Abstract
Rosmarinic acid is a hydroxycinnamic acid ester commonly found in the Boraginaceae and Lamiaceae plant families. It exhibits various biological activities, including antioxidant, anti-inflammatory, antibacterial, antiallergic, and antiviral properties. Rosmarinic acid is used as a food and cosmetic ingredient, and several pharmaceutical applications have been suggested as well. Rosmarinic acid is currently produced by extraction from plants or chemical synthesis; however, due to limited availability of the plant sources and the complexity of the chemical synthesis method, there is an increasing interest in producing this compound by microbial fermentation. In this study, we aimed to produce rosmarinic acid by engineered baker’s yeast Saccharomyces cerevisiae. Multiple biosynthetic pathway variants, carrying only plant genes or a combination of plant and Escherichia coli genes, were implemented using a full factorial design of experiment. Through analysis of variances, the effect of each enzyme variant (factors), together with possible interactions between these factors, was assessed. The best pathway variant produced 2.95 ± 0.08 mg/L rosmarinic acid in mineral medium with glucose as the sole carbon source. Increasing the copy number of rosmarinic acid biosynthetic genes increased the titer to 5.93 ± 0.06 mg/L. The study shows the feasibility of producing rosmarinic acid by yeast fermentation.
Keywords: metabolic engineering, Saccharomyces cerevisiae, rosmarinic acid, cytochrome P450
Rosmarinic acid (RA) is a polyphenolic antioxidant that occurs in some members of the Lamiaceae family, and is one of the active compounds in medical herbs such as rosemary Rosmarinus officinalis, sage Salvia officinalis, and lemon balm Melissa officinalis. It displays numerous antioxidant, anti-inflammatory, antitumor, anticancer, and antimicrobial activities. The antioxidant activity of RA is higher than that of vitamin E.1 Several clinical trials found RA effective in the treatment of asthma, allergies, and reactive airway diseases.2−5 Additionally, rosmarinic acid is a neuroprotective and cognitive-enhancer molecule, as shown in treatment of Parkinson and prevention of Alzheimer’s diseases in rat models.6,7
Similar to most of the plant-derived bioactive compounds, RA is currently obtained from plant cells. The highest content of 31% of dry weight (6.4 g/L) was reported for cell cultures of S. officinalis by feeding 0.1 g/L L-phenylalanine after 25 days.8 Considering that a suspension cell culture of plants is difficult to scale up, lacks the specificity for RA production, and requires long-term cultivation, there is a need to develop less expensive and simpler processes for RA production. Microbial cell factories, such as Saccharomyces cerevisiae, have the potential to serve as a host for heterologous production of plant metabolites and are well-amenable for engineering and large scale fermentation on inexpensive medium.9,10
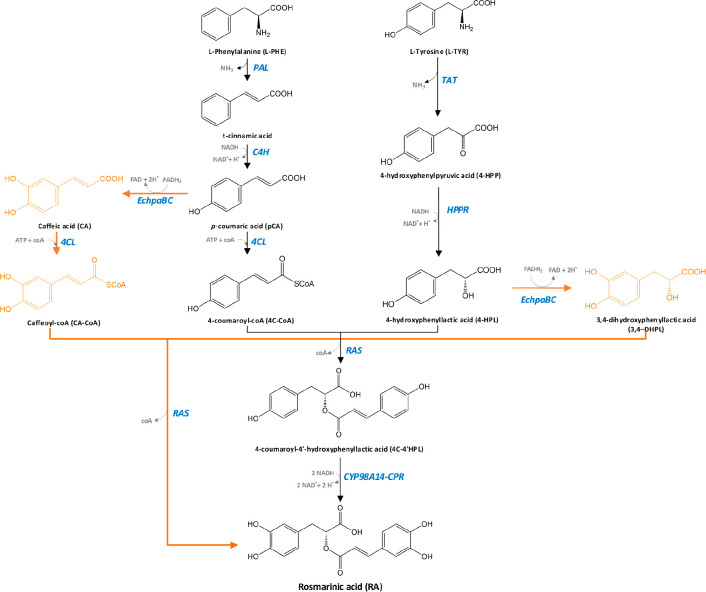
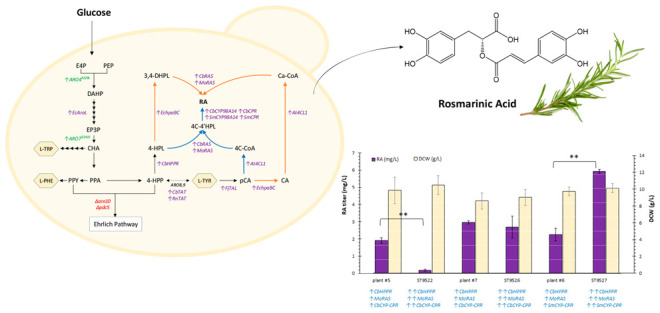
Rosmarinic acid consists of two aromatic rings with two hydroxyl groups on each ring (Figure 1), which give the free radical scavenging ability against superoxide anions and hydroxyl radicals.11 The RA biosynthetic enzymes were studied in suspension cell cultures of Plectranthus scutellarioides (Coleus blumei) by Petersen et al.12 (Figure 1). In “plant pathway”, the two parts of RA molecule, caffeic acid (CA) and 3,4-dihydroxyphenyllactic acid (3,4-DHPL), are synthesized separately from L-phenylalanine and L-tyrosine, respectively. The first step is the deamination of both amino acids, to form t-cinnamic acid from L-phenylalanine, and 4-hydroxyphenylpyruvic acid (4-HPP) from L-tyrosine. Next, t-cinnamic acid is hydroxylated in positon 4 of the phenolic ring by cytochrome P450 monooxygenase cinnamate 4-hydroxylase (C4H) to form p-coumaric acid (pCA). The following step is the activation of pCA by 4-coumaryl CoA-ligase (4CL). In the other branch, 4-HPP is reduced to 4-hydroxyphenyllactic acid (4-HPL). Rosmarinic acid synthase (RAS) catalyzes the coupling of 4-coumaroyl-CoA and 4-HPL with concurrent release of coenzyme A. This enzyme requires unimolar amounts of both substrates to form one mole of product, 4-coumaroyl-4′-hydroxyphenyllactic acid. This compound is then hydroxylated in positions 3 and 3′ by cytochrome P450 hydroxycinnamoyl-hydroxyphenyllactate 3- and 3′-hydroxylase (CYP98A14). These final hydroxylation steps are considered to be the main flux-limiting steps in the production of rosmarinic acid.13 The final reaction of rosmarinic acid production by the pathway described (and shown in black in Figure 1) can be summarized as:L-tyrosine + L-phenylalanine + 4NADH + ATP → rosmarinic acid + 2NH3 + 4NAD+
Figure 1.
Biosynthetic plant pathway to rosmarinic acid in plant cell C. blumei(14) (black compounds and arrows), and chimeric pathway (orange compounds and arrows). The enzymes involved are abbreviated in blue, PAL = phenylalanine ammonia lyase, C4H = cinnamic acid 4-hydroxylase, 4CL = 4-coumaryl CoA-ligase, TAT = tyrosine aminotransferase, HPPR = hydroxyphenylpyruvate reductase, RAS = “rosmarinic acid synthase”, hydroxycinnamoyl-CoA/hydroxyphenyllactate hydroxycinnamoyltransferase, CYP98A14 = 4-coumaroyl-4′-hydroxyphenyllactate 3/3′-hydroxylases, EchpaBC = E. coli 4-hydroxyphenylacetate 3-hydroxylase complexes B and C.
