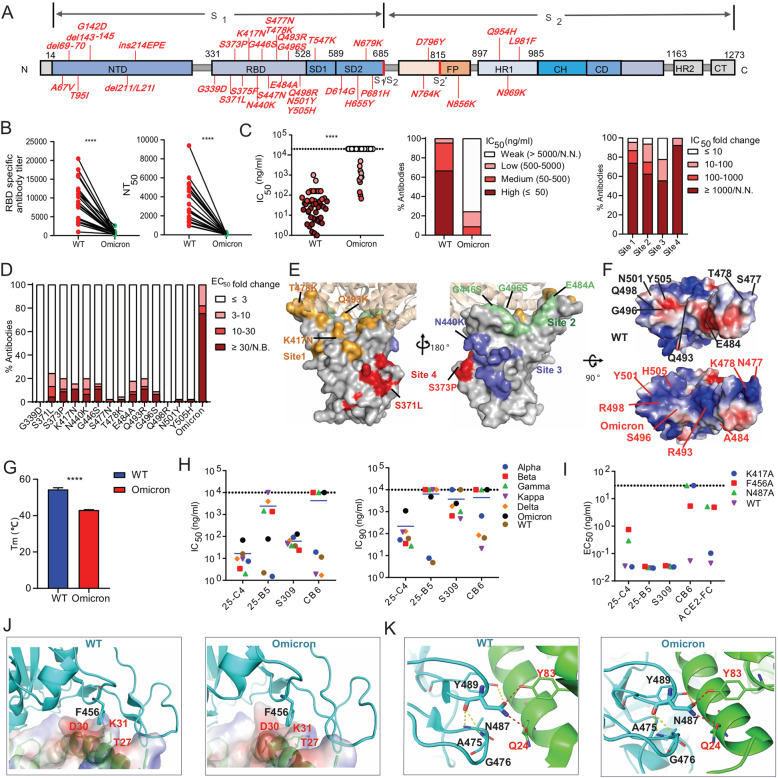
The newly emerged Omicron (B.1.1.529) variant of SARS-CoV-2 is quickly overtaking the Delta variant and becoming the dominant strain around the world due to its enhanced transmissibility and high immune escape potential [1, 2]. Compared to the Wuhan strain, the Omicron variant carries 37 spike mutations, of which 15 are within the receptor-binding domain (RBD) (Fig. 1A). A high mutation rate is associated with remarkable resistance to current vaccines and RBD-specific antibody therapeutics [2–4]. It is urgent to comprehensively understand the impacts of these RBD mutations on the neutralization capability of RBD-specific neutralizing antibodies (NAbs) targeting diverse epitopes and search for highly potent and broadly reactive NAbs against SARS-CoV-2 variants, including the Omicron strain. This information will be critical for understanding the evolutionary mechanisms that govern SARS-CoV-2 immune escape and for guiding vaccine design.
Fig. 1.
Immune evasion analysis of the Omicron variant and identification of a cross-variant neutralizing antibody targeting conserved RBD amino acids. A The key spike mutations present in the Omicron variant (B.1.1.529) are highlighted. B The anti-RBD antibody titer and neutralizing activity of the plasma from COVID-19 convalescent individuals (n = 21) against Wuhan-Hu-1 (wild-type, WT) and the Omicron variant. RBD-specific IgG antibody (Ab) titers were calculated as the endpoint dilution that remained positively detectable for the wild-type and Omicron RBD. Fifty percent neutralizing antibody titers (NT50) were calculated against wild-type and Omicron pseudoviruses. C Neutralization capability of Omicron pseudovirus by a panel of 45 RBD-specific mAbs. The left panel shows the IC50 values of the tested antibodies against wild-type and Omicron pseudoviruses. The middle panel shows the percentage of antibodies with the indicated neutralization activities. The right panel shows the percentage of different degrees of IC50 fold change for antibodies targeting four antigenic sites. The IC50 fold change is the ratio of the IC50 of antibodies against Omicron compared with wild-type pseudovirus. D The percentage of different degrees of antibody binding EC50 fold change of the Omicron RBD or single-amino acid substitutions present in Omicron. The EC50 fold change was calculated as the EC50 of the mutant RBD /the EC50 of the wild-type RBD. E The key immune escape substitutions found in Omicron are highlighted at four distinct antigenic sites in the RBD region. The color-coding scheme: site 1 (orange), site 2 (green), site 3 (slate blue), site 4 (red); ACE2 (wheat). Left and right are shown from different angles. F Electrostatic potential surface representation of the RBD from wild-type and Omicron. Electrostatic surface potentials are colored red and blue for negative and positive charges, respectively; white represents neutral residues. The wild-type positions are shown in black font and Omicron in red. G The melting temperature (Tm) of the recombinant RBD of wild-type