Abstract
Combining potential diagnostics markers might be necessary to achieve sufficient diagnostic test performance in a complex state such as cancer. Applying this philosophy, we have identified a 13-protein, blood-based classifier for the detection of colorectal cancer. Using mass spectrometry, we evaluated 187 proteins in a case-control study design with 274 samples and achieved a validation of 0.91 receiver operating characteristic area under the curve.
Introduction:
Colorectal cancer (CRC) testing programs reduce mortality; however, approximately 40% of the recommended population who should undergo CRC testing does not. Early colon cancer detection in patient populations ineligible for testing, such as the elderly or those with significant comorbidities, could have clinical benefit. Despite many attempts to identify individual protein markers of this disease, little progress has been made. Targeted mass spectrometry, using multiple reaction monitoring (MRM) technology, enables the simultaneous assessment of groups of candidates for improved detection performance.
Materials and Methods:
A multiplex assay was developed for 187 candidate marker proteins, using 337 peptides monitored through 674 simultaneously measured MRM transitions in a 30-minute liquid chromatography-mass spectrometry analysis of immunodepleted blood plasma. To evaluate the combined candidate marker performance, the present study used 274 individual patient blood plasma samples, 137 with biopsy-confirmed colorectal cancer and 137 age- and gender-matched controls. Using 2 well-matched platforms running 5 days each week, all 274 samples were analyzed in 52 days.
Results:
Using one half of the data as a discovery set (69 disease cases and 69 control cases), the elastic net feature selection and random forest classifier assembly were used in cross-validation to identify a 15-transition classifier. The mean training receiver operating characteristic area under the curve was 0.82. After final classifier assembly using the entire discovery set, the 136-sample (68 disease cases and 68 control cases) validation set was evaluated. The validation area under the curve was 0.91. At the point of maximum accuracy (84%), the sensitivity was 87% and the specificity was 81%.
Conclusion:
These results have demonstrated the ability of simultaneous assessment of candidate marker proteins using high-multiplex, targeted-mass spectrometry to identify a subset group of CRC markers with significant and meaningful performance.
Keywords: Classification, Colorectal cancer, Machine learning, Mass spectrometry, Multiple reaction monitoring
Introduction
Colorectal cancer (CRC) remains a major cause of morbidity and mortality in the United States, with 142,820 new cases and 50,830 deaths reported in 2013.1 In 2014, the American Cancer Society reported that despite the establishment of CRC testing and prevention guidelines2–4 and the demonstration of the efficacy of such programs,5 only 59.1% of those recommended to participate in testing do so either by endoscopy (56.4%) or guaiac fecal occult blood test (gFOBT; 8.8%).6
Recent methods have been proposed to improve the CRC detection rates, including stool- and blood-based methods.7,8 Stool-based methods such as gFOBT and fecal immunochemical test (FIT) have focused on the detection of blood released to the lumen from cancerous lesions. Improvements in these methods have combined additional markers (eg, methylated DNA) to improve the performance.9 These methods have displayed varying performance, with 70% sensitivity and 93% specificity and 70% sensitivity and 95% specificity for gFOBT and FIT,10 respectively, and 92% sensitivity and 87% specificity for ColoGuard.9 Although FIT has been suggested as an effective detection assay,11 a wide variation in test performance has been observed, most likely resulting from varying test cutpoints and sample handling conditions.12 Blood-based tests have long been sought that will combine the assay performance of colonoscopy with the ease of plasma or serum collection and handling. When patients refusing colonoscopy are offered alternative, noninvasive assay methods, the vast majority select blood-based tests.13 Although some assay performance has been demonstrated with certain blood-based markers such as SEPT9,14 the clinical performance has not yet been sufficient to displace colonoscopy or FIT or gFOBT as a part of standard clinical practice.
Many studies have attempted to identify new CRC markers with clinical utility in either the blood plasma or serum15–18; however, none has identified a single marker with sufficient performance to develop a clinically useful test. A few studies have attempted to combine multiple markers for improved performance.19–21 However, these studies have suffered from technical limitations regarding the number of analytes that can be combined in standard methods such as immunoassays. Targeted mass spectrometry (MS) leverages the multiplex properties of MS to simultaneously measure tens or hundreds of target proteins.22–25 This approach has achieved renewed recognition as a valuable analytical tool for protein measurement.26,27 In the present study, we have implemented a workflow that includes abundant protein immunodepletion and targeted MS and real-time monitoring as a method to rapidly evaluate 187 candidate CRC marker proteins, enabling the evaluation of biomarker groups with significantly better performance than when used as single components. Using a collection of 274 CRC and control, age- and gender-matched patient plasma samples, divided into discovery and validation sets, we validated a 13-protein and 15-multiple reaction monitoring (MRM) transition classifier with significant performance (area under the curve [AUC] 0.91; 87% sensitivity and 81% specificity). The present study demonstrates the potential for high-multiplex, targeted MS to play a useful role in biomarker panel discovery.
