Abstract
Fluoroquinolone-resistant mutants, selected from a wild-type Escherichia coli K-12 strain and its Mar mutant by exposure to increasing levels of ofloxacin on solid medium, were analyzed by Northern (RNA) blot analysis, sequencing, and radiolabelled ciprofloxacin accumulation studies. Mutations in the target gene gyrA (DNA gyrase), the regulatory gene marR, and additional, as yet unidentified genes (genes that probably affect efflux mediated by the multidrug efflux pump AcrAB) all contributed to fluoroquinolone resistance. Inactivation of the acrAB locus made all strains, including those with target gene mutations, hypersusceptible to fluoroquinolones and certain other unrelated drugs. These studies indicate that, in the absence of the AcrAB pump, gyrase mutations fail to produce clinically relevant levels of fluoroquinolone resistance.
Fluoroquinolone (FQ) resistance in Escherichia coli can be caused by mutations in the target topoisomerases of the drugs, DNA gyrase (e.g., in gyrA) (11, 30) and topoisomerase IV (e.g., in parC) (10, 13, 14). Mutations that affect regulatory genes such as marA (3, 5) or soxS (2) also lead to FQ resistance. The latter genes regulate intracellular drug concentrations by producing decreased uptake and/or increased efflux of the drug (1). In E. coli, overexpression of MarA causes decreased expression of the OmpF porin (6) as well as increased expression of the multidrug efflux pump AcrAB (23), thereby conferring resistance to a large number of antimicrobial agents (for a review, see reference 1). Studies with mutants selected in vitro have been confined to descriptions of phenotypic differences (MICs, outer membrane profiles, accumulation of FQs) or have focused on mutations in the quinolone resistance-determining regions (QRDRs) of gyrA and/or parC (10, 25, 27, 29). In this in vitro study we sought to understand the role of the AcrAB efflux pump in the FQ resistance mediated by topoisomerase mutations acquired during selection on ofloxacin. The AcrAB efflux pump was found to be critical to the FQ resistance level.
(This study was presented in part at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy, 24 to 27 September 1998, San Diego, Calif.)
MATERIALS AND METHODS
Antibiotics, chemicals, and media.
Ofloxacin (OFL) was kindly donated by Hoechst, Frankfurt, Germany. Radiolabelled [14C]ciprofloxacin ([14C]CIP) was a generous gift of Bayer AG, Leverkusen, Germany. Carbonyl cyanide m-chlorophenylhydrazone (CCCP) was purchased from Sigma Chemical Co., St. Louis, Mo., and organic solvents were purchased from Aldrich, Milwaukee, Wis. Strains were grown in Luria-Bertani (LB) broth (10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl per liter) unless otherwise noted.
Bacterial strains and plasmids.
All strains were derivatives of plasmid-free E. coli K-12 strain AG100 (8) and its Mar mutant AG112. The latter was selected on tetracycline in two steps (this study). Wild-type E. coli GC4468, its derived Sox mutant JTG1078 (soxR105 [9]), and plasmid pSXS bearing the soxS gene on a 432-bp fragment (2) were a generous gift of B. Demple. Strain JZM120 (JC7623ΔacrAB::Tn903 Kanr) (17) was kindly supplied by H. Nikaido.
Selection of FQ-resistant mutants.
Mutants 1-AG100, 2-AG100, 3*-AG100, and 4*-AG100 and mutants 1-AG112, 2-AG112, and 3-AG112 were sequential step mutants derived from strains AG100 and AG112, respectively, on solid media. For each step, about 1011 cells of an overnight culture were plated on several LB agar plates supplemented with increasing concentrations of OFL (0.5 to 16 μg/ml). Single colonies were further purified on OFL-supplemented agar plates. Mutants 1-AG100 and 2-AG100 came out of one series, while mutants 3*-AG100 and 4*-AG100 came from a second series.
Susceptibility testing.
