Abstract
Evaluating patient criticality is the foremost step in administering appropriate COVID-19 treatment protocols. Learning an Artificial Intelligence (AI) model from clinical data for automatic risk-stratification enables accelerated response to patients displaying critical indicators. Chest CT manifestations including ground-glass opacities and consolidations are a reliable indicator for prognostic studies and show variability with patient condition. To this end, we propose a novel attention framework to estimate COVID-19 severity as a regression score from a weakly annotated CT scan dataset. It takes a non-locality approach that correlates features across different parts and spatial scales of the 3D scan. An explicit guidance mechanism from limited infection labeling drives attention refinement and feature modulation. The resulting encoded representation is further enriched through cross-channel attention. The attention model also infuses global contextual awareness into the deep voxel features by querying the base CT scan to mine relevant features. Consequently, it learns to effectively localize its focus region and chisel out the infection precisely. Experimental validation on the MosMed dataset shows that the proposed architecture has significant potential in augmenting existing methods as it achieved a 0.84 R-squared score and 0.133 mean absolute difference.
Keywords: COVID-19 severity, Non-local attention, Squeeze, Deep learning, 3D CNN
1. Introduction
The SARS-Cov-2 coronavirus (COVID-19) created a global medical emergency in 2020 and its variants continue to spread in many countries, impacting 435 million people worldwide with 5.9 million deaths. Over this period, the health sector has faced a major setback in the allocation of medical resources. Access to ventilators, life support, and intensive care has to be prioritized for severely affected patients exhibiting critical symptoms. Therefore, accurate assessment of severity plays a crucial factor in the precise triage of COVID-19 patients. From the medical perspective, several biochemical and clinical parameters are studied as potential biomarkers for predicting patient outcomes and disease progression. Feng et al. associated COVID-19 with inflammatory levels expressed in terms of neutrophil count, lactic dehydrogenase, and C-reactive protein [1]. Similarly, biological variables like oxygen flow rate, diastolic pressure, and comorbidities investigated in clinical trials can help forecast severity conversion rate [2]. Scoring indicators like Pneumonia Severity Index (PSI), CURB-65, A-DROP show good sensitivity for use as prognostic variables [3]. Thus, medical history and clinical findings are effective factors in the creation of triage tools to identify high-risk patients [4].
While manual patient triage from clinical reports and lab tests can be very labor-intensive, automated data-driven methods are actively explored. Currently, the use of chest Computed Tomography (CT) scan imaging in prognostic studies has motivated the development of imaging-based Artificial Intelligence (AI) solutions that can analyze these findings to quantify severity. An international level consensus was reached about the applicability of imaging predictors as a primary tool in the clinical decision making and triage of patients suspected of COVID-19 [5]. Radiological features statistically distinguish between severe and non-severe patient groups [6]. Lieveld et al. statistically proved the associations between CT scoring and hospital/ICU admissions and 30-day mortality, suggesting that CT directly offers cues for prognosis [7]. Clinical features in the CT are taken as the reference even for affirming the diagnostic nature of other modalities including lung ultrasound, biochemical tests [8]. Common CT features attributed to COVID-19 include posterior and peripheral ground-glass opacities with or without consolidation, which are leveraged to facilitate contextual information for AI learning.
Challenges in learning an AI model from CT arise from the large inter-case variabilities in the data acquired from different scanners which have different parameters. Furthermore, the presence of certain CT biomarkers like ground-glass opacities, septal thickening, crazy paving patterns may not always contribute to severity, thus the assessment depends on the individual case and the adjoining context. It is also observed that patients with similar CT patterns might experience different extents of severity. Further, the CT manifestations responsible for COVID-19 are often presented with intricate morphological structures. From exhibits low contrast against background noise and lesser discriminability from similar manifestations. AI that learns to assess severity should be guided by a mechanism that aligns its receptivity to this lesion volume.
Recent works in CT severity assessment employ statistical, machine learning, and deep learning approaches to learn from CT images. In CNN, predominant methods include transfer learning from pre-trained models, multi-pathway branching networks, spatial/channel attention, and multiple instance learning. Section 2 discusses the pros and cons of these techniques in relationship with the advantages presented by the proposed work. To this end, we propose a novel weakly supervised attention framework that simultaneously stratifies COVID-19 risks and extracts infected volumes. Deep attention models especially in medical vision tasks, selectively enhance the properties of the infected tissues while diminishing other structures. Specifically, the use of the non-locality based attention in this work helps to capture long-range connectivity between lesion spread in different parts of the CT scan. Further, as attention networks develop a self-guided mechanism to adjust their weights, they are finely tuned in response to feedback from other layers. The attention modules proposed in this work are designed to fully analyze the semantic cues in deep voxel features on a range of scales and complexity. It comprehensively correlates findings across the scan to recognize salient CT features responsible for severity.
2. Related works
Statistical analysis, machine learning, and deep learning models are mainly studied for COVID-19 severity assessment. Several works utilize multi-modal learning that fuses predictor variables from not only CT image features but also patient medical history, signs and symptoms, demographic data, comorbidities, other clinical characteristics, and diagnostic biomarkers. Thus, the severity could be explained through any of these modes.
