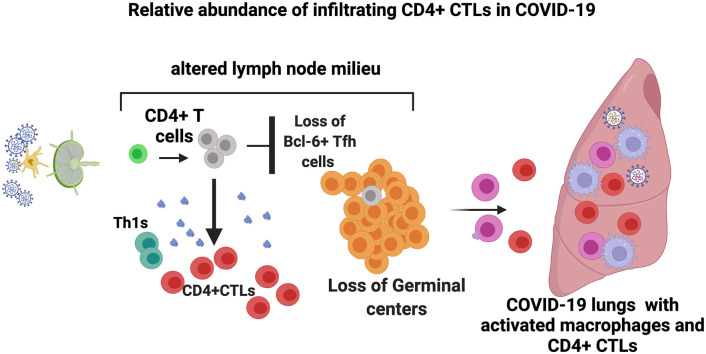
Abstract
Many studies have been performed in severe COVID-19 on immune cells in the circulation and on cells obtained by bronchoalveolar lavage. Most studies have tended to provide relative information rather than a quantitative view, and it is a combination of approaches by various groups that is helping the field build a picture of the mechanisms that drive severe lung disease. Approaches employed to date have not revealed information on lung parenchymal T cell subsets in severe COVID-19. Therefore, we sought to examine early and late T cell subset alterations in the lungs and draining lymph nodes in severe COVID-19 using a rapid autopsy protocol and quantitative imaging approaches. Here, we have established that cytotoxic CD4+ T cells (CD4 + CTLs) increase in the lungs, draining lymph nodes and blood as COVID-19 progresses. CD4 + CTLs are prominently expanded in the lung parenchyma in severe COVID-19. In contrast CD8+ T cells are not prominent, exhibit increased PD-1 expression, and no obvious increase is seen in the number of Granzyme B+ CD8+ T cells in the lung parenchyma in severe COVID-19. Based on quantitative evidence for re-activation in the lung milieu, CD4 + CTLs may be as likely to drive viral clearance as CD8+ T cells and may also be contributors to lung inflammation and eventually to fibrosis in severe COVID-19.
Keywords: COVID-19, CD8+ T cells, Cytotoxic CD4+ T cells
Graphical abstract
1. Introduction
When hospitalized patients with acute COVID-19 were traced back to establish whether they had a previous mRNA vaccine course in the previous 3 to 6 months or had been naturally infected, the Odds Ratio supporting a protective effect of vaccination over natural infection ranged from 5 to 7 depending on the vaccine taken. However, when patients were segregated by age in those 64 and older the Odds Ratio favoring prior mRNA vaccination over natural infection rose to 19.5 [1]. Persistence of SARS-CoV-2 in the gut for many months after a lung infection has also been documented by biopsy studies [2]. A recent study further suggests that SARS-CoV-2 persists in pulmonary and extra-pulmonary sites including the brain for up to seven months after infection [3]. These studies suggest that adaptive immunity against this virus is sub-optimal at least in the first six to eight months or so after infection, and that viral persistence might contribute to the Post-Acute Sequelae of SARS-CoV-2 (PASC) [4,5].
Why might prior infection result in sub-optimal adaptive immunity? In severe COVID-19, rapid autopsy studies revealed a loss of T follicular helper cells and of germinal centers. We showed that this phenomenon was linked to a prominent extrafollicular B cell response in the lymph nodes and the blood and switched memory phenotype B cells were readily observed in the lymph nodes and were shown to be antigen-specific in the blood [6]. It is now well established that the humoral immune response is sub-optimal roughly during the first six months of COVID-19, with switched memory B cells largely of extra-follicular origin, a lack of neutralization breadth, and limited somatic hypermutation [[7], [8], [9]]. While the altered cytokine milieu in the lungs and in draining lymph nodes likely impedes T follicular helper cell differentiation, we sought to ask if quantitative evidence supported the attenuation of cellular immunity as well in COVID-19, that might possibly help explain why there is viral persistence in this disease.
Lymphocyte populations have been extensively analyzed in patients with COVID-19. Lymphopenia has been widely documented, especially in severe disease, but the underlying basis for this phenomenon remains poorly understood [10,11]. Flow cytometry including high dimensional analyses of peripheral blood T lymphocytes has demonstrated the heterogeneity of the immune response in patients hospitalized with COVID-19 and has helped identify several immune signatures that have been linked to distinct clinical outcomes [12]. Crucial studies on circulating lymphocytes have established the existence of antigen-specific T cells in COVID-19 [[13], [14], [15], [16], [17], [18], [19]]. Single cell RNA-sequencing studies have also been performed on circulating T cells in severely ill patients [17,20,21], and one study showed the expansion of antigen-specific TPH (T peripheral helper)-like and cytotoxic CD4+ T cells [17]. Expansion in the blood of antigen specific CD8+ T cells that contribute to virus elimination has been documented in other viral infections [22,23], but interestingly in COVID-19 although antigen-specific CD8+ T cells have been seen in the blood these cells have not revealed evidence either for quantitative temporal increases in the acute stages or evidence of antigen-specific CD8+ T cell clonal expansion in acute COVID-19.
Many studies have been performed on bronchoalveolar lavage exudates from severely ill COVID-19 patients [[24], [25], [26], [27]], and different studies have revealed dysfunctional CD8+ T cells and expansion of Th17 cells [28,29]. An autopsy study using a spatial transcriptomic imaging approach revealed few detectable T cells in the lungs in severe COVID-19 and no information on T cell subsets [30]. Two studies using single nuclear RNA sequencing on rapid autopsies of the lung (single cell sequencing was not technically possible) suggested that T cells are not abundant in the lungs, but no details about T cell subsets were elicited by this approach as well [31,32]. One study on autopsies used targeted RNA-sequencing examining the expression of selected genes and examined gene signatures to quantify immune cells and indirectly concluded that CD8+ T cells were increased late in severe disease, though they had a dysfunctional signature [33]. An examination of antigen-specific T cell responses has been undertaken in convalescent subjects undergoing lung resection for other reasons, and revealed SARS-CoV-2 specific T cell responses in the lung during convalescence [34]. No study to date has directly quantified T cell subsets in the lung parenchyma or in the draining thoracic lymph nodes in severe COVID-19 patients.
We show here, however, using a rapid autopsy protocol and quantitative imaging approaches, that in the later stages of severe COVID-19 there was no increase in CD8+ T cells or of re-activated Granzyme B (GZMB) + CD8+ T cells (CD8 + CTLs) in the lungs compared to the earlier stages of this disease. However, these CD8+ T cells expressed high levels of PD-1. Cytotoxic CD4+ T cells (CD4 + CTLs) were the only CD4+ T cell subset that exhibited significant expansion in the lungs in severe COVID-19 and the numbers of GZMB + CD8+ T cells and GZMB+ CD4 + CTLs in this end-organ were roughly equivalent. These data suggest that CD8+ T cells in severe COVID-19 may be dysfunctional, possibly due to exhaustion, and CD4+ CTLs likely complement remaining functional CD8 + CTLs in order to limit or eliminate the virus. These quantitative findings provide the only picture of T cell subset alterations in the lung parenchyma induced in the acute stages of severe SARS-CoV-2 infection.
