradical form at the beginning of our measurement and the concentration of the mono-radical form remained constant over the first hour while the concentration of the biradical form decreased. For cells that were pre-treated with 1 mM NEM, 40% of the AMUPol was in the mono-radical form at the beginning and the concentration of the monoxide form remained constant while the concentration of the biradical form decreased. On the other hand, for cells pre-treated with 2.5 mM NEM, all the AMUPol was in the biradical form at the beginning of our measurement, although about 20% of the AMUPol was in the mono-radical form after an hour of room temperature incubation. However, there was no contribution to the EPR spectra from the mono-radical form of AMUPol for cells that were pre-treated incubated with 10 mM NEM, even at long incubation times.
Figure 4:
Effect of NEM pre-treatment on AMUPol reduction, cellular viability, and DNP performance. A) Pre-treatment of cells with NEM slows cellular reduction of AMUPol. HEK293 cells were incubated with varying concentrations of NEM in PBS which was removed before the introduction of AMUPol by electroporation. Total nitroxide concentration was monitored by EPR for cells. B) AMUPol EPR spectra for cells that were pre-treated with 2.5 mM NEM (orange) or PBS (gray) at the earliest time point and after an hour of room temperature incubation. C) Pre-treatment of cells with NEM compromises cellular membrane integrity. The trypan blue permeability decreased from 90 ± 11 % for cells incubated with 0 or 1 mM NEM to 11 ± 5% after incubation in 2.5 mM NEM. This change was significant (student’s t-test, * p < 0.05, *** p < 0.001). D) Cells were unable to grow after pre-treatment with NEM. E) The DNP performance for control cells was similar across the major biomass components. Peaks for the major biomass components are indicated by arrow heads for protein, nucleotide, and lipids (green, blue, and pink, respectively) for the microwaves on and microwaves off 13C CP spectra. F) Despite protection of the radical from reduction by 2.5 mM NEM pre-treatment, while DNP performance was similar across the major biomass components, the DNP enhancements were low. G) NEM pre-treatment does not result in differences in the 13C CP spectra for the cellular biomass.
Pre-treatment with NEM is toxic to cells
Although NEM prevented reduction of AMUPol by cellular components, NEM is a general oxidizing agent that will interact with many cellular factors, which may affect cellular integrity43. We assessed cellular integrity using two complementary approaches. We assessed membrane integrity using trypan blue exclusion and we assessed toxicity by quantitative regrowth assays 21,36. Pre-treatment with 1 mM NEM did not affect membrane integrity while pre-treatment with 2.5 mM and 10 mM NEM reduced membrane integrity by more than 80% (Fig 4C). Since trypan dye exclusion assay reports only on membrane permeability, which is imperfectly correlated with viability, we also assessed the ability of cells to grow after exposure to NEM. Despite the maintenance of membrane integrity after exposure to 1 mM NEM, after exposure to 1 mM NEM, cells were unable to propagate. Cells exposed to 2.5 mM and 10 mM of NEM were likewise unable to propagate (Figure 4D). Thus, because exposure to the concentration of NEM required to stop reduction of AMUPol compromised membrane integrity and eliminated cellular propagative ability, the use of NEM to improve AMUPol stability in biological environments is not appropriate in cases where cellular viability is required.
NEM reduces the polarization efficiency of AMUPol
Slowing the reduction of AMUPol by pre-treatment with NEM may be a useful approach to increase the sensitivity of DNP experiments if cellular viability is not a concern. Thus, we assessed the DNP performance of AMUPol in cells that had been pre-treated with 2.5 mM NEM and of cells that had not. After incubation in either 2.5 mM NEM or PBS for 30 minutes, the incubation buffer was removed, and cells were electroporated in the presence of AMUPol. Cells were allowed to recover for 10 minutes and then washed twice to remove extracellular AMUPol before cryoprotection and DNP analysis. We collected 13C cross-polarization (CP) spectra with and without microwave irradiation to determine the DNP enhancement for cells prepared with and without NEM pre-treatment. We determined DNP enhancements for peaks in the 13C CP spectra that are representative of the major biomass components of HEK293 cells; proteins, nucleotides and lipids21. We found that for cells pre-treated with NEM, the DNP enhancements for proteins, nucleotides and lipids were 8, 10 and 7, respectively while for cells that were not pre-treated with NEM, the DNP enhancements for proteins, nucleotides and lipids were 32, 24, and 27. Although the EPR measurements indicated that most of the AMUPol was in the biradical DNP-active form when the cells were pre-treated with NEM, the AMUPol in this sample did not enhance the cellular components as well as it did in untreated cells. The DNP enhancements for cells pre-treated with NEM were 70% lower than those for cells without NEM pre-treatment. We next assessed DNP build up times (TB,on) for these samples and found that the TB,on values for cells pre-treated with NEM for proteins, nucleotides and lipids were 3.4, 3.8 and 3.7 seconds, respectively while for cells that were not pre-treated with NEM, the DNP enhancements for proteins, nucleotides and lipids were 6.3, 6.4, and 7.1 seconds (Table S3)44. TB,on values of NMR signals are shorter when radical concentrations increase. The TB,on values for cells pre-treated with NEM were half of the TB,on value for cells without NEM pre-treatment, consistent with the observation that treatment with NEM prevents radical reduction. To assess the homogeneity of the AMUPol distribution throughout the sample, we used both the regression error of the fit of the TB,on data to a mono-exponential equation as well as the β-factor from a stretched exponential fit45. If the concentration distribution of AMUPol is heterogenous, there will be a mixture of underlying TB,on values in the same, which will increase the regression error and decrease the β-factor. For both samples, the regression errors were 1.6 ± 0.6% and the β-factors were 0.92 ±0.1 across all three representative biomass components, indicating a homogenous dispersion of AMUPol throughout the biomass, as expected when AMUPol is introduced into cells by electroporation. Thus, although pre-treatment with NEM preserves AMUPol in biradical form and delivery by electroporation results in homogenous dispersion through cells, pre-treatment of cellular biomass with NMR is not an effective approach to increase sensitivity of in-cell DNP experiments.