Materials and Methods
Candidate Marker Proteins
A search of the published data was performed to compile a list of candidate marker proteins with some degree of individual evidence for CRC detection. The proteins considered for inclusion on the list generally needed to be detectable in human blood serum or plasma and to have been validated with some degree of CRC assay performance in a reasonably sized human clinical study. An upper limit of approximately 200 proteins was set, given the initial estimates on the instrument limitations for concurrent data collection in a 30-minute scheduled MRM assay. The selected proteins are listed in Supplemental Table 1 (available in online). The assembled list was not intended to be exhaustive or rigorously systematic but, rather, to be a reasonable starting point for a discovery project evaluating the potential for high multiplex-targeted MS to combine individual analyte measurements into a higher performing group.
Targeted MS Assays
The peptide selection process for targeted MS using the MRM system described in the present study (Figure 1) follows the guidelines established in published MS standards24,28 and the selection criteria outlined in the Clinical and Laboratory Standards Institute and National Committee for Clinical Laboratory Standards guidelines.29–31 The initial 187 distinct proteins selected are represented by a total of 310 known isoform variants as annotated in the Ensembl database. In silico tryptic digestion was performed on this list of proteins, resulting in 77,772 total peptides. Common peptide selection strategies were used to reduce the number to 9447 candidate peptides, represented by 5904 unique sequences.32 The interim list of unique peptides was further evaluated by in silico models that predict the responsiveness in liquid chromatography (LC)-MS applications33; 5 to 6 peptides per protein, total of 1056, were selected for synthesis by New England Peptide (Gardner, MA) and empirical evaluation of MS performance. This analysis eliminated 430 peptides because of poor ionization or excessive charge state distributions. A total of 3130 transitions from the remaining 626 peptides were evaluated using triplicate 12-point dilution curves (1/2 log10 steps) in neat and digested plasma matrices. Each transition’s dilution profile was assessed for linearity and accuracy of the fit line to the dilution response data and the precision of measurement at each dilution step. Standard methods for the calculation of these metrics were used, for which an acceptable peptide had to have ≥ 2 transitions that passed the criteria for each metric.28 The acceptance criteria were as follows: linearity (adjusted R2 values of > 0.95); accuracy (relative residual values of < 0.80); and precision, coefficient of variation values of < 0.25. This resulted in a multiplexed, targeted MS-MRM assay with a total of 337 peptides with 2 transitions each that were then synthesized as high purity (> 95%) stable isotope peptides (all C13) arginine (R) or lysine (K; New England Peptide). Together with the C13-labeled reference peptides, this yielded a final assay with 1348 distinct analytes in a single 30-minute LC-MS injection. This qualifies as a tier 2 MRM research assay design.34
Figure 1.
Flow Diagram Depicting the Steps Involved to Reduce an Initial List of Candidate Protein Biomarkers to a Viable Multiple Reaction Monitoring Assay. In Brief, Target Proteins Underwent In Silico Tryptic Digestion From Which Peptides Were Down selected by Both In Silico Modeling and Empirical Measurements to an Interim List of Candidate Peptides. These Candidate Peptides Each Have 5 Transitions Optimized for Instrument Response and Evaluated for Matrix Interference. Additional Down selection for the Final Assay, Based on Performance Metrics, Resulted in 337 Peptides, Having 2 Representative Transitions per Each Peptide
Abbreviations: CRC = colorectal cancer; LCMS = liquid chromatography-mass spectrometry.
CRC Samples
For the present initial discovery study, the CRC and control plasma samples were obtained from 3 different commercial sample repositories for a total study collection of 274 age- and gender-matched patients. A summary of the sample cohort characteristics is listed in Table 1. The 3 repositories, CapitalBio (Gaithersburg, MD), Asterand Bioscience (Detroit, MI), and ProteoGenex (Culver City, CA), had previously collected samples from Russian populations using their own institutionally approved protocols and procedures. All patients with CRC had initially presented with colon cancer, diagnosed by colonoscopy and subsequent pathologic examination. The CapitalBio samples were collected from 3 sites immediately before colonoscopy in advance of any procedure medications. Blood samples were collected in 10-mL K2EDTA tubes, processed to plasma by 1300g centrifugation within 30 minutes of sampling, and stored in polypropylene tubes at −70°C within 4 hours of collection. The Asterand Bioscience samples were collected from 2 sites between the colonoscopy and resection surgery. These blood samples were collected in K3EDTA tubes, processed by double-centrifugation at 1500g, and frozen in 2-mL cryovials at −70°C within 4 hours of collection. The controls samples for this group were collected after colonoscopy using the same processing protocol after procedure confirmation of the absence of pathologic findings. The ProteoGenex samples were collected from 2 sites on the day of resection before any preoperative medications. The blood samples were collected in K2EDTA tubes, processed to plasma by 1300g centrifugation, and stored in 2-mL cryovials. The control samples for this group were collected using the same processing protocol from healthy visits to a practitioner at the same site, with the proviso that a gastrointestinal condition was not the reason for the visit. The varied nature of the sample collection for each of these cohorts raised the concern that any one cohort might contain systematic bias incidental to the target pathologic features. Therefore, the samples from all 3 of these cohorts were pooled to mitigate any bias that any one collection might contribute to the discovery process (detailed further in the Results section). This collective pool was randomly divided in half, preserving the age and gender matching, to create a 138-sample discovery set (69 CRC and 69 control) to be used for classifier training and a 136-sample validation set to be used for final testing.
