Abstract
Ever since the outbreak of COVID-19, the entire world is grappling with panic over its rapid spread. Consequently, it is of utmost importance to detect its presence. Timely diagnostic testing leads to the quick identification, treatment and isolation of infected people. A number of deep learning classifiers have been proved to provide encouraging results with higher accuracy as compared to the conventional method of RT-PCR testing. Chest radiography, particularly for using X-ray images, is a prime imaging modality for detecting the suspected COVID-19 patients. However, the performance of these approaches still needs to be improved. In this paper, we propose a capsule network called COVID-WideNet for diagnosing COVID-19 cases using Chest X-ray (CXR) images. Experimental results have demonstrated that a discriminative trained, multi-layer capsule network achieves state-of-the-art performance on the COVIDx dataset. In particular, COVID-WideNet performs better than any other CNN based approaches for the diagnosis of COVID-19 infected patients. Further, the proposed COVID-WideNet has the number of trainable parameters that is 20 times less than that of other CNN based models. This results in a fast and efficient diagnosing COVID-19 symptoms, and with achieving the 0.95 of Area Under Curve (AUC), 91% of accuracy, sensitivity and specificity, respectively. This may also assist radiologists to detect COVID and its variant like delta.
Keywords: Capsule Networks, CNN, COVID-19, COVID-19: Virus variants, Deep learning, X-Rays, RT-PCR
1. Introduction
The upsurge of the infectious coronavirus disease 2019 (COVID-19) has caused a state of a global health crisis. Being a new strain, which has not been previously recognized by humans, it is necessary to detect the disease in order to prevent its spread. Timely diagnosis of infected and suspected people on the onset of symptoms related to COVID-19, can help in their immediate isolation.
Presently, intensive research is being done for the fast detection of the SARS-CoV-2 virus, which is responsible for the spread of COVID-19 disease. There exist a very popular test known as Polymerase Chain Reaction (PCR) which is widely used for easy detection of respiratory viruses. Therefore, RT-PCR is being considered as one of the front-line testing methods for detection of COVID-19 symptoms [1]. Additionally, scientists all around the world are working meticulously on potential treatments, vaccines, and many other technological advancements, Artificial Intelligence (AI), and Deep Learning-based methods for the fast detection and protection from COVID-19.
At present, deep learning techniques like Convolutional Neural Networks (CNNs) are being used widely to extract the various features from image-based datasets. However, the presence of adversarial examples in real-time can be hard to predict by using a CNN approach as it fails to identify the same [2]. These examples are images that contains a small amount of added noise which can cause a neural network to incorrectly classify the images. This is because CNN uses a large amount of augmented training data that enables to use modified versions of original input images. In comparison to CNN, a much finer approach of capsules [3], [4] has been used which employs dynamic routing to mutually agree on different orientations of the same object, thus making it performs well on adversarial examples as well. In this work, we have used Capsule Networks to overcome the various disadvantages of CNN. Capsule networks provide a strong mechanism for an improved detection of features in images. Capsule Networks, consisting of Capsules, introduce the concept of inverse rendering to solve the problem of providing spatial information, and make up for the shortcomings of CNN model. Initially, capsules were introduced by Hinton et al. [4]. They condense all the relevant information about the state of the feature which they aim to detect, and store it in a vector form. The probability of the presence of a feature is encoded as the length of their output vector, while the state of the feature is encoded as the direction of the vector. In a survey by [5], a comprehensive survey of the architecture and performance of Capsule Networks has been done. After a comparison with the existing robust architectures of CNN, it is suggested that Capsule Networks can prove to be a boon in the field of Computer Vision. [6] have experimentally showed the effectiveness of using Capsule Networks on Medical images as compared to CNNs. Its applications have been significantly seen in real-world scenarios such as its use on Medical Images. For instance, classification of brain tumor using capsule networks have been explained by [7].
The benchmark CNNs have a major drawback, which is the loss of spatial information. The inefficient process of max pooling in CNNs results in a loss of valuable spatial information as only the most active neurons are chosen to be moved to the next layer during max pooling [3], [4]. Some of the major drawbacks of CNNs are given below:
-
•
In CNNs, there is a chance of losing the information about the composition and position of the components present in an image due to max-pooling layers which throws away information about the precise position. This limitation is reduced by data augmentation which increases the number of training dataset images that are required to train the neural network. Thus, it increases the computational cost significantly.
-
•
CNNs may fail when they are encountered with adversarial examples like noisy images to learning models. Thus, when CNNs take images containing noise, they recognize the image as a completely different image, whereas, human visual systems can easily identify it as the same original image.
-
•
CNNs do not have coordinate frames, which are considered as the fundamental part of human vision.
-
•
CNNs are Invariant of Translation, since they fail to identify spatial information, whereas capsule networks are able to identify the position of the object as well.
-
•
CNNs may consist of several layers which result in an increased time for training the network on a large dataset and also increased computation cost in a real-time scenario.
To overcome these major challenges in CNNs, Sabour et al. [3] proposed a robust method known as Dynamic Routing which encodes this spatial information present in images into features. While CNNs use replicas of learned feature detectors, the process of dynamic routing use vector-output capsules. Max-pooling of CNN is replaced with a routing-by-agreement mechanism, such that information about the precise position of an entity within a region of the image is preserved. Using dynamic routing as a backbone, Capsule Networks are implemented to provide improved results in the field of Computer Vision and overcome the short-comings of CNNs.
In the present work, we have proposed COVID-WideNet, a capsule network based model for the detection of COVID-19 positive patients in Chest X-rays(CXR). Proposed model eliminates the shortcoming of CNN based approach while diagnosing of COVID-19 symptoms from CXR images. The main contributions of this research work can be summarized as follows:
-
1.
We propose COVID-WideNet which is based on capsule network and deep-learning based model and comprises of 2 Convolutional layers and 3 Capsule layers.
-
2.
We have proposed ‘DiseaseCapsule’, a layer consisting of ‘2 8D’ capsules, here ‘2 8D’ refers to 2 capsules of 8-dimensions each.
-
3.
A wider ‘PrimaryCapsule’ layer is also introduced consisting of ‘32 16D’ capsules, where ‘32 16D’ refers to thirty-two capsules of 16-Dimensions each.
-
4.
COVID-WideNet achieves the state-of-the-art accuracy with 20 times less number of trainable parameters. Thus, it decreases the execution speed and computation cost in a real-time COVID-19 detection.
-
5.
Proposed model achieves 91% of sensitivity and specificity. As a result, the AUC of the model is 0.95 and represents a good rank of classification.
-
6.
We have presented a discriminative approach towards COVID-19 X-ray detection which eliminates the various limitations of CNN based model.
The remaining paper is organized into the following sections. Section 2 reviews with critical analysis of different deep learning/AI approaches to detect the COVID-19. Section 3 presents the proposed model COVID-WideNet and its Algorithm 1 for fast and efficient detection of COVID-19 positive cases. Section 4 describes the core experimental steps. Section 5 presents the experimental results obtained and the related discussion. Finally, Section 6 concludes this paper.