Cytochrome P450s are heme-containing monoxygenases. Using molecular oxygen as a substrate, they insert one oxygen atom into an aliphatic position of an organic substrate and reduce the other oxygen atom to water.15 The successful expression and activation of CYP enzymes in cells relies on two main factors: (i) coupling the P450 enzyme to electron-donating protein, cytochrome P450 reductase (CPR), and (ii) colocalizing of both CYP and CPR to the membrane of endoplasmic reticulum (ER) anchored via their N-terminus.16 The expression of a functioning CYP–CPR enzyme pairs in bacterial cell factories such as Escherichia coli, where CPR and ER membrane are absent, is problematic. Hence, for RA production, Bloch and Schmidt-Dannert developed a “chimeric pathway” for E. coli, by 3-hydroxylation of the substrate precursors prior to RA ester formation.17 In this method, they used the E. coli 4-hydroxyphenylacetate 3-hydroxylase complex (EchpaBC) to hydroxylate pCA to caffeic acid, and 4-HPL to 3,4-DHPL.18 RA synthase can act on caffeic acid and 3,4-DHPL, producing RA (Figure 1). But the enzyme shows almost 10 times lower affinity for 3,4-DHPL as substrate compared to 4-HPL.19 Also, the enzyme complex EchpaBC is reported to catalyze the hydroxylation of L-tyrosine to L-DOPA (L-(3,4-dihydroxy)phenylalanine)20 reducing the L-tyrosine pool as the main amino acid precursor for RA production. The optimized E. coli strain of Bloch and Schmidt-Dannert was shown to produce 0.65 ± 0.11 mg/L of rosmarinic acid from glycerol.17 The main issue with this strain is that the catalytic efficiency of the EchpaBC enzyme complex is probably not so high for 4-HPL, as isorinic acid (the ester conjugate of 4-HPL and caffeic acid) was detected in much higher concentration than rosmarinic acid. To increase this catalytic activity and achieve higher concentration of 3,4-DHPL, Yao et al. used a variant of lactate dehydrogenase enzyme (LDHY52A) from Lactobacillus pentosus and coexpressed it together with EchpaBC.21 By feeding the precursor caffeic acid, they achieved the highest titer of 130 mg/L.22,23 Further engineering the strain to aim for higher L-tyrosine titer is reported by Li et al., who in their work reached an RA titer of 4.5 mg/L from glucose.24 In the same study, 172 mg/L RA was obtained by coculturing three E. coli strains on complex medium. The genetic modifications of these three strains were similar to that in the previous works.22,23 The difference was in segregation of the production of precursors and RA into three different strains: the first strain producing caffeic acid from glucose, the second strain produced 3,4-DHPL from glucose, and the third strain conjugated caffeic acid and 3,4-DHPL to rosmarinic acid.24 It will however be difficult to scale up a process based on coculturing of several strains, as it will require maintaining a stable strain-to-strain ratio throughout the fermentation.
As the basic step in engineering a heterologous pathway, one needs to find the optimal combination of enzyme homologues in the pathway, in order to have the highest conversion of intermediates to the final product of interest. Owing to investigations of the key enzymes involved in biosynthesis of rosmarinic acid in several plant species of the family Lamiaceae during the past decade, there are several candidates for the genes CYP98A14, RAS, and TAT, with coding DNA sequences available.25−29 In this case, the design of experiments (DOE) serves as an efficient tool for systematic screening of the combinatorial space of enzyme homologues and interpretation of the results.30,31 With the full factorial design, all possible combinations of the levels across all the factors are taken into account to analyze the effects of each factor, evaluating these effects at several levels and, more importantly, the possible interactions between the factors. The obtained main effects and interaction plots together with the magnitude of effect for each single factor or interaction can provide an insight into the biological systems. In the case of large combinatorial libraries, fractional factorial DOE was shown to be a powerful tool for the algorithmic design of yeast strains to improve itaconic acid titer.32
Eukaryotic cells such as S. cerevisiae have been shown to be an ideal host for production of plant-derived bioactive compounds. The native cytochrome P450 reductase of S. cerevisiae (NCP1 gene, accession code P16603) has 31% identity to protein sequence of CPR from two rosmarinic acid producer cells, S. officinalis and P. scutellarioides, making S. cerevisiae a potential host for production of rosmarinic acid. We herein provide the first description of rosmarinic acid production from glucose in mineral media using the plant originated enzymes expressed in a microbial host.
Results and Discussion
Investigation of Rosmarinic Acid Degradation in Fermentation Medium
A major issue with microbial production of bioactive phenolic compounds such as RA is the degradation of these compounds in the presence of oxygen, heat, and light.33,34 Previous electrochemical studies have shown that RA is oxidized in a pH-dependent two-step reaction, withdrawing two electrons from each aromatic ring, four electrons in total.35 Additionally, when produced in a cell, RA can be hydrolyzed into caffeic acid and 3,4-DHPL by the action of reversible rosmarinic acid synthase.36 We tested the method for RA extraction with ethyl acetate described by Bloch and Schmidt-Dannert,17 but could not extract RA added to a liquid medium. Extraction with a more polar solvent (acetonitrile) also gave a very low efficiency (1%). As RA solubility is reported to increase in binary solvent mixtures of ethanol and water, we mixed the aqueous sample of RA with ethanol in 1:1 v/v ratio and in this way could detect 80–90% of the added RA. To prevent the oxidation of RA during the sample preparation, we added 0.01% butylated hydroxytoluene as an antioxidant, and in this way, we could detect 99% of the added RA (Supporting Information, Table S1). Next, we tested the stability of RA in yeast cultures. For this, we supplemented the mineral medium (pH 6.0) with 1 g/L RA and incubated it for 3 days at 30 °C with or without yeast cells. To our surprise, we could recover 90% of RA at the end of the cultivation from the cells-containing broth, but less than 3% from the medium without cells. We hypothesized that RA was degraded less in the cell culture due to pH decrease over the course of fermentation because of the production of organic acids by yeast. Indeed, when we repeated the experiment with the medium without cells set to pH 3.0, no degradation of RA was observed. To prevent the degradation of RA in the course of cultivation, we tested the addition of 10 mM ascorbic acid as antioxidant. The recovery of RA after 3 days was 94% and 91% in the absence or presence of yeast cells, respectively (Supporting Information, Figure S1). We could thus conclude that nonengineered S. cerevisiae does not degrade RA and that RA can be stabilized in the medium by the addition of ascorbic acid, which was applied in the subsequent cultivations in this work.