One of the popular approaches towards clinical risk estimation is the statistical hypothesis validation on lab experimental studies. These trials and multi-variate analysis aim to identify useful parameters that indicate the extent of severity in the lungs. For instance, Yang et al. developed a clinical framework that attributes the COVID-19 severity score to 20 different regions of the lungs depending on the parenchymal opacification involved in that region [9]. Sun et al. demonstrated that CT parameters including lesion, ground-glass opacities, consolidations have strong correlations with laboratory inflammatory biomarkers, which are observable indicators of severity [10]. The severity scores calculated for these parameters were statistically different between severe and non-severe groups. In a similar research, Feng et al. combinedly analyzed the CT lung abnormalities and clinical characteristics as risk factors to develop a severity score [1]. These parameters accurately reflected the extent of lesion involvement in the lungs. Evaluating standard risk prediction tools, such as Pneumonia Severity Index (PSI), quick Sepsis Related Organ Failure Assessment (qSROFA) renders calibration of the COVID-19 associated risks in terms of these variables. Fan et al. compared the accuracy of these indices to identify a reliable risk stratification system [3]. Neto et al. employed these indices and statistically confirmed their significance in differentiating severely affected populations [11]. Similarly, Zhang et al. developed a scoring system that uses a multi-variate analysis to select certain risk factors of severe pneumonia [12]. The usefulness of D-dimer levels as a reliable prognostic marker for mortality rate determination and hospitalization was demonstrated by Yao et al. [13]. In a clinical study to confirm the correlation of CAD-based quantification of lung parenchyma with other clinical findings, Durhan et al. decisively highlighted CT scoring utility in predicting severe pneumonia and ICU admissions [14]. Ebrahimian et al. investigated the closeness between standard Radiographic Assessment of Lung Edema (RALE) score and the severity determined by a commercial AI algorithm that scores proportionate to COVID-19 related findings [15]. In the clinical trial, it was observed that the difference between these scores was statistically insignificant and strongly correlated with patient outcomes. Overall, these statistical risk scoring techniques define a set of known variables, such as opacities, consolidations, and express severity as a heuristic function of these terms. In contrast, the deep learning model proposed in this work automatically performs feature engineering from the CT scans to determine useful abnormal attributes.
Machine learning techniques model the severity as a function of radiographical features and other clinical, biochemical parameters from Electronic Health Records (EHR). The combined feature-set is an effective modality for predicting clinical outcomes, triaging, and early identification of symptoms. Feature engineering aims to build effective descriptors out of these variables. As an example, Ye et al. combined the infection morphological features, and the texture attributes like coarseness, contrast, roughness, and entropy, to create a fusion assessment descriptor for severity estimation [16]. Along similar lines, Wu et al. developed a hierarchy of features encompassing radiological findings and clinical attributes [6]. Learning from quantitative CT lung-lesion features and clinical parameters, Zhang et al. employed a light gradient boosting machine regression model to assess severe or non-severe clinical stages [17]. Bagged trees recursive feature elimination was tried for feature selection followed by a logistic regression model to discriminate between the severe and non-severe groups. Cai et al. investigated histogram texture features of the CT lesion volumes to build random forest models for severity classification [18]. Leveraging the EHR data, Schoning et al. trained logistic regression and decision trees to classify severe and non-severe patients into distinct groups depending upon different severity levels [19]. Similarly, Bats et al. designed a framework of clinical characteristics consisting of 26 variables to learn a probability distribution for severe and risk-free patients [20]. Selection of the best predictors was made using the Akaike information criterion and the classifier was learnt as logistic regression. The utility of biochemical tests as a prognostic indicator of COVID-19 severity was demonstrated by Cobre et al. [21]. The trained neural networks, decision trees, and K-nearest neighbors showed the ability to predict positivity and relative importance order of the biomarkers in clinical decision making. Quiroz came up with a ML approach (gradient boosted trees) that utilizes the CT imaging features such as ground-glass volume, consolidate volume, effusion volume for assessing the severity [22]. Clinical and laboratory indices have been shown to enhance the learnability of ML for severity estimation. Tang et al. trained a random forests model over these fusion features [23]. The significance of each CT feature or laboratory index was obtained as a correlation of that variable to the predicted severity band. Though ML methods offer explainability of important clinical and imaging variables, they still do not directly derive features from CT images. They demand domain awareness and also extensive parameter fine-tuning for optimal performance. Deep learning on the other hand performs end-to-end feature extraction and learning. Also, the proposed CNN encodes key lesion information in its layers which can be incrementally transferred to further prognostic tasks/datasets, unlike ML design that specializes in a target application.