The MICs of selected antimicrobial agents were determined by a standard broth microdilution procedure with cation-adjusted Mueller-Hinton broth (Becton Dickinson, Cockeysville, Md.) and an inoculum of 5 × 105 CFU/ml according to National Committee for Clinical Laboratory Standards (NCCLS) performance and interpretive guidelines (20). The antimicrobial agents in the commercially available microtiter plates (Merlin Diagnostics GmbH, Bornheim, Germany) included the FQs CIP, enoxacin, fleroxacin, norfloxacin, OFL, pefloxacin, sparfloxacin (SPX), and trovafloxacin (TVA). They also included tetracycline (TET), chloramphenicol (CML), trimethoprim, cefoxitin (CFOX), cefaclor, cefixime, and loracarbef. The MICs of bile salts were determined by a broth macrodilution procedure: sodium cholate and sodium deoxycholate were serially diluted in twofold steps in LB broth, and tubes were inoculated with 5 × 105 CFU/ml. The MICs of both bile salts (data not shown) and antibiotics were determined twice in independent experiments, with reproducible results.
P1 transduction.
acrAB-deleted strains were constructed by P1 transduction (26) of ΔacrAB::Tn903 Kanr from strain JZM120 into AG100, AG112, and all derived FQ-resistant mutants (18). The AcrAB gene-deleted strains were designated with the suffix “AK”, e.g., AG100AK, 1-AG100AK, and AG112AK. Two independent transductants were saved for each recipient. Deletion of the gene for AcrAB was confirmed by the absence of intact target DNA in a PCR assay and by greatly increased susceptibility to bile salts (data not shown), as reported previously (29). Besides positive and negative controls in each reaction, the gapA gene was used as the internal standard for determination of the validities of the PCR assays.
DNA sequencing.
The QRDRs of gyrA (nucleotides 123 to 366) or parC (nucleotides 145 to 492) in the FQ-resistant mutants were amplified by PCR and were purified by use of Qiaquick spin columns (Qiagen, Hilden, Germany), as described previously (7; S. Conrad, L. Scheit, M. Oethinger, G. Klotz, R. Marre, and W. V. Kern, Abstr. 36th Intersci. Conf. Antimicrob. Agents Chemother, abstr. C9, 1996). marOR (which encodes the operator [marO] and repressor [marR] of the Mar operon) was amplified from base pairs 1311 to 1858 with primer pair ORAB2 and RK3 as described earlier (22). Direct cycle sequencing in both directions was performed with the same primers in an automatic 373A DNA Sequencer (Applied Biosystems).
RNA extraction and Northern blot analysis.
Northern blot analysis was performed as described previously (22). In brief, RNA was harvested by a cesium chloride method from mid-logarithmic-phase cultures grown at 30°C. For assessment of the state of the marRAB operon or the soxRS operon, cultures were split and half the cultures were induced with 5 mM sodium salicylate (marRAB operon) or 1.3 mM paraquat (soxRS operon). The level of transcription from both operons was assessed by hybridization of radiolabelled DNA probes (marA or soxS) to the membrane-bound RNA (20 μg/lane), exposure on a PhosphoImager screen, and visualization with Image-Quant Software (Molecular Dynamics, Sunnyvale, Calif.), as described recently (22).
Accumulation of [14C]CIP in whole cells.