2. Materials and methods
2.1. Human subjects
2.1.1. Tissue analysis cohort
Thoracic lymph nodes and lung samples from COVID-19 patients were obtained through the Brigham and Women's Hospital Department of Pathology and included 22 patients (information on age and gender in Table S1and S2) including 16 with laboratory confirmed COVID-19 who underwent autopsy in 2020. All patients had tested positive for SARS-CoV-2 by RT-PCR of nasopharyngeal swabs in a laboratory during hospital admission. All cases were divided into two groups; early (less than ten days from respiratory symptoms onsets to death, hospitalization of up to ten days), and late (hospitalized for 15–36 days prior to death). Patients who had been on trials for known immunosuppressive agents were excluded. Controls were age- and gender-matched individuals with lung disease who were exhaustively investigated when hospitalized but had no evidence for SARS-CoV-2 infection (Table S2; Kaneko et al., 2020).
2.1.2. Peripheral blood cohort
Peripheral blood samples were drawn from both Outpatients and Inpatients with COVID-19 at Massachusetts General Hospital and fresh blood was analyzed for flow cytometry using a multi- color panel (information in Table S3). Data is presented on T cell populations from 62 patients, including moderately ill, severely ill and convalescent patients. Convalescence was defined as a clinically asymptomatic state on the date of blood draw, either from a baseline asymptomatic state or recuperated from moderate clinical symptoms of COVID- 19. Moderate disease was defined as active clinical symptoms of COVID-19 on the date of blood draw that did not necessitate a hospital admission. Severe disease was defined as active clinical symptoms of COVID-19 on the date of blood draw that did necessitate a hospital admission. Severe disease was further subdivided by maximum CRP level during hospital admission as CRP < 200 mg/L classified as ‘CRP intermediate (int)’ and CRP > 200 mg/L as ‘CRP high (hi)’.
2.2. Study approval
This study was conducted with the approval of the Institutional Review Boards at the Massachusetts General Hospital and the Brigham and Women's Hospital.
2.3. Multi-color immunofluorescence staining
Tissue samples were fixed in formalin, embedded in paraffin, and sectioned. These specimens were incubated with the following antibodies: anti-CD3 (A045229–2; DAKO), anti-CD4 (ab133616; Abcam), anti-Pan-CK (ab27988; Abcam), anti-CD31 (3528; Cell Signaling Technology), anti-SLAMF7 (HPA055945; Sigma-Aldrich), anti-CX3CR1 (ab8021; Abcam), anti-T-bet (ab150440; Abcam), anti-GATA3 (MA1028; Invitrogen), anti-Ror gamma (ab212496; Abcam), anti-CXCR5 (clone: MAB190; R&D Systems), anti-FoxP3 (clone: 98377; Cell Signaling Technology), anti-CD8 (ab85792; Abcam), anti-PD1 (B13300; Lifespan Bioscience), anti-GZMB (ab4095; Abcam), anti-HLA-DR (ab20181; Abcam), anti-TGF-β (ab27969; Abcam) and anti-cleaved caspase-3 (9664; Cell Signaling Technology) followed by incubation with a secondary antibody using an OpalTM Multiplex Kit (Perkin Elmer). The samples were mounted with ProLongTM Diamond Antifade mountant containing DAPI (Invitrogen).
2.4. Microscopy and quantitative image analysis
Images of the tissue specimens were acquired using the TissueFAXS platform (TissueGnostics). For quantitative analysis, the entire area of the tissue was acquired as a digital grayscale image in five channels with filter settings for FITC, Cy3, Cy5 and AF75 in addition to DAPI. Both in the lungs and in lymph nodes, we analyzed the entire area including airspaces such as alveoli, blood vessels and lymph vessels. Cells of a given phenotype were identified and quantitated using the TissueQuest software (TissueGnostics), with cut-off values determined relative to the positive controls. This microscopy-based multicolor tissue cytometry software permits multicolor analysis of single cells within tissue sections similar to flow cytometry. StrataQuest (TissueGnostics) software was also used to quantify cell-to-cell contact. In the StrataQuest cell-to-cell contact application, masks of the nuclei based on DAPI staining establish the inner boundary of the cytoplasm and the software “looks” outwards towards the plasma membrane boundary. Overlap of at least 3 pixels of adjacent cell markers is required to establish a “contact” criterion. Although the software has been developed and validated more recently, the principle of the method and the algorithms used have been described in detail elsewhere [35].
2.5. Quantification and statistical analysis
For tissue studies, flow cytometry and clinical correlations, data are shown as median [interquartile range: 25th–75th percentiles]. GraphPad Prism version 8 was used for statistical analysis, curve fitting and linear regression. A two-tailed Mann-Whitney U test was used to calculate p values for continuous, non-parametric variables. The paired t-test was also performed to calculate p-values between two variables in the same subject. For comparing more than one population, Kruskal-Wallis testing was used with Dunn's multiple comparison testing. A p value of <0.05 was considered significant.
2.6. Flow cytometry
1 million fresh PBMCs were stained within 2 h of isolation. Prior to antibody staining, Fc receptors were blocked using human FcR blocking reagent (Miltenyi) at a concentration of 1:50 at 4 °C for 10 min. Cells were surface stained at 4 °C, protected from light, using optimized concentrations of fluorochrome-conjugated primary antibodies for 30 min as well as live/dead fixable blue stain (Thermo Fisher) at a concentration of 1:20 using the following antibody panels (clone, manufacturer): CD14 (HCD14, Biolegend), CD19 (HIB19, Biolegend), CD34 (581, Biolegend), CD94 (HP-3D9, BD Biosciences), CD11c (Bu15, Biolegend), CD3 (UCHT1, BD Biosciences), CD4 (SK3, BD Biosciences), CD8 (RPA-T8, BD Biosciences), CD38 (HIT2, BD Biosciences), HLA-DR (L243, Biolegend), CD57 (NK-1, BD Biosciences), CD25 (2A3, BD Biosciences), CXCR3 (G025H7, Biolegend), CCR6 (G034E3, Biolegend), CCR4 (L291H4, Biolegend), CX3CR1 (2A9–1, Biolegend), PD-1 (EH12.1, BD Biosciences), CXCR5 (RF8B2, BD Biosciences), CD62L (DREG-56, Biolegend), CD45RA (HI100, BD Biosciences), CD127 (A019D5, Biolegend), CD16 (3G8, BD Biosciences), CD56 (HCD56, Biolegend).
Cells were then washed in PBS and fixed using 4% paraformaldehyde for 30 min at at 4 °C. Flow cytometry was performed on a BD Symphony (BD Biosciences, San Jose, CA) and rainbow tracking beads (8 peaks calibration beads, Fisher) were used to ensure consistent signals between flow cytometry batches. FCS files were analyzed, and B cell subsets were quantified using FlowJo software (version 10).