Deep learning is seen as a major future trend in COVID-19 prognosis [24]. The models like CNN, fully connected neural networks, sequence networks develop a perception of the severity through a layered stack of learnable units. They rigorously analyze the CT features in different levels of the processing hierarchy. Irmak et al. extracted the ground-glass opacities and extent of lung involvement through a feedforward CNN to quantify COVID-19 severity [25]. Lassau et al. improved the prognosis performance of the deep learning model by composite training on the multi-contextual dataset of deep CT features, clinical parameters, and biomarkers [2]. Different from these methods, our proposed work operates directly on raw CT images, progressively filtering the salient regions with attention. To optimize 3D CNN complexity for fine-grained severity assessment, Li et al. introduced an input slicing based on multi-view slicing [26]. Furthermore, the performance was enhanced by utilizing dual-Siamese channels and clinical metadata for prior knowledge transfer. In contrast to Li et al. that considers nine fixed slices in a 3D scan, the proposed CNN comprehensively attends to every 3D voxel at multiple resolutions. Karthik et al. presented a filter optimization module in CNN that can capture characteristic patterns of COVID-19 [27]. Similarly in our work, the attention module identifies the COVID-19 indicators by minimizing an additional infection segmentation error term. Karthik et al. introduced a contour enhancement to CNN for locating COVID-19 infected tissues [28]. Samala et al. leveraged the severity map generated by a pre-trained GoogLeNet to form intensity-based global descriptors from the image and classified them with logistic regression [29]. Naeem et al. used SIFT, GIST features to build a CNN-LSTM fusion model for severity prediction [30]. Aboutalebi et al. designed a projection–expansion CNN that generates enhanced representations for predicting the airspace severity of COVID-19 [31]. In a similar fashion, to adaptively recalibrate feature responses at each encoder level our proposed CNN explores a squeeze and channel-attention combination. Wang et al. explored dual parallel branching neural networks to share and co-learn features on joint segmentation tasks, which mirrors the setup of severity prediction and infection localization in our proposed approach [32]. Zhao et al. proposed an image registration and knowledge-aided CNN approach to effectively learn from limited segmentation labels in training data [33]. The proposed deep learning model also builds upon the concept of weak supervision that guides the attention head to learn lesion localization.
Deep transfer learning from off-the-shelf CNNs including Inception V3, ResNet, and DenseNet was attempted by Yu et al. [34]. The COVID-19 cases were assessed as severe or not from exploring these CT features on multiple ML classifiers. Aswathy et al. applied transfer learning from the fusion of ResNet50 and DenseNet201 [35]. Goncharov et al. proposed a residual U-Net model for severity assessment [29]. Exploiting both diagnostic and prognostic information evident from a CT scan, Feng et al. designed a U-Net based encoder network for extracting lesion features [36]. The encoded sequence was collectively classified to a set of severity bands, at the same time explicitly supervised to predict disease progression from mild to severe. Along similar lines, Goncharov et al. designed an encoder–decoder CNN to process severe lesions in the CT scan [37]. Lessmann et al. trained multiple pre-trained CNNs for assigning severity scores based on the degree of parenchymal involvement in the lung pulmonary lobes [38]. While transfer learning from standard CNN architectures exploits prior knowledge, the model design elements are not tailored to handle specific aspects of COVID-19 lesions. For instance, the CNN developed in this work inspired non-locality based attention to correlating lesions spread across the spatial and axial dimensions of the scan.
Applying CAD-based tools to quantify severity scores is another robust approach. Huang et al. tracked the CT lung opacification percentages of infected patients between initial assessment and follow-up treatments. These scores generated by a commercial deep learning software significantly varied amongst mild, severe, critical patients, thus validating the sensitivity of CT features [39]. Pu et al. designed a CAD system that utilizes elastic registration of lung boundaries and vessels, between two consecutive CT scans to assess severity and disease progression [40]. Different from statistical techniques and CAD tools that rely on biochemical variables and risk indices, the proposed approach learns solely from the chest CT modality. Inclusion of these other clinical parameters into our attention modeling would help determine their effect on the severity quantification.
Zhou et al. proposed an attention-based multi-modality feature fusion learning for severity prediction [41]. Mohammed et al. used spatial-channel attention CNN to detect infected regions with weak supervision [42]. Compared to these spatial/channel attention models, the proposed CNN is customized as a multi-stage analyzer that encodes a range of cues into hierarchical attention layers, such as global contextual awareness, cross-channel interdependencies, and variably-sized receptive field information across different feature sizes.
Multiple Instance Learning (MIL) on CT patches aids in severity estimation of different lung regions at a finer granularity. The patch-wise scores are collectively pooled to express the severity of the CT scan. The work by Li et al. proposed an instance-level attention model to weigh the severity of the cropped patch instances and combine them for bag-level classification [43]. The MIL was enhanced by performing a virtual bag-based data augmentation and learning a self-supervised pretext task ahead of actual training. He et al. developed an embedding-level MIL, to encode the patch instances into feature embeddings [44]. A global contrast pooling was applied over these instance embeddings in the bag to estimate severity. In a similar work, Xue et al. used MIL to generate deep representations from lung ultrasound [45]. These ultrasound features were fused with the encoded clinical information and assessed for the likelihood of being severe or not. Although MIL offers an effective means to gather severity from different parts of the CT image, the pooling function can dilute the quality of embeddings, as key activations might get diminished. While dense aggregations or attention-based MIL (as in [43]) overcome to an extent, the features are not robustly tuned at the micro-level. Our proposed CNN on the other hand, exploits non-local attention operations to comprehensively correlate fine-grained voxel features across the scan. Unlike the self-guided MIL that takes longer to converge, we employ a weakly-supervised refinement technique to auto-correct the attention focus. Though this model uses pooling layers, these effects are compensated by considering a range of scales and resolutions.