Cultures were grown to the logarithmic phase in LB broth at 30°C, washed in 50 mM potassium phosphate–0.2% glucose (pH 7.4), and resuspended in the same buffer to an optical density at 600 nm (OD600) of 5 to 7. [14C]CIP (specific activity, 59 mCi/mmol) was added to 10 μM. Accumulation was measured at equilibrium after 5 and 15 min at 30°C by dilution of 50 μl of the cell-labelling suspension into 5 ml of 100 mM LiCl–50 mM KPO4 (pH 7.4), collection of cells immediately on Metricel mixed-cellulose ester membrane filters (pore size, 0.45 μm; Gelman Sciences Inc., Ann Arbor, Mich.), and washing with 5 ml of the same buffer. The filters were dried, and the radioactivity was assayed with a liquid scintillation counter by using Betafluor (National Diagnostics, Somerville, N.J.). The counting efficiency was 90%. The amount of radiolabel that bound to filters in the absence of cells was subtracted. When used, CCCP, which destroys the proton motive force, was added at 20 min to a final concentration of 200 μM, and the levels of accumulation of CIP were assayed 5 and 15 min thereafter. The assay was designed in a way to investigate up to five strains in one experiment and to include, in addition, AG112 as a control strain. Results were calculated as accumulation of CIP in picomoles per OD600 unit, where 1 OD600 unit represented the number of cells in 1 ml when the OD600 was equal to 1 (approximately 109 E. coli cells; about 0.3 mg of protein). The ratio (expressed as percent) of CIP accumulation of energized cells divided by that of deenergized (CCCP-treated) cells was used as an indirect measure of active efflux (15). The values used to obtain this ratio were the separate averages of accumulation before (5 and 15 min) and after (25 min and 35 min) the addition of CCCP.
RESULTS AND DISCUSSION
Selection of FQ-resistant mutants in vivo.
The number of mutants observed for the total number of cells plated at the different levels of OFL ranged between 8 × 10−8 and 1 × 10−10, which agrees well with previous data (10, 27, 29). None of the mutants was defective in growth. The first mutation step increased resistance to OFL by eightfold in both AG100 and its Mar mutant AG112 (Table 1). Subsequent increases were two- to fourfold in all steps (Table 1 and Table 2), with the highest MIC being that of OFL for 3-AG112 (MIC, 8 μg/ml). The MICs of all FQs increased in parallel, with the order of MICs being OFL > CIP > TVA = SPX (Table 2).
TABLE 1.
FQ resistance mutations in E. coli AG100 and its Mar mutant AG112a
| Strain or mutant | OFL MIC (μg/ml) | Fold increase in OFL MICb | Substitution in gyrA | Mutation in marOR | marA expression | CIP uptake (% of deenergized accumulation) |
|---|---|---|---|---|---|---|
| AG100 | 0.03 | None | None | Wild type | 51 | |
| 1-AG100 | 0.25 | 8 | D87G | None | Wild type | 43 |
| 2-AG100 | 1 | 32 | D87G | Frameshift at aa 14 (position 1485 + 1 bp) | Overexpression | 30 |
| 3*-AG100 | 2 | 64 | D87G | Asp67Tyr (1643G→T) | Overexpression | 9 |
| 4*-AG100 | 4 | 128 | S83L, D87G | Asp67Tyr (1643G→T) | Overexpression | 9 |
| AG112 | 0.125 | 4 | None | 5-bp deletion (Δ1481–1485) | Overexpression | 25 |
| 1-AG112 | 1 | 32 | D87G | 5-bp deletion (Δ1481–1485) | Overexpression | 24 |
| 2-AG112c | 4 | 128 | D87G | 5-bp deletion (Δ1481–1485) | Overexpression | 12 |
| 3-AG112c | 8 | 256 | D87G | 5-bp deletion (Δ1481–1485) | Overexpression | 9 |
Abbreviations and symbols: aa, amino acid; Δ, deletion of the bases at the indicated positions; *, mutants were not derived from 2-AG100.
Fold change in MIC compared to the MIC for AG100 (wild type).
Strains have putative mutations at additional unknown loci that affect acrAB.
TABLE 2.