2. Related work on AI approaches for COVID-19 detection
An Intelligence-based techniques have been widely accepted in the mainstream of computer vision as they have abilities to perform many tasks, such as object detection and image classification. Similarly, its applications are seen significantly in medical image analysis and disease diagnosis [8], [9], [10], [11]. Until now, a large number of research articles have been published using AI-based approaches in context to COVID-19 [12]. This section summarizes the various techniques used for detection of COVID-19 using medical images such as CXR and CT-Scan images, and the application of capsule network while diagnosing COVID-19.
2.1. Detection of COVID-19 using chest X-rays (CXR) and CT- scans
Due to the speedy spread of COVID-19, fast and efficient methods for testing COVID-19 patients is being done. Timely diagnosis of the disease is crucial for control and treatment. Recent studies show that Chest CT-scan Imaging may offer a faster and reliable diagnosis of the novel COVID-19, even better than the RT-PCR testing [13], [14], [15], [16]. Ai et al. [17] have even compared Chest CT to the RT-PCR evaluation by investigating the diagnostic value on 1014 patients in Wuhan, who had gone through both the types of testing. They concluded that Chest CT could be considered as the foremost method for detection of COVID-19 due to their high sensitivity. Similarly in a study done by Caruso et al. [18] on 158 COVID-19 pneumonia participants in Rome, the results were sensitive (97%) when Chest CT was used but not specific (56%). As stated by Kim [19], CT scans are recommended to people with suspicious lung abnormality, and leveraging diagnostic AI models for CT Scans may help in assuaging the burden of radiologists and clinicians by enhancing rapid testing and treatment. To identify the changes in lungs during COVID-19, Chest CT-Scans can be used intensively. Pan et al. [20] have stated that the abnormalities on the Chest CT scans are the most severe after 10 days of initial onset of a COVID-19 infected person. Wang et al. [21] also state that CT may play a prime role in the control of coronavirus disease, especially in patients with COVID-19 pneumonia.
Since CXR and CT scans play a significant role in the detection of COVID-19 patients, numerous deep learning-based approaches have been proposed recently. In Huang et al. [22], a deep learning approach has been proposed for the quantitative Chest CT assessment on 126 COVID-19 patients. In another study by Shan et al. [23], segmentation network to classify the regions of lungs obtained by CT images. It is claimed to provide the accurate quantitative information, track the disease progression and longitudinal changes during the treatment process. However, this method is only for the segmentation of the COVID-19 infected regions in the lungs, and not for the detection of the disease itself. Li et al. [24] proposed a deep learning method to detect COVID-19 and further differentiate it from the other chest CT images of pneumonia and other non-pneumonic lung diseases using 4356 images. The deep learning model used by them consists of a ResNet50 as the backbone that takes Region of Interest of CT images as input and generates features. However, the test set used by them to evaluate their model is from the same hospitals from which the train set came. Thus, various techniques have been proposed to detect COVID-19 in images of Chest CT-Scans, and our work also focuses on the same.
Using CXR images, Narin et al. [25] have proposed three convolutional neural network-based models that are : InceptionResNetV2, ResNet50 and InceptionV3,for the detection of coronavirus pneumonia infected patients. They achieved the highest performance for classification with an accuracy of 98% using the pre-trained ResNet50 model. In a similar study, Apostolopoulos and Mpesiana [26] have used and highlighted the notion of COVID-19 detection using CXR images based on transfer learning approach from public medical repositories. They used VGG19 and MobileNetV2 models to implement transfer learning and achieved a high accuracy of 96.72%.
Thus, recently a number of studies have been done on Chest X-ray and CT Scan images using deep learning based methods for detection of COVID-19. To overcome the drawbacks of CNN-based approaches, a survey of Capsule Networks is also done.
2.2. Applications of capsule networks to detect COVID-19
Off late, Capsule Networks have also been applied for the automated detection of COVID-19. In Afshar et al. [27], a Capsule Network based framework is proposed which when implemented on X-ray images is able to classify images with COVID-19. They achieved 0.97 area under the ROC curve, and an accuracy of 95.7%. Similarly, in another study by Toraman et al. [28], a Capsule Network based model was successful in the binary classification into ‘COVID-19’ and ‘No Findings’ of X-ray images with an accuracy of 97.24%, and multi classification into ‘Pneumonia’, ‘COVID-infected’ and ‘No Findings’ with an accuracy of 84.22%. Afshar et al. [29] developed an Artificial Intelligence (AI)-based framework using a collected dataset of LDCT/ULDCT (low-dose and ultra-low-dose) scans protocols that reduce the radiation exposure close to that of a single X-ray. They used a pre-trained U-Net-based lung segmentation model, referred to as “U-net (R231CovidWeb)”, to segment lung regions and discard irrelevant information. The model uses a two stage capsule network architecture to classify COVID-19, community-acquired pneumonia (CAP), and normal cases, with the sensitivity of 89.5% for COVID-19, 95% for CAP, 85.7 for normal, and overall accuracy is 90%. Tiwari and Jain [30] proposed CNN-CapsNet and VGGCapsNet decision support system based on the X-ray image to diagnose the presence of the COVID-19 virus. The proposed model presents 97% of accuracy, 92% of sensitivity respectively. Quan et al. [31] proposed DenseCapsNet using CNN with capsule network and applied on 750 CXR lung images of healthy patients pneumonia with coronavirus, and obtained the accuracy of 90.7% and an F1 score of 90.9%, and sensitivity of 96%. Heidarian et al. [32] proposed ‘CT-CAPS’ a fully automated Capsule network-based framework, it extracts distinctive features of chest CT scans via Capsule Networks to identify COVID19 cases in a coarsely-labeled dataset of COVID-19, CAP, and normal cases. The experimental results indicate the capability of the CT-CAPS to automatically analyze volumetric chest CT scans and distinguish different cases with the accuracy of 90.8%, sensitivity of 94.5%, and specificity of 86.0%. Aksoy and Salman [33] analyzed chest X-ray images of 1019 patients using Capsule Networks (CapsNet) model, designed can detect COVID-19 disease with an accuracy rate of 98.02%. Saha et al. [34] proposed GraphCovidNet model to detect COVID-19 from CT-scans and CXRs of the affected patients & evaluated this model on four standard datasets: SARS-COV-2 Ct-Scan dataset, COVID-CT dataset, combination of covid-chestxray-dataset, Chest X-ray Images (Pneumonia) dataset and CMSC-678-MLProject dataset. The model shows an impressive accuracy of 99% for all the datasets and its prediction capability becomes 100% accurate for the binary classification problem of detecting COVID-19 scans.