2.1. Research gaps and motivation
The key motivations behind the proposed design are as follows,
-
•
Prevalent AI methods treat severity assessment as a classification task into one of mild, moderate, severe, and fatal categories. Since such a rigid stratification has blurred boundaries, learning to separate these classes can be limiting the key information desired in precise triage.
-
•
While attention-based design enhancements are generally used to augment the CNN capabilities, there exists a methodological possibility of exploring fusion systems with different architectural elements that can complement and fully harness the potential of attention learning for the given task/dataset at hand. Although several works have researched multi-level attention structures, access to the interpretability of predictions at various levels can offer better penetration into the decision process which better informs the clinicians.
-
•
As it is commonly not feasible to enable large-scale fully annotated medical image databases for diagnostic studies, there is a significant shift towards AI architectures that can effectively exploit weakly labeled datasets to provide insights. Navigating the CNN focus areas in response to weakly-guided learning amplifies contextuality in features.
2.2. Research contributions
Motivated by the above research opportunities, the major contributions put forth by the proposed work are listed below:
-
•
The proposed AI framework learns the severity scoring as a regression model on a continuous 0–1 scale encompassing five bands of criticality. Different from existing works, this formulation helps the model respond well to variations in the CT manifestations that influence the degree of severity.
-
•
The proposed model employs a non-local attention scheme to correlate severe structures encoded in different parts of the 3D scan. Weakly supervising this attention head from a limited set of infection annotated samples guides the CNN to refine its focus towards severely infected areas. Designing explicit guidance to fine-tune the non-locality based attention module is a novelty of this work.
-
•
The proposed model builds an information channel across the multi-scale features, and attention modules by encapsulating them into a layer-wise attention encoder–decoder hierarchy that is tunable at a micro-level of granularity.
-
•
Salient knowledge transfer from individual modules in the attention unit is designed to facilitate information discovery and complement learning at other modules in the unit. The Squeeze and Channel-attention layers encode large receptive fields and long-range connectivity that can be globally queried at the attention Decoder to extract infections.
-
•
The proposed CNN intrinsically highlights the CT hotspots influencing the severity prediction, as pointed by the forward attention tended to individual 3D voxels.
3. Proposed work
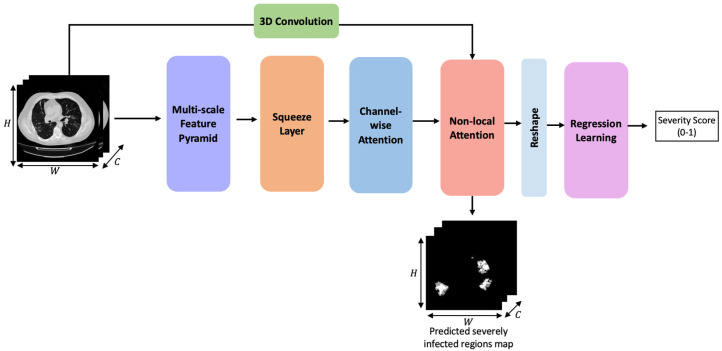
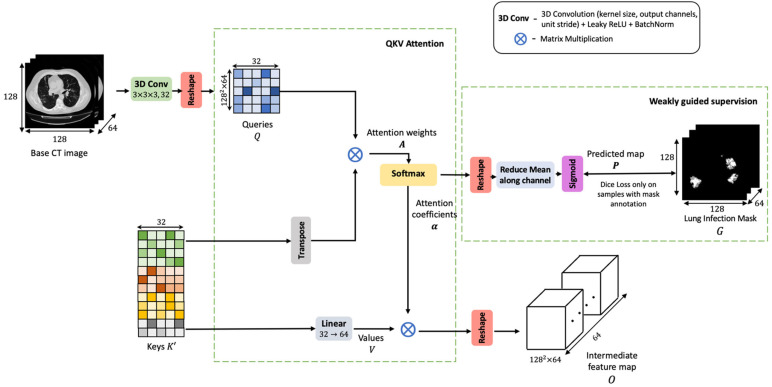
The proposed network is modeled as a 3D CNN that performs attention fusion over diverse feature sets. A high-level architectural sketch of the proposed CNN framework is presented in Fig. 1. The first three blocks make up the encoder phase. It is designed to robustly analyze the CT scan and encode adequate contextual information that would facilitate the non-local attention layer. The non-locality based attention module extracts relevant cues from this encoded map and correlates these features with voxels in the base CT scan to decode the severely infected areas. Weak supervision from a limited set of infection-labeled samples is used to guide this attention learning in the refinement of key focus areas and further quantify severity. Finally, the learning objective is defined on dual loss functions applied both to lesion localization and severity regression.
Fig. 1.
Architecture sketch of the proposed non-local squeeze attention 3D CNN framework for severity assessment.