Effect of deletion of acrAB on susceptibility of FQ-resistant mutants of E. coli AG100 and AG112a
| Strain | MICa (μg/ml)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OFL
|
CIP
|
TVA
|
SPX
|
TET
|
CML
|
CFOX
|
||||||||
| + | Δ | + | Δ | + | Δ | + | Δ | + | Δ | + | Δ | + | Δ | |
| AG100 | 0.03 | ≤0.015 | ≤0.015 | ≤0.015 | 0.06 | ≤0.03 | ≤0.015 | ≤0.015 | 1 | 0.5 | 4 | 1 | 4 | 0.5 |
| 1-AG100 | 0.25 | 0.06 | 0.25 | 0.06 | 0.25 | 0.06 | 0.125 | ≤0.015 | 2 | 1 | 4 | 1 | 4 | 0.5 |
| 2-AG100 | 1 | 0.06 | 0.5 | 0.06 | 0.25 | 0.06 | 0.25 | ≤0.015 | 2 | 0.5 | 16 | 1 | 16 | 0.5 |
| 3*-AG100 | 2 | 0.06 | 1 | 0.06 | 0.5 | 0.06 | 0.5 | ≤0.015 | 8 | 1 | 32 | 1 | 32 | 0.5 |
| 4*-AG100 | 4 | 0.125 | 2 | 0.125 | 2 | 0.125 | 2 | ≤0.015 | 8 | 1 | 64 | 1 | 32 | 1 |
| AG112 | 0.125 | ≤0.015 | 0.06 | ≤0.015 | 0.125 | ≤0.03 | 0.06 | ≤0.015 | 4 | 1 | 16 | 1 | 32 | 1 |
| 1-AG112 | 1 | 0.06 | 0.5 | 0.03 | 0.5 | 0.06 | 0.25 | ≤0.015 | 4 | 0.5 | 32 | 1 | 16 | 1 |
| 2-AG112 | 4 | 0.06 | 2 | 0.03 | 1 | 0.06 | 1 | ≤0.015 | 16 | 1 | 64 | 1 | 32 | 0.5 |
| 3-AG112 | 8 | 0.06 | 4 | 0.03 | 2 | 0.06 | 2 | ≤0.015 | 8 | 0.5 | 64 | 1 | 64 | 0.5 |
Determined by the broth microdilution method according to the standards of the National Committee for Clinical Laboratory Standards (20). Results are representative of experiments done in duplicate. +, acrAB present; Δ, acrAB was deleted.
Identification of chromosomal mutations in structural and regulatory genes and susceptibilities to unrelated antibiotics.
Sequencing of the QRDRs of gyrA revealed that an identical point mutation at codon 87 (substitution of glycine for aspartate) occurred during the first mutation step in both AG100- and AG112-derived mutants (Table 1). These mutations led to an increase in the MICs of FQs without an additional multiple resistance (Mar) phenotype. There are several reports that a gyrA mutation is the first “visible” mutation in the chain of events that leads to higher levels of FQ resistance during stepwise in vitro mutagenesis (10, 25; W. V. Kern, M. Oethinger, A. S. Ritter, S. Conrad, R. Marre, and S. B. Levy, Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-182, 1997). That the first gyrA mutations in both AG100 and AG112 were identical may be coincidental.
The second-step mutation in the AG100 background was a Mar mutation, as shown by overexpression of marRAB by Northern blot analysis of 2-AG100. Such overexpression was found in three independently selected second-step mutants of AG100 (data not shown). Constitutive overexpression of marA was associated with increased levels of resistance to TET, CML, and CFOX (Table 2). Sequencing of marOR of a third-step mutant, mutant 3*-AG100, from a different series showed a different mar mutation but the same gyrA mutation.
As expected (9), marRAB was derepressed in Mar mutant AG112 and all subsequently derived mutants. Sequencing of marOR of the parental strain AG112 identified a 5-bp deletion after the codon for amino acid 12 of MarR, resulting in deletion of one amino acid and a change in the complete protein sequence thereafter (Table 1).