Table 1.
Summary of Patient Demographics and Clinical Annotations for 138 Discovery Set and 136 Validation Set Samples
| Variable | Discovery (n = 138) | Validation (n = 136) | ||
|---|---|---|---|---|
|
| ||||
| Control | CRC | Control | CRC | |
| Total | 69 | 69 | 68 | 68 |
| ProteoGenex | 24 | 24 | 24 | 24 |
| Asterand | 24 | 24 | 24 | 24 |
| CapitalBio | 21 | 21 | 20 | 20 |
| Gender | ||||
| Male | 29 | 29 | 28 | 28 |
| Female | 40 | 40 | 40 | 40 |
| Mean age (years) | 56.8 | 60.5 | 58.0 | 62.0 |
| CRC stage | NA | NA | ||
| I | 13 | 16 | ||
| II | 35 | 35 | ||
| III | 15 | 14 | ||
| IV | 6 | 3 | ||
| CRC lesion location | NA | NA | ||
| Colon | 33 | 39 | ||
| Rectum | 34 | 26 | ||
| Rectosigmoid junction | 2 | 3 | ||
Abbreviations: CRC = colorectal cancer; NA = not applicable.
Primary Data Acquisition
The patient plasma samples were prepared for MRM LC-MS measurement as follows. The plasma samples were thawed at 4°C for 30 minutes, followed by a 20-fold dilution of 25 μL of plasma with 475 μL of multiple affinity removal system (MARS) buffer A (Agilent Technologies). The diluted plasma was filtered through a 0.22-μm filter (Agilent Technologies), followed by a 5K molecular weight cutoff (Agilent Technologies) filtration step for lipid removal. The retentate was reconstituted to 950 μL with MARS buffer A and transferred to an autosampler vial for immunoaffinity depletion using a 10-mm × 100-mm MARS-14 LC column (Agilent Technologies). The flow-through peak of the immunoaffinity column was collected into a 2-mL, 96-well plate (Eppendorf). The entire collected sample volume was transferred to a new 5K molecular weight cutoff filter to exchange the MARS A buffer with 100 mM ammonium bicarbonate before a total protein assay (total protein assay, Life Technologies). The sample was transferred to a 2-mL, 96-well plate and lyophilized in a proteomic CentriVap system (Labconco). The plate was transferred to a Tecan EVO150 liquid handler for denaturation with 50% 2,2,2-trifluoroethanol in 100 mM ammonium bicarbonate, reduction with 200 mM DL-dithiothreitol (Sigma-Aldrich), alkylation with 200 mM iodoacetamide (Arcos), and enzymatic digestion with trypsin (Promega) for 16 hours at 37°C. The digestion was quenched with 10 μL of neat formic acid and transferred to a 330-μL, 96-well plate (Costar; Sigma-Aldrich) for lyophilization.
The LC-MS data for the samples were obtained using 6490 triple quadrupole mass spectrometers coupled to 1290 ultra high pressure liquid chromatography (UHPLC) instruments (Agilent Technology), with a capillary flow electron ionization source used for ionization. The LC flow rate was optimized at 450 μL/min and remained stable around 800 bar. High-purity nitrogen gas was used for collisionally activated dissociation at energies optimized individually for each MRM transition. Agilent 1290 autosamplers were used to deliver a 10-μL injection volume of 3 μg/μL digested plasma, reconstituted to contain all stable isotope-labeled standard peptides at 100 fmol/mL, for chromatographic separation on a ZORBAX rapid resolution high definition Eclipse Plus C18 column (Agilent Technologies) with dimensions of 2.1 × 150 mm and 1.8-μm particle size. LC mobile phase A was composed of 0.1% formic acid in water and mobile phase B was composed of 0.1% of formic acid in acetonitrile. A 30-minute UHPLC linear segment gradient was used to separate the analytes with the following segments: 3% B for the first 0.5 minute, 3% to 6% for 0.5 minute, 6% to 10% for 2 minutes, 10% to 30% for 18.75 minutes, 30% to 40% for 5 minutes, 40% to 80% for 1.25 minutes, and held at 80% for 1.25 minutes, before returning to 3% B for 0.75 minute.