The subsequent Sections 3.1 to 3.4 elucidate the stages involved in the CNN framework. Lastly, Section 3.5 presents the multi-task learning setup and loss functions for training the model.
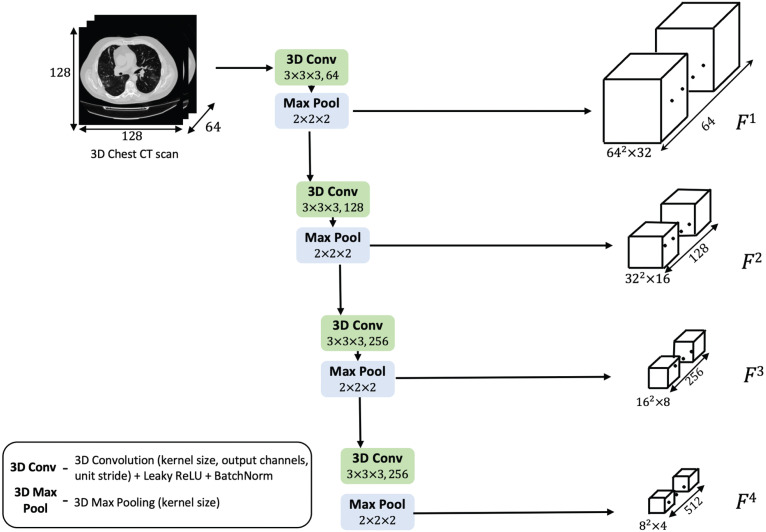
3.1. Multi-scale feature pyramid
The input 3D CT scan is processed by a stack of 3D convolutions as shown in Fig. 2. The feature pyramid robustly extracts salient features in four different spatial scales. The shallower layers extract low-level information, structural details such as lesion edge, boundaries, morphology, intensity range, and contrast variations with other manifestations. In the inner layers, more semantic features with objectness information are derived. Building an encoder representation with such broad-ranging contextual details is central to assessing severity of the COVID-19 infection present in the scan.
Fig. 2.
Schematic diagram of the multi-scale feature pyramid.
In Fig. 2, the convolutional layer consists of a 3D convolution operation, followed by a Leaky ReLU activation and batch-wise Z-normalization. Every pyramid level emits double the number of channels from the previous level. The activated feature map is downsized using a 3D max-pooling layer which reduces the dimensions by half. Thus, with each layer, the resolution of the volume gets halved, and the voxel feature depth is doubled. The pyramid feature maps are denoted by , , , in the decreasing order of resolution.
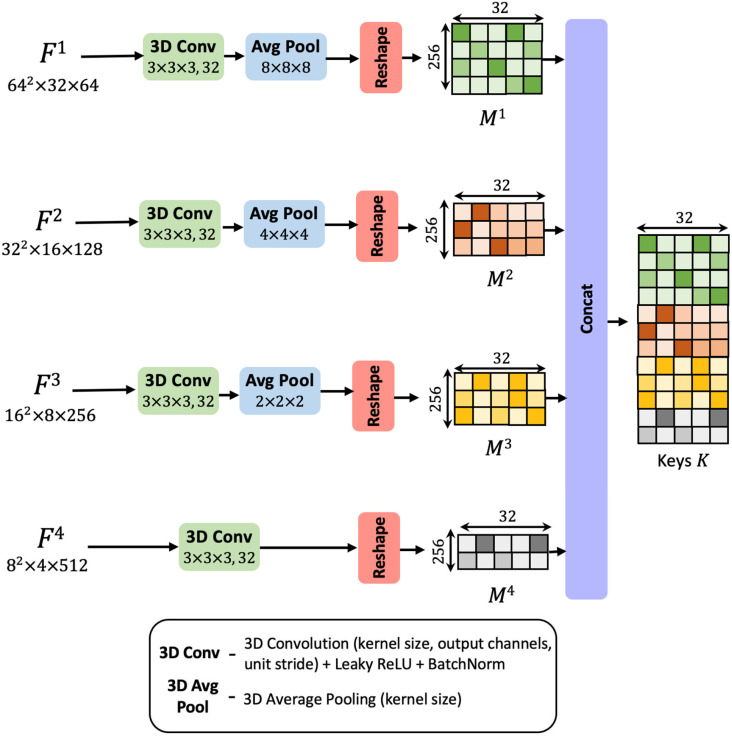
3.2. Squeeze layer
This feature pyramid encodes rich semantic features in the deep layers while preserving the low-level structures in the initial layers. Exploiting such multi-scale, varied contextual information plays an important role in precisely assessing the criticality of different regions in the CT scan. Multi-scale feature learning is a key ingredient in the design of neural networks for medical image processing and has led to state-of-the-art results in several tasks. Particularly, leveraging this information enables weak localization of the infected CT voxels.