2.7. Peptide stimulation of PBMCs
PBMCs from 3 hospitalized subjects with acute COVID-19 were used for the activation-induced marker assay. Briefly, cryopreserved PBMCs were thawed and rested at 37 °C prior to peptide stimulation. Cells were quantified and plated at a concentration of approximately 2.5 million cells per mL in AIM-V media. Cells were then incubated for 16 h at 37C, 5% CO2 with or without the addition of an overlapping peptide pool of Spike antigen (JPT Peptide Technologies; PM- WCPV-S) according to the manufacturer's recommended protocol. Cells were harvested, washed, and underwent standard Fc block and cell staining for flow cytometric analysis. Additional antibodies used in this assay not included in the other blood studies included mouse anti-human CD69-BB700 (BD Bioscience, clone FN50, stained at 1:50 at 4C), mouse anti-human CD38- BUV661 (BD Bioscience, clone HIT2, stained at 1:400 at 37C), mouse anti-human CX3CR1-APC (BioLegend, clone 2A9–1, stained at 1:50 at 4C), and mouse anti-human CD28-BV480 (BD Bioscience, clone CD28.2, stained at 1:100 at 37C) .
3. Results
3.1. A paucity of activated CD8+ T cells in the lungs late in severe COVID-19
Staining of epithelial and endothelial cells confirmed that lung architecture is severely distorted in severe COVID-19 (Fig. S1A).Given the rampant viral pneumonia and severe lung inflammation seen in severe COVID-19, the expectation was that there would be large-scale infiltration of the lungs by activated CD8+ T cells, and that these cells would be re-activated in the lungs [[36], [37], [38]]. As evidence for recent re-activation, we examined GZMB expression in CD8+ T cells. There was little discernible infiltration of CD8+ T cells in the lungs in severe COVID-19 and no increase in CD8+ T cells expressing GZMB as compared with age-matched control lung tissues from rapid autopsies on non-COVID-19 patients collected in the same time window (Fig. 1A, B). Indeed, the total numbers of activated lung parenchymal CD8+ T cells in severe COVID-19 averaged as little as 10 to 15 cells per sq. mm of lung parenchyma. Moreover, the numbers of activated CD8+ T cells in a range of age-matched controls with no clinically established lung infection was higher than in the COVID-19 patients. Although there was also no evidence for CD8+ T cell lymphopenia in the draining thoracic lymph nodes in severe COVID-19, no expansion of CD8+ T cells was observed in these draining lymph nodes in both the acute and resolving phases of severe disease (Fig. 2A). In addition to the high levels of TNF-α in draining lymph nodes, we noted the induction of a potentially immunosuppressive milieu that is TGF-β rich (Fig. S1B, S1C).
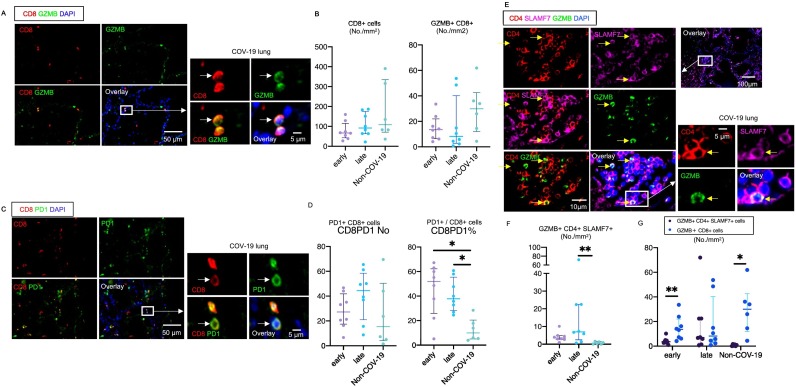
Fig. 1.
A paucity of activated CD8+ T cells and infiltration of SLAMF7+ Granzyme B+ CD4+ T cells in the lungs in severe COVID-19
(A) Representative multi-color immunofluorescence images of CD8 (red), GZMB (green) and DAPI (blue) staining in a lung from a COVID (COV) -19 patient. Arrows indicate GZMB expressing CD8+ T cells. (B) Absolute numbers of CD8+ T cells (left) and GZMB+CD8+ T cells (right) in lungs from early (purple) (n = 8) and late (blue) (n = 8) COV-19 and non-COVID-19 (non-COV-19) patients (green) (n = 6). (C) Representative multi-color immunofluorescence images of CD8 (red), PD1 (green) and DAPI (blue) staining in a lung from a COV-19 patient. Arrows indicate PD1+ CD8+ T cells. (D) Absolute numbers and relative proportions of PD1 + CD8+ T cells in lungs from early (purple) (n = 8) and late (blue) (n = 8) COV-19 and non-COV-19 patients (green) (n = 6). (E) Representative multi-color immunofluorescence images of CD4 (red), SLAMF7 (purple), GZMB (green) and DAPI (blue) staining in a lung from a COV-19 patient. Arrows indicate GZMB+ SLAMF7+ CD4 + CTLs. (F) Absolute numbers of GZMB+ SLAMF7+ CD4+ T cells in lungs from early (purple) (n = 8) and late (blue) (n = 8) COV-19 and non-COV-19 patients (green) (n = 6). (G) Absolute numbers of GZMB+ SLAMF7+ CD4 + CTLs (purple) and GZMB+ CD8+ T cells (blue) in lungs from early (left) (n = 8) and late (middle) (n = 8) COV-19 patients and non-COV-19 patients (right) (n = 6). Multiple comparisons are controlled for by Kruskal-Wallis test. P values are calculated by paired t-test. *p < 0.05; **p < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
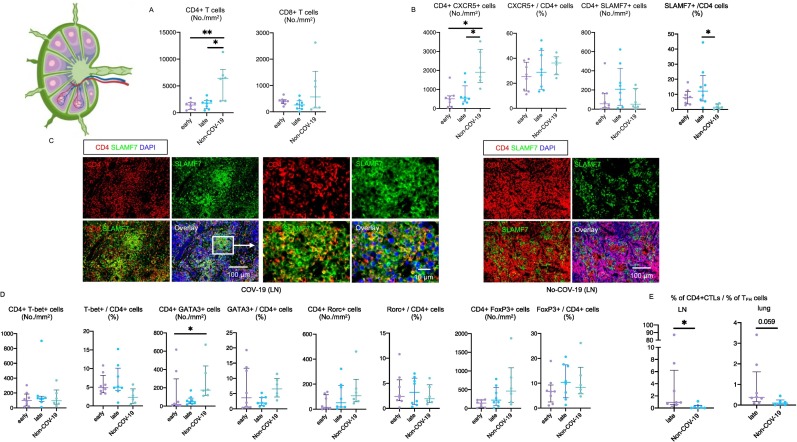
Fig. 2.
Relative increase in CD4 + CTLs but no expansion of CD8+ T cells in COVID-19 lymph nodes.