A single mutation in the gyrA gene conferred a somewhat higher level of FQ resistance than overexpression of marA by itself did: the OFL MICs were 0.25 μg/ml (eightfold increase) and 0.125 μg/ml (fourfold increase) for 1-AG100 and AG112, respectively (Table 1). The two mutations are multiplicative (OFL MICs, 1 μg/ml [32-fold increase] for 2-AG100 and 1-AG112; Table 1). During further steps to higher levels of FQ resistance, a mutant in another series, mutant 4*-AG100, acquired a second mutation in gyrA, at codon 83, substituting leucine for serine (Table 1). No additional mutations in gyrA, parC, or marOR could be identified in any of the more resistant mutants, nor did they constitutively overexpress soxS at any step (data not shown). Mutant 2-AG112, derived from gyrA marR double mutant 1-AG112, displayed increased levels of resistance to multiple drugs (Table 2), with no further mutation in marOR. The additional mutation possibly led to upregulation of acrAB (see below). In contrast, the next step mutant, 3-AG112, had increased levels of resistance to only FQs and CFOX (Table 2). The genetic basis for this resistance is also unknown.
Accumulation of [14C]CIP in whole cells.
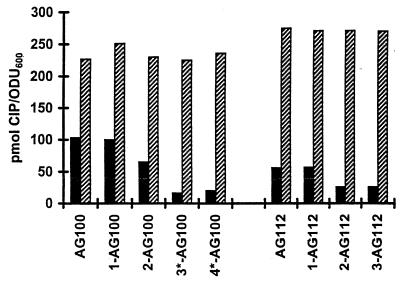
The accumulation of CIP in whole energized cells reached a plateau by 5 min and averaged 103 pmol of CIP/OD600 unit in AG100. When the proton motive force was dissipated by adding 200 μM CCCP, the level of accumulation of [14C]ciprofloxacin doubled (201 pmol of CIP/OD600 unit). These findings confirmed earlier studies with norfloxacin (4) that FQ-susceptible E. coli cells use energy to reduce the level of FQ accumulation; i.e., they show active efflux. This phenomenon was also observed for Proteus vulgaris with OFL (12). In comparison, accumulation in AG112 was 54% of that in the wild-type strain, averaging 56 pmol of CIP/OD600 unit, and increased to 226 pmol CIP/OD600 unit after the addition of CCCP. The rapid accumulation of CIP (this work) or norfloxacin (4) to the level in the AG100 parental strains upon deenergization of the cells rules out downregulation of outer membrane porins as the major mechanism of reduced levels of drug accumulation in Mar mutants. The amount of CIP accumulated by energized first-step (gyrA) mutants 1-AG100 and 1-AG112 and the increase in the level of drug uptake following deenergization were virtually identical to those for parental strains AG100 and AG112, respectively (data not shown). In second-step Mar mutant 2-AG100, the level of accumulation decreased to 65 pmol CIP/OD600 unit (63% of that for the wild-type strain; Fig. 1). Independently isolated mutants 3*-AG100 and 4*-AG100 showed even lower levels of accumulation (16 and 19% of that for the wild-type strain, respectively; Fig. 1). Similarly, mutants 2-AG112 and 3-AG112 accumulated considerably less CIP than Mar mutants AG112 and 1-AG112 (Fig. 1). With less drug accumulation, the cells can survive in the presence of higher external concentrations of FQ. The mutations also increased the cells' resistance to TET, CML, and CFOX.
FIG. 1.
Cellular accumulation of CIP. [14C]CIP accumulation by FQ-resistant mutants derived in vitro from E. coli K-12 strain AG100 and its Mar mutant AG112 was assayed at 30°C at equilibrium after addition of 10 μM CIP, as described in Materials and Methods. Cells carried either the wild-type acrAB gene (solid bars) or had an acrAB deletion (hashed bars). ODU600, OD600 units.
Effects of deletion of acrAB.