In the squeeze layer shown in Fig. 3, the four differently sized maps , , , are parallelly reduced to a uniform channel dimension of 32. The reduction achieved by 3D convolutional layers introduces a bottleneck representation. These 3D convolutional transformations analyze the channel-wise dependencies, squeeze and consolidate the filters information into a lower-dimensional space, comprising of 32 channels. The average pooling layers down-sample the spatial resolution to a uniform size of 16 × 16. The resultant feature maps are reshaped into a matrix form. Here the rows represent voxels in the 3D volume, and the columns correspond to the 32-dimensional feature embedding associated with the voxel. Let be the transformed matrix corresponding to feature map . These feature matrices infuse contextual information from differently sized receptive fields. They encapsulate the semantic details drawn from various scales.
Fig. 3.
Diagrammatic representation of stages involved in the squeeze layer.
The matrices are linearly densely stacked along the voxels dimension to yield an aggregated feature set. This matrix entity, denoted by is termed as the ‘keys’ (refer to Fig. 3). The densely fused contextual map, , comprehensively encodes the local spatial descriptions from multiple scales and captures the channel relationships as well. It will be analyzed by the subsequent layers to decode the lesion tissues.
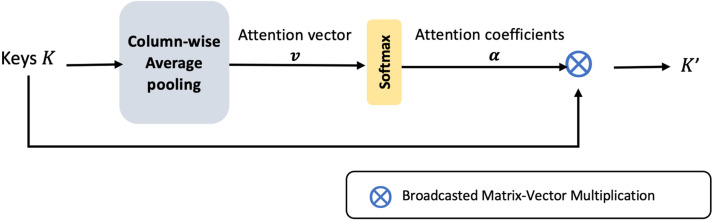
3.3. Channel-wise attention layer
Learning a mechanism to weigh the channel information, enables the CNN to adaptively recalibrate its parameters and converge on significant features. The keys, generated in the previous step are pooled from the multi-scale feature pyramid. Thus, the merged 32 number of filter channels in are a result of convolutional layers with different parameters. Therefore, a channel-wise attention model at this step would normalize the effects of fusing divergent channel information. It clearly highlights the relevant channels for the subsequent layers. Also, it constrains the upstream convolutional layers to align the feature extraction and produce complementary features that enhance the dense fusion.
Aggregation of voxel features along the channel dimension leads to global channel-wise statistics. The attention layer applies a global average pooling of columns in , to form 32 channel descriptors (presented in Fig. 4). This operation squeezes the global voluminal information encoded by each channel into a scalar value. A vector of such 32 channel descriptors is called the attention vector, denoted by . The attention vector is softmax normalized and masked along the columns of . The attention gating function results in optimal weighing of the channel-wise features.
Fig. 4.
Block diagram of the channel-wise attention layer.
3.4. Non-local attention block
Let denote the transformed set of keys emitted at the channel-attention block. is used as context to guide the decoding of COVID-19 infections and criticality scoring from the base CT scan. The non-local attention block learns a comprehensive attention function, where voxels in the base CT scan attend to the information contained in the encoded contextual map. With such learning, a voxel’s receptive field is not restricted to its local spatial neighborhood but is set to the entire context of the scan. Since the voxel features can tune in response to the global view of the CT, its representational capabilities are enhanced. Global attention to multi-scale features boosts the CNN’s discriminatory learning and precise localization of the severely infected tissues.
Fig. 5 depicts the steps involved in non-local attention. First, the CT scan is processed into a feature map, termed as queries, . Queries are an entity used to search and extract relevant details from the encoded context. On the other hand, the keys, are utilized to serve the required information as queried by . The attention map, is computed as a matrix product between the query and keys, as presented in Eq. (1).
| (1) |
refers to the degree of correlation between voxel and the encoding . The resulting matrix is row-wise softmax normalized and transformed into a unified attention coefficient map alpha (refer Eq. (2)).
| (2) |
For facilitating feature adaptation and easing the convergence of attention parameters, the attention weighing is performed over a contextual map, . This entity, termed as values, is derived as a linear projection of . The attention coefficients, are combined over as given in Eq. (3). Combining on V offers the CNN a learnable mechanism to refine its output feature map in response to the attention feedback.
| (3) |
The final representation emitted at the non-local attention, given by O has selectively infused salient details from all areas in the CT and suppressed insignificant signals.
Fig. 5.
Schematic diagram of the non-local attention block.
To enable sharp refinement of the attention focus towards lesion-rich areas, this attention coefficient map, is further subjected to supervision from a subset of infection annotated samples. This matrix is reshaped into a 4D tensor and averaged along the channel dimension to yield . Then given sample with infection mask and predicted attention map , the localization loss is defined as follows,
| (4) |
From Eq. (4) it is clear that this dice loss function is only optimized for samples with mask.