The final assay was built to minimize the sparse sampling effects owing to the high frequency in the concurrent analytes measured, targeting ≥ 12 points across a peak for each analyte. The average number of points across the peak was 16.2 ± 5.4. Within the 30-minute chromatography profile, each analyte was allocated a 42-second window for data acquisition with the MS instrument in dynamic MRM acquisition mode. Minimizing the data acquisition window allowed for a maximum single-injection analyte capacity of approximately 1500. Figure 2 shows a plot of concurrency by LC time with a maximum concurrency of just 100 transitions. The minimum and maximum dwell times for the described dynamic MRM acquisition method were 3.19 and 123.75 ms, respectively.
Figure 2.
Plot of Concurrent Assay Transitions Across Mass Spectrometry (MS) Elution Time. Median Chromatography Full Width at Half Maximum for Heavy Peptides Was 3.4 Seconds, 8.6 Seconds at Baseline. Within the 30-Minute Chromatography Profile, Each Analyte Was Allocated a 42-Second Window for Data Acquisition With the MS Instrument in Dynamic Multiple Reaction Monitoring Acquisition Mode Resulting in an Number of Points Across Each Peak of 16.2 ± 5.4
Robustness tests for chromatographic drift indicated approximately 100 LC-MS injections could be accomplished without needing to readjust the targeted retention times or replacing reverse phase LC-MS columns. Figure 3 shows the trend for the retention time drift over the duration of the experiment. Column exchanges were triggered when the lower 97.5 quartile in deviation from the expected retention time was < −21 seconds, representing a loss of approximately 18 heavy peptide transitions.
Figure 3.
Box Plots (Whiskers at 95% Confidence Interval [CI]) of Differences of Measured Heavy Peptide Retention Times From Expected Times for Each Sample Injection. The Close Monitoring of Retention Time Drift Was Used to Justify the Exchange of the Main Chromatography Column Owing to Significant Risk to Losing Peak Measurements (A). Events for Column Exchange Were Triggered by the Lower 95% CI at 21 Seconds or a Loss of Approximately 17 Heavy Peptide Transitions. Additionally, a Chromatography Column Was Exchanged Owing to Risk of Liquid Chromatography Over Pressure (B)
Data Reduction
The raw MS data were extracted using the data conversion module in ProteomeWizard 2.135 and subject to peak picking and quantitative assessment through proprietary software developed at Applied Proteomics, Inc. A real-time analytical pipeline was also developed to archive and process the data files immediately after acquisition. The data files were processed through a series of operations that included moving the file to a central server, extraction of raw data, data reduction, calculation of metrics, and uploading of data to a SQL server. An internal web client, accessing the SQL server, allowed researchers and technicians to monitor the progress, assess trends, review traces, and download data for offline analyses. In addition, algorithms were used to monitor the trends in analyte retention times and changes in signal abundance, distributing automated electronic mail alerts when the trends deviated > 2 standard deviations from the expected distribution.
Classifier Discovery and Validation
The classifier discovery and validation data sets consisted of relative feature concentrations, calculated as the ratio of the unlabeled peptide peak area to the associated labeled standard peptide peak area for each transition. No other normalization of the transitions’ relative abundance was applied before classifier analysis, because the labeled peptides provided a sufficient internal control. The missing values for any transition were imputed as the minimum value for each particular transition. Before model building, the transitions were log2-transformed and scaled (0 mean, unit variance) across the patients within each sample cohort. The total number of transition values used for the classifier analysis was 532 after filtering for assay performance.
To reduce the total number of predictor candidates in the classifier models, an initial transition filtering step was performed on the discovery set using 11 different methods provided by the FSelector R package36 (correlation selection, χ2 filtering, consistency filtering, linear correlation filtering, rank correlation filtering, information gain filtering, gain ratio filtering, symmetric uncertainty filtering, OneR filtering, random forest filtering, and RReliefR filtering). A total set of 43 transitions was obtained by retaining all transitions selected by ≥ 1 of the feature selection methods. Because this initial transition-filtering step used only the discovery set data, the holdout validation set provided a completely independent assessment of the transitions’ classification performance.
The classifier models were assessed using a 10-by-10-fold cross-validation procedure. For each single 10-fold cross-validation, the 138 paired samples in the discovery set were randomly assigned to 1 of 10 folds. Nine of these folds were pooled together as a training set, and the remaining fold was used as the test set. This method was repeated 10 times, such that each fold was held out once for cross-validation testing. Within each fold cycle, transition selection was first applied to the training set using elastic net regularization implemented in the GLMNet R package.37 In this process, elastic net models were built, and the model coefficients were used to select the top n transitions, usually ranging from 2 to 20 transitions. A classifier model was built with the selected transitions using one of several different algorithms, including support vector machines, random forests, elastic network models, logistic regression, and k-nearest neighbors models. After construction of the classifier model on the given fold’s training set, the model was directly applied, without modification, to that fold’s test set. Test set performance was evaluated using its receiver operating characteristic (ROC) curve and its associated AUC. After these 10 internal cycles, the total discovery set was once again randomly divided into 10 folds, and the procedure was repeated for a total of 10 outer cycles. The transition selection and model assembly process was performed using only the data from each individual fold’s training set. At the completion of this process, the top-performing models, as assessed by the discovery set cross-validation AUC values, were selected for validation. These models were directly applied to the validation set data, and AUC performance was determined. Despite the evaluation of a large grid of feature selection and classifier assembly parameters, multiple testing correction concerns were not an issue because of the hold out of a completely independent validation set (n = 136).