3. COVID-WideNet
3.1. Capsule network
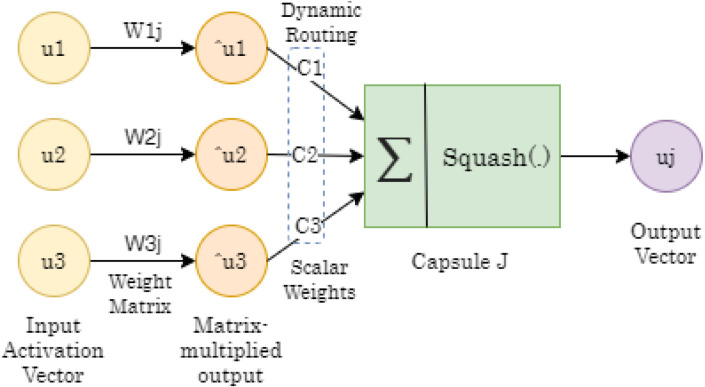
In this section, we present COVID-WideNET by starting with introduction of capsule network. A capsule is an activation vector that basically executes on its inputs some complex internal computations. Length of these activation vectors signifies the probability of availability of a feature. Furthermore, the condition of the recognized element is encoded as the direction in which the vector is pointing. In traditional, CNN uses Max pooling for invariance activities of neurons, which is nothing except a minor change in input and the neurons of output signal will remains same. The Max pooling loses all the significant data and furthermore does not encode relative spatial connections between features and capsules. It encapsulate all the significant data about the condition of the features that can be recognized with the help of a vector as presented in Fig. 1. It consist of three layers, input activation vector defined by . is a weight matrix between input activation vector and matrix multiplied output. is also known as prediction vector. Matrix multiplied output is . The predictions, however, are taken into account based on a coefficient, through the “Routing by Agreement” process, that determine the actual output of the Capsule , denoted by . The output vector is refers the classification of class values like COVID and non-COVID.
Fig. 1.
A schematic diagram of internal working of capsule network .
In the capsule, input weights are computed using “dynamic routing”, which is a new technique to decide where each capsule’s output goes Sabour et al. [3] and Hinton et al. [4]. The essence of the dynamic routing algorithm is that the lower level capsule needs to “choose” the capsule at a more significant level where it will send the output. Lower level capsules have a method of estimating which upper level capsule better fits its outcomes and will consequently alter its weight that will multiply this capsule’s output before sending it to higher level capsules. Capsule network introduces a new nonlinear activation function called Squash function that takes a vector, and afterwards “squashes” it to have length close 1, however it does not change its course Sabour et al. [3] and Hinton et al. [4].
| (1) |
Here, the total number of inputs is indicated by , and refers to the capsule’s vector output, calculated from Eq. (1). For every capsule except the lower level capsules i.e., it denotes the prediction of for capsule .
As shown in Eq. (2), is produced by multiplying a weight matrix with the output of lower level capsule.
| (2) |
| (3) |
where the are score of the predictions calculated using Eq. (3) that are decided by the repetitive dynamic routing process.
Capsule network uses a separate loss function called Margin loss. The length of the instantiating vector shows the probability of the detected objects present in an image. A top-level capsule has a long vector if and only if its closely associate with the object present in an image. To consider multiple objects in an image, a separate margin loss is calculated for each capsule. Down weighting the loss for missing objects stops the learning from shrinking the vector lengths for all objects. The total marginal loss is the sum of the losses of all entities. The Margin loss function is calculated from Eq. (4), where is associated with Capsule ,
| (4) |
Eq. (4) explains the final loss that considers the value of , whenever the object is present, otherwise . The hyper parameters , , and are set before the process initiates so the length does not collapse. Here, the main function of is down-weighting for absent digit classes that stops the initial learning process from shrinking the lengths of the activity vectors of all the digit capsules (see Fig. 3).
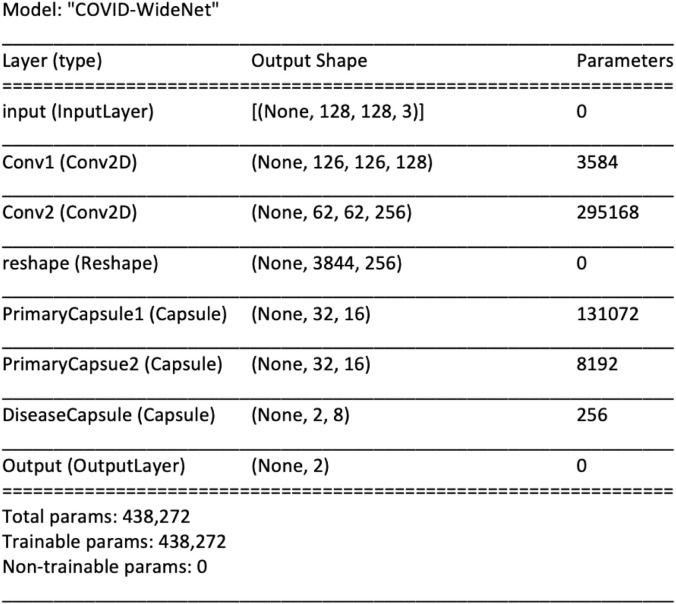
Fig. 3.
Model description of the proposed COVID-WideNet with 2 Convolutional layers, 3 Capsule Layers and 438,272 parameters .
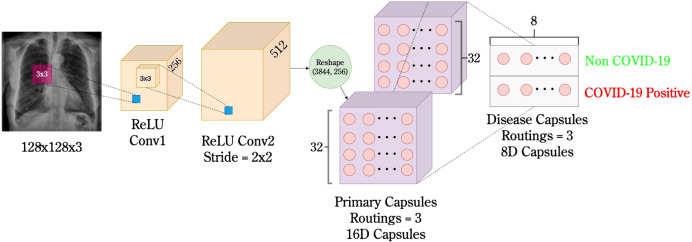
3.2. Proposed model - COVID-WideNet
The proposed architecture of COVID-WideNet is shown in Fig. 2. It is a shallow architecture with only 3 Convolution layers and a fully connected layer. The first convolution layer Conv1 is a simple convolution layer with a filter size of 256, 3 × 3 convolution kernels, and ReLU activation [35]. Whereas, Conv2 layer has a filter size of 512, 3 × 3 convolution kernels with a stride of 2 and RelU activation. The first two convolution layers are used to extract the basic features from the pixel intensities and are then used as input to the primary capsules. There are two Primary Capsule layers each having 32 ‘16D’ capsules. Here ‘32’ signifies the number of capsules in each layer and ‘16D’ represent the dimension of each capsule. Each dimension corresponds to a unique feature. The last layer is the Disease Capsule which has 2 ‘8D’ capsules. Here, ‘2’ signifies the number of capsules in the layer and 8D represent the dimension of each capsule.
Fig. 2.
Architecture of the proposed COVID-WideNet .