Discussion:
In this study, we investigate the reduction of AMUPol in both lysed and intact HEK293 cells. We find that nitroxide radicals are reduced with first order reduction kinetics by cell lysates at a rate of ~12% of the added nitroxide radical concentration per hour. Electroporation delivered a consistent amount of AMUPol to intact cells and that nitroxide radicals are reduced just slightly more rapidly (~15% per hour) by intact cells than by cell lysates. The two nitroxide radicals of AMUPol are reduced independently and this leads to considerable accumulation of the DNP-silent mono-radical form of AMUPol, particularly in preparations of intact cells where nearly half of the AMUPol is already reduced to the DNP silent mono-radical form at the earliest experimental time points. This confirms that the loss of DNP-active bi-radical form of AMUPol is faster than the nitroxide reduction rate. Finally, we investigate the effect of adding N-ethyl maleimide, a well-known inhibitor of thiol (-SH) group-based reduction of nitroxide bi-radical in cells, on AMUPol reduction, cellular viability, and DNP performance. Although pre-treatment of cells with NEM effectively inhibited the reduction of AMUPol, exposure to NEM compromised cellular viability and, surprisingly, did not improve DNP performance. Collectively, these results indicate that currently, the most effective strategy to obtain high DNP enhancements for DNP-assisted in-cell NMR is to minimize room temperature contact times with cellular constituents because pre-incubation with NEM was toxic to cells and results in reduced DNP performance despite radical stabilization.
Although addition of NEM effectively inhibits reduction of nitroxide radicals by cells, stabilization of the radicals does not translate to any improvement in DNP performance. Poor DNP performance of NEM-stabilized nitroxide radical based polarization agents in cellular samples appears to be independent of cell type (bacterial vs mammalian), polarization agent (TOTAPOL vs AMUPol) and choice of cryoprotectant (10% DMSO vs 15% glycerol). In the prior study of polarization stability in biological samples of NEM-stabilized TOTAPOL mixed with bacterial cells, the poor DNP performance was suggested to possibly result from poor permeability of the polarization agent into bacterial cells35. In this work, we eliminated permeability as a confounding factor by introduction of the polarization agent into the cells by electroporation, yet still find that the DNP performance in the presence of NEM is poor. DNP enhancements are dependent upon the concentration of polarization agent; high concentrations of polarization agents decrease DNP enhancements by bleaching the NMR signal. However, the experimental concentrations used in these experiments are well below the concentrations that result in bleaching in mammalian cells21. The values of TB,on in this work are consistent with low micromolar concentrations of AMUPol in cellular samples21 and analysis of the fits of TB,on do not indicate inhomogeneity in the distribution of the AMUPol throughout the sample. Our current hypothesis is that the decrease in DNP efficiency may be due to a product of the interaction between NEM and cellular constituents that compromises DNP performance, especially because a large proportion of NEM is removed by a wash step before DNP. Interestingly, in carefully matched standard samples, the inclusion of 10 mM NEM in a standard proline sample in DNP juice decreased the enhancements on both proline and glycerol by 15% with no changes of the TB,on, indicating that even the presence of NEM in a non-redox active sample compromises DNP performance. Overall, although the mechanism for the loss of DNP performance is frustratingly unclear, the empirical observation is unequivocal. While NEM prevents reduction of the radicals in polarization agents, exposure of the sample to NEM not only kills mammalian cells but also results in poor DNP performance. Thus, although pre-treatment with NEM preserves AMUPol in biradical form and delivery by electroporation results in homogenous dispersion through cells, pre-treatment of cellular biomass with NMR is not an effective approach to increase sensitivity of in-cell DNP experiments.