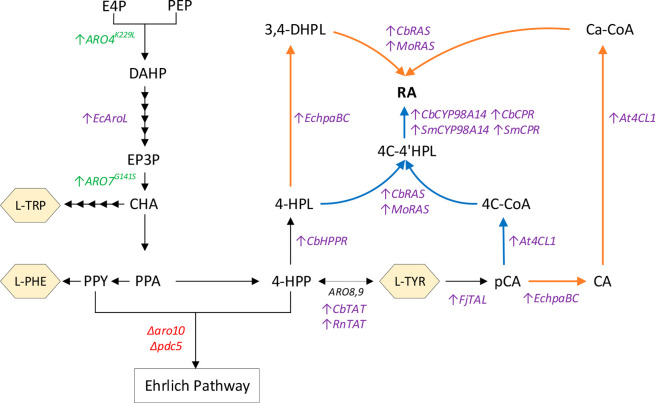
The precursor molecules for RA biosynthesis in plants are L-tyrosine and L-phenylalanine (Figure 1). These amino acids also serve as the precursor for numerous biosynthesis pathways of commercial interest, including flavonoids,37 stilbenoids,38 and fusel alcohols,39 to name a few. The engineering strategies to increase the production of these amino acids have been extensively studied on yeast before. The most common strategies are the alleviation of the allosteric feedback regulation of the DAHP synthase and chorismate mutase genes, by expressing mutated version of ARO4 (ARO4K229L) and ARO7 (ARO7G141S), overexpression of heterologous shikimate kinase AroL from E. coli (EcAroL), and deletion of ARO10 and PDC5 to block the aromatic amino acids degradation pathway (Ehrlich pathway).40,41 We implemented these modifications into S. cerevisiae CEN.PK113-7D strain42 to create the platform strain for the production of RA (Figure 2). The next step, was to convert aromatic amino acids into precursors for rosmarinic acid production. As L-tyrosine has one hydroxyl group in the aromatic ring, replacing L-phenylalanine with L-tyrosine as the sole precursor for RA production can save a hydroxylation step. To do this, we used the tyrosine ammonia lyase from F. johnsoniae (FjTAL) to synthesize pCA from L-tyrosine.43 By overexpressing tyrosine aminotransferase (TAT) from either plant P. scutellarioides (CbTAT) or from rat R. norvegus (RnTAT), and hydroxyphenylpyruvate reductase from P. scutellarioides (CbHPPR) we were able to synthesize 4-HPL from L-tyrosine as well. The activation of p-coumaric acid to 4-comaroyl-coA was achieved by overexpression of 4CL1 of A. thaliana, which was shown to have better performance than the other isoenzymes, 4CL2 and 4CL3.44 By expressing rosmarinic acid synthase originating from either M. officinalis (MoRAS) or from P. scutellarioides (CbRAS), followed by overexpression of CYP98A14 hydroxylase and the corresponding reductase genes from either S. miltiorrhiza (SmCYP98A14-CPR) or from P. scutellarioides (CbCYP98A14-CPR), we successfully assembled the plant pathway for RA biosynthesis. For high expression of the pathway enzymes, the enzyme homologues were expressed under the control of a strong constitutive promoters Ptef1 (transcriptional elongation factor EF-1α) and Ppgk1 (phosphoglycerate kinase) as shown in Figure 3 and Figure S3. These promoters are already characterized to have high constant activity at different glucose concentrations.45 For terminator selection, bidirectional terminators Tadh1 and Tcyc1 were used.46
Figure 2.
Engineered route for rosmarinic acid (RA) production in S. cerevisiae. Native genes are indicated in black, overexpressed yeast genes in green, overexpressed heterologous genes in purple, and deleted genes in red. The arrows in orange show the chimeric pathway, and the arrows in blue are for plant pathway. The aromatic amino acids are shown in hexagons: E4P, erythrose-4-phosphate; PEP, phosphoenolpyruvate; DAHP, 3-deoxy-d-arabino-heptulosonate-7-phosphate; EP3P, 5-enolpyruvylshikimate-3-phosphate; CHA, chorismate; L-TRP, L-tryptophan; PPA, prephenate; PPY, phenylpyruvate; L-PHE, L-phenylalanine; 4-HPP, 4-hydroxyphenylpyruvate; L-TYR, L-tyrosine; 4-HPL, 4-hydroxyphenyllactate; pCA, p-coumaric acid; CA, caffeic acid; 4C-CoA, 4-coumaroyl-coA; 4C-4′HPL, 4-coumaroyl-4′-hydroxyphenyllactic acid; Ca-CoA, caffeoyl-coA; 3,4-DHPL, 3,4-dihydroxyphenyllactate; RA, rosmarinic acid.
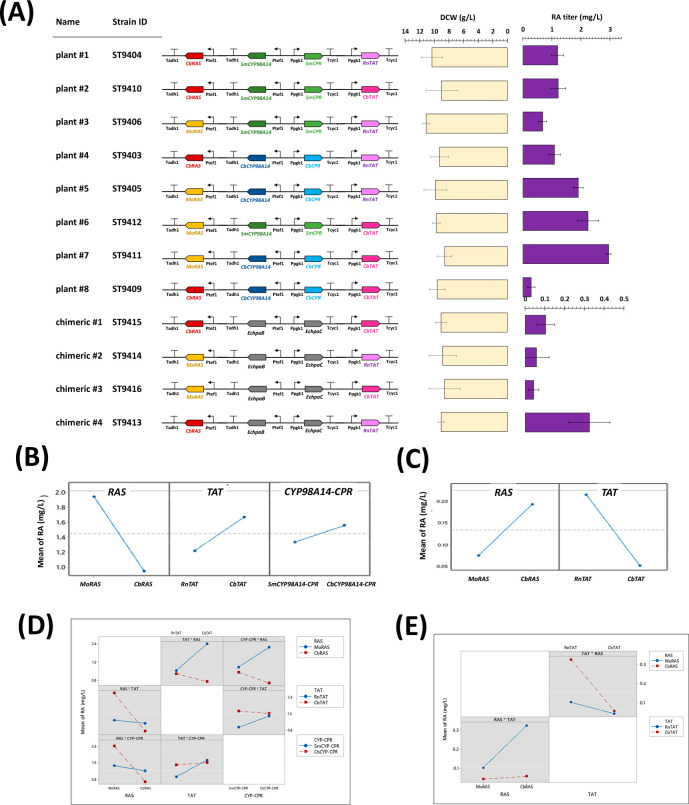
Figure 3.
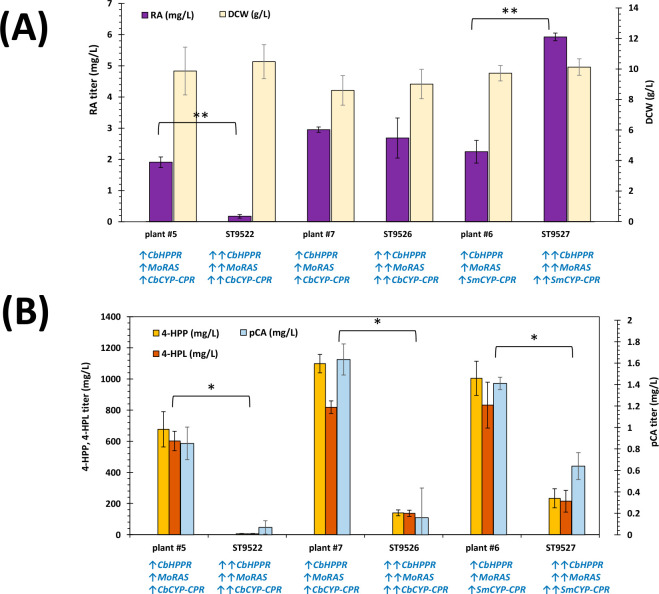
(A) The full factorial DOE strain construction with implemented heterologous plant and chimeric pathways for RA production, along with the promoters and terminators used to control the expression of genes. The phenotype of the constructed strains as rosmarinic acid titer and DCW (g/L) is shown for each construct. The mean value of response (RA titer) versus levels of each factor is shown in panel B for plant pathway strains, and panel C for chimeric strains. The 2-way interaction plot for factors affecting fitted means of RA (mg/L) is also shown for plant strains (D), and for chimeric strains (E). The values for RA titer and DCW measurements represent the mean of n = 3 biologically independent samples, and error bars show standard deviation.