Statistical Analysis
Data analysis was performed using R.38 ROC analysis and the graphic data were generated using the ROCR R package.39
Results
Assay Performance
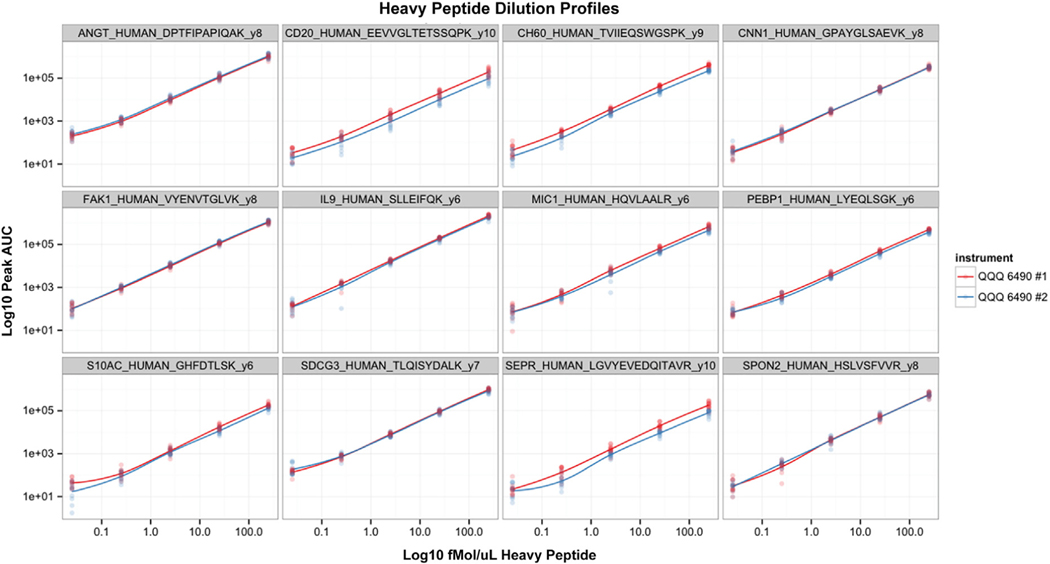
The performance of the MS-MRM assays for the 187 targeted proteins was assessed after LC-MS data collection by the ability to detect the presence of endogenous peptides in ≥ 50% of the discovery set samples. The criterion for detection was defined by the observation of a chromatographic peak of approximately Gaussian shape with a 4- to 8-second full-width at half-maximum. In addition, the peak center of the endogenous analyte was required to have been within 2 seconds of the peak center for the internal heavy peptide standard. By this definition of assay performance, 424 transitions, 260 peptides, and 168 proteins of the initial list of 187 targets were quantitatively measured, with an assay development success rate of 90%. For the 674 stable isotope peptides used in the present study, the median coefficient of variation for both instruments was 0.214 and 0.228. Figure 4 shows the 5-point dilution profile run on each instrument for every day of study collection. The overall instrument dynamic range was determined to be approximately 2.5 to 3 orders of magnitude, with good stability and linearity between both instruments.
Figure 4.
Calibration Curves for a Randomly Selected Set of Heavy Peptide Transitions, Showing the 5-Point Daily Calibration Curve Covering Individual Peptide Concentrations of 250 fmol/μL to 0.025 fmol/μL. All 12 Days, on Each Instrument, Are Represented in the Point Cluster. A Loess Smooth Line Was Plotted to Guide the Eye
Abbreviations: AUC = area under the curve; QQQ = triple quadropole.
Classifier Performance
After assessment of the discovery set classifier models, a classifier that used the random forest ensemble approach (random forest R package, version 4.6–7),40 with default parameters and 15 transitions selected by elastic net regularization selected for final validation. A final random forest classifier model was built on the entire discovery set data and locked down before application to the validation set data. This model was composed of 15 transitions from 13 proteins: A1AG1, A1AT, AMY2B, CLUS, CO9, ECH1, FRIL, GELS, OSTP, SBP1, SEPR, SPON2, and TIMP1. The names of the proteins, peptides, and transitions selected for the classifier are listed in Table 2. The ROC plot for the discovery set cross-validation is shown in Figure 5, with the error bars representing the distribution of values from the 10 rounds of the 10-fold cross-validation. The average AUC from these 10 rounds was 0.82. After final classifier assembly, the performance in the validation set was 0.91 (Figure 6). The gray curves represent the individual classifier performance for each of the component transitions in the validation set. At the point of maximum accuracy on the validation ROC curve (84%), the sensitivity was 87% and the specificity was 81%. Overall, 90% of the stage I and II cancers were correctly classified (12 of 16 for stage I and 34 of 35 for stage II), suggesting that early CRC detection with this classifier could be possible.