The observation that mono-radical forms of AMUPol accumulate inside cells provides insight into a confusing observation in prior work. In prior work, electroporation of HEK293 cells in the presence of 20 mM AMUPol resulted in the best DNP performance, although the amount of AMUPol that was actually delivered into the cells was not quantified 19. In this study, we found that electroporation consistently delivers approximately 10% of the concentration of the AMUPol in the buffer during electroporation into the cells and that at the earliest possible starting point for experimental measurements, about half of the AMUPol has already been reduced to the DNP-silent mono-radical form. Thus, in these intact cell samples, the concentration of DNP-competent AMUPol that resulted in the best DNP performance is ~2 mM. In contrast, in this work, we found that the starting concentration of AMUPol corresponded to the amount of AMUPol added to the sample and was all in the bi-radical form at the start of the experiment. In prior work, addition of 5 mM AMUPol to lysed cells followed by flash freezing resulted in best DNP performance with a DNP enhancement is 93, which is considerably higher than the maximal DNP enhancement of 60 in intact cells 21. This suggested that delivery of higher concentrations of DNP-active AMUPol to cells should result in higher DNP enhancements. However, although electroporation in the presence of 30 mM AMUPol results in the delivery of more radical to the cells -- the TB,on values decrease to 2.6 ± 1 seconds from 4.0 ± 0.6 seconds -- the DNP enhancements decrease to 53 ± 2 from 60 ± 6 21. The quantitative deconvolution of the EPR spectra undertaken in this work confirmed that the DNP-inactive mono-radical form of AMUPol accumulates to significant levels in intact cells, even at the earliest possible measurement point. Accumulation of the mono-radical form of AMUPol can compromise the DNP efficiency due to paramagnetic relaxation effects46-49. Therefore, the reduction of the biradical form of AMUPol by cellular environments to the mono-radical form limits the sensitivity of in-cell DNP NMR; the experimental sensitivity of in-cell DNP NMR is not limited by radical delivery but rather by the reduction of the polarization agent by cellular environments that results in the accumulation of DNP-inactive mono-radical species that compromise the DNP performance.
Because even relatively slow reduction rates of polarization agents that rely upon the cross-effect can limit the maximal enhancement through the build-up of DNP-inactive mono-radical intermediate forms, development of bio-resistant radicals could improve experimental sensitivity in complex biological systems. Indeed, in this example, improved bio-stability could potentially double the DNP enhancement in the samples discussed here, a change that would translate into either detection of biomolecules with half the abundance in the same experimental time or more facile collection of spectra with lower efficiency (such as heteronuclear experiments) or higher dimensionality. Moreover, development of bio-resistant polarization agents could result in dramatic increases in the sensitivity of DNP-assisted in-cell NMR, even if the modifications that confer biostability result in polarization agents that are less efficient in purified settings. Development of bio-resistant polarization agents could lengthen the sample manipulation time to accommodate studies that seek to target the polarization agent to specific subcellular compartments or investigate time-dependent processes particularly for radicals that are covalently attached to a biomolecule of interest9,50-56. Tri-radical based polarization agents could be more effective polarization agents in cellular environments since reduction of one nitroxide will not immediately kill the cross-effect mechanism57-59. Collectively, the work here indicates that the most effective strategy to obtain high DNP enhancements for DNP-assisted in-cell NMR is to minimize room temperature contact times with cellular constituents and suggests the development of bio-resistant polarization agents for DNP could considerably increase the sensitivity of DNP-assisted in-cell NMR experiments.
Materials and Methods
Sample Preparation
HEK293 cells were grown in 10 cm culture dish in complete media, DMEM (Gibco, Cat# 10569) with 10% FBS and 1% Pen-Strep at 37 °C humidified incubation containing 5% CO2. Upon reaching 80%-90% confluency, cells were washed with 10 mL of 1X PBS (pH 7.4, Gibco, Cat#10010) and un-attached using 2 mL of Tryp-LE (Gibco, Cat#12605). Trypsin was inactivated by addition of 8 mL of complete media. Cell suspension was centrifuged at 233 x g for 5 min to pellet the cells. Trypsin containing media was removed and the cell pellet was washed 1X with PBS.
For electroporation, 50 μL of cell pellet was mixed with 50 μL electroporation buffer (Amaxa SF cell line 4D nucleofactor X kit L, Lonza, Cat# V4XC-2024) containing 20 mM AMUPol (CortecNet). Cells were mixed thoroughly and electroporated using Lonza electroporator (Lonza 4D-Nucleofactor) using CM130 pulse. After electroporation, cells were allowed to recover for 10 min at room temperature and washed twice with 1x PBS to remove the extracellular AMUPol. Then, cells were mixed with DNP buffer (perdeuterated PBS with 15% (v/v) d8-glycerol) and centrifuged at 233 x g for 1-2 min to pellet the cells. The supernatant was removed, and the cell pellet was transferred into 0.94-mm-inner diameter EPR capillary tube (VWR) and sealed. Samples contained ~10 million cells in the 27.5 μL volume of the rotor.
For lysates, 50 μL of cell pellet was mixed with 50 μL DNP buffer perdeuterated PBS with 15% (v/v) d8-glycerol and AMUPol. Cells were lysed by four flash freeze-thaw cycles. Lysed cells were transferred into EPR capillary tube and sealed. Lysates contained half the number of cells per volume relative to intact cell samples.
For NEM treated cells, harvested cells were washed 1X with PBS to remove residual media. 50 μL of cells were suspended into 50 μL of NEM (Sigma, Cat# 04259) dissolved in PBS and incubated at room temperature for 30 minutes. 50 μL of cells suspended in 50 μL of 1x PBS were used as untreated control. After incubation, cells were washed with PBS once and then mixed with 50 μL of electroporation buffer containing 40 mM AMUPol so that the final concentration of AMUPol in the cell suspension is 20 mM. Cells were electroporated as described, allowed to recover for 10 minutes at room temperature, and washed twice with 1x PBS to remove extracellular AMUPol. Cells were then transferred into EPR capillary tube and sealed for EPR experiments.