As we used two homologues from different origins for the three enzymes tyrosine aminotransferase (RnTAT/CbTAT), rosmarinic acid synthase (MoRAS/CbRAS), and CYP98A14 hydroxylase and reductase (SmCYP98A14-CPR/CbCYP98A14-CPR), we used the full factorial DOE for strain design and data analysis. The number of enzymes (termed as Factors of DOE) and the homologues (termed as Levels of DOE) are 3 and 2, respectively, hence a full factorial design would result in 23 = 8 genotypes (termed as Experiments of DOE) for plant pathway assembly in yeast. In the same manner, with two homologues for enzymes tyrosine aminotransferase and rosmarinic acid synthase for the chimeric pathway, a two-level two-factor DOE of full factorial would result in 22 = 4 experiments. A schematic overview of the plant and chimeric pathways is indicated in Figure 2. As the total number of combinatorial strain designs is feasible to construct and phenotype experimentally, we constructed the whole library of strains accordingly and did not make any fractional factorials to reduce the number of combinatorial designs.
With DOE full factorial strain construction, we ended up with eight strains expressing plant pathways and four strains with chimeric pathways, each design being a unique genetic modification. The gene expression levels for different Easyclone sites have already been measured by Jessob-Fabre et al.47 with fluorescence protein GFP as the indicator. However, a small variation in GFP-expression level was also reported.47 Therefore, to have a fair comparison, the gene homologues were expressed at similar loci (Easyclone sites), with similar promoter and terminator sets (Figure S3), to ensure that the observed difference in RA titer is not a result of difference in gene expression level. Next, we screened the constructed strains for the highest rosmarinic acid production (the desired phenotype), using feed-in-time media with glucose as the sole carbon source. The DOE full factorial design with screening results for RA titer and dry cell weight (DCW, g/L) is shown in Figure 3. For plant–pathway strains, the model for the prediction of response was fitted with a quadratic polynomial, with an R-square of 99.78%. Regression analysis (Supplementary Table S2) showed that of all factors, only the origin of the RAS enzyme had significant influence on rosmarinic acid titer (F-value = 175, p-value < 0.05). Also the 2-way interaction between the enzymes rosmarinic acid synthase (RAS) and tyrosine aminotransferase (TAT) with an F-value of 126 had an impact on the RA titer, while not rejecting null hypothesis (p-value = 0.057). To select for the best combinatorial design of plant strains, the interaction plot of factors shown in Figure 3D is used that shows that the RA mean titer (mg/L) for a plant pathway is the highest when RAS is from M. officinalis, TAT is from P. scutellarioides, and CYP98A14-CPR is from P. scutellarioides. The resulting strain plant #7 showed the highest RA titer for plant-pathway strains, with 2.95 ± 0.08 mg/L.
For chimeric-pathway strains, the linear model was fitted with R-square of 79.03%, while the regression analysis (Table S3) of factors did not show any factor as an important variant in RA titer (p-value > 0.1, F-values close to 1). The optimal design is again selected based on the interaction plot of factors shown in Figure 3E, which shows the highest RA mean titer (mg/L) for the chimeric pathway is in the design with RAS from P. scutellarioides and TAT from R. norvegus. The resulting strain chimeric #4 produced RA in a titer of 0.33 ± 0.10 mg/L, which is ca. half of the titer reported for RA producing E. coli with the same pathway.17 Heterologous enzymes originating from plants are reported to be organized in protein complexes, in which the proteins involved in the pathway can interfere in spatial colocalization of other proteins. These protein–protein interactions are also the driving force for coevolution and regulation of the protein structure of these enzymes.48 Therefore, the origin of these proteins plays a significant role in obtained RA titers (Figure 3A), which also highlights the fact that these interactions have to be taken into account in developing cell factories for plant-derived compounds.
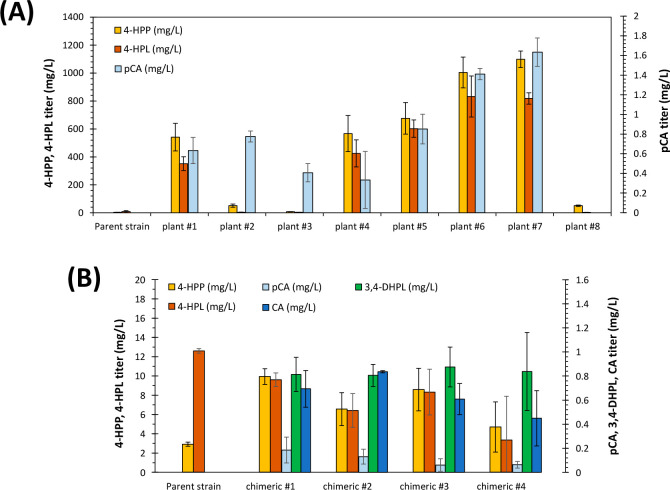
Apart from RA titer in the engineered strains, we also measured the titers of RA biosynthetic intermediates in the extracted samples (Figure 4). A major difference between the plant and chimeric strains, as seen from this graph, was the titer of 4-HPP, that is almost a hundred times higher in plant strains (Figure 4A) compared to the chimeric ones (Figure 4B). As the difference between plant and chimeric strains is in the hydroxylase enzyme that is beyond 4-HPP, then one should assume that the titers of 4-HPP and 4-HPL must be comparable between the two types of strains. The observed difference is due to the promiscuity of enzyme EchpaBC, as this enzyme is reported to hydroxylate quite a broad range of aromatic compounds, including L-tyrosine hydroxylation to L-DOPA.20,49 In principal, the expression of TAL enzyme should catalyze the conversion of L-DOPA to caffeic acid,20 which would contribute to the RA synthesis pathway. However, we could observe a darker color of chimeric strains at the end of 72 h cultivation, compared to the parent strain (Figure S4). Indeed, the produced L-DOPA was partly oxidized to dark colored-melanin pigment,50 which probably drained the L-tyrosine pool and resulted in lower titers of 4-HPP and 4-HPL in chimeric strains. The parent strain (ST7574) without precursor supply optimization accumulated 2.91 ± 0.22 mg/L of 4-HPP (Figure 4B), while the best RA-producing strain plant #7 had 1099 ± 59 mg/L 4-HPP. Similarly, the concentration of the reduced derivative of 4-HPP, 4-HPL, was 97-fold higher in the precursor-engineered strain (plant #7) compared to the parent strain. Considering that CoA-ligase enzyme (At4CL1) is similar in both plant and chimeric strains, we assumed from titers of pCA and CA in Figure 4 that the amount of 4C-CoA and Ca-CoA are the limiting substrates for RA production in plant and chimeric strains, respectively. Based on km values for the enzyme RAS with 4-HPL or 3,4-DHPL as substrate, obtained from The Comprehensive Enzyme Information System (http://www.brenda-enzymes.org/), there is a higher affinity of RAS enzyme (almost 10 times) to 4-HPL compared to 3,4-DHPL.19 This seems to be the main reason for the observed difference in RA synthesis through the plant and chimeric pathways. Moreover, a comparison of the precursors’ accumulation in plant #7 and plant #8 in Figure 4A shows a higher titer of intermediates for plant #7, while the only difference in the two designs is the RAS homologue, the second last step of RA biosynthesis that apparently should not affect precursors accumulation. However, as mentioned earlier, the protein–protein interactions for MoRAS and CbRAS with the other proteins of the pathway might be different, influencing the performance of other enzymes that have role in precursors production. In addition, as concluded from Figure 3, MoRAS gives higher titers of RA than CbRAS, so in plant #7, this enzyme can drive the conversion of intermediates to RA faster and reduce the accumulation of these compounds.
Figure 4.
Precursors titer measured for yeast strains with (A) plant pathway and (B) chimeric pathway at the end of production experiment. The designs of strains plant #1 to plant #8 and chimeric #1 to chimeric #4 is shown in Figure 3: 4-HPP, 4-hydroxyphenylpyruvate; 4-HPL, 4-hydroxyphenyllactate; pCA, p-coumaric acid; 3,4-DHPL, 3,4-dihydroxyphenyllactate; CA, caffeic acid. The values represent the mean of n = 3 biologically independent samples, and error bars show standard deviation.