Table 2.
Proteins, Peptides, and Transitions Constituting 13-Protein/15-Transition Validated Classifier for CRC Detection Using Targeted MS-MRM
| Protein Description | Protein ID | Peptide | Transition |
|---|---|---|---|
| α1-Acid glycoprotein 1 | A1AG1_HUMAN | NWGLSVYADK | y7 |
| α1-Acid glycoprotein 1 | A1AG1_HUMAN | SDVVYTDWK | y5 |
| α1-Antitrypsin | A1AT_HUMAN | SVLGQLGITK | y7 |
| α-Amylase 2B | AMY2B_HUMAN | LVGLLDLALEKDYVR | b3 |
| Clusterin | CLUS_HUMAN | EPQDTYHYLPFSLPHR | y3 |
| Complement component C9 | CO9_HUMAN | TEHYEEQIEAFK | y2 |
| Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | ECH1_HUMAN | LRDLLTR | b3 |
| Ferritin light chain | FRIL_HUMAN | GGRALFQDIK | b3 |
| Gelsolin | GELS_HUMAN | AGALNSNDAFVLK | b4 |
| Gelsolin | GELS_HUMAN | AGALNSNDAFVLK | y7 |
| Metalloproteinase inhibitor 1 | TIMP1_HUMAN | GFQALGDAADIR | b4 |
| Osteopontin | OSTP_HUMAN | AIPVAQDLNAPSDWDSR | y9 |
| Selenium-binding protein 1 | SBP1_HUMAN | EPLGPALAHELR | y6 |
| Seprase | SEPR_HUMAN | LGVYEVEDQITAVR | y8 |
| Spondin-2 | SPON2_HUMAN | HSLVSFVVR | y8 |
Abbreviations: CRC = colorectal cancer; MRM = multiple reaction monitoring; MS = mass spectrometry.
Figure 5.
Average Receiver Operating Characteristic (ROC) Curve From the 15-Transition and 13-Protein Classifier Model Applied to the Discovery Set Data in Cross-Validation Assessment. The Plot Represents the Average of the 10 ROC Curves Obtained by Combining Model Predictions for All Test Set Samples Across the 10 Folds of Each Inner Replicate of the Cross-Validation Procedure. The Mean Area Under the Curve of These 10 ROCs Was 0.82
Figure 6.
Validation Set Receiver Operating Characteristic (ROC) Curve for the Locked Discovery Set Model Applied to the Validation Set (Black Line). The Associated Area Under the Curve Was 0.91. The ROC Curves From the Individual Transition Components of the Classifier Model Are Shown in Light Gray for Comparative Assessment Against the Combined Marker Panel Performance
As described in the Materials and Methods section, to rule out the potential that a collection bias in 1 of the 3 combined cohorts used in the present study might influence classifier assembly and performance, a permutation analysis was performed. In the present analysis, the data from each protein in the 15-transition and 13-protein classifier model were randomly permuted among the samples and cohorts, 1 protein at a time, leaving the data for the other proteins intact. For each protein permutation, the classifier model was applied to the new data set, and the number of samples correctly and incorrectly classified by sample cohort was tabulated, assessed at the point of maximum accuracy. This resulted in a 2 × 3 table of the correct and incorrect classification versus the 3-sample cohorts. Fisher’s exact test was then applied to the 2 × 3 table to assess the possibility of association of sample misclassification with any of the sample cohorts. None of the resulting P values reached significance (α = 0.05, Bonferroni corrected; Table 3), suggesting no association was present in the sample cohorts with misclassification and that any one particular protein in the classifier was not selected because of a cohort-specific bias.
Table 3.
Results for Tests for Significance of Incorrectly Called Samples as Function of Individual Cohort as Assessed by Panel Component Permutation Analysis
| Protein | Samples With Incorrect Results | Samples With Correct Result | P Value (Fisher’s Exact Test; Bonferroni) | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Asterand | CapitalBio | ProteoGenex | Asterand | CapitalBio | ProteoGenex | ||
| A1AG1 | 4 | 9 | 6 | 44 | 31 | 42 | 1.00 |
| A1AT | 4 | 11 | 7 | 44 | 29 | 41 | 0.88 |
| AMY2B | 4 | 10 | 9 | 44 | 30 | 39 | 1.00 |
| CLUS | 4 | 12 | 8 | 44 | 28 | 40 | 0.48 |
| CO9 | 10 | 10 | 10 | 38 | 30 | 38 | 1.00 |
| ECH1 | 4 | 13 | 6 | 44 | 27 | 42 | 0.14 |
| FRIL | 4 | 11 | 7 | 44 | 29 | 41 | 0.88 |
| GELS | 7 | 9 | 10 | 41 | 31 | 38 | 1.00 |
| OSTP | 4 | 12 | 6 | 44 | 28 | 42 | 0.29 |
| SBP1 | 5 | 11 | 7 | 43 | 29 | 41 | 1.00 |
| SEPR | 2 | 10 | 8 | 46 | 30 | 40 | 0.20 |
| SPON2 | 3 | 13 | 7 | 45 | 27 | 41 | 0.06 |
| TIMP1 | 5 | 11 | 8 | 43 | 29 | 40 | 1.00 |
Discussion
Research efforts in blood-based marker proteins for CRC have, to date, demonstrated little success in the identification of markers with sufficient performance to be clinically useful. In the present initial study, using a highly multiplexed approach to measure proteins by targeted MS, we have rapidly evaluated the combined discovery performance of candidate CRC markers and identified ≥ 1 group of markers that merit further study in the appropriate patient subsets.