Cell viability assay using Trypan Blue dye
10 μL of cells suspended in PBS were mixed with 10 μL of 2X Trypan Dye. Total cell viability was measured using automated cell counting machine (Countess II FL, Life Technologies).
Cellular Growth Assay
0.2 million cells were plated in 10 cm culture dish containing 10 mL of complete media (DMEM with 10% FBS and 1% Pen-Strep). Cells were allowed to attach overnight. Media was changed after cells had adhered and growth was monitored for 7-8 days and images were taken. Overall confluency as assess by visual inspection of three fields of view from the tissue culture plate was plotted against time (in days) for determining growth.
DNP Sample preparation
Samples for DNP analysis were prepared as described 21. Briefly, 50 μL of cells (with or without NEM pre-treatment) were mixed with 50 μL of 40 mM AMUPol in electroporation buffer. Cells were electroporated using the pulse program CM130 in Lonza electroporator, allowed to recover for 10 minutes at room temperature and then washed twice with PBS to remove extracellular AMUPol. Next, cells were suspended 50 μL of perdeuterated PBS (85% D2O: 15% H2O) and 18 μL of d8-glycerol for a final percentage of cryoprotectant in the matrix of 15% (v/v). The cell suspension was centrifuged at 233 x g for 2 minutes into a 3.2 mm sapphire rotor and the supernatant was removed. A silicone plug was inserted into the rotor and capped. The rotor was frozen at a rate of in 1 °C/min cooler to −80 °C before transferring into liquid nitrogen storage. Frozen samples cryogenically transferred into the NMR spectrometer 36.
NMR spectroscopy
Rotors were transferred in liquid nitrogen directly into the NMR probe that was pre-equilibrated at 100 K15. All dynamic nuclear polarization magic angle spinning nuclear magnetic resonance (DNP MAS NMR) experiments were performed on a 600 MHz Bruker Ascend DNP NMR spectrometer/7.2 T Cryogen-free gyrotron magnet (Bruker), equipped with a 1H, 13C, 15N triple-resonance, 3.2 mm low temperature (LT) DNP MAS NMR Bruker probe (600 MHz). The sample temperature was 104 K and the MAS frequency was 12 kHz. The DNP enhancement for the instrumentation set-up for a standard sample of 1.5 mg of uniformly 13C, 15N labeled proline (Isotech) suspended in 25 mg of 60:30:10 d8-glycerol: D2O:H2O containing 10 mM AMUPol was between 130 and 140 and a TB,on of 4.6 s. For 13C cross-polarization (CP) MAS experiments, the 13C radio frequency (RF) amplitude was linearly swept from 75 kHz to 37.5 kHz with an average of 56.25 kHz. 1H RF amplitude was 68~72 kHz for CP, 83 kHz for 90-degree pulse, and 85 kHz for 1H TPPM decoupling with phase alternation of ± 15° during acquisition of 13C signal. The DNP enhancements were determined by comparing 1D 13C CP spectra collected with and without microwaves irradiation using a recycle delay of 10 s. For TB,on measurements, recycle delays ranged from 0.1 s to 300 s. To determine the TB,on, the dependence of the recycle delay using saturation recovery on both 13C peak intensity or volume was fit to the mono-exponential equation and the stretched-exponential equation , respectively. For 1D experiments, the data were processed using NMRPipe60. The real part of the processed spectrum was exported using pipe2txt.tcl command. Peaks were integrated, and the time constants were obtained by least-squares fitting with a single-exponential function. DNP enhancements were determined by peak intensity.
EPR experiments
CW EPR data was acquired using a Bruker EMXnano X-band EPR spectrometer at a frequency of 9.6 GHz with a center field of 3424 G and sweep width of 172 G. Unless otherwise indicated, the following instrumental parameters were used: modulation frequency, 100 kHz; modulation amplitude, 1.00 G; conversion time, 5 ms; microwave power, 0.32 mW; scan time, 8.6 s. All samples were loaded into 50 μL microcapillary tubes, and their EPR spectra were recorded at room temperature.
The concentrations of the biradical and mono-radical species of AMUPol were obtained using Bruker’s Xenon software to subtract the spectrum of an AMUPol standard from that of the bi-radical/mono-radical mixture at different time points. For the samples of intact cells (electroporated with AMUPol) and the lysate mixed with 1 mM AMUPol, a standard of 0.5 mM AMUPol in water was used for subtraction. For the lysate sample mixed with 10 mM AMUPol, a standard of 5 mM AMUPol was used for subtraction. The spectra of the standards were shifted, and the gain was adjusted during subtraction. The total nitroxide concentration and the concentration of mono-radical were determined by double integration and the concentration of bi-radical and the totally reduced forms were determined by subtraction.
The nitroxide radical reduction curves for electroporated samples and lysates were fit to a mono-exponential model with an offset (y=a*e^(−k*x)+c) using a least squares regression algorithm in MATLAB. In this equation, y is either the total nitroxide concentration in mM or the normalized fractional signal intensity, x is the time, and k is the rate constant. The terms a and c both describe the offset and were fit as independent parameters. The sum of a and c equaled one in our fits.