The high amounts of residual unreacted metabolites in the RA producing strains in Figure 4A suggested a limitation in the activity of HPPR, RAS, and CYP98A14 associated with CPR enzymes. Hence, we chose three of the high RA producing strains in Figure 3, plants #5, #6, and #7, and integrated the second copy of HPPR, RAS, and CYP98A14-CPR. The results of this modification are shown in Figure 5A, where the highest RA titer of 5.93 ± 0.06 mg/L was achieved for ST9527, 164% higher than in the parent strain plant #6 (p = 0.0007). Doubling the copy number of HPPR, RAS, and CYP98A14 genes also decreased the concentrations of RA intermediates 4-HPP, 4-HPL, and pCA in Figure 5B (p < 0.01). However, this reduction in accumulation of intermediates did not result in higher titer of RA in ST9526 with double-integration of MoRAS, CbHPPR, and CbCYP-CbCPR when compared to its parent strain plant #7 (p = 0.27), and even caused a 91% decrease in RA synthesis of ST9522 compared to the parent plant #5 (p = 0.0006).
Figure 5.
Rosmarinic acid titer and DCW (A), and the precursor titers (B) for the strains with integrated second copy of HPPR, RAS, and CYP98A14-CPR genes. Statistical analysis was performed for RA titer by using Student’s t test (one-tailed, two-sample unequal variances, *p < 0.01, **p < 0.001). The values represent the mean of n = 3 biologically independent samples, and error bars show standard deviation.
We speculated if the low RA titer can be related to the toxicity of the compound to the host organism. We cultivated nonproducing and RA-producing strains with different concentrations of RA (0.1–5 g/L) and monitored the growth by OD600 measurement (Figure S2). The final OD600 was only decreased by ca. 25% when RA was added at the highest level of 5 g/L, hence the compound is not strongly toxic to S. cerevisiae.
Despite the fact that the RA titer of 5.93 ± 0.06 mg/L in strain ST9527 is the highest titer achieved for production of rosmarinic acid by a single heterologous microbe using a defined medium, this titer is still well below the required titer for a competitive production of RA, and further strain engineering is hence required. The future strain engineering efforts can comprise the further optimization of precursor and cofactor supply in the engineering of RA pathway enzymes, optimizing their expression and localization, and fermentation optimization.
Conclusion
Rosmarinic acid was produced by expressing biosynthetic enzymes from plants in yeast S. cerevisiae. The strain was optimized by selection of optimal enzyme variants, increasing the gene copy number of the pathway genes, and by improving the precursor supply through the aromatic amino acids pathway. Up to 5.93 ± 0.06 mg/L rosmarinic acid was produced by engineered yeasts on mineral medium with glucose as the sole carbon source. This proof-of-concept study shows the possibility of developing a fermentative process for rosmarinic acid using a S. cerevisiae cell factory.
Materials and Methods
Strains and Media
E. coli strain DH5α was used for cloning and plasmid propagation. Lysogeny Broth (LB) liquid medium or LB solid medium supplemented with 20 g/L agar containing 100 mg/L ampicillin was used for E. coli cultivation at 37 °C.
The strain CEN.PK 113-7D42 was used as the parent strain in this study, and the following strain ST7574 was derived by transforming the episomal vector for expression of Cas9 protein, with a kanamycin resistance marker (Table S4). To maintain the selection for Cas9, G418 (Sigma-Aldrich) was supplemented into media at 200 mg/L. Further metabolic engineering of the yeast cells was carried out by the EasyClone Markerfree toolkit.47 Briefly, the integrative vectors (Figure S3A–M) were linearized by digestion with NotI enzyme (New England BioLabs), and together with the corresponding gRNA helper vector (Figure S3, N) were transformed into the host yeast strain carrying Cas9 expression plasmid. Yeast transformation was performed using the standard lithium acetate method, and the transformants were plated on YPD medium (10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose) supplemented with antibiotics: nourseothricin (Jena Bioscience GmbH) at 100 mg/L and G418 (Sigma-Aldrich) at 200 mg/L. The chemicals were all obtained from Sigma-Aldrich, unless otherwise mentioned.
To verify the correct integration of DNA fragment into yeast cells, colony PCR with OneTaq Master Mix (New England BioLabs) and the set of primers shown and described in Table S8 was used. Each colony PCR reaction contained 1 μL of each of the primers 1, 2, and 3 (Table S8), 10 μL of OneTaq Master Mix and a small amount of yeast colony as template, with water to the final volume of 20 μL. The thermal program used is initial denaturation at 95 °C for 2 min, 30 cycles of denaturation at 95 °C for 20 s, annealing at 55 °C for 20 s, and extension for 72 °C 1 min/kb. The correct tranformants are then cured to lose the gRNA vector.
Plasmids
All the integrative vectors together with gRNA vectors are listed in Table S5, and the plasmid maps of integrative and gRNA vectors are shown in Figure S3. The integrative vectors were assembled by USER cloning according to Table S5. Generally, 0.5 μL of CutSmart buffer (NEB) was mixed with 0.5 μL of USER enzyme and 100–200 ng of each BioBrick or linearized parent vector to a final volume of 5 μL. The parent vectors contain a USER cloning site surrounded by bidirectional terminators Tadh1 and Tcyc1. These vectors were linearized by digestion with FastDigest SfaAI (Life Technologies) and sticky ends obtained by nicking with Nb.BsmI (New England Biolabs). The USER reaction was then incubated for 25 min at 37 °C, 15 min at 25 °C, and 10 min at 15 °C, followed by transformation to E. coli. Correct assembly was verified by DNA sequencing (Eurofins Genomics, Germany).47,51
All BioBricks with templates and primers used for USER cloning are listed in Table S6, with primer sequences listed in Table S7. For amplification of BioBricks, Phusion U polymerase chain reaction (PCR) from Thermo Fisher Scientific was implemented using the following thermal program: Initial denaturation at 98 °C for 5 min, denaturation at 98 °C for 10 s, 30 cycles of annealing at 52 °C for 10 s, extension at 72 °C for 30 s/kb.51
Enzyme Genes
The heterologous pathway for rosmarinic acid production from l-tyrosine was constructed in yeast by implementing the genes 4-coumaryl coA-ligase from Arabidopsis thaliana (At4CL1), tyrosine ammonia lyase from Flavobacterium johnsoniae (FjTAL), tyrosine aminotransferase from P. scutellarioides (CbTAT) and from Rattus norvegus (RnTAT), hydroxyphenylpyruvate reductase from P. scutellarioides (CbHPPR), rosmarinic acid synthase from P. scutellarioides (CbRAS) and from M. officinalis (MoRAS), 4-hydroxyphenylacetate 3-monooxygenase complex B and C from E. coli (EchpaB, EchpaC), cytochrome P450-dependent hydroxylase from S. miltiorrhiza (SmCYP98A14) and from P. scutellarioides (CbCYP98A14), and the corresponding cytochrome P450 reductases (SmCPR and CbCPR). It should be noted that throughout this study, for the nomenclature of the genes originated from P. scutellarioides, the former name of this plant Coleus blumei (Cb) is used. Synthetic genes were ordered in codon-optimized variants for S. cerevisiae from GeneArt (Thermo Fisher Scientific). The codon-optimized nucleotide sequences for all the synthetic genes are listed in Table S9.