From a technical perspective, we have demonstrated that targeted MS is a viable approach to quickly establish assays for the relative abundance of many a priori interesting proteins that can then be measured simultaneously in many samples. Of the 187 candidate CRC marker proteins selected for multiplex targeted MS-MRM, 90% yielded evaluable data after a simple workflow according to abundant protein immunodepletion. No analyte-specific affinity reagents (eg, antibodies, aptamers) were used. The total assay development time was approximately 2 months, and the greatest expense was for the synthetic, stable-isotope peptide controls. The rapidity and productivity of this approach suggests that in this and many other clinical research areas, the ability to combine the performance of previously insufficient marker proteins might produce useful assays. We have shown the ability to rapidly evaluate and select from a very large group of initial candidates, using relative quantification by MS, and found ≥ 1 group of proteins that merits further development. Conversion of the identified marker panel to more specific assay formats, either analyte-specific enrichment mass spectrometry or traditional multiplex immunoassay, might further improve precision and accuracy. Such refinement would also increase assay throughput and reduce the costs. Although some studies have endeavored to identify and combine the markers to improve performance with more standard approaches (eg, enzyme-linked immunosorbent assay), the challenges of running many individual assays on limited amounts of sample material or the technical limitations of these approaches have kept these studies from achieving better performance.23−25
Conclusion
From a clinical research perspective, we have demonstrated the feasibility of the development of a panel of candidate proteins for the detection of CRC from a blood plasma sample. Our assay performance of 87% sensitivity and 81% specificity at the point of maximum accuracy (84%) has demonstrated the power of identifying and combining proteins that individually might be not clinically relevant, but, as a group, have significant clinical performance. The results of the present initial study have demonstrated the potential to discover a sufficiently performing, noninvasive, blood-based biomarker panel that could help to improve compliance for CRC testing in populations ineligible for colonoscopy.
Supplementary Material
Clinical Practice Points.
Patients directed in accordance with current guidelines for CRC screening (eg, endoscopy, stool-based tests) have not been fully compliant, in large part because of the inconvenience or sample format of the currently available tests.
Despite a long history of attempts, a blood-based, single-protein test for CRC with sufficient clinical performance has been not be demonstrated.
The present study has demonstrated the feasibility of identifying combinations of candidate protein markers, using high-multiplex MS, to define diagnostic tests with superior performance.
Blood tests for CRC will soon be developed with sufficient clinical performance and utility to aid in overall CRC detection and diagnosis.
Acknowledgments
Disclosure
The indicated authors are employees of, and have an equity interest in, Applied Proteomics Inc (JJ, BW, RB, NB, GB, TB, EC, LC, PC, RD, SK, AK, RP, SS, HS, WS, JY, JB). No outside source of funding was used for this study.
Footnotes
The remaining authors declare that they have no competing interests.
Supplemental Data
Supplemental table accompanying this article can be found in the online version at http://dx.doi.org/10.1016/j.clcc.2016.02.004.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63:11–30. [DOI] [PubMed] [Google Scholar]
- 2.Lieberman DA, Rex DK, Winawer SJ, Giardiello FM. Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US multi-society task force on colorectal cancer. Gastroenterology 2012; 143:844–57. [DOI] [PubMed] [Google Scholar]
- 3.Maisonneuve P, Botteri E, Lowenfels AB. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps. Gastroenterology 2008; 135: 1570–95. [DOI] [PubMed] [Google Scholar]
- 4.Pignone M, Sox HC. Screening for colorectal cancer: U.S. preventive services task force recommendation statement. Ann Intern Med 2008; 149:627–37. [DOI] [PubMed] [Google Scholar]
- 5.Zauber AG, Winawer SJ, O’Brien MJ, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med 2012; 366:687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.American Cancer Society. Colorectal Cancer Facts & Figures 2014–2016. Atlanta: American Cancer Society; 2014. [Google Scholar]
- 7.Vu HT, Burke CA. Advances in colorectal cancer screening. Curr Gastroenterol Rep 2009; 11:406–12. [DOI] [PubMed] [Google Scholar]
- 8.Imperiale TF. Noninvasive screening tests for colorectal cancer. Dig Dis 2012; 30(suppl 2):16–26. [DOI] [PubMed] [Google Scholar]
- 9.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014; 370:1287–97. [DOI] [PubMed] [Google Scholar]