and Omicron measured by nano differential scanning fluorimetry (nanoDSF). H IC50 and IC90 values of four RBD-specific NAbs against different SARS-CoV-2 VOC pseudoviruses. I Binding EC50 values of the tested NAbs or ACE2 with the F456A, N487A and K417A RBD mutants. J Structural interaction between F456 on RBD from wild-type and omicron with ACE2. The binding surface of ACE2 is shown by electrostatic surface representations. K Structural interaction between N487 on the RBD of wild-type and omicron with ACE2. Yellow dashed lines, intramolecular hydrogen bond within the RBD; red dashed lines, polar interactions between the RBD and ACE2. All experimentally obtained data represent one of two independent experiments. Nonneutralizing IC50 values were set to over 10 μg/ml, and nonbinding EC50 values were set to over 5 μg/ml. The structures of RBD-ACE2 complexes were obtained from PDB (6m0j for wild-type [12] and 7 WBP for Omicron [10]). P values shown in the figures were determined using paired two-tailed Student’s t tests (****P < 0.0001)
First, we found that the Omicron RBD retains high binding affinity with human ACE2 and that Omicron pseudovirus is able to infect target cells more efficiently than the Wuhan strain (Supplementary Fig. 1A, B). To investigate the degree of immune evasion mediated by Omicron, the RBD-specific antibody titer and neutralization activity of 21 convalescent plasma samples collected at the early stage of the pandemic (early 2020) were measured [5]. Striking reductions in both plasma RBD-specific antibody titers (4.7- to 42.9-fold reductions) and neutralization titers (9.7- to 123.4-fold reductions) against Omicron compared to the Wuhan strain were observed, which is in line with previous studies (Fig. 1B, Supplementary Fig. 2 and Supplementary Table 1) [2, 3].
To understand the resistance profile of the Omicron strain, we tested a panel of 45 neutralizing mAbs reported by our group and other groups that efficiently neutralize the Wuhan strain by targeting diverse antigenic sites within the RBD [5–8]. Nearly all the NAbs exhibited reduced binding and neutralizing activity against the Omicron RBD and pseudovirus (Supplementary Table 2, Supplementary Fig. 3 and Fig. 1C). We noted that the neutralizing activity of ~ 75% of antibodies tested, including several SARS-CoV-2 and SARS-CoV cross-reactive NAbs, was either abolished or sharply reduced. Omicron was found to be able to evade all four of the antigenic site-targeting NAbs (Fig. 1C right panel, Supplementary Fig. 3C). Most NAbs targeting antigenic sites 1 and 2, which overlap with the receptor-binding motif (RBM), lacked in vitro neutralizing activity against Omicron. Site 3 targeting antibodies showed less resistance, which may be attributed to the relatively conserved amino acid sequences. Although a fraction of SARS-CoV-2 and SARS-CoV cross-reactive NAbs that target site 4 showed moderate reduction of binding activity with the Omicron RBD, they displayed higher neutralization resistance and failed to neutralize Omicron pseudovirus (Supplementary Table 2).