(A) Absolute numbers of total CD4+ T cells (left) and CD8+ T cells (right) in lymph nodes from early (purple) (n = 8) and late (blue) (n = 8) COV-19 patients and non-COV-19 patients (green) (n = 6). (B) Absolute numbers and relative proportions of TFH cells (left) and CD4 + CTLs (right) in lungs from early (purple) (n = 8) and late (blue) (n = 8) COV-19 patients and non-COV-19 patients (green) (n = 6). (C) Representative multi-color immunofluorescence images of CD4 (red), SLAMF7 (green) and DAPI (blue) staining in lungs from a COV-19 patient (left) and non- COV-19 patient (right). Yellow cells with red (CD4) and green (SLAMF7) merged are CD4 + CTLs and are frequently seen in COV-19 patients. In contrast, red and green can still be separately identified, and yellow cells are relatively sparse in non-COV-19 patients. (D) Absolute numbers and relative proportions of TH1 (left), TH2 (middle right), TH17 (middle right) and Treg (right) cells in lungs from early (purple) (n = 8) and late (blue) (n = 8) COV-19 patients and non-COV-19 patients (green) (n = 6). (E) Ratios of percentage of CD4 + CTLs to that of TFH cells in total CD4+ T cells in lymph nodes (LN) (left) and lungs (right) from late COV-19 patients (purple) (n = 8) and non-COV-19 patients (blue) (n = 6). Mann-Whitney U test was used to calculate p value. *p < 0.05; **p < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
PD-1 expression per se on CD8+ T cells (studied in the blood) does not necessarily imply dysfunction [39], but a relatively high proportion of the limited numbers of CD8+ T cells found in the lungs in severe COVID-19 do express PD-1 (Fig. 1C, D). Since TGF-β signaling can also contribute to PD-1 expression in T cells, we suspect that the altered lymph node milieu contributes to this process [40].
Although only a small fraction of CD8+ T cells express GZMB, the numbers of CD4+ SLAMF7+ T cells that express GZMB increased late in the disease and at this time the absolute numbers of GZMB expressing CD4 + CTLs and CD8+ T cells were comparable (Fig. 1E-G). In summary, we did not observe any increase in cytotoxic CD8+ T cells in the lungs in severe COVID-19 compared to controls, most of these cells expressed PD-1 and there was a paucity of re-activated GZMB expressing CD8+ T cells in this end-organ in acute disease.
3.2. Loss of TFH cells and an increase in CD4 + CTLs in draining lymph nodes in severe COVID- 19
Examination of the draining lymph nodes revealed CD4+ T cell lymphopenia and a late increase in cells with a CD4 + CTL phenotype (Fig. 2A-D). Most of the CD4+ T cell lymphopenia could be attributed to the loss of CXCR5 + CD4+ TFH cells in the lymph nodes (Fig. 2B). The increase in CD4 + CTL phenotype cells in draining lymph nodes was also validated by CX3CR1 staining (Fig. S2A, S2B). Relative attenuation was seen of TH2 cells.
Interestingly there is an inverse relationship between the decrease in T follicular helper cells in draining lymph nodes and the lungs in severe COVID-19 and an increase in CD4 + CTLs (Fig. 2E). This phenomenon may represent an evolutionary conserved immune deviation phenomenon that occurs in a secondary lymphoid organ milieu, but only in the context of severe intracellular infections, driving activated CD4+ T cells away from a humoral TFH cell function towards a more cellular TH1 or CD4 + CTL immune context [[41], [42], [43]].
3.3. Increased infiltration of the lungs by cytotoxic CD4+ T cells is relatively prominent as severe COVID-19 progresses
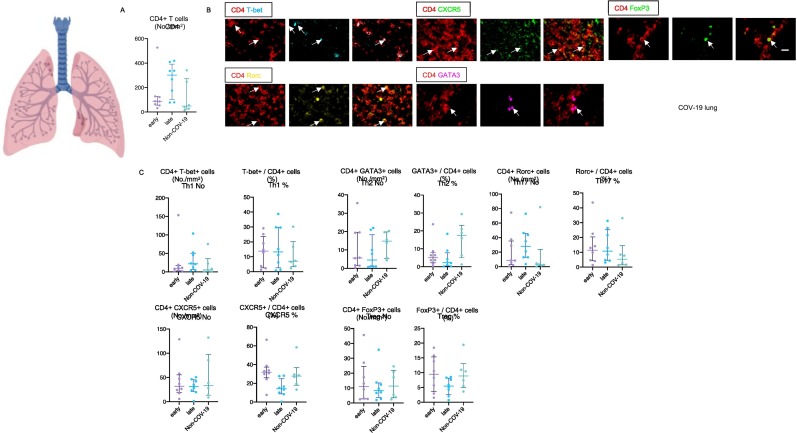
We systematically quantitated prominent CD4+ T cell subsets in patients with COVID-19 in the lung and in draining thoracic lymph nodes. In spite of documented CD4+ T cell lymphopenia in draining lymph nodes in severe COVID-19 (see also Fig. 2A), infiltration of the lungs by CD4+ T cells was comparable in absolute numbers between COVID-19 patients and age-matched controls (Fig. 3A) [6]. The absolute numbers and relative proportions of T-bet expressing TH1 cells, GATA3 expressing TH2 cells, RORC expressing TH17 cells, CXCR5 expressing TFH cells and FOXP3 expressing Tregs in the lungs in COVID-19 were comparable to the numbers seen in controls (Fig. 3B, C).
Fig. 3.
CD4+ T cell subsets in the lungs in severe COVID-19.
(A) Absolute numbers of total CD4+ T cells in lungs from early (purple) (n = 8) and late (blue) (n = 8) COV-19 and non-COV-19 patients (green) (n = 6). (B) Representative multi-color staining showing TH1, TH2, TH17, TFH and Treg cells in lungs from COV-19 patients. [TH1: CD4+ (red) T-bet+ (light blue)] [TH2: CD4+ (red) GATA3+ (purple)] [TH17: CD4+ (red) RORγ+ (yellow)] [TFH: CD4+ (red) CXCR5+ (green)] [Treg: CD4+ (red) FoxP3+ (green)]. (C) Absolute numbers and relative proportions of TH1 (upper left), TH2 (upper middle), TH17 upper right), TFH (lower left) and Treg (lower middle) cells in lungs from early (purple) (n = 8) and late (blue) (n = 8) COV-19 patients and non-COV-19 patients (green) (n = 6). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
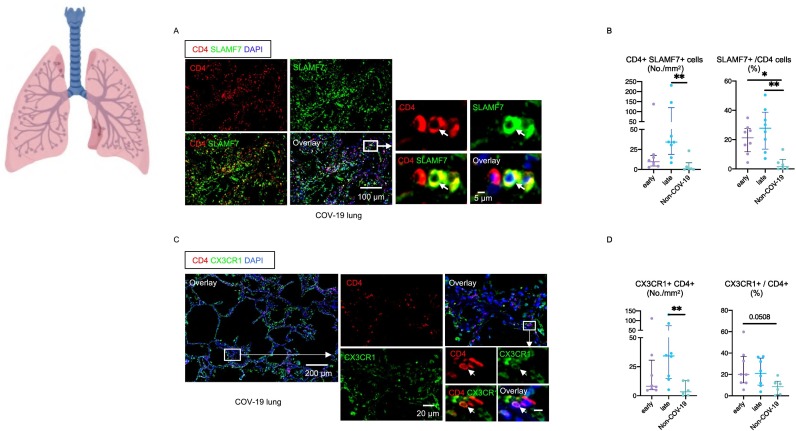
The validity of using CX3CR1 and SLAMF7 as markers for CD4 + CTLs has been previously established and we have re-examined this issue and revalidated it in the COVID-19 context by multi-color flow cytometry and the intracellular analysis of granzymes and perforin (Fig. S3) [44,45]. The one CD4+ T cell subset that was clearly expanded in the lungs of COVID-19 patients was the CD4 + SLAMF7+ CD4 + CTL population (Fig. 4A, B). A very similar increase was observed using CX3CR1 instead of SLAMF7 as a marker for CD4 + CTLs (Fig. 4C, D). CD4 + CTLs were increased both in terms of absolute numbers as well as proportions and this increase was most significant in late or resolving patients. Because CD4 + CTLs induce cell death in a class II restricted manner, we examined the expression of HLA Class II in the lungs. In this milieu of inflammation, many endothelial cells and about 60% of epithelial cells expressed HLA Class II (Fig. 5A, B). CD4 + CTLs expand in many human fibrotic diseases and secrete cytokines [[46], [47], [48], [49]]. The late influx of CD4 + CTLs and the clearance of virus may be linked to both an increase in apoptotic death of infected cells (Fig. 5C, D) and enhanced inflammation in the lungs.