Supplementary Material
Highlights.
AMUPol is reduced at a modest rate by intact mammalian cells and lysates
In cells, half the AMUPol is in the DNP-silent monoradical form at earliest timepoint
Electroporation delivers consistent quantities of AMUPol to intact cells
Reduction is prevented by 2.5mM N-ethlymaleimide but viability is compromised
No improvement in DNP enhancements post reduction inhibition with NEM
Acknowledgements:
R.G. is supported by a fellowship from the O'Donnell Brain Institute Neural Science Training Program. This work was supported by grants from the National Institute of Health (NS-111236), the Welch Foundation (I-1923-20200401, the Lupe Murchison Foundation and the Kinship Foundation (Searle Scholars Program) to K.K.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Competing Financial interests:
The authors declare no competing financial interests.
References
- (1).Narasimhan S; Scherpe S; Lucini Paioni A; van der Zwan J; Folkers GE; Ovaa H; Baldus M DNP-Supported Solid-State NMR Spectroscopy of Proteins Inside Mammalian Cells. Angew Chem Int Ed Engl 2019, 58 (37), 12969–12973. 10.1002/anie.201903246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ackermann BE; Debelouchina GT Heterochromatin Protein HP1α Gelation Dynamics Revealed by Solid-State NMR Spectroscopy. Angew Chem Int Ed Engl 2019, 58 (19), 6300–6305. 10.1002/anie.201901141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Frederick KK; Michaelis VK; Corzilius B; Ong T-C; Jacavone AC; Griffin RG; Lindquist S Sensitivity-Enhanced NMR Reveals Alterations in Protein Structure by Cellular Milieus. Cell 2015, 163 (3), 620–628. 10.1016/j.cell.2015.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Schlagnitweit J; Friebe Sandoz S; Jaworski A; Guzzetti I; Aussenac F; Carbajo RJ; Chiarparin E; Pell AJ; Petzold K Observing an Antisense Drug Complex in Intact Human Cells by In-Cell NMR Spectroscopy. Chembiochem 2019, 20 (19), 2474–2478. 10.1002/cbic.201900297. [DOI] [PubMed] [Google Scholar]
- (5).Viennet T; Viegas A; Kuepper A; Arens S; Gelev V; Petrov O; Grossmann TN; Heise H; Etzkorn M Selective Protein Hyperpolarization in Cell Lysates Using Targeted Dynamic Nuclear Polarization. Angew Chem Int Ed Engl 2016, 55 (36), 10746–10750. 10.1002/anie.201603205. [DOI] [PubMed] [Google Scholar]
- (6).Etzkorn M; Martell S; Andronesi OC; Seidel K; Engelhard M; Baldus M Secondary Structure, Dynamics, and Topology of a Seven-Helix Receptor in Native Membranes, Studied by Solid-State NMR Spectroscopy. Angew Chem Int Ed Engl 2007, 46 (3), 459–462. 10.1002/anie.200602139. [DOI] [PubMed] [Google Scholar]
- (7).Daube D; Vogel M; Suess B; Corzilius B Dynamic Nuclear Polarization on a Hybridized Hammerhead Ribozyme: An Explorative Study of RNA Folding and Direct DNP with a Paramagnetic Metal Ion Cofactor. Solid State Nucl Magn Reson 2019, 101, 21–30. 10.1016/j.ssnmr.2019.04.005. [DOI] [PubMed] [Google Scholar]
- (8).Narasimhan S; Pinto C; Lucini Paioni A; van der Zwan J; Folkers GE; Baldus M Characterizing Proteins in a Native Bacterial Environment Using Solid-State NMR Spectroscopy. Nat Protoc 2021, 16 (2), 893–918. 10.1038/s41596-020-00439-4. [DOI] [PubMed] [Google Scholar]
- (9).Yang Y; Yang F; Gong Y-J; Chen J-L; Goldfarb D; Su X-C A Reactive, Rigid GdIII Labeling Tag for In-Cell EPR Distance Measurements in Proteins. Angew Chem Int Ed Engl 2017, 56 (11), 2914–2918. 10.1002/anie.201611051. [DOI] [PubMed] [Google Scholar]
- (10).Akbey Ü; Oschkinat H Structural Biology Applications of Solid State MAS DNP NMR. J Magn Reson 2016, 269, 213–224. 10.1016/j.jmr.2016.04.003. [DOI] [PubMed] [Google Scholar]
- (11).Costello WN; Xiao Y; Frederick KK DNP-Assisted NMR Investigation of Proteins at Endogenous Levels in Cellular Milieu. Methods Enzymol 2019, 615, 373–406. 10.1016/bs.mie.2018.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Renault M; Tommassen-van Boxtel R; Bos MP; Post JA; Tommassen J; Baldus M Cellular Solid-State Nuclear Magnetic Resonance Spectroscopy. Proc Natl Acad Sci U S A 2012, 109 (13), 4863–4868. 10.1073/pnas.1116478109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hänsel R; Luh LM; Corbeski I; Trantirek L; Dötsch V In-Cell NMR and EPR Spectroscopy of Biomacromolecules. Angew Chem Int Ed Engl 2014, 53 (39), 10300–10314. 10.1002/anie.201311320. [DOI] [PubMed] [Google Scholar]