- 10.Zauber AG. Cost-effectiveness of colonoscopy. Gastrointest Endosc Clin N Am 2010; 20:751–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allison JE. FIT: a valuable but underutilized screening test for colorectal cancer— it’s time for a change. Am J Gastroenterol 2010; 105:2026–8. [DOI] [PubMed] [Google Scholar]
- 12.Lee JK, Liles EG, Bent S, Levin TR, Corley DA. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann Intern Med 2014; 160:171–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adler A, Geiger S, Keil A, et al. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC Gastroenterol 2014; 14:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson DA, Barclay RL, Mergener K, et al. Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: a prospective multicenter study. PLoS One 2014; 9:e98238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hundt S, Haug U, Brenner H. Blood markers for early detection of colorectal cancer: a systematic review. Cancer Epidemiol Biomarkers Prev 2007; 16:1935–53. [DOI] [PubMed] [Google Scholar]
- 16.Jimenez CR, Knol JC, Meijer GA, Fijneman RJ. Proteomics of colorectal cancer: overview of discovery studies and identification of commonly identified cancer-associated proteins and candidate CRC serum markers. J Proteomics 2010; 73: 1873–95. [DOI] [PubMed] [Google Scholar]
- 17.Newton KF, Newman W, Hill J. Review of biomarkers in colorectal cancer. Colorectal Dis 2012; 14:3–17. [DOI] [PubMed] [Google Scholar]
- 18.Ma Y, Zhang P, Wang F, Qin H. Searching for consistently reported up- and down-regulated biomarkers in colorectal cancer: a systematic review of proteomic studies. Mol Biol Rep 2012; 39:8483–90. [DOI] [PubMed] [Google Scholar]
- 19.Shimwell NJ, Wei W, Wilson S, et al. Assessment of novel combinations of biomarkers for the detection of colorectal cancer. Cancer Biomark 2010; 7:123–32. [DOI] [PubMed] [Google Scholar]
- 20.Wild N, Andres H, Rollinger W, et al. A combination of serum markers for the early detection of colorectal cancer. Clin Cancer Res 2010; 16:6111–21. [DOI] [PubMed] [Google Scholar]
- 21.Lumachi F, Marino F, Orlando R, Chiara GB, Basso SM. Simultaneous multi-analyte immunoassay measurement of five serum tumor markers in the detection of colorectal cancer. Anticancer Res 2012; 32:985–8. [PubMed] [Google Scholar]
- 22.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics 2006; 5:573–88. [DOI] [PubMed] [Google Scholar]
- 23.Whiteaker JR, Zhao L, Anderson L, Paulovich AG. An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein biomarkers. Mol Cell Proteomics 2010; 9:184–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzyk MA, Parker CE, Domanski D, Borchers CH. Development of MRM-based assays for the absolute quantitation of plasma proteins. Methods Mol Biol 2013; 1023:53–82. [DOI] [PubMed] [Google Scholar]
- 25.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using stable isotope standards and capture by anti-peptide antibodies (SISCAPA). J Proteome Res 2004; 3:235–44. [DOI] [PubMed] [Google Scholar]
- 26.Gillette MA, Carr SA. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat Methods 2013; 10:28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marx V. Targeted proteomics. Nat Methods 2013; 10:19–22. [DOI] [PubMed] [Google Scholar]
- 28.Chace DH. Mass spectrometry in the clinical laboratory: general principles and guidance. Approved guideline. Clinical and Laboratory Standards Inst, Wayne, PA USA, 2007. [Google Scholar]
- 29.Tholen DW. Evaluation of the linearity of quantitative analytical methods: proposed guideline: EP6eP2. Clinical and Laboatory Standards Inst, Wayne, PA USA, 2001. [Google Scholar]
- 30.EP6, CLSI proposed guideline. Evaluation of the linearity of quantitative analytical methods. Wayne, PA USA: National Committee for Clinical Laboratory Standards; 1986. [Google Scholar]
- 31.EP7, CLSI proposed guideline. Testing in clinical chemistry. Wayne, PA USA: National Committee for Clinical Laboratory Standards; 1986. [Google Scholar]
- 32.EP5, CLSI proposed guideline. User evaluation of precision performance of clinical chemistry devices. Wayne, PA USA: National Committee for Clinical Laboratory Standards; 1984. [Google Scholar]
- 33.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol 2008; 4:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fusaro VA, Mani DR, Mesirov JP, Carr SA. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat Biotechnol 2009; 27:190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carr SA, Abbatiello SE, Ackermann BL, et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteomics 2014; 13:907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambers MC, MacLean B, Burke R, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 2012; 30:918–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010; 33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 38.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. Available at: http://www.R-project.org. Accessed January 15, 2015. [Google Scholar]
- 39.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics 2005; 21:3940–1. [DOI] [PubMed] [Google Scholar]
- 40.Liaw A, Wiener M. Classification and regression by randomForest. R News 2002; 2:18–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.