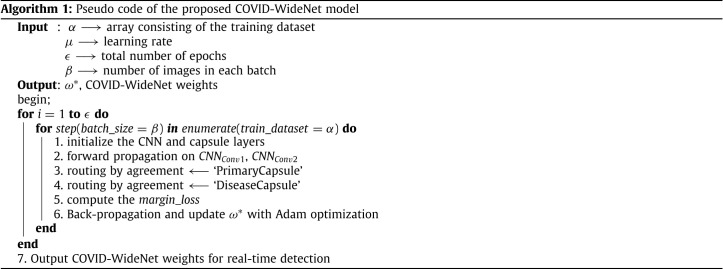
The working of proposed Algorithm 1, COVID-WideNet is explained in the following steps:
-
1.
Generate the array using the training images from the COVIDx dataset having the shape (None, 128, 128, 3) where ‘None’ refers to the number of images in and is replaced depending on . The input image dimension is changed into 128 × 128 × 3 for every image in .
-
2.
Initialize the hyperparameters – , and . For our study we have used , , and . Here, is the learning rate using Adam optimizer, is the epoch length, and is the batch size Afshar et al. [27].
-
3.
The outer loop corresponds to iteration over . Here, one epoch is equal to one iteration of the model over the COVIDx dataset training array . The inner loop corresponds to iteration over the COVIDx dataset training array with a regular step of batch size . For example, if and , then the first iteration of training over the first 64 samples from will occur and the weights will be updated. Once each sample Chest X-ray in is at least iterated over once, inner outloop ends and the second iteration () is started.
-
4.
Initialize the two Convolution layers and which input a tensor of rank 4 with shape (number of samples, height, width, filters). As seen in Fig. 2, has shape (None, 126, 126, 128) while has shape (None, 62, 62, 256) where ‘None’ can be replaced by the number of images in the training set.
-
5.
The input batch propagates through the architecture where the Convolution operation is applied in the first two layers, then the output of is reshaped to the target shape reducing the 4D tensor into a 3D tensor as per the requirement of the capsule layers.
-
6.
‘PrimaryCapsule’ can be compared to inverting the rendering process. Thus, forming and inverse a graphics perspective. The primary capsule has ‘32 16D’ capsules with 3 routing iterations. In total there are two primary capsules which perform routing-by-agreement process.
-
7.
‘DiseaseCapsule’ consists of complex entities and thus each capsule in this layer has eight dimensions. In total, ‘DiseaseCapsule’ layer has ‘2 8D’ capsules. Each capsule in this layer represents the type of disease i.e., COVID-19 or Non-COVID-19 with each dimension representing a separate and unique feature.
-
8.
The model is trained by calculating the margin loss and optimized by using the Adam optimization technique. The weight matrix is then updated using back propagation.
-
9.
Once the model is trained on iterations the final weights are saved and now can be used to detect COVID-19 in real-time.
4. Experiments
4.1. Dataset collection
Data acquisition is the first step in developing any diagnostic tool and AI has been able to significantly contribute in working with medical data images. We have used COVIDx [36] as a benchmark dataset in this study, which is a compilation of five different datasets containing CXR images—COVID-chest X-ray [37], Fig. 1 COVID-19-chest X-ray [38], ActualMed COVID-19-chest X-ray [39], COVID-19 Radiography [40], Chest X-ray Images (Pneumonia) [41]. Their detailed description has been provided here.
-
(i)
COVID-chest X-ray-dataset- This dataset comprises CXR images of COVID-19 cases and is publicly available Cohen et al. [37], [42]. It is constantly updated with new images. At the time of this study, the dataset consists 930 images of COVID-19 positive patients.
-
(ii)
Fig. 1 COVID-19-chest X-ray-dataset- This is an open source data set of COVID-19 CXR images. This data set is compiled by Chung [38].
-
(iii)
ActualMed COVID-19-chest X-ray-dataset- This is a public data set of CXR images of patients infected by COVID-19. This data set is compiled by Chung [39] and is collected by different research groups from Canada and Vision and Image Processing Research Group, DarwinAI Corp., Canada. It also comprises COVID-19 CXR images that have both positive and negative cases.
-
(iv)
COVID-19 Radiography Database- This is a open data set of X-ray images of chests for COVID-19 positive patients along with Normal and Viral Pneumonia case images publicly available on Kaggle [40]. This data set is compiled by a team of researchers from the University of Dhaka, Bangladesh and Qatar University, Doha, Qatar with the help of medical doctors. In their current release, this data set consists of 219 images of COVID-19 positive patients, 1341 images of normal patients and 1345 images of patients having viral pneumonia.
-
(v)
Chest X-ray Images (Pneumonia)- This is an open source data set of chest radiograph (CXR) for diagnosis of pneumonia. This data set is compiled by RSN [41] in collaboration with the US National Institutes of Health, The Society of Thoracic Radiology, and MD.ai and is made publicly available on Kaggle.
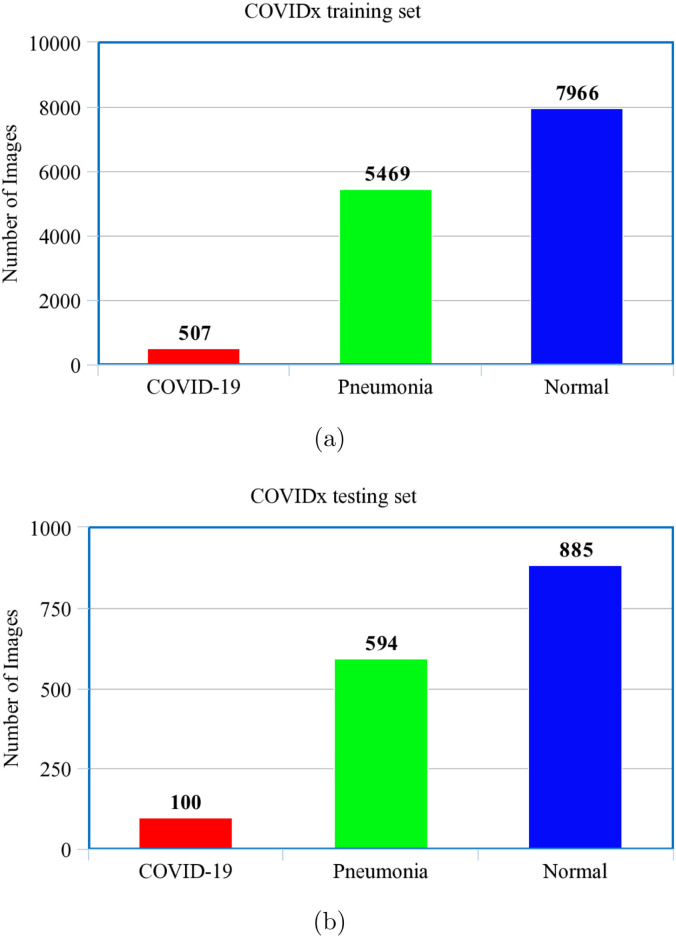
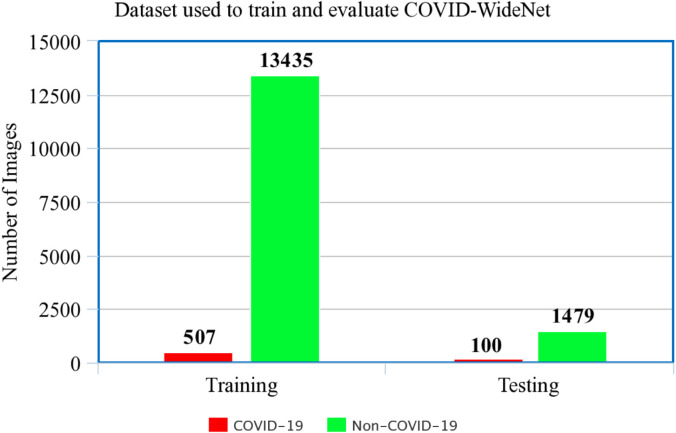
The compiled COVIDx dataset is shown in Fig. 4, Fig. 4. Since the main purpose of our study is to detect COVID-19 patients we have combined the Normal and Pneumonia CXR Images present in COVIDx as shown in Fig. 5. Thus, for training and testing COVID-WideNet we have used two class values: ‘COVID-19’ and ‘Non-COVID-19’ trained on 13,942 training images including 507 CXR images for ‘COVID-19’ and 13,435 for ‘Non-COVID-19’. As shown in Fig. 5 CXR images of COVID-19 positive are significantly less than the Non-COVID-19 images. This may cause class imbalance problem which is handled by modifying the loss function inspired by Heidarian et al. [43] and Saif et al. [44] as shown in Eq. (5).