- (14).Reckel S; Lopez JJ; Löhr F; Glaubitz C; Dötsch V In-Cell Solid-State NMR as a Tool to Study Proteins in Large Complexes. Chembiochem 2012, 13 (4), 534–537. 10.1002/cbic.201100721. [DOI] [PubMed] [Google Scholar]
- (15).Kuppusamy P; Li H; Ilangovan G; Cardounel AJ; Zweier JL; Yamada K; Krishna MC; Mitchell JB Noninvasive Imaging of Tumor Redox Status and Its Modification by Tissue Glutathione Levels. Cancer Res 2002, 62 (1), 307–312. [PubMed] [Google Scholar]
- (16).Albert BJ; Gao C; Sesti EL; Saliba EP; Alaniva N; Scott FJ; Sigurdsson ST; Barnes AB Dynamic Nuclear Polarization Nuclear Magnetic Resonance in Human Cells Using Fluorescent Polarizing Agents. Biochemistry 2018, 57 (31), 4741–4746. 10.1021/acs.biochem.8b00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hu K-N; Yu H; Swager TM; Griffin RG Dynamic Nuclear Polarization with Biradicals. J Am Chem Soc 2004, 126 (35), 10844–10845. 10.1021/ja039749a. [DOI] [PubMed] [Google Scholar]
- (18).Can TV; Ni QZ; Griffin RG Mechanisms of Dynamic Nuclear Polarization in Insulating Solids. J Magn Reson 2015, 253, 23–35. 10.1016/j.jmr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Hu K-N Polarizing Agents and Mechanisms for High-Field Dynamic Nuclear Polarization of Frozen Dielectric Solids. Solid State Nucl Magn Reson 2011, 40 (2), 31–41. 10.1016/j.ssnmr.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Sauvée C; Rosay M; Casano G; Aussenac F; Weber RT; Ouari O; Tordo P Highly Efficient, Water-Soluble Polarizing Agents for Dynamic Nuclear Polarization at High Frequency. Angew Chem Int Ed Engl 2013, 52 (41), 10858–10861. 10.1002/anie.201304657. [DOI] [PubMed] [Google Scholar]
- (21).Ghosh R; Xiao Y; Kragelj J; Frederick KK In-Cell Sensitivity-Enhanced NMR of Intact Viable Mammalian Cells. J Am Chem Soc 2021, 143 (44), 18454–18466. 10.1021/jacs.1c06680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Mentink-Vigier F; Barra A-L; van Tol J; Hediger S; Lee D; De Paëpe G De Novo Prediction of Cross-Effect Efficiency for Magic Angle Spinning Dynamic Nuclear Polarization. Phys Chem Chem Phys 2019, 21 (4), 2166–2176. 10.1039/c8cp06819d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Mentink-Vigier F Optimizing Nitroxide Biradicals for Cross-Effect MAS-DNP: The Role of g-Tensors’ Distance. Phys Chem Chem Phys 2020, 22 (6), 3643–3652. 10.1039/c9cp06201g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Azarkh M; Okie O; Eyring P; Dietrich DR; Drescher M Evaluation of Spin Labels for In-Cell EPR by Analysis of Nitroxide Reduction in Cell Extract of Xenopus Laevis Oocytes. J Magn Reson 2011, 212 (2), 450–454. 10.1016/j.jmr.2011.07.014. [DOI] [PubMed] [Google Scholar]
- (25).Belkin S; Mehlhorn RJ; Hideg K; Hankovsky O; Packer L Reduction and Destruction Rates of Nitroxide Spin Probes. Arch Biochem Biophys 1987, 256 (1), 232–243. 10.1016/0003-9861(87)90441-3. [DOI] [PubMed] [Google Scholar]
- (26).Saphier O; Silberstein T; Shames AI; Likhtenshtein GI; Maimon E; Mankuta D; Mazor M; Katz M; Meyerstein D; Meyerstein N The Reduction of a Nitroxide Spin Label as a Probe of Human Blood Antioxidant Properties. Free Radic Res 2003, 37 (3), 301–308. 10.1080/1071576021000050410. [DOI] [PubMed] [Google Scholar]
- (27).Schmidt MJ; Fedoseev A; Bücker D; Borbas J; Peter C; Drescher M; Summerer D EPR Distance Measurements in Native Proteins with Genetically Encoded Spin Labels. ACS Chem Biol 2015, 10 (12), 2764–2771. 10.1021/acschembio.5b00512. [DOI] [PubMed] [Google Scholar]
- (28).Kveder M; Sentjurc M; Schara M Spin Probe Reduction in Cells and Tissues. Magn Reson Med 1988, 8 (3), 241–247. 10.1002/mrm.1910080302. [DOI] [PubMed] [Google Scholar]