Statistical Analysis
The regression and statistical analysis of variances (ANOVA) of the model was performed using Minitab19.2 software. To construct the strains, a two-level full factorial design was used in order to evaluate the effect of the combination of gene homologues in such a way to maximize RA titer as the response. Full factorial design allows for studying all possible combinations of the levels in the factors in the strain construction and also evaluate the joint effect of the factors on the response.52 The factors and levels are described in Table 1. As such, 23 = 8 strains for plant pathway, and 22 = 4 strains for chimeric pathway were constructed according to this experimental design.
Table 1. Factors and Levels Used for Full-Factorial DOE of Strain Construction.
| levels |
||
|---|---|---|
| factors | 1 | 2 |
| rosmarinate synthase (RAS) homologues | M. officinalis (MoRAS) | P. scutellarioides (CbRAS) |
| tyrosine aminotransferase (TAT) homologues | R. norvegus (RnTAT) | P. scutellarioides (CbTAT) |
| cytochrome P450-dependent hydroxylase (CYP) and reductases (CPR) homologues | S. miltiorrhiza (SmCYP98A14-CPR) | P. scutellarioides (CbCYP98A14-CPR) |
As part of the ANOVA analysis, the F-test was used to determine the statistical significance of the effect of each factor (the genes RAS, TAT, and CYP98A14-CPR) on the final response (rosmarinic acid titer). The F-value (and also p-value) of each factor in the DOE design is shown in Tables S2 and S3. It should be noted that a larger F-value for a factor means that the variation of the two mean production titers at 2 levels (gene origins) is significant, and the change in this factor has a large effect on the RA titer.31 The F-value is calculated as described by Zhou et al.31 The p-value is also calculated using Minitab19.2 software, with a significance of α = 0.05.
Rosmarinic Acid Production
The screening of constructed yeast strains for RA production was carried out in 24-deep-well plates with air-penetrable metal lids (EnzyScreen, The Netherlands) containing 2 mL of fresh mineral medium, which contains 7.5 g/L (NH4)2SO4, 14.4 g/L KH2PO4, 0.5 g/L MgSO4·7H2O, 2 mL/L trace metal (3.0 g/L FeSO4·7H2O, 4.5 g/L ZnSO4·7H2O, 4.5 g/L CaCl2·2H2O, 0.84 g/L MnCl2·2H2O, 0.3 g/L CoCl2·6H2O, 0.3 g/L CuSO4·5H2O, 0.4 g/L Na2MoO4·2H2O, 1.0 g/L H3BO3, 0.1 g/L KI, and 19.0 g/L Na2EDTA·2H2O), and 1 mL/L vitamin solutions (0.05 g/L D-biotin, 1.0 g/L D-pantothenic acid hemicalcium salt, 1.0 g/L thiamin–HCl, 1.0 g/L pyridoxin–HCl, 1.0 g/L nicotinic acid, 0.2 g/L p-aminobenzoic acid, and 25.0 g/L myo-inositol).53 To mimic fed-batch fermentation, 60 g/L polysaccharide powder (EnPump Substrate, Enpresso GmbH, Germany) with an enzyme mix of final concentration of 0.3% v/v (Reagent A, Enspresso GmbH, Germany) was added to the mineral media immediately before use. To prevent RA degradation, 10 mM filter-sterile ascorbic acid was supplemented to the media right before inoculation.
Metabolite Extraction
The preculture of each yeast strain was grown in 5 mL of YPD medium in 13 mL tubes overnight at 30 °C and 250 rpm. The inoculation of all strains was done with an adequate amount of preculture to obtain the starting OD600 = 0.1. The plates were light-protected, and incubated at 30 °C with shaking at 300 rpm at 5 cm orbit cast. Samples were taken after 72 h and analyzed for cell growth and metabolite extraction. The optimized extraction method was as follows: 800 μL of the culture was mixed 1:1 (v:v) with absolute ethanol supplemented with 0.01% w/v 3,5-di-tert-4-butylhydroxyltoluene (BHT). The mixture was then shaken vigorously using the Precellys T 24 homogenizer (Bertin Corp.) in three cycles of 6500 rpm for 45 s to disrupt the cells. After the disruption, the cell debris was separated by centrifuging for 30 min at 21 000g, and the supernatant was moved to amber-colored HPLC vials for metabolite analysis.
Analytical Methods
Cell growth was monitored by measuring the optical density at 600 nm (OD600) using a NanoPhotometer (Implen GmbH, Germany). The values were then used to calculate dry cell weight (DCW) of the culture in g/L using the following empirical equation:
For metabolite quantification, the standards for rosmarinic acid (RA), 3,4-dihydroxyphenyllactic acid (3,4-DHPL), p-coumaric acid (pCA), caffeic acid (CA), 4-hydroxyphenyllactic acid (4-HPL), and 4-hydroxyphenylpyruvic acid (4-HPP) with certified purities greater than 96% were purchased from Sigma-Aldrich (St. Louis, Missouri, USA). The LC system consisted of an Advance liquid chromatograph (Bruker, Fremont, CA, USA) equipped with a solvent delivery compartment with high-pressure mixing, a column compartment, and a CTC auto sampler. The injection volume was 1 μL. The separation of compounds was performed on an ACQUITY UPLC HSS T3, 100 mm × 2.1 mm, 1.8 μm particle size (Waters, Ireland). In front of the separation column was a Phenomenex krudkatcher filter, 0.5 μm. The total flow rate of eluent A (Milli-Q water with 0.1% formic acid) and B (acetonitrile with 0.1% formic acid) was 0.5 mL min–1. The initial gradient was 100% and held for 1 min, decreasing to 5% after 4 min and held for 1 min, then back to 100% after 0.1 min and held at 100% for 0.9 min. The total run time was 6 min. The column temperature was set at 45 °C. The MS/MS detection was performed on an EVOQ triple quadrupole instrument (Bruker, Fremont, CA, USA) equipped with an atmospheric pressure ionization (API) interface. The mass spectrometer was operated with electrospray both in the negative-ion mode (ESI–). The capillary voltage was set to 3000 V. The heated probe temperature was 350 °C, and the heated probe gas flow was 40 L h–1. The cone gas flow and temperature were 20 L/h and 350 °C, respectively. Nebulizer gas flow was set at 40 L/h, and the exhaust gas was turned on. Argon was used as the collision gas at a pressure of 1.5 mTorr. Detection was performed in multiple reacting monitoring (MRM) mode. The collision energies were optimized for the different compounds.
Acknowledgments
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No. 814408 (SHIKIFACTORY100 project). IB also acknowledges the financial support from the Novo Nordisk Foundation (Grant Agreement No. NNF10CC1016517) and from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (YEAST-TRANS project, Grant Agreement No. 757384). The authors wish to thank Diogo Romano for assistance with experimental work, and Peter Kötter for the use of the CEN.PK strain.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.0c00048.