Fig. 4.
CD4 + CTLs are the most expanded CD4 + T cell subset in COVID-19 lungs.
(A) Representative multi-color immunofluorescence images of CD4 (red), SLAMF7 (green) and DAPI (blue) staining in a lung from a COV-19 patient. Arrows indicate SLAMF7+ CD4 + CTLs. (B) Absolute numbers and relative proportions of SLAMF7+ CD4 + CTLs in lungs from early (purple) (n = 8) and late (blue) (n = 8) COV-19 patients and non-COV-19 patients (green) (n = 6). (C) Representative multi-color immunofluorescence images of CD4 (red), CX3CR1 (green) and DAPI (blue) staining in a lung from a COV-19 patient. (D) Absolute numbers and relative proportions of CX3CR1+ CD4 + CTLs in lungs from early (purple) (n = 8) and late (blue) (n = 8) COV-19 patients and non-COV-19 patients (green) (n = 6). Multiple comparisons are controlled for by Kruskal-Wallis test. *p < 0.05; **p < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
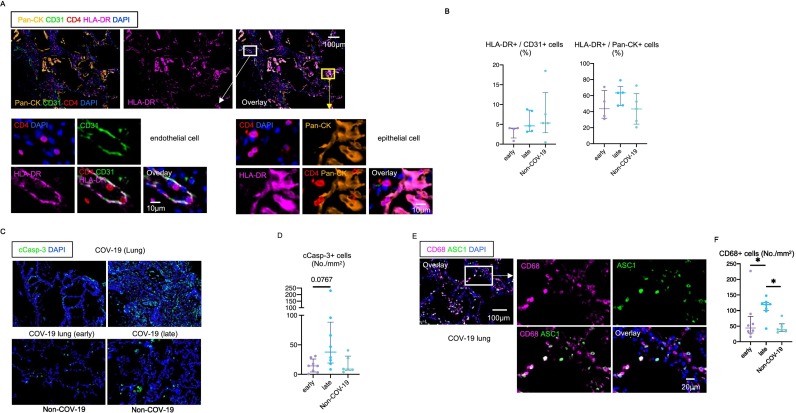
Fig. 5.
Epithelial cells and endothelial cells express HLA class II in severe COVID-19.
(A) Representative multi-color immunofluorescence images of Pan-CK (orange), CD31 (green), CD4 (red), HLA-DR (purple) and DAPI (blue) staining in a lung from a COV-19 patient. White and yellow boxes highlight HLA-DR+ CD31+ endothelial cells (left) and HLA-DR+ Pan-CK+ epithelial cells (right). CD4+ T cells are observed in close proximity to HLA-DR+ endothelial cells and epithelial cells. (B) Relative proportions of HLA-DR+ endothelial cells (left) and epithelial cells (right) in lungs from early (purple) (n = 4) and late (blue) (n = 5) COV-19 and non-COV-19 patients (green) (n = 5). (C) Representative multi-color immunofluorescence images of cleaved-caspase3 (c-Casp3) (green) and DAPI (blue) staining in lungs from COV-19 patients (upper) and non-COV-19 patients (lower). (D) Absolute numbers of total c-Casp3+ cells in lungs from early (purple) (n = 8) and late (blue) (n = 8) COV-19 patients and non-COV-19 patients (green) (n = 6). (E) Representative multi-color immunofluorescence images of CD68 (purple), ASC1 (green) and DAPI (blue) staining in a lung from a COVID-19 patient. (F) Absolute numbers of CD68+ cells in lungs from early (purple) (n = 8) and late (blue) (n = 8) COVID-19 and non-COVID-19 patients (green) (n = 6). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Macrophage infiltration is a hallmark of COVID-19, and the macrophages and T cells likely collaborate to promote inflammation in the lungs in COVID-19 [50,51]. A recent study utilized multipe modalities including tissue single-cell genomics described the accumulation of macrophages expressing profibrotic genes in COVID-19 patinets with acute respiratory distress syndrome (ARDS) and pulmonary fibrosis [52]. We therefore investigated whether CD4 + CTLs coexist with macrophages in COVID-19 lung lesions. Lung infiltrating macrophages with abundant ASC-1 specks (a hallmark of inflammasome activation) were abundant early and even more abundant later in the disease, indicative of ongoing inflammation at a time when SARS-CoV-2 may have largely been eliminated from the lungs (Fig. 5E, F).
Overall, these data indicated that CD4 + CTLs represent a prominent CD4+ T cell subset that infiltrates and/or expands in the major affected organ in severe COVID-19, and this infiltration/expansion is most prominent in the resolving phase of the disease (in spite of persisting CD4+ T cell lymphopenia in draining lymph nodes). Macrophages also infiltrate the lung, and that is more pronounced in the late phase as well as CD4 + CTLs, which may contribute to the pulmonary fibrosis.
3.4. Blood “TFH-like” cells do not mirror the loss of TFH cells in lymph nodes in severe COVID-19
Although T cell subsets in the lungs reflect changes initiated in draining lymph nodes, many immune cells seen in lymph nodes and in the lungs are localized to tissue sites and T cells in the blood and the lungs do not follow parallel paths for the most part. The blood does however more readily permit the examination of antigen-specific cells and it is currently not technically possible to efficiently examine antigen-specific T cells in autopsy tissues. CD4 + CTLs have been studied in the blood of COVID-19 patients by examining SARS-CoV-2 peptide-specific CD4+ T cells using an assay in which the authors gated on antigen- activated CD4+ T cells in vitro and then performed single-cell RNA-sequencing [17]. Though it is not possible to gauge the magnitude of the specific response in those reported blood studies, leave alone determine whether these cells do or do not infiltrate tissues, the demonstration of antigen specific cells in blood does provide valuable information regarding immunity being virus-directed.