Fig. 4.
COVIDx dataset consisting of CXR images and compiled from the five different datasets for (a) Training, and (b) Testing purpose.
Fig. 5.
Training set compiled from the five datasets.
Here, is the number of positive samples, is the number of negative samples, denotes the loss associated with positive samples, and denotes the loss associated with negative samples.
| (5) |
4.1.1. Data pre-processing
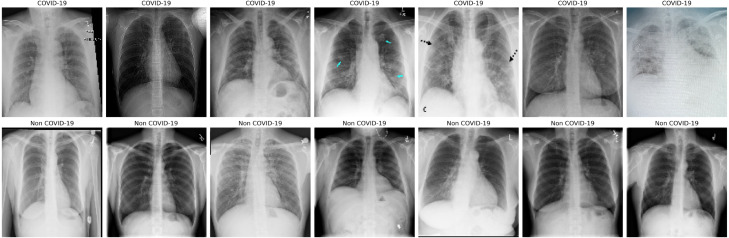
Data compiled for the purpose of this study consists of X-ray images which are not uniform in size. The images are resized to a size of 128 × 128 pixels to input uniform images to the model. The images have 3 channels—RGB(Red, Blue, Green) and the shape—128 × 128 × 3. The images are converted into an array of shape (13,942, 128, 128, 3). Since, capsule networks focus on the spatial information and orientation of pixels in an image, data augmentation is not applied in our study, unlike the commonly adopted method in case of CNNs. A sample from the dataset after data pre-processing is shown in Fig. 6.
Fig. 6.
Examples of CXR images from COVIDx dataset (first row: COVID-19; second row: Non-COVID-19.
4.1.2. Data leakage
In medical imaging, usually the data used is prone to large amount of data leakage. It is probable that a single patient has more than one sample in the dataset, and while splitting the dataset into training and testing the sample images of that patient can be appended to both training and testing sets thus causing data leakage. The direct effect of data leakage is visible on the loss associated with training and testing of the model. If the testing loss is smaller than the training set we can say that there may have been some sort of data leakage. To prevent this problem, we have considered 75 unique images of X-rays of patients with COVID-19 in the testing set, resulting into 100 samples (refer to Fig. 4(b)). This can be considered as a benchmark and is referred to as COVIDx dataset proposed by Wang et al. [36].
4.2. Performance evaluation
The performance of COVID-WideNet has been evaluated using confusion matrix, sensitivity, specificity and accuracy. Further, we also calculated the Area Under the Curve (AUC) by plotting the Receiver Operating Characteristics (ROC).
4.2.1. Confusion matrix
Typically, confusion matrix is a 2 × 2 matrix, each cell of the matrix represents the model detection rate which are represented by—True Positive (TP), True Negative (TN), False Positive(FP), and False Negative(FN). These four parameters can be explained with an example scenario of a hospital [45], [46].
-
1.
True Positive (TP): If a person is actually COVID positive and also predicted/detected as COVID-19.
-
2.
True Negative (TN): If a person is normal, and also correctly predicted/detected as non-COVID.
-
3.
False Positive (FP): It represents an incorrect prediction/ detection, when a normal person is predicted/detected as COVID positive.
-
4.
False Negative (FN): It also represents an incorrect detection, when a COVID positive person predicted/detected as normal one.
Using these four performance evaluation parameters, Eqs. (4), (6) and (7) are calculated each representing the accuracy, sensitivity and specificity of the model respectively.
| (6) |
| (7) |
| (8) |
4.2.2. Receiver operating characteristic (ROC)
ROC is an evaluation metric used to speculate the performance of a model at each classification threshold. Initially, it was used in radar identification during World War II to differentiate the signal from noise [47]. Due to its fine evaluation performance, it is notably used in numerous medical laboratory tests such as radiology, epidemiology, and endocrinology [48], [49], [50]. The task of data analysis with predictive knowledge on research related to health informatics can be simplified with the calculation of ROC. Therefore, it is highly recommended in tomography scans for COVID-19 detection [51], [52]. We can also say that, it is the trade-off between sensitivity and specificity [53].
In this research work, the performance of COVID-19 dataset has been evaluated using ROC plots. This has been done by calculating the area under the ROC curve, that is known as Area under the curve (AUC). This calculated AUC value of the curve is in between 0 and 1. An AUC value being close to 1.0 indicates that the model has a high sensitivity and specificity [54]. The AUC of the ROC is estimated by the Trapezoidal formula as shown in Eq. (9).
| (9) |
Trapezoid (T) is the integration of to values from a functional configuration that is further divided into equal vertical segments. The summation of forms the trapezium of each vertical segment, where the upper end is followed by a chord and their approximate summation provide the promising result of the AUC in the numerical form. This trapezoidal rule shows a definite integral function , and the points of domain subdivision of the integration (p, q) are labeled as ; where . Further, the function is defined by using Eq. (9) [55]. Thus, AUC is a potent method to compute the accuracy of ROC obtained from the model.