Optimization of rosmarinic acid extraction method; regression statistics; lists of strains, plasmids, BioBricks, and primers; additional figures (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Park S. U.; Uddin R.; Xu H.; Kim Y. K.; Lee S. Y. (2008) Biotechnological applications for rosmarinic acid production in plant. Afr. J. Biotechnol. 7, 25. [Google Scholar]
- Takano H.; Osakabe N.; Sanbongi C.; Yanagisawa R.; Inoue K.; Yasuda A.; Natsume M.; Baba S.; Ichiishi E.; Yoshikawa T. (2004) Extract of Perilla frutescens enriched for rosmarinic acid, a polyphenolic phytochemical, inhibits seasonal allergic rhinoconjunctivitis in humans. Exp. Biol. Med. 229, 247–254. 10.1177/153537020422900305. [DOI] [PubMed] [Google Scholar]
- Stansbury J. (2014) Rosmarinic Acid as a Novel Agent in the Treatment of Allergies and Asthma*. J. Restor. Med. 3, 121–126. 10.14200/jrm.2014.3.0109. [DOI] [Google Scholar]
- Jiang W.-L.; Chen X.-G.; Qu G.-W.; Yue X.-D.; Zhu H.-B.; Tian J.-W.; Fu F.-H. (2009) Rosmarinic acid protects against experimental sepsis by inhibiting proinflammatory factor release and ameliorating hemodynamics. Shock 32, 608–613. 10.1097/SHK.0b013e3181a48e86. [DOI] [PubMed] [Google Scholar]
- Sanbongi C.; Takano H.; Osakabe N.; Sasa N.; Natsume M.; Yanagisawa R.; Inoue K.-I.; Sadakane K.; Ichinose T.; Yoshikawa T. (2004) Rosmarinic acid in perilla extract inhibits allergic inflammation induced by mite allergen, in a mouse model. Clin. Exp. Allergy 34, 971–977. 10.1111/j.1365-2222.2004.01979.x. [DOI] [PubMed] [Google Scholar]
- Wang J.; Xu H.; Jiang H.; Du X.; Sun P.; Xie J. (2012) Neurorescue effect of rosmarinic acid on 6-hydroxydopamine-lesioned nigral dopamine neurons in rat model of Parkinson’s disease. J. Mol. Neurosci. 47, 113–119. 10.1007/s12031-011-9693-1. [DOI] [PubMed] [Google Scholar]
- Hamaguchi T.; Ono K.; Murase A.; Yamada M. (2009) Phenolic Compounds Prevent Alzheimer’s Pathology through Different Effects on the Amyloid-β Aggregation Pathway. Am. J. Pathol. 175, 2557–2565. 10.2353/ajpath.2009.090417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippolyte I.; Marin B.; Baccou J. C.; Jonard R. (1992) Growth and rosmarinic acid production in cell suspension cultures of Salvia officinalis L. Plant Cell Rep. 11, 109–112. 10.1007/BF00232160. [DOI] [PubMed] [Google Scholar]
- Li M.; Borodina I. (2014) Application of synthetic biology for production of chemicals in yeast Saccharomyces cerevisiae.. FEMS Yeast Res. 15, 1–12. 10.1111/1567-1364.12213. [DOI] [PubMed] [Google Scholar]
- Stovicek V.; Holkenbrink C.; Borodina I. (2017) CRISPR/Cas system for yeast genome engineering: advances and applications. FEMS Yeast Res. 17, fox030 10.1093/femsyr/fox030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordán M. J.; Lax V.; Rota M. C.; Lorán S.; Sotomayor J. A. (2012) Relevance of carnosic acid, carnosol, and rosmarinic acid concentrations in the in vitro antioxidant and antimicrobial activities of Rosmarinus officinalis (L.) methanolic extracts. J. Agric. Food Chem. 60, 9603–9608. 10.1021/jf302881t. [DOI] [PubMed] [Google Scholar]
- Petersen M.; Häusler E.; Karwatzki B.; Meinhard J. (1993) Proposed biosynthetic pathway for rosmarinic acid in cell cultures of Coleus blumei Benth. Planta 189, 10–14. 10.1007/BF00201337. [DOI] [Google Scholar]
- Petersen M. (1997) Cytochrome P450-dependent hydroxylation in the biosynthesis of rosmarinic acid in Coleus. Phytochemistry 45, 1165–1172. 10.1016/S0031-9422(97)00135-0. [DOI] [Google Scholar]
- Petersen M.; Abdullah Y.; Benner J.; Eberle D.; Gehlen K.; Hücherig S.; Janiak V.; Kim K. H.; Sander M.; Weitzel C.; Wolters S. (2009) Evolution of rosmarinic acid biosynthesis. Phytochemistry 70, 1663–1679. 10.1016/j.phytochem.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Bak S.; Beisson F.; Bishop G.; Hamberger B.; Höfer R.; Paquette S.; Werck-Reichhart D. (2011) Cytochromes P450. Arab. Book Am. Soc. Plant Biol. 9, e0144. 10.1199/tab.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault H.; Bassard J.-E.; Hamberger B.; Werck-Reichhart D. (2014) Cytochrome P450-mediated metabolic engineering: current progress and future challenges. Curr. Opin. Plant Biol. 19, 27–34. 10.1016/j.pbi.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Bloch S. E.; Schmidt-Dannert C. (2014) Construction of a chimeric biosynthetic pathway for the de novo biosynthesis of rosmarinic acid in Escherichia coli. ChemBioChem 15, 2393–2401. 10.1002/cbic.201402275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.; Yan Y. (2012) Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex. Microb. Cell Fact. 11, 42. 10.1186/1475-2859-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M.; Petersen M. (2011) Distinct substrate specificities and unusual substrate flexibilities of two hydroxycinnamoyltransferases, rosmarinic acid synthase and hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl-transferase, from Coleus blumei Benth. Planta 233, 1157–1171. 10.1007/s00425-011-1367-2. [DOI] [PubMed] [Google Scholar]
- Huang Q.; Lin Y.; Yan Y. (2013) Caffeic acid production enhancement by engineering a phenylalanine over-producing Escherichia coli strain. Biotechnol. Bioeng. 110, 3188–3196. 10.1002/bit.24988. [DOI] [PubMed] [Google Scholar]
- Yao Y.-F.; Wang C.-S.; Qiao J.; Zhao G.-R. (2013) Metabolic engineering of Escherichia coli for production of salvianic acid A via an artificial biosynthetic pathway. Metab. Eng. 19, 79–87. 10.1016/j.ymben.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Jiang J.; Bi H.; Zhuang Y.; Liu S.; Liu T.; Ma Y. (2016) Engineered synthesis of rosmarinic acid in Escherichia coli resulting production of a new intermediate, caffeoyl-phenyllactate. Biotechnol. Lett. 38, 81–88. 10.1007/s10529-015-1945-7. [DOI] [PubMed] [Google Scholar]
- Zhuang Y.; Jiang J.; Bi H.; Yin H.; Liu S.; Liu T. (2016) Synthesis of rosmarinic acid analogues in Escherichia coli. Biotechnol. Lett. 38, 619–627. 10.1007/s10529-015-2011-1. [DOI] [PubMed] [Google Scholar]
- Li Z.; Wang X.; Zhang H. (2019) Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering. Metab. Eng. 54, 1–11. 10.1016/j.ymben.2019.03.002. [DOI] [PubMed] [Google Scholar]
- Huang B.; Yi B.; Duan Y.; Sun L.; Yu X.; Guo J.; Chen W. (2008) Characterization and expression profiling of tyrosine aminotransferase gene from Salvia miltiorrhiza (Dan-shen) in rosmarinic acid biosynthesis pathway. Mol. Biol. Rep. 35, 601–612. 10.1007/s11033-007-9130-2. [DOI] [PubMed] [Google Scholar]
- Eberle D.; Ullmann P.; Werck-Reichhart D.; Petersen M. (2009) cDNA cloning and functional characterisation of CYP98A14 and NADPH:cytochrome P450 reductase from Coleus blumei involved in rosmarinic acid biosynthesis. Plant Mol. Biol. 69, 239–253. 10.1007/s11103-008-9420-7. [DOI] [PubMed] [Google Scholar]