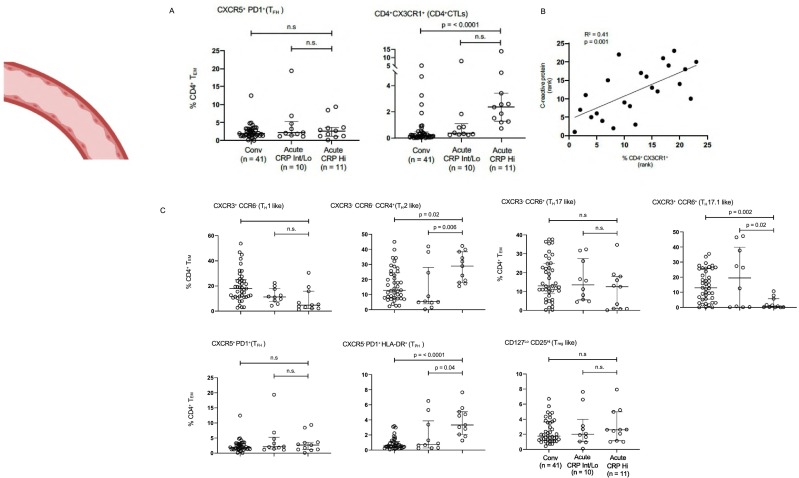
We used 24 color-flow cytometry to analyze effector CD4+ T cell subsets from 62 convalescent and acute COVID-19 patients. CD4+ CD62Llo CD45RAlo effector T cells were gated on and eight major non-overlapping CD4+ T cell subsets (or their close equivalents) were interrogated using surface markers. As seen in Fig. 6A and C, the only CD4+ T cell subsets that were expanded in acute disease compared with convalescence were CD4+ CX3CR1+ T cells (CD4 + CTLs) and CD4 + CXCR5-PD-1+ HLA-DR+ T peripheral helper (TPH) cells. CXCR5 + CD4 + TFH-like cells in the blood were neither expanded nor decreased. The correlation of both CD4 + CTL and TPH cell expansion with disease severity is also depicted in Fig. 6B. Limited AIM assays also confirm that blood CD4 + CTLs are SARS-CoV-2 specific (Fig. S4). These data, although they cannot be extrapolated even in a semi-quantitative way to the lungs in severe COVID-19, indirectly support our findings, based on the examination of the lung parenchyma, that CD4 + CTLs likely represent a functionally relevant CD4+ T cell subset in severe COVID-19.
Fig. 6.
Blood TFH-like cells do not mirror the loss of TFH cells in lymph nodes in severe COVID-19.
(A) TFH cells (left) and CD4 + CTLs (right) were defined by surface markers including chemokine receptors and cytokine receptors. Dot plots display circulating T cell subsets as proportions of total CD4+ effector-memory T cells. Multiple comparisons are controlled for by Kruskal-Wallis test. *p < 0.05; **p < 0.01. CRP = C-reactive protein. Conv = convalescent. (B) CD4 + CTL accumulation correlates with disease severity as assessed by CRP levels. (C) CD4+ T cell subsets in the peripheral blood of patients with COVID-19 at states of convalescence (n = 39), severe illness with an intermediate maximum CRP level during hospitalization (< 200 mg/L; severe (CRP int); n = 10), and severe illness with a high maximum CRP level during hospitalization (> 200 mg/L; severe (CRP hi); n = 11) as defined by the clinical criteria listed in supplementary Information Table 3. A. TH1, TH2, TH17 and Treg cells were defined by surface markers including chemokine receptors and cytokine receptors and not on the basis of transcription factors, hence the nomenclature with a suffix “-like”. Dot plots display circulating T cell subsets as proportions of total CD4+ effector-memory T cells. All p-values were calculated using the Kruskal-Wallis test to control for multiple comparisons. CRP = C-reactive protein. Conv = convalescent.
4. Discussion
When an organ is infected, lymphocyte populations are activated in draining lymph nodes and home to the site of infection. The degree to which CD8+ T cells in particular, can be clonally expanded, has long been established to be enormous, and indeed this was first established in a human viral context in 1996 [53]. In many murine models of respiratory infections including by respiratory syncytial virus and influenza, expansion of CD8+ T cells is seen in the draining lymph nodes and in the lungs [[54], [55], [56]].
In order to connect specific T cells to disease pathology in humans, T cell subsets in the affected end organ require to be quantitatively analyzed; ideally data should be obtained in parallel from draining lymph nodes. Synthesizing quantitative data on T cell subsets in the lung parenchyma and lymph nodes obtained from rapid autopsies with other non-quantitative data, including published reports on antigen-specific T cell subsets in the blood of living patients, may help build a more accurate picture of the adaptive T cell response in the lungs to SARS-CoV-2. Comprehensively following changes in individual T cell subsets in any end-organ and the relevant draining lymph nodes has rarely been undertaken in any human infection, though newer technologies may make this possible. In COVID-19 there is no ethical basis by which antigen-specific T cells could have been studied at the site of active disease (namely the lungs) in severely ill patients, but we have nevertheless successfully interrogated both the lungs and draining lymph nodes for major T cell subsets and established the lack of any discernible increase in activated CD8+ T cells and the relative expansion of CD4 + CTLs. We consider it likely that this milieu-dependent transient impairment of CD8+ T cell immunity is not antigen-specific and might partially contribute to viral and fungal superinfections described in severe COVID-19 [57]. It is also the likely reason for the long-term tissue persistence of SARS-CoV-2 in many infected individuals.
We show here that in severe COVID-19 there is no discernible increase of CD8+ T cells in the draining lymph nodes or in the lungs. We did not gauge proliferation or activation directly and can only comment on absolute cell numbers in the lung parenchyma and in draining lymph nodes. One likely biological explanation for this striking lack of CD8+ T cell reactivation and expansion has emerged from studies that indicate that the ORF-6 and ORF-8 proteins of SARS CoV-2 may contribute to the downregulation of HLA class I expression in infected cells. ORF-6 has been shown to both inhibit the transcriptional induction and nuclear entry of NLRC5, a key transcriptional regulator of HLA class I expression [58]. SARS-CoV-2 ORF-8 is a resident endoplasmic reticulum protein that can physically associate with newly synthesized HLA class I molecules and promote their entry into autophagosomes as a prelude to lysosomal degradation [59]. However, other mechanisms might impair the initial activation of CD8+ T cells in draining lymph nodes as well. The severity of the viral infection and possibly excessive DAMP generation in severe COVID-19 significantly alter the milieu of the draining lymph nodes, inducing very high levels of TNF-α as we have previously reported [6]. TNF-α can influence CD8+ T cells in a number of positive and negative ways but high levels of this cytokine are known to cause the elimination of CD8+ effectors [60]. Type I interferons have also been reported to enhance the cytolytic activity of recruited CD8+ effector/memory T cells and thus control early viral replication in a lung viral infection setting [61]. The reported presence of autoantibodies to Type I interferons as a pre-disposing factor for severe COVID-19 pneumonia [62], the functional lack of Type I interferons could contribute to CD8+ T cell dysfunctionality in some patients with severe COVID-19.