5. Experimental results and discussion
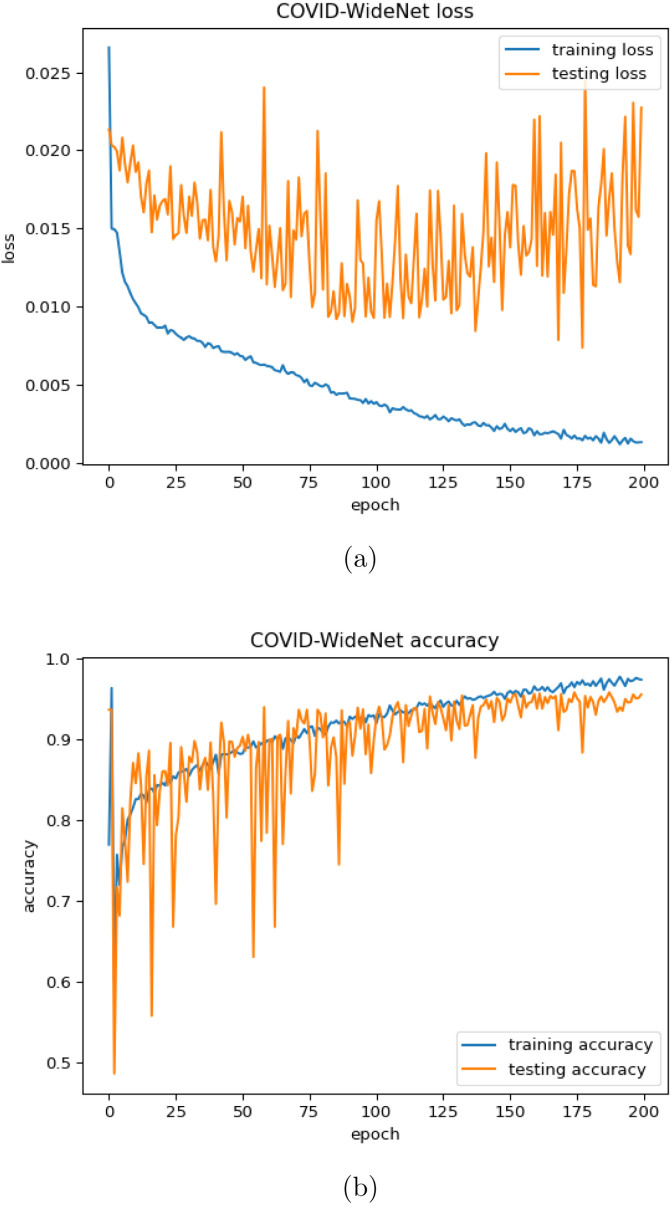
The proposed COVID-WideNet is trained on the COVID-19 dataset having 200 epochs with the learning rate as and a batch size of 64. Both training and testing accuracy are shown in Fig. 8(b), and it can be observed that the best training accuracy is 97.78%, which is obtained at the epoch. Same has also been observed in Fig. 8, Fig. 8, that the testing loss is greater than the training loss while testing accuracy is almost always lesser than the training accuracy. Since the model has never been trained on the testing set the same behavior was expected. This observation removes suspicion of any kind related to data leakage which might have occurred during splitting the dataset in the training set and testing set.
Fig. 8.
Training and testing for the proposed COVID-WideNet (a) Loss (b) Accuracy.
Further Confusion Matrix shown in Table 1 is used to calculate the performance metrices—accuracy, sensitivity and specificity of COVID-WideNet based TP, FP, TN and FN. From the test set, we obtained both sensitivity and specificity approximately equal 91% and 91.14% respectively. Here, sensitivity indicates whether the model is able to accurately detect the COVID-19 positive cases. On the contrary, specificity indicates the accurate detection of the COVID-19 negative cases. Since, our model has achieved high sensitivity and specificity. So, we can confidently say that the proposed model is both highly sensitive and specific.
Table 1.
Confusion matrix for COVID-WideNet trained on COVIDx dataset with two classes.
| Predicted class |
|||
|---|---|---|---|
| COVID-19 | Non-COVID-19 | ||
| Actual class | COVID-19 | ||
| Non-COVID-19 | |||
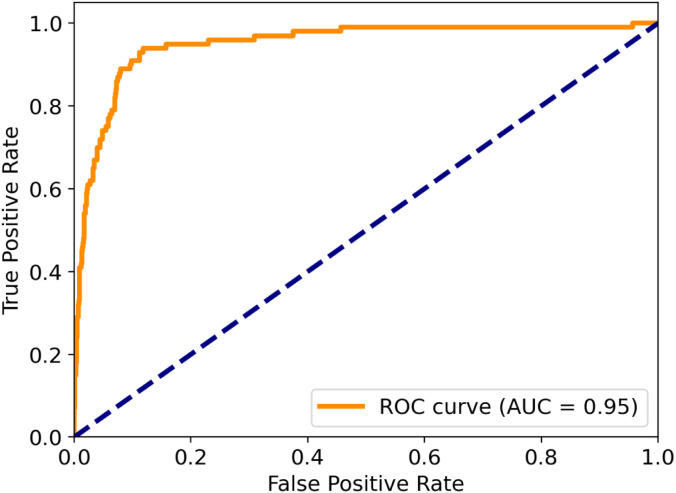
The AUC of ROC of propose model is 0.95 and is depicted in Fig. 7, which is much higher than the threshold level (i.e., 0.5). This is clear indication of the least trade-off between sensitivity and specificity of the model and can be considered as a good grade of classification. This is also considered to be ‘outstanding’ in the domain of medical disease diagnosis [33], [58].
Fig. 7.
ROC and AUC for the proposed COVID-WideNet.
We have compared the results of our proposed model with seven other state-of-the-art models keeping the COVIDx dataset as a benchmark since it comprises a fixed testing set. As seen in Table 2, VGG-19 [36], ResNet-50 [36] and COVID-Net [36] having 20 times more parameters than our proposed model-COVID-WideNet and uses data augmentation. Because of the small number of parameters in our model we can deploy the model in real-time and reduce the diagnosis time and the computational cost which is the need of the hour. COVID-CAPS [27] on the other hand has not used data-augmentation since the model is based on capsule networks but uses the transfer learning based approach and is trained on 112,120 X-ray images for 14 thorax abnormalities [56]. Due to pre-training on a larger dataset their model has a lower sensitivity of 80%. Through this study, we establish a novel approach for the fusion of medical imaging and neural network models, focusing more on the information provided by each pixel of a positive and negative sample. Thus, eliminating the traditional CNN based approach which uses pooling layers, data augmentation and transfer learning making the model less sensitive to positive samples.
Table 2.