- (29).Bobko AA; Kirilyuk IA; Grigor’ev IA; Zweier JL; Khramtsov VV Reversible Reduction of Nitroxides to Hydroxylamines: Roles for Ascorbate and Glutathione. Free Radic Biol Med 2007, 42 (3), 404–412. 10.1016/j.freeradbiomed.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Khramtsov VV; Yelinova VI; Weiner LM; Berezina TA; Martin VV; Volodarsky LB Quantitative Determination of SH Groups in Low- and High-Molecular-Weight Compounds by an Electron Spin Resonance Method. Anal Biochem 1989, 182 (1), 58–63. 10.1016/0003-2697(89)90718-5. [DOI] [PubMed] [Google Scholar]
- (31).Głebska J; Skolimowski J; Kudzin Z; Gwoździński K; Grzelak A; Bartosz G Pro-Oxidative Activity of Nitroxides in Their Reactions with Glutathione. Free Radic Biol Med 2003, 35 (3), 310–316. 10.1016/s0891-5849(03)00306-x. [DOI] [PubMed] [Google Scholar]
- (32).Kasazaki K; Yasukawa K; Sano H; Utsumi H Non-Invasive Analysis of Reactive Oxygen Species Generated in NH4OH-Induced Gastric Lesions of Rats Using a 300 MHz in Vivo ESR Technique. Free Radic Res 2003, 37 (7), 757–766. 10.1080/1071576031000103069. [DOI] [PubMed] [Google Scholar]
- (33).Keana JF; Pou S; Rosen GM Nitroxides as Potential Contrast Enhancing Agents for MRI Application: Influence of Structure on the Rate of Reduction by Rat Hepatocytes, Whole Liver Homogenate, Subcellular Fractions, and Ascorbate. Magn Reson Med 1987, 5 (6), 525–536. 10.1002/mrm.1910050603. [DOI] [PubMed] [Google Scholar]
- (34).Linden AH; Lange S; Franks WT; Akbey U; Specker E; van Rossum B-J; Oschkinat H Neurotoxin II Bound to Acetylcholine Receptors in Native Membranes Studied by Dynamic Nuclear Polarization NMR. J Am Chem Soc 2011, 133 (48), 19266–19269. 10.1021/ja206999c. [DOI] [PubMed] [Google Scholar]
- (35).McCoy KM; Rogawski R; Stovicek O; McDermott AE Stability of Nitroxide Biradical TOTAPOL in Biological Samples. J Magn Reson 2019, 303, 115–120. 10.1016/j.jmr.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Ghosh R; Kragelj J; Xiao Y; Frederick KK Cryogenic Sample Loading into a Magic Angle Spinning Nuclear Magnetic Resonance Spectrometer That Preserves Cellular Viability. J Vis Exp 2020, No. 163. 10.3791/61733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Sözer EB; Pocetti CF; Vernier PT Transport of Charged Small Molecules after Electropermeabilization - Drift and Diffusion. BMC Biophys 2018, 11, 4. 10.1186/s13628-018-0044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Zu Y; Huang S; Lu Y; Liu X; Wang S Size Specific Transfection to Mammalian Cells by Micropillar Array Electroporation. Sci Rep 2016, 6, 38661. 10.1038/srep38661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).DiFranco M; Quinonez M; Capote J; Vergara J DNA Transfection of Mammalian Skeletal Muscles Using in Vivo Electroporation. J Vis Exp 2009, No. 32, 1520. 10.3791/1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Lund A; Casano G; Menzildjian G; Kaushik M; Stevanato G; Yulikov M; Jabbour R; Wisser D; Renom-Carrasco M; Thieuleux C; Bernada F; Karoui H; Siri D; Rosay M; Sergeyev IV; Gajan D; Lelli M; Emsley L; Ouari O; Lesage A TinyPols: A Family of Water-Soluble Binitroxides Tailored for Dynamic Nuclear Polarization Enhanced NMR Spectroscopy at 18.8 and 21.1 T. Chem Sci 2020, 11 (10), 2810–2818. 10.1039/c9sc05384k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Eaton SS; Woodcock LB; Eaton GR Continuous Wave Electron Paramagnetic Resonance of Nitroxide Biradicals in Fluid Solution. Concepts Magn Reson Part A Bridg Educ Res 2018, 47A (2), e21426. 10.1002/cmr.a.21426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Torricella F; Pierro A; Mileo E; Belle V; Bonucci A Nitroxide Spin Labels and EPR Spectroscopy: A Powerful Association for Protein Dynamics Studies. Biochim Biophys Acta Proteins Proteom 2021, 1869 (7), 140653. 10.1016/j.bbapap.2021.140653. [DOI] [PubMed] [Google Scholar]
- (43).Fuchs J; Mehlhorn RJ; Packer L Free Radical Reduction Mechanisms in Mouse Epidermis Skin Homogenates. J Invest Dermatol 1989, 93 (5), 633–640. 10.1111/1523-1747.ep12319780. [DOI] [PubMed] [Google Scholar]
- (44).Pinon AC; Schlagnitweit J; Berruyer P; Rossini AJ; Lelli M; Socie E; Tang M; Pham T; Lesage A; Schantz S; Emsley L Measuring Nano- to Microstructures from Relayed Dynamic Nuclear Polarization NMR. J. Phys. Chem. C 2017, 121 (29), 15993–16005. https://doi.org/June 21, 2017. [Google Scholar]