We speculate that during severe intracellular infections perhaps provide the conditions for the expansion of CD4+ CTLs, a CD4+ sub-lineage that may possibly have evolved to compensate for CD8+ T cell dysfunction that is sometimes observed in severe or chronic infections, CD4 + CTLs have previously been only studied in the blood in the context of COVID-19 and this previous description of the relatively selective expansion of CD4 + CTLs in the blood in severe COVID-19 are consistent with our current results on T cells in the lung parenchyma, draining lymph nodes and the blood [17].
Although there have been many qualitative studies of T cell in COVID-19 in blood and bronchoalveolar lavage specimens, no other quantitative assessments of CD8+ T cells or CD4+ T cell subsets in the lung parenchyma during acute severe COVID-19 have been reported. It has been established that immune changes in the circulation in COVID-19 poorly mirror immunity in the lungs, underscoring the importance of quantifying immune cells in the lung parenchyma [63,64].
The study described here remains the only quantitative study of lung parenchymal T cell subsets in patients with severe acute COVID-19. A study using targeted RNA-sequencing on autopsy tissues from patients who succumbed to severe COVID-19 has suggested relative CD8+ T cell expansion later in severe disease and the signature also suggested CD8+ T cell dysfunction [33].
The increase in inflammation that accompanies the late surge in CD4 + CTLs may potentially contribute to viral clearance, but this surge may also have negative consequences. CD4 + CTLs can, in some disease contexts, possibly provide the adaptive immune trigger for fibrosis [[46], [47], [48], [49]]. Given the abundance of macrophages in the lungs, the studies on bronchoalveolar lavage derived cells that suggest that infected macrophages induce chemoattractant to attract T cells and the potential contribution of both these cell types to the fibrosis seen in COVID-19, the possibility that these cells might also contribute in some patients to the Post-Acute Sequelae of SARS-CoV-2 (PASC) should be kept in mind. Indeed, progressive lung fibrosis has been reported even after less severe acute COVID-19 [52,65].
5. Conclusions
A quantitative imaging approach identified temporal changes in T cell subsets, including the expansion of CD4 + CTLs and no obvious increase of activated CD8+ T cells in the lungs in severe COVID-19. A schematic model from our previous and present study for altered lymph node milieu resulting in sub-optimal immune pressure in the lungs in COVID-19 is presented in graphical abstract. The mechanisms by which an immunosuppressive draining lymph node milieu is generated in the disease and the exact molecular mechanisms that drive what appears to be an immune deviation event causing developing TFH cells to possibly branch off into a CD4 + CTL fate, require further investigation.
Ethics approval
This study was conducted with the approval of the Institutional Review Boards at the Massachusetts General Hospital and the Brigham and Women's Hospital.
Author contributions
Conceptualization (SP, RFP, NK, JB, HK, CP, BDW, JL, GG, XY, ML); Methodology (NK, JB, CP, VSM, JF, HL, TD, KP, GG); Investigation (NK, JB, HK, CP, HL, KL, VSM, RFP); Resources (RFP, BDW, XY, ML, GG, JL, VSM, AT, KL, MW); Original Draft (SP, NK, CP); Review/Editing (SP, NK, CP, BDW, JL, ML); Supervision (SP).
Funding
This work was supported by NIH U19 AI110495 to SP and the Massachusetts Consortium on Pathogen Readiness. NK was supported by JSPS KAKENHI (Grant Number: 21KK0163), Mochida foundation and the QR program from Kyushu University.
Data availability statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Declaration of Competing Interest
None.
Acknowledgments
We thank members of the Massachusetts consortium on Pathogen Readiness Specimen Working Group for blood samples, and Doug Kwon and Brooke Spencer for access to Tissue Core specimens. The graphical abstract was prepared using Biorender.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.clim.2022.108991.
Appendix A. Supplementary data
Supplementary Figures 1-4, Supplementary Figure legends and Supplementary Tables 1-3
Supplementary material 2
References
- 1.Bozio C.H., et al. Laboratory-confirmed COVID-19 among adults hospitalized with COVID-19-like illness with infection-induced or mRNA vaccine-induced SARS-CoV-2 immunity - nine states, January-September 2021. MMWR Morb. Mortal. Wkly Rep. 2021;70(44):1539–1544. doi: 10.15585/mmwr.mm7044e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaebler C., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591(7851):639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniel C., et al. 2022. Nature Portfolio. [Google Scholar]
- 4.Al-Aly Z., Xie Y., Bowe B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature. 2021;594(7862):259–264. doi: 10.1038/s41586-021-03553-9. [DOI] [PubMed] [Google Scholar]
- 5.Crook H., et al. Long covid-mechanisms, risk factors, and management. Bmj. 2021;374:n1648. doi: 10.1136/bmj.n1648. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko N., et al. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183(1):143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodruff M.C., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21(12):1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark S.A., et al. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell. 2021;184(10):2605–2617.e18. doi: 10.1016/j.cell.2021.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robbiani D.F., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584(7821):437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan W.-J., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X., et al. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583(7816):437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 12.Mathew D., et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508) doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grifoni A., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiskopf D., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5(48) doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni L., et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52(6):971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng Y., et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21(11):1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meckiff B.J., et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4(+) T cells in COVID-19. Cell. 2020;183(5):1340–1353.e16. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kusnadi A., et al. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8(+) T cells. Sci. Immunol. 2021;6(55) doi: 10.1126/sciimmunol.abe4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saini S.K., et al. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8(+) T cell activation in COVID-19 patients. Sci. Immunol. 2021;6(58) doi: 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilk A.J., et al. Multi-omic profiling reveals widespread dysregulation of innate immunity and hematopoiesis in COVID-19. J. Exp. Med. 2021;218(8) doi: 10.1084/jem.20210582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephenson E., et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021;27(5):904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Callan M.F., et al. Direct visualization of antigen-specific CD8+ T cells during the primary immune response to Epstein-Barr virus in vivo. J. Exp. Med. 1998;187(9):1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bree G.J., et al. Selective accumulation of differentiated CD8+ T cells specific for respiratory viruses in the human lung. J. Exp. Med. 2005;202(10):1433–1442. doi: 10.1084/jem.20051365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chua R.L., et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38(8):970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 25.Liao M., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 26.Ren L.L., et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin. Med. J. 2020;133(9):1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szabo P.A., et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity. 2021;54(4):797–814.e6. doi: 10.1016/j.immuni.