Results comparisons of the proposed COVID-WideNet to other state-of-the-art models.
| Architecture | Accuracy | Sensitivity | Specificity | AUC | Parameters (in Millions) |
Pre-trained on | Data augmentation |
|---|---|---|---|---|---|---|---|
| COVID-CAPS [27] | 98.3% | 80% | 98.6% | 0.99 | 0.29M | NIH Chest X-ray dataset [56] | False |
| VGG-19 [36] | 83.0% | 58.7% | – | – | 20.37M | ImageNet [57] | True |
| ResNet-50 [36] | 90.6% | 83.0% | – | – | 24.97M | ImageNet [57] | True |
| COVID-Net [36] | 93.3% | 91% | – | – | 11.75M | ImageNet [57] | True |
| VGG-CapsNet [30] | 97% | – | – | 0.96 | – | X rays and CTS | – |
| DenseCapsNet [31] | 90.7% | – | 95.3% | 0.93 | – | CXR images | – |
| CT-Caps [32] | 90.8% | 94.5% | 86% | – | – | COVID-CT-MD | – |
| COVID-WideNet | 91% | 91% | 91.14% | 0.95 | 0.43M | No pre-training | False |
6. Conclusion
There is a dire need to develop fast methods for diagnosis of COVID-19, a capsule networks based detection model called COVID-WideNet has been presented in this paper. The proposed COVID-WideNet consists of two convolutional layers i.e. ‘PrimaryCapsule’ layer with ‘32 16D’ capsules, and ‘DiseaseCapsule’ layer with ‘2 8D’ capsules (here ‘32 16D’ refers to thirty-two capsules of 16-Dimensions each and ‘2 8D’ refers the two capsules each of them is 8-dimensions; where each dimension of capsule represents a unique feature). After replacing the scalar-output CNN-based approach with the vector-output capsule networks and substituting max-pooling with a routing-by-agreement method, we have achieved outstanding performance results that comprises 0.95 AUC along with sensitivity and specificity respectively. Obtained results during performance evaluations of the proposed model, indicate significant sensitivity and specificity both while diagnosing the COVID-19 symptoms. Further, the total number of trainable parameters is decreased to up to 20 times in comparison to any other CNN-based models. Therefore, (COVID-WideNet) can act as an alternative testing approach for faster detection of COVID-19 with the reduced diagnosis time and computational cost. With the use of capsule networks, we have overcome the various shortcomings of CNN based models caused by the use of pooling layers, data augmentation and transfer learning. Overall, this work presents a discriminative approach towards binary image classification in COVID-19 diagnosis using medical imaging.
In future, the proposed model will be tuned accordingly and implement with the help of big data analytic tools on cloud computing services this will help to monitor and analyze the patterns of the recent COVID-19 variants like delta, omicron, etc.
CRediT authorship contribution statement
P.K. Gupta: Research concept and team supervision. Mohammad Khubeb Siddiqui: Team supervision, Neural network algorithm, methodology, and write-up, Writing – review & editing. Xiaodi Huang: Neural network algorithm, methodology, and write-up, Writing – review & editing. Ruben Morales-Menendez: Writing – original draft. Harsh Panwar: Acquisition of data, analysis and/or interpretation of data. Hugo Terashima-Marin: Review. Mohammad Saif Wajid: Acquisition of data, analysis and/or interpretation of data.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
All authors approved the version of the manuscript to be published.
References
- 1.Olofsson S., Brittain-Long R., Andersson L.M., Westin J., Lindh M. PCR for detection of respiratory viruses: seasonal variations of virus infections. Expert Rev. Anti-Infect. Ther. 2011;9(8):615–626. doi: 10.1586/eri.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.X. Li, F. Li, Adversarial examples detection in deep networks with convolutional filter statistics, in: Proceedings of the IEEE International Conference on Computer Vision, 2017, pp. 5764–5772.
- 3.Sabour S., Frosst N., Hinton G.E. 2017. Dynamic routing between capsules. arXiv:1710.09829. [Google Scholar]
- 4.Hinton G.E., Krizhevsky A., Wang S.D. International Conference on Artificial Neural Networks. Springer; 2011. Transforming auto-encoders; pp. 44–51. [Google Scholar]
- 5.Patrick M.K., Adekoya A.F., Mighty A.A., Edward B.Y. Capsule networks–a survey. J. King Saud Univ.-Comput. Inf. Sci. 2019 [Google Scholar]
- 6.Jiménez-Sánchez A., Albarqouni S., Mateus D. Intravascular Imaging and Computer Assisted Stenting and Large-Scale Annotation of Biomedical Data and Expert Label Synthesis. Springer; 2018. Capsule networks against medical imaging data challenges; pp. 150–160. [Google Scholar]
- 7.Afshar P., Mohammadi A., Plataniotis K.N. 2018 25th IEEE International Conference on Image Processing (ICIP) IEEE; 2018. Brain tumor type classification via capsule networks; pp. 3129–3133. [Google Scholar]
- 8.Saurabh S., Gupta P. 2021 Thirteenth International Conference on Contemporary Computing (IC3-2021) 2021. Functional brain image clustering and edge analysis of acute stroke speech arrest MRI; pp. 234–240. [Google Scholar]
- 9.Bhardwaj P., Gupta P., Panwar H., Siddiqui M.K., Morales-Menendez R., Bhaik A. Application of deep learning on student engagement in e-learning environments. Comput. Electr. Eng. 2021;93 doi: 10.1016/j.compeleceng.2021.107277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siddiqui M.K., Islam M.Z., Kabir M.A. A novel quick seizure detection and localization through brain data mining on ECoG dataset. Neural Comput. Appl. 2019;31(9):5595–5608. [Google Scholar]
- 11.Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Bhardwaj P., Sharma S., Sarker I.H. AquaVision: Automating the detection of waste in water bodies using deep transfer learning. Case Stud. Chem. Environ. Eng. 2020;2 [Google Scholar]
- 12.Chakraborty K., Bhatia S., Bhattacharyya S., Platos J., Bag R., Hassanien A.E. Sentiment analysis of COVID-19 tweets by deep learning classifiers—A study to show how popularity is affecting accuracy in social media. Appl. Soft Comput. 2020;97 doi: 10.1016/j.asoc.2020.106754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Bhardwaj P., Singh V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-scan images. Chaos Solitons Fractals. 2020 doi: 10.1016/j.chaos.2020.110190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., Chen J., Wang R., Zhao H., Chong Y., et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021;18(6):2775–2780. doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., Xie Y., Li Y., Shen C., Xia Y. 2020. Covid-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv preprint arXiv:2003.12338. [Google Scholar]
- 16.Siddiqui M.K., Morales-Menendez R., Gupta P.K., Iqbal H.M., Hussain F., Khatoon K., Ahmad S. Correlation between temperature and COVID-19 (suspected, confirmed and death) cases based on machine learning analysis. J. Pure Appl. Microbiol. 2020;14 [Google Scholar]