To investigate how Omicron RBD mutations contribute to antibody resistance, we expressed wild-type RBD with single-amino acid substitutions that are present in Omicron and tested their binding activity with the same panel of 45 NAbs. We identified eight major immune escape substitutions, which are located within antigenic site 1 (K417N and Q493R), site 2 (G446S, E484A, G496S), site 3 (N440K), and site 4 (S371L and S373P). Except for the known escape positions K417 and E484 found in other variants of concern (VOCs), such as Beta and Gamma, the Omicron variant carries another six new major escape mutations within the RBD. Interestingly, Omicron mutations S477N, T478K, N501Y, and Y505H also resulted in resistance to a limited number of NAbs, whereas the mutations G339D and Q498R did not affect the binding activity of any of the Abs tested to the RBD (Fig. 1D, E). In addition, we found some mAbs were tolerant to any of single mutation but resistant to the mutant bearing all Omicron RBD mutations (Supplementary Fig. 4). Structural analysis demonstrated that the multiple mutations present in Omicron lead to an unusually high positive electrostatic potential at the contact surface of site 1- and site 2-targeting antibodies (Fig. 1F). The marked change in electrostatic potential of Omicron may create an unfavorable environment for the binding of antibodies but is more likely to facilitate ACE2 interaction due to the negative electrostatic surface potential of the receptor [9, 10], suggesting another immune escape mechanism. A dramatic decline in the melting temperature of the Omicron RBD was also observed (Fig. 1G and Supplementary Fig. 5), indicating that the substantial mutations present in Omicron may alter the local antigenic conformation and stability.
Although the Omicron variant exhibits striking antibody evasion, 2 of our previously isolated mAbs, 25-C4 and 25-B5, retained neutralizing potency against Omicron, with an IC50 of approximately 100 ng/ml (Supplementary Table 2). It is worth noting that 25-C4 was also able to neutralize five other SARS-CoV-2 variants of concern, including Alpha, Beta, Kappa, Delta, and Gamma. Moreover, based on the IC50 and IC90 values, 25-C4 exhibited a higher level of neutralizing activity on average than the previously described broadly neutralizing antibody S309 (Supplementary Fig. 6 and Fig. 1H).
Similar to CB6, 25-C4 belongs to VH3-53/3-66-encoded NAbs and exhibits a high level of receptor-blocking activity [5]. Our mutagenesis results provide clues as to why 25-C4 exhibits a broader neutralizing profile than CB6 (Fig. 1I and Supplementary Fig. 4). According to our RBD alanine scanning mutagenesis [5], F456 and N487 are the key epitope residues for 25-C4 and CB6 recognition but not for 25-B5 and S309 (Supplementary Fig. 7). Mutation at spike residue K417 might lead to resistance to CB6 but not to 25-C4. F456 and N487 are essential for ACE2 binding with spike and are conserved among all VOCs, which is well illustrated by the structure of RBDs from different variants complexed with ACE2 (Supplementary Fig. 8 and Fig. 1J, K). F456 forms a tight hydrophobic interaction with ACE2, and N487 interacts with ACE2 by forming a hydrogen bond with Q24 and Y83 of ACE2. In addition, N487 plays an important role in the formation of intramolecular hydrogen bonds to maintain the stability of the large loop of the RBD, which contains residues Ile468 to Pro491 at the ACE2 contact surface. K417 is a known key immune escape position of CB6 due to the critical salt bridge interaction between K417 and a negatively charged residue [5, 6]. Moreover, we found that 25-C4 functionally mimics ACE2 to enhance S2’ cleavage of wild-type spike, resulting in the formation of more S2’ fragments after protease treatment compared to S309 with no receptor-binding inhibition activity (Supplementary Fig. 9). The key epitope residues of 25-C4 are involved in ACE2-spike interface and are strictly conserved among VOCs, indicating that targeting conserved receptor-binding residues might lead to cross-variant neutralization.
In summary, the extensive mutations found in the Omicron spike protein are associated with unprecedented immune evasion of SARS-CoV-2, which has been proven by our results and other studies [2–4]. Our study reveals that the Omicron variant not only carries previously identified immune escape mutations found in other VOCs, such as substitutions at K417 and E484 (Supplementary Fig. 8), but has also evolved new resistance mutations at all four antigenic sites, resulting in resistance to NAbs targeting diverse RBD epitopes. The change in electrostatic potential at the antibody contact surface and local antigenic conformation within the RBM may also contribute to NAb evasion. Although Omicron is resistant to the majority of NAbs, it is still able to recognize ACE2 in a manner comparable to the Wuhan strain (Supplementary Fig. 1A). The RBM of the SARS-CoV-2 spike protein is important both for binding with ACE2 and inducing neutralizing Abs [5]. According to our study and Omicron structural studies [10], the ACE2 binding pattern of Omicron has been partially changed by the compromise of affinity-enhancing (S477N, Q493R, Q498R and N501Y) and affinity-decreasing (K417N, G446S and E484A) substitutions within the RBM (Supplementary Fig. 10). Therefore, accumulating comutations that are compatible with viral fitness and enhanced viral resistance are the evolutionary mechanisms that govern SARS-CoV-2 immune escape [5, 11].