- (45).Rankin AGM; Trébosc J; Pourpoint F; Amoureux J-P; Lafon O Recent Developments in MAS DNP-NMR of Materials. Solid State Nucl Magn Reson 2019, 101, 116–143. 10.1016/j.ssnmr.2019.05.009. [DOI] [PubMed] [Google Scholar]
- (46).Tran NT; Mentink-Vigier F; Long JR Dynamic Nuclear Polarization of Biomembrane Assemblies. Biomolecules 2020, 10 (9), E1246. 10.3390/biom10091246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Rogawski R; Sergeyev IV; Li Y; Ottaviani MF; Cornish V; McDermott AE Dynamic Nuclear Polarization Signal Enhancement with High-Affinity Biradical Tags. J Phys Chem B 2017, 121 (6), 1169–1175. 10.1021/acs.jpcb.6b09021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Jaroniec CP Solid-State Nuclear Magnetic Resonance Structural Studies of Proteins Using Paramagnetic Probes. Solid State Nucl Magn Reson 2012, 43–44, 1–13. 10.1016/j.ssnmr.2012.02.007. [DOI] [PubMed] [Google Scholar]
- (49).Liao SY; Lee M; Wang T; Sergeyev IV; Hong M Efficient DNP NMR of Membrane Proteins: Sample Preparation Protocols, Sensitivity, and Radical Location. J Biomol NMR 2016, 64 (3), 223–237. 10.1007/s10858-016-0023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Qi M; Gross A; Jeschke G; Godt A; Drescher M Gd(III)-PyMTA Label Is Suitable for in-Cell EPR. J Am Chem Soc 2014, 136 (43), 15366–15378. 10.1021/ja508274d. [DOI] [PubMed] [Google Scholar]
- (51).Yang Y; Yang F; Li X-Y; Su X-C; Goldfarb D In-Cell EPR Distance Measurements on Ubiquitin Labeled with a Rigid PyMTA-Gd(III) Tag. J Phys Chem B 2019, 123 (5), 1050–1059. 10.1021/acs.jpcb.8b11442. [DOI] [PubMed] [Google Scholar]
- (52).Karthikeyan G; Bonucci A; Casano G; Gerbaud G; Abel S; Thomé V; Kodjabachian L; Magalon A; Guigliarelli B; Belle V; Ouari O; Mileo E A Bioresistant Nitroxide Spin Label for In-Cell EPR Spectroscopy: In Vitro and In Oocytes Protein Structural Dynamics Studies. Angew Chem Int Ed Engl 2018, 57 (5), 1366–1370. 10.1002/anie.201710184. [DOI] [PubMed] [Google Scholar]
- (53).Gauto D; Dakhlaoui O; Marin-Montesinos I; Hediger S; De Paëpe G Targeted DNP for Biomolecular Solid-State NMR. Chem Sci 2021, 12 (18), 6223–6237. 10.1039/d0sc06959k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Fleck N; Heubach CA; Hett T; Haege FR; Bawol PP; Baltruschat H; Schiemann O SLIM: A Short-Linked, Highly Redox-Stable Trityl Label for High-Sensitivity In-Cell EPR Distance Measurements. Angew Chem Int Ed Engl 2020, 59 (24), 9767–9772. 10.1002/anie.202004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Juliusson HY; Sigurdsson ST Reduction Resistant and Rigid Nitroxide Spin-Labels for DNA and RNA. J Org Chem 2020, 85 (6), 4036–4046. 10.1021/acs.joc.9b02988. [DOI] [PubMed] [Google Scholar]
- (56).Jagtap AP; Krstic I; Kunjir NC; Hänsel R; Prisner TF; Sigurdsson ST Sterically Shielded Spin Labels for In-Cell EPR Spectroscopy: Analysis of Stability in Reducing Environment. Free Radic Res 2015, 49 (1), 78–85. 10.3109/10715762.2014.979409. [DOI] [PubMed] [Google Scholar]
- (57).Yau W-M; Jeon J; Tycko R Succinyl-DOTOPA: An Effective Triradical Dopant for Low-Temperature Dynamic Nuclear Polarization with High Solubility in Aqueous Solvent Mixtures at Neutral PH. J Magn Reson 2020, 311, 106672. 10.1016/j.jmr.2019.106672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Li Y; Chaklashiya R; Takahashi H; Kawahara Y; Tagami K; Tobar C; Han S Solid-State MAS NMR at Ultra Low Temperature of Hydrated Alanine Doped with DNP Radicals. J Magn Reson 2021, 333, 107090. 10.1016/j.jmr.2021.107090. [DOI] [PubMed] [Google Scholar]
- (59).Mentink-Vigier F; Mathies G; Liu Y; Barra A-L; Caporini MA; Lee D; Hediger S; G Griffin R; De Paëpe G Efficient Cross-Effect Dynamic Nuclear Polarization without Depolarization in High-Resolution MAS NMR. Chem Sci 2017, 8 (12), 8150–8163. 10.1039/c7sc02199b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Delaglio F; Grzesiek S; Vuister GW; Zhu G; Pfeifer J; Bax A NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J Biomol NMR 1995, 6 (3), 277–293. 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.