Upon deletion of the AcrAB multidrug efflux pump, the ability to actively efflux CIP was completely lost by all strains (data not shown). Energized AG100 cells bearing the acrAB deletion accumulated CIP to more than twice the level seen for energized acrAB-positive cells (Fig. 1). No difference in the levels of CIP uptake was noted between any acrAB-deleted strains, irrespective of their mutations in marRAB or gyrA (Fig. 1). Although all ΔacrAB strains became profoundly hypersusceptible to all FQs, mutants with newly acquired mutations in gyrA (Table 1) still retained some gyrA-mediated FQ resistance (Table 2), although this was therefore well below the level of clinical significance. The OFL MIC for the gyrA double mutant 4*-AG100 ΔacrAB, for instance, was only 0.125 μg/ml. Interestingly, the differences in MICs of different FQs were no longer observed for ΔacrAB strains, e.g., for 2-AG100 ΔacrAB, the OFL, CIP, and TVA MICs were 0.06 μg/ml, while for acrAB-positive strain 2-AG100, the OFL MIC was 1 μg/ml, the CIP MIC was 0.5 μg/ml, and the TVA MIC was 0.25 μg/ml (Table 2). The SPX MICs for all ΔacrAB mutants were below the detection limit (MICs, ≤0.015 μg/ml). Deletion of acrAB also eliminated the multidrug resistance conferred by overexpression of marA, thus underlining previous findings that the AcrAB multidrug efflux pump plays a major role in the antibiotic resistance phenotype of Mar mutants (23). Effects of additional, as yet unknown mutations were also completely abolished by deletion of acrAB (Table 2). We conclude, therefore, that the effect of one or more additional mutations on drug resistance is also affected by acrAB.
Concluding remarks.
In view of the large number of pumps in E. coli (21, 24), it is remarkable that none of the other pumps actively effluxes CIP in the absence of the AcrAB efflux pump. Thus, the AcrAB multidrug efflux pump appears to be the only, or at least the most important, pump in E. coli which uses CIP as a substrate.
Our findings correspond with recent data on the triclosan susceptibility of E. coli, which was greatly affected by loss of the AcrAB pump (18). While overexpression of acrAB, marA, or soxS increased the cells' resistance to triclosan about twofold, a mutation in the target of triclosan, enoyl reductase (encoded by fabI), rendered the cell about 100-fold more resistant (19). However, deletion of acrAB reduced the level of resistance 10-fold in all strains, rendering the fabI mutation less effective, similar to the decrease in the effectiveness of topoisomerase mutations in the case of FQs.
The prominent finding of our work is that the AcrAB efflux pump has a powerful role in both the intrinsic and the acquired level of FQ resistance in E. coli. As previously reported by others for Pseudomonas (21), we show that efflux mechanisms significantly decrease the action of FQs also in E. coli, even though in the latter organism the hydrophilic drugs presumably traverse the outer membrane more rapidly through the more permeable porin channels (21). Mutations in unidentified chromosomal loci, not marOR or soxRS, modulate the level of resistance apparently by increasing efflux via AcrAB. Blockage of the AcrAB efflux pump would increase the potencies of drugs such as FQs even in the face of topoisomerase mutations. While this work was in progress, a report on the importance of the Mex multidrug efflux pumps for FQ resistance in Pseudomonas aeruginosa drew a parallel conclusion (16).
ACKNOWLEDGMENTS
This study was supported in part by a research grant from the U.S. Public Health Service (grant GM 51661 to S.B.L. and L.M.M.), the Deutsche Forschungsgemeinschaft (grant Oe 195/1-1 to M.O.), and the University of Ulm (grants Ke700/1-1 and P172/1994 to M.O. and W.V.K.).