- 17.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caruso D., Zerunian M., Polici M., Pucciarelli F., Polidori T., Rucci C., Guido G., Bracci B., de Dominicis C., Laghi A. Chest CT features of COVID-19 in Rome, Italy. Radiology. 2020 doi: 10.1148/radiol.2020201237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H. Outbreak of novel coronavirus (COVID-19): What is the role of radiologists? Eur Radiol. 2020;30(6):3266–3267. doi: 10.1007/s00330-020-06748-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., Zheng D., Wang J., Hesketh R.L., Yang L., et al. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 2020 doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Dong C., Hu Y., Li C., Ren Q., Zhang X., Shi H., Zhou M. Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology. 2020 doi: 10.1148/radiol.2020200843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang L., Han R., Ai T., Yu P., Kang H., Tao Q., Xia L. Serial quantitative chest ct assessment of covid-19: Deep-learning approach. Radiol.: Cardiothorac. Imaging. 2020;2(2) doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shen D., Shi Y. Abnormal lung quantification in chest CT images of COVID-19 patients with deep learning and its application to severity prediction. Med. Phys. 2021;48(4):1633–1645. doi: 10.1002/mp.14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021:1–14. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. Covid-caps: A capsule network-based framework for identification of covid-19 cases from x-ray images. Pattern Recognit. Lett. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toraman S., Alakus T.B., Turkoglu I. Convolutional capsnet: A novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110122. URL: http://www.sciencedirect.com/science/article/pii/S0960077920305191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Afshar P., Rafiee M.J., Naderkhani F., Heidarian S., Enshaei N., Oikonomou A., Fard F.B., Anconina R., Farahani K., Plataniotis K.N., et al. 2021. Human-level COVID-19 diagnosis from low-dose CT scans using a two-stage time-distributed capsule network. arXiv preprint arXiv:2105.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiwari S., Jain A. Convolutional capsule network for COVID-19 detection using radiography images. Int. J. Imaging Syst. Technol. 2021;31(2):525–539. doi: 10.1002/ima.22566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quan H., Xu X., Zheng T., Li Z., Zhao M., Cui X. DenseCapsNet: Detection of COVID-19 from X-ray images using a capsule neural network. Comput. Biol. Med. 2021;133 doi: 10.1016/j.compbiomed.2021.104399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heidarian S., Afshar P., Mohammadi A., Rafiee M.J., Oikonomou A., Plataniotis K.N., Naderkhani F. ICASSP 2021-2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE; 2021. Ct-caps: Feature extraction-based automated framework for covid-19 disease identification from chest ct scans using capsule networks; pp. 1040–1044. [Google Scholar]
- 33.Aksoy B., Salman O.K.M. Detection of COVID-19 disease in chest X-Ray images with capsul networks: application with cloud computing. J. Exp. Theor. Artif. Intell. 2021:1–15. [Google Scholar]
- 34.Saha P., Mukherjee D., Singh P.K., Ahmadian A., Ferrara M., Sarkar R. GraphCovidNet: A graph neural network based model for detecting COVID-19 from CT scans and X-rays of chest. Sci. Rep. 2021;11(1):1–16. doi: 10.1038/s41598-021-87523-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Nair V., Hinton G.E. Proceedings of the 27th International Conference on International Conference on Machine Learning. Omni Press; Madison, WI, USA: 2010. Rectified linear units improve restricted boltzmann machines; pp. 807–814. (ICML’10). [Google Scholar]
- 36.Wang L., Lin Z.Q., Wong A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen J.P., Morrison P., Dao L. 2020. COVID-19 image data collection. arXiv preprint arXiv:2003.11597. [Google Scholar]
- 38.Chung A. 2020. Figure 1 COVID-19 chest x-ray data initiative. URL: https://github.com/agchung/Figure1-COVID-chestxray-dataset. [Google Scholar]
- 39.Chung A. 2020. Actualmed covid-19 chest x-ray data initiative. URL: https://github.com/agchung/Actualmed-COVID-chestxray-dataset. [Google Scholar]
- 40.Chowdhury M.E., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Al Emadi N., et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 41.Radiological Society of North America . 2019. RSNA pneumonia detection challenge. URL: https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data. [DOI] [PubMed] [Google Scholar]
- 42.Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. 2020. Covid-19 image data collection: Prospective predictions are the future. arXiv preprint arXiv:2006.11988. [Google Scholar]
- 43.Heidarian S., Afshar P., Enshaei N., Naderkhani F., Rafiee M.J., Fard F.B., Samimi K., Atashzar S.F., Oikonomou A., Plataniotis K.N., et al. Covid-fact: A fully-automated capsule network-based framework for identification of covid-19 cases from chest ct scans. Front. Artif. Intell. 2021;4 doi: 10.3389/frai.2021.598932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saif A., Imtiaz T., Rifat S., Shahnaz C., Ahmad M.O., Zhu W.-P. CapsCovNet: A modified capsule network to diagnose Covid-19 from multimodal medical imaging. IEEE Trans. Artif. Intell. 2021 doi: 10.1109/TAI.2021.3104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siddiqui M.K., Huang X., Morales-Menendez R., Hussain N., Khatoon K. Machine learning based novel cost-sensitive seizure detection classifier for imbalanced EEG data sets. Int. J. Interact. Des. Manuf. (IJIDeM) 2020;14(4):1491–1509. [Google Scholar]
- 46.Siddiqui M.K., Morales-Menendez R., Huang X., Hussain N. A review of epileptic seizure detection using machine learning classifiers. Brain Inform. 2020;7:1–18. doi: 10.1186/s40708-020-00105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lusted L.B. Signal detectability and medical decision-making. Science. 1971;171(3977):1217–1219. doi: 10.1126/science.171.3977.1217. [DOI] [PubMed] [Google Scholar]
- 48.Obuchowski N.A. Receiver operating characteristic curves and their use in radiology. Radiology. 2003;229(1):3–8. doi: 10.1148/radiol.2291010898. [DOI] [PubMed] [Google Scholar]
- 49.Siddiqui M.K., Morales-Menendez R., Ahmad S. Application of receiver operating characteristics (ROC) on the prediction of obesity. Braz. Arch. Biol. Technol. 2020;63 doi: 10.1590/1678-4324-2020190736. [DOI] [Google Scholar]
- 50.Aljumah A.A., Siddiqui M.K., Ahamad M.G. Application of classification based data mining technique in diabetes care. J. Appl. Sci. 2013;13(3):416–422. doi: 10.3923/jas.2013.416.422. [DOI] [Google Scholar]
- 51.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of COVID-19 in X-Rays using nCOVnet. Chaos Solitons Fractals. 2020 doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Streiner D.L., Cairney J. What’s under the ROC? An introduction to receiver operating characteristics curves. Can. J. Psychiatry. 2007;52(2):121–128. doi: 10.1177/070674370705200210. [DOI] [PubMed] [Google Scholar]
- 54.Lalkhen A.G., McCluskey A. Clinical tests: sensitivity and specificity. Contin. Educ. Anaesth. Crit. Care Pain. 2008;8(6):221–223. [Google Scholar]
- 55.S.-T. Yeh, et al. Using trapezoidal rule for the area under a curve calculation, in: Proceedings of the 27th Annual SAS® User Group International (SUGI’02), 2002.
- 56.X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri, R.M. Summers, Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 2097–2106.
- 57.Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. 2009 IEEE Conference on Computer Vision and Pattern Recognition. Ieee; 2009. Imagenet: A large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 58.Hosmer D.W., Jr., Lemeshow S., Sturdivant R.X. John Wiley & Sons; 2013. Applied Logistic Regression, Vol. 398. [Google Scholar]