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wauters E., et al. Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages. Cell Res. 2021;31(3):272–290. doi: 10.1038/s41422-020-00455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y., et al. Clonal expansion and activation of tissue-resident memory-like Th17 cells expressing GM-CSF in the lungs of severe COVID-19 patients. Sci. Immunol. 2021;6(56) doi: 10.1126/sciimmunol.abf6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desai N., et al. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat.Commun. 2020;11:6319. doi: 10.1038/s41467-020-20139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delorey T.M., et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595(7865):107–113. doi: 10.1038/s41586-021-03570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melms J.C., et al. A molecular single-cell lung atlas of lethal COVID-19. Nature. 2021;595(7865):114–119. doi: 10.1038/s41586-021-03569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nienhold R., et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat. Commun. 2020;11(1):5086. doi: 10.1038/s41467-020-18854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grau-Expósito J., et al. Peripheral and lung resident memory T cell responses against SARS-CoV-2. Nat. Commun. 2021;12(1):3010. doi: 10.1038/s41467-021-23333-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ecker R.C., Steiner G.E. Microscopy-based multicolor tissue cytometry at the single-cell level. Cytometry A. 2004;59(2):182–190. doi: 10.1002/cyto.a.20052. [DOI] [PubMed] [Google Scholar]
- 36.Heidema J., et al. CD8+ T cell responses in bronchoalveolar lavage fluid and peripheral blood mononuclear cells of infants with severe primary respiratory syncytial virus infections. J. Immunol. 2007;179(12):8410–8417. doi: 10.4049/jimmunol.179.12.8410. [DOI] [PubMed] [Google Scholar]
- 37.Heidema J., et al. Dynamics of human respiratory virus-specific CD8+ T cell responses in blood and airways during episodes of common cold. J. Immunol. 2008;181(8):5551–5559. doi: 10.4049/jimmunol.181.8.5551. [DOI] [PubMed] [Google Scholar]
- 38.Jozwik A., et al. RSV-specific airway resident memory CD8+ T cells and differential disease severity after experimental human infection. Nat. Commun. 2015;6(1):10224. doi: 10.1038/ncomms10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rha M.S., et al. PD-1-expressing SARS-CoV-2-specific CD8(+) T cells are not exhausted, but functional in patients with COVID-19. Immunity. 2021;54(1):44–52.e3. doi: 10.1016/j.immuni.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park B.V., et al. TGFβ1-mediated SMAD3 enhances PD-1 expression on antigen-specific T cells in Cancer. Cancer Discov. 2016;6(12):1366–1381. doi: 10.1158/2159-8290.CD-15-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryg-Cornejo V., et al. Severe malaria infections impair germinal center responses by inhibiting T follicular helper cell differentiation. Cell Rep. 2016;14(1):68–81. doi: 10.1016/j.celrep.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Popescu M., Cabrera-Martinez B., Winslow G.M. TNF-α contributes to lymphoid tissue disorganization and germinal center B cell suppression during intracellular bacterial infection. J. Immunol. 2019;203(9):2415–2424. doi: 10.4049/jimmunol.1900484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elsner R.A., Shlomchik M.J. IL-12 blocks Tfh cell differentiation during Salmonella infection, thereby contributing to germinal center suppression. Cell Rep. 2019;29(9):2796–2809.e5. doi: 10.1016/j.celrep.2019.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiskopf D., et al. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc. Natl. Acad. Sci. U. S. A. 2015;112(31):E4256–E4263. doi: 10.1073/pnas.1505956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perugino C.A., et al. CD4+ and CD8+ cytotoxic T lymphocytes may induce mesenchymal cell apoptosis in IgG4-related disease. J. Allergy Clin. Immunol. 2021;147:368–382. doi: 10.1016/j.jaci.2020.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattoo H., et al. Clonal expansion of CD4(+) cytotoxic T lymphocytes in patients with IgG4-related disease. J. Allergy Clin. Immunol. 2016;138(3):825–838. doi: 10.1016/j.jaci.2015.12.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maehara T., et al. Cytotoxic CD4+ T lymphocytes may induce endothelial cell apoptosis in systemic sclerosis. J. Clin. Invest. 2020;130:2451–2464. doi: 10.1172/JCI131700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., et al. A novel CD4+ CTL subtype characterized by chemotaxis and inflammation is involved in the pathogenesis of Graves’ orbitopathy. Cell. Mol. Immunol. 2021;18(3):735–745. doi: 10.1038/s41423-020-00615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allard-Chamard H., et al. CD4(+)CTLs in fibrosing mediastinitis linked to histoplasma capsulatum. J. Immunol. 2021;206(3):524–530. doi: 10.4049/jimmunol.2000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rendeiro A.F., et al. The spatial landscape of lung pathology during COVID-19 progression. Nature. 2021;593(7860):564–569. doi: 10.1038/s41586-021-03475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grant R.A., et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590(7847):635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wendisch D., et al. SARS-CoV-2 infection triggers profibrotic macrophage responses and lung fibrosis. Cell. 2021;184(26):6243–6261.e27. doi: 10.1016/j.cell.2021.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Callan M.F., et al. Large clonal expansions of CD8+ T cells in acute infectious mononucleosis. Nat. Med. 1996;2(8):906–911. doi: 10.1038/nm0896-906. [DOI] [PubMed] [Google Scholar]
- 54.Hufford M.M., et al. The effector T cell response to influenza infection. Curr. Top. Microbiol. Immunol. 2015;386:423–455. doi: 10.1007/82_2014_397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim T.H., Lee H.K. Differential roles of lung dendritic cell subsets against respiratory virus infection. Immune Netw. 2014;14(3):128–137. doi: 10.4110/in.2014.14.3.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McDermott D.S., et al. Central role of dendritic cells in shaping the adaptive immune response during respiratory syncytial virus infection. Futur. Virol. 2011;6(8):963–973. doi: 10.2217/fvl.11.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu X., et al. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285 doi: 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoo J.S., et al. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat. Commun. 2021;12(1):6602. doi: 10.1038/s41467-021-26910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y., et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc. Natl. Acad. Sci. U. S. A. 2021;118(23) doi: 10.1073/pnas.2024202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mehta A.K., Gracias D.T., Croft M. TNF activity and T cells. Cytokine. 2018;101:14–18. doi: 10.1016/j.cyto.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kohlmeier J.E., et al. Type I interferons regulate cytolytic activity of memory CD8(+) T cells in the lung airways during respiratory virus challenge. Immunity. 2010;33(1):96–105. doi: 10.1016/j.immuni.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bastard P., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370(6515) doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daamen A.R., et al. Comprehensive transcriptomic analysis of COVID-19 blood, lung, and airway. Sci. Rep. 2021;11(1):7052. doi: 10.1038/s41598-021-86002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sposito B., et al. The interferon landscape along the respiratory tract impacts the severity of COVID-19. Cell. 2021;184(19):4953–4968.e16. doi: 10.1016/j.cell.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doane J.J., et al. Progressive pulmonary fibrosis after non-critical COVID-19: a case report. Am. J. Case Rep. 2021;22 doi: 10.12659/AJCR.933458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-4, Supplementary Figure legends and Supplementary Tables 1-3
Supplementary material 2
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.