REFERENCES
- 1.Alekshun M N, Levy S B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amábile-Cuevas C F, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen S P, Hächler H, Levy S B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen S P, Hooper D C, Wolfson J S, Souza K S, McMurry L M, Levy S B. Endogenous active efflux of norfloxacin in susceptible Escherichia coli. Antimicrob Agents Chemother. 1988;32:1187–1191. doi: 10.1128/aac.32.8.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen S P, McMurry L M, Hooper D C, Wolfson J S, Levy S B. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989;33:1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen S P, McMurry L M, Levy S B. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conrad S, Oethinger M, Kaifel K, Klotz G, Marre R, Kern W V. gyrA mutations in high-level fluoroquinolone-resistant Escherichia coli clinical isolates. J Antimicrob Chemother. 1996;38:443–455. doi: 10.1093/jac/38.3.443. [DOI] [PubMed] [Google Scholar]
- 8.George A M, Levy S B. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid determined efflux of tetracycline. J Bacteriol. 1983;155:531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberg J T, Chou J H, Monach P A, Demple B. Activation of oxidative stress genes by mutations at the soxQ/cfxB/marA locus in Escherichia coli. J Bacteriol. 1991;173:4433–4439. doi: 10.1128/jb.173.14.4433-4439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40:879–885. doi: 10.1128/aac.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooper D C, Wolfson J S, Ng E Y, Swartz M N. Mechanisms of action of and resistance to ciprofloxacin. Am J Med. 1987;82:12–20. [PubMed] [Google Scholar]
- 12.Ishii H, Sato K, Hoshino K, Sato M, Yamaguchi A, Sawai T, Osada Y. Active efflux of ofloxacin by a highly quinolone-resistant strain of Proteus vulgaris. J Antimicrob Chemother. 1991;28:827–836. doi: 10.1093/jac/28.6.827. [DOI] [PubMed] [Google Scholar]
- 13.Khodursky A B, Zechiedrich E L, Cozzarelli N R. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumagai Y, Kato J, Hoshino K, Akasaka T, Sato K, Ikeda H. Mutants of Escherichia coli DNA topoisomerase IV parC gene. Antimicrob Agents Chemother. 1996;40:710–714. doi: 10.1128/aac.40.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy S B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992;36:695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren M S, Boyer E, Chamberland S, Lee V J. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:1340–1346. doi: 10.1128/aac.43.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 18.McMurry L M, Oethinger M, Levy S B. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol Lett. 1998;166:305–309. doi: 10.1111/j.1574-6968.1998.tb13905.x. [DOI] [PubMed] [Google Scholar]
- 19.McMurry L M, Oethinger M, Levy S B. Triclosan targets lipid synthesis. Nature. 1998;394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility. Tests for bacteria that grow aerobically. 4th ed. 1997. Approved standard. NCCLS document M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 21.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oethinger M, Podglajen I, Kern W V, Levy S B. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob Agents Chemother. 1998;42:2089–2094. doi: 10.1128/aac.42.8.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piddock L J V, Hall M C, Walters R N. Phenotypic characterization of quinolone-resistant mutants of Enterobacteriaceae selected from wild type, gyrA type and multiply-resistant (marA) type strains. J Antimicrob Chemother. 1991;28:185–198. doi: 10.1093/jac/28.2.185. [DOI] [PubMed] [Google Scholar]
- 26.Provence D L, Curtiss R., III . Gene transfer in gram-negative bacteria. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 317–347. [Google Scholar]
- 27.Tenney J H, Maack R W, Chippendale G R. Rapid selection of organisms with increasing resistance on subinhibitory concentrations of norfloxacin in agar. Antimicrob Agents Chemother. 1983;23:188–189. doi: 10.1128/aac.23.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thanassi D G, Cheng L W, Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe M, Kotera Y, Yosue K, Inoue M, Mitsuhashi S. In vitro emergence of quinolone-resistant mutants of Escherichia coli, Enterobacter cloacae, and Serratia marcescens. Antimicrob Agents Chemother. 1990;34:173–175. doi: 10.1128/aac.34.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida J, Kojima T, Yamagishi J, Nakamura S. Quinolone-resistant mutations of gyrA gene of Escherichia coli. Mol Gen Genet. 1988;211:1–7. doi: 10.1007/BF00338386. [DOI] [PubMed] [Google Scholar]