A small number of broadly neutralizing NAbs targeting evolutionarily conserved non-RBM spike epitopes among SARS-CoV-2 variants, even SARS-CoV, such as S309, have been identified [7]. Alternatively, our study suggests that targeting the conserved epitope that mimics the spike-binding interface on ACE2 may constitute a new solution to induce cross-variant neutralizing antibodies. Newly identified 25-C4 represents another class of cross-variant antibodies that targets the conserved key residues F456 and N487, which are essential for interaction between spike and ACE2. This class of antibodies retains the potential to combat the emerging Omicron variant and possible new variants. More importantly, how to induce 25-C4-like cross-variant broadly neutralizing antibodies by rational vaccine design remains a question to be solved in the future.
Supplementary information
Materials and methods, Supplemental Figures
Acknowledgements
We thank the Core Facilities of Chemical Biology, Cell Biology, and Molecular Biology of the Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences for their technical support. This work was supported by the Ministry of Science and Technology of China (2018YFA0507402) and the National Natural Science Foundation of China (32100123, 32100751, and 82041015).
Author contributions
B.S., X.Y.S., H.Z.L., and C.Y.Y. initiated, designed, and supervised the study. C.Y.Y., Z.Y.L., X.L., and X.Y.S. designed and performed all the experiments. Y.D.F, S.N.W.M., Y.G.Z., L.Y.M., and W.P.G. provided materials and helped with some experiments. S.F.C. analyzed the data. Z.Y. performed the structural analysis. Z.Y.L., C.Y.Y., X.Y.S., and B.S. wrote the paper. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Chunyan Yi, Zhiyang Ling, Xiao Lu.
Contributor Information
Hongzhou Lu, Email: luhongzhou@fudan.edu.cn.
Xiaoyu Sun, Email: xysun@sibcb.ac.cn.
Bing Sun, Email: bsun@sibs.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-022-00853-6.
References
- 1.Choi SJ, Kim DU, Noh JY, Kim S, Park SH, Jeong HW, et al. T cell epitopes in SARS-CoV-2 proteins are substantially conserved in the Omicron variant. Cell Mol Immunol. 2022;19:447–8. doi: 10.1038/s41423-022-00838-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu L, Iketani S, Guo Y, Chan JF, Wang M, Liu L, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2021 doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 3.Cameroni E, Bowen JE, Rosen LE, Saliba C, Zepeda SK, Culap K, et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2021 doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2021 doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi C, Sun X, Lin Y, Gu C, Ding L, Lu X, et al. Comprehensive mapping of binding hot spots of SARS-CoV-2 RBD-specific neutralizing antibodies for tracking immune escape variants. Genome Med. 2021;13:164. doi: 10.1186/s13073-021-00985-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–4. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 7.Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–5. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 8.Hansen J, Baum A, Pascal KE, Russo V, Giordano S, Wloga E, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–4. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascarella S, Ciccozzi M, Bianchi M, Benvenuto D, Cauda R, Cassone A. The electrostatic potential of the Omicron variant spike is higher than in Delta and Delta-plus variants: a hint to higher transmissibility? J Med Virol. 2021 doi: 10.1002/jmv.27528. [DOI] [PubMed] [Google Scholar]
- 10.Han P, Li L, Liu S, Wang Q, Zhang D, Xu Z, et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell. 2022 doi: 10.1016/j.cell.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi C, Sun X, Ye J, Ding L, Liu M, Yang Z, et al. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell Mol Immunol. 2020;17:621–30. doi: 10.1038/s41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–20. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and methods, Supplemental Figures