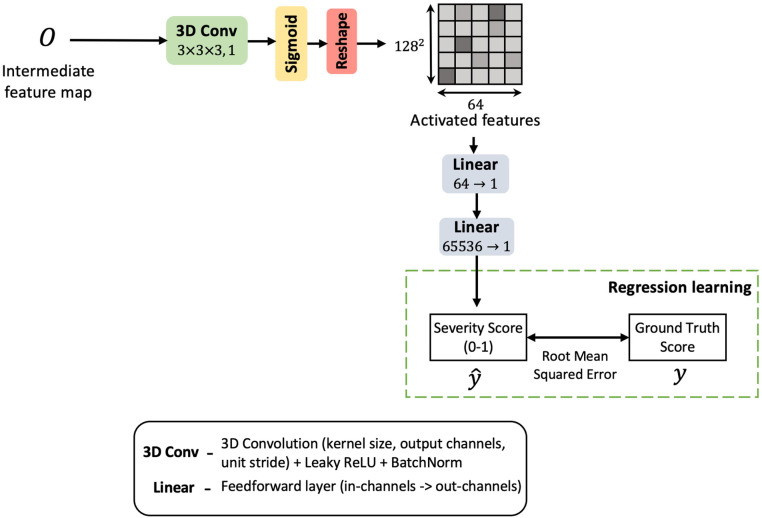
3.5. Regression learning
The proposed CNN is trained for severity regression as shown in Fig. 6. The severity score is regressed within a range of 0–1. The dataset offers five bands of criticality observed in the COVID-19 patients based on their degree of lung abnormalities. These bands are set at 0.00, 0.25, 0.50, 0.75 and 1.00. The regression head predicts a severity measure closest to one of these marks. The decoded feature map from the previous layer is convolved and linear projected to a regression score, (refer Fig. 6). The regressor head is trained against target score on mean squared loss, which is given in Eq. (5).
| (5) |
The joint loss function applied to both the localizer and regressor during training is presented in Eq. (6). As a result, during prediction, the model is not only able to a generate criticality score but also renders a rough estimation of the affected volume.
| (6) |
Fig. 6.
Schematic sketch of the CNN head for regression.
4. Dataset description
The MosMed CT scans dataset was used to train and validate the proposed method for severity assessment. The reason for the choice of this severity dataset is because it also comes with infection labeling annotated for a subset of 3D scans. These expert annotated samples are leveraged through weak supervision to tune the attention head to roughly localize the severely infected regions.
The dataset contains 1110 3D CT volumes, collected from individual patients in the municipal hospitals of Moscow, Russia. The scans are stratified into five bands of severity. Table 1 describes the severity thresholds and sample counts under these bands. Based on the criticality range, a suitable regression target score between 0 to 1 is assigned to the samples. Out of all scans in the dataset, infection masks are available only for a subset of 50 scans which are used to train the attention module.
Table 1.
Distribution of CT scan samples based on severity of the COVID-19 infection.
| Severity band | Pulmonary parenchymal involvement | Number of 3D scans | Regression target score |
|---|---|---|---|
| Zero | Absent | 254 | 0.00 |
| Mild | 25% | 684 | 0.25 |
| Moderate | 25% to 50% | 125 | 0.50 |
| Severe | 50% to 75% | 45 | 0.75 |
| Critical | 75% | 2 | 1.00 |
4.1. Data pre-processing
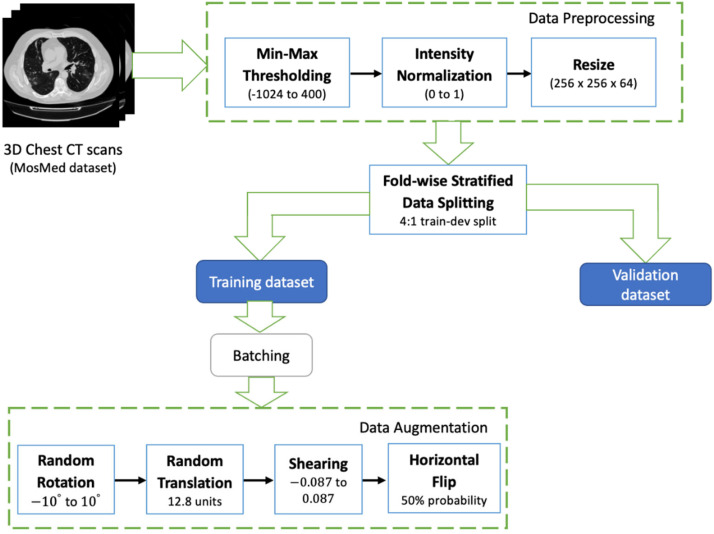
The data pipeline consists of the steps described in Fig. 7. First, the raw 3D scans are intensity normalized. The CT voxel intensities are saved in Hounsfield Units (HU). For scans in the MosMed dataset, this ranges from –1024 to 2000. 400HU is chosen as the upper bound since the radio-intensities above this threshold indicate bones. Therefore, the scans are min–max thresholded to the range bounded between –1024 and 400. Post this step, they are normalized between 0 to 1. Normalized scans are then resized to a uniform spatial resolution of 128 ×128 and depth of 64 slices. Interpolating the scan intensities to fit these dimensions facilitates consistent data batching and tensor processing across different CNN layers.
Fig. 7.
Stages involved in the data preparation and augmentation of the 3D CT scans.