- Weitzel C.; Petersen M. (2010) Enzymes of phenylpropanoid metabolism in the important medicinal plant. Planta 232, 731–742. 10.1007/s00425-010-1206-x. [DOI] [PubMed] [Google Scholar]
- Prabhu P. R.; Hudson A. O.; Pike R. (2010) Identification and Partial Characterization of an L-Tyrosine Aminotransferase (TAT) from Arabidopsis thaliana. Biochem. Res. Int. 2010, 549572. 10.1155/2010/549572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove J. L.; Scoble H. A.; Mathews W. R.; Baumstark B. R.; Biemann K. (1989) The structure of tyrosine aminotransferase. Evidence for domains involved in catalysis and enzyme turnover. J. Biol. Chem. 264, 45–53. [PubMed] [Google Scholar]
- Smanski M. J.; Zhou H.; Claesen J.; Shen B.; Fischbach M.; Voigt C. A. (2016) Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 14, 135–149. 10.1038/nrmicro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.; Vonk B.; Roubos J. A.; Bovenberg R. A. L.; Voigt C. A. (2015) Algorithmic co-optimization of genetic constructs and growth conditions: application to 6-ACA, a potential nylon-6 precursor. Nucleic Acids Res. 43, 10560–10570. 10.1093/nar/gkv1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E. M.; Zhao Z.; Gielesen B. E. M.; Wu L.; Benjamin Gordon D.; Roubos J. A.; Voigt C. A. (2018) Iterative algorithm-guided design of massive strain libraries, applied to itaconic acid production in yeast. Metab. Eng. 48, 33–43. 10.1016/j.ymben.2018.05.002. [DOI] [PubMed] [Google Scholar]
- Oh H. K. F.; Siow L. F.; Lim Y. Y. (2019) Approach to preserve phenolics in Thunbergia laurifolia leaves by different drying treatments. J. Food Biochem. 43, e12856 10.1111/jfbc.12856. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Smuts J. P.; Dodbiba E.; Rangarajan R.; Lang J. C.; Armstrong D. W. (2012) Degradation study of carnosic acid, carnosol, rosmarinic acid, and rosemary extract (Rosmarinus officinalis L.) assessed using HPLC. J. Agric. Food Chem. 60, 9305–9314. 10.1021/jf302179c. [DOI] [PubMed] [Google Scholar]
- Gil E. de S.; Enache T. A.; Oliveira-Brett A. M. (2013) Redox behaviour of verbascoside and rosmarinic acid. Comb. Chem. High Throughput Screening 16, 92–97. 10.2174/1386207311316020003. [DOI] [PubMed] [Google Scholar]
- Petersen M.; Simmonds M. S. J. (2003) Rosmarinic acid. Phytochemistry 62, 121–125. 10.1016/S0031-9422(02)00513-7. [DOI] [PubMed] [Google Scholar]
- Rodriguez A.; Strucko T.; Stahlhut S. G.; Kristensen M.; Svenssen D. K.; Forster J.; Nielsen J.; Borodina I. (2017) Metabolic engineering of yeast for fermentative production of flavonoids. Bioresour. Technol. 245, 1645–1654. 10.1016/j.biortech.2017.06.043. [DOI] [PubMed] [Google Scholar]
- Li M.; Schneider K.; Kristensen M.; Borodina I.; Nielsen J. (2016) Engineering yeast for high-level production of stilbenoid antioxidants. Sci. Rep. 6, 36827. 10.1038/srep36827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassing E.-J.; de Groot P. A.; Marquenie V. R.; Pronk J. T.; Daran J.-M. G. (2019) Connecting central carbon and aromatic amino acid metabolisms to improve de novo 2-phenylethanol production in Saccharomyces cerevisiae. Metab. Eng. 56, 165–180. 10.1016/j.ymben.2019.09.011. [DOI] [PubMed] [Google Scholar]
- Rodriguez A.; Kildegaard K. R.; Li M.; Borodina I.; Nielsen J. (2015) Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab. Eng. 31, 181–188. 10.1016/j.ymben.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Liu Q.; Yu T.; Li X.; Chen Y.; Campbell K.; Nielsen J.; Chen Y. (2019) Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat. Commun. 10, 4976. 10.1038/s41467-019-12961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K.-D.; Kötter P.; Stansfield I.; Stark M. J. (2007) 25 Yeast Genetic Strain and Plasmid Collections. Methods Microbiol. 36, 629–666. 10.1016/S0580-9517(06)36025-4. [DOI] [Google Scholar]
- Jendresen C. B.; Stahlhut S. G.; Li M.; Gaspar P.; Siedler S.; Förster J.; Maury J.; Borodina I.; Nielsen A. T. (2015) Highly Active and Specific Tyrosine Ammonia-Lyases from Diverse Origins Enable Enhanced Production of Aromatic Compounds in Bacteria and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 81, 4458–4476. 10.1128/AEM.00405-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Kildegaard K. R.; Chen Y.; Rodriguez A.; Borodina I.; Nielsen J. (2015) De novo production of resveratrol from glucose or ethanol by engineered. Metab. Eng. 32, 1–11. 10.1016/j.ymben.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Partow S.; Siewers V.; Bjørn S.; Nielsen J.; Maury J. (2010) Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast 27, 955–964. 10.1002/yea.1806. [DOI] [PubMed] [Google Scholar]
- Uwimana N.; Collin P.; Jeronimo C.; Haibe-Kains B.; Robert F. (2017) Bidirectional terminators in Saccharomyces cerevisiae prevent cryptic transcription from invading neighboring genes. Nucleic Acids Res. 45, 6417–6426. 10.1093/nar/gkx242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop-Fabre M. M.; Jakočiu̅nas T.; Stovicek V.; Dai Z.; Jensen M. K.; Keasling J. D.; Borodina I. (2016) EasyClone-MarkerFree: A vector toolkit for marker-less integration of genes into Saccharomyces cerevisiae via CRISPR-Cas9. Biotechnol. J. 11, 1110–1117. 10.1002/biot.201600147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J.; Guiping L.; Xiujun L.; Xincai H.; Hongmei L. (2009) Influence of growth regulators and sucrose concentrations on growth and rosmarinic acid production in calli and suspension cultures of Coleus blumei. Nat. Prod. Res. 23, 127–137. 10.1080/14786410801890338. [DOI] [PubMed] [Google Scholar]
- Yao J.; He Y.; Su N.; Bharath S. R.; Tao Y.; Jin J.-M.; Chen W.; Song H.; Tang S.-Y. (2020) Developing a highly efficient hydroxytyrosol whole-cell catalyst by de-bottlenecking rate-limiting steps. Nat. Commun. 11, 1515. 10.1038/s41467-020-14918-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordjour E.; Adipah F. K.; Zhou S.; Du G.; Zhou J. (2019) Metabolic engineering of Escherichia coli BL21 (DE3) for de novo production of L-DOPA from D-glucose. Microb. Cell Fact. 18, 74. 10.1186/s12934-019-1122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkenbrink C.; Dam M. I.; Kildegaard K. R.; Beder J.; Dahlin J.; Belda D. D.; Borodina I. (2018) EasyCloneYALI: CRISPR/Cas9-Based Synthetic Toolbox for Engineering of the Yeast. Biotechnol. J. 13, 1700543. 10.1002/biot.201700543. [DOI] [PubMed] [Google Scholar]
- Antony J. (2014) Full Factorial Designs, in Design of Experiments for Engineers and Scientists (Second ed.) (Antony J., Ed.) 2nd ed., Chapter 6, pp 63–85, Elsevier, Oxford. [Google Scholar]
- Verduyn C.; Postma E.; Scheffers W. A.; Dijken J. P. V. (1992) Effect of benzoic acid on metabolic fluxes in yeasts: A continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8, 501–517. 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.