4.2. Dataset augmentation
To improve generalizability of the model and expose to new orientations during training, the samples were augmented in an online fashion. In each randomly sampled mini-batch, the samples were dynamically subjected to a series of data transformations on the fly at the time of training, as opposed to creating a static augmented set (refer Fig. 7). The parameter ranges for the affine transformations are as follows: (1) angle of rotation is randomly drawn from the range of to , (2) translation along x-y directions are within 10% of the image’s height and width, (3) horizontal and vertical shears factors are randomized between and for a shear angle of , (4) horizontal flipping is performed with a 50% probability. These augmentations are carried out on the raw data in the training set. Samples in the validation or testing sets are directly considered for evaluation.
5. Results and discussions
This section presents the performance analysis of the proposed CNN through multiple experiments. In the ablation study, the effectiveness of each individual building block of the model is investigated in isolation from other components. The proposed architecture is robustly evaluated on the K-fold cross-validation scheme. In addition, we present a quantitative comparison of the proposed work’s efficacy with state-of-the-art methods.
5.1. Experimental setup
The K-fold cross-validation approach is chosen to evaluate CNN performance and carry out model training/validation. The training-validation splits are generated in a stratified manner, i.e., the ratio between the five categories of samples is the same in both sets. When creating data batches, an equal number of samples from the five severity bands are drawn into a single batch. The weighted-class data sampling technique ensures that this percentage of severity classes is equally balanced within the batch.
The proposed CNN was trained on a 32 GB NVIDIA V100 GPU in an Ubuntu VM instance on the Google Cloud. The model was implemented in PyTorch. Adam was used as the default gradient descent optimization algorithm in all the experiments. The initial learning rate was set to 0.01. The rate of gradient updates is controlled by a multiplicative learning rate decay scheduler. The learning rate drops by a factor of 0.1 when no improvement is observed in the validation accuracy for a span of 10 epochs. Considering the GPU memory limitations, a batch size of 32 was chosen for training the CNN.
5.2. Results of CNN training
The proposed model was empirically analyzed on the 5-fold cross-validation framework. To measure convergence of the dice loss applied at the attention head, it was ensured that the CT samples with infection mask were split in a 4:1 ratio between the training and validation sets in each fold. The model was combinedly trained for severity regression and weakly supervised attention localization. The trained model was evaluated on regression metrics that included Mean Squared Error (MSE), Root Mean Squared Error (RMSE), Mean Absolute Error (MAE), and Coefficient of Determination (R2 score). The Dice Similarity Coefficient (DSC) was used to evaluate the degree of closeness between the predicted and actual infection masks at the attention head.
Results of evaluating the proposed CNN on 5-fold cross-validation are presented in Table 2. The model converged with a residual error of 0.13. Since the difference between any consecutive two severity levels is 0.25, a confidence interval of 0.13 on unseen data reasonably demarcates the boundary between these bands. It suggests that the model has learnt characteristic features for different severity ranges and exploits these cues to distinguish the degree of infection between two adjacent bands. It has learnt to correlate similarity of CT manifestations within a severity level and differentiate the patterns across the levels. A mean squared error of 0.019 further asserts the good degree of fit achieved by the model. R2 score projects the extent of variability of the severity scoring that can be explained by the model. With a coefficient of 84%, the model reliably fits the distribution of severity values, by capturing most of the variance inherent to that data. It accurately forecasts the criticality measure, as a dependent variable of the CT scan data with only a low margin of error.
Table 2.
Fold-wise computation of the trained model metrics for severity score regression, recorded on the validation set from each fold.
| Folds | MSE | RMSE | MAE | R score |
|---|---|---|---|---|
| Fold 1 | 0.017 | 0.131 | 0.126 | 0.860 |
| Fold 2 | 0.019 | 0.140 | 0.136 | 0.840 |
| Fold 3 | 0.021 | 0.148 | 0.139 | 0.824 |
| Fold 4 | 0.022 | 0.149 | 0.146 | 0.820 |
| Fold 5 | 0.016 | 0.126 | 0.116 | 0.873 |
| Average | 0.019 | 0.139 | 0.133 | 0.843 |
The trendline variation in the R2 score and decay of mean square error are visualized in Table 3. The values are tabulated in Table 4. The R2 curve shows an increasing trend that saturates in about 14 epochs. In all folds, the model converged close to 20 epochs. The minimal gap observed between training and validation curves suggests that the CNN generalized well for the data distribution. The decay in mean squared error follows a decreasing trend, where the validation loss initially staggers but stabilizes in the later phase. Fact that the mean difference in MSE across the folds is only 0.003, proves the model’s robustness to perform alike on different splits of unseen data. An optimal mean dice loss of 0.199 at the attention module suggests that it has significantly complemented the regression learning, by guiding the model’s focus towards infection areas. Being able to implicitly associate this severity score to specific regions in the CT scan using attention also highlights the explainability aspects of the model design.
Table 3.
Learning curves recorded during training. Mean squared error is the criteria for severity regression, while reduction dice loss is monitored for the attention head. The coefficient of determination is computed on the train/dev sets after every epoch.
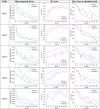 |
Table 4.
Metrics recorded for regression and attention localization on the 5-fold cross-validation scheme.
| Folds | Mean squared error |
R score |
Dice loss at attention head |
|||
|---|---|---|---|---|---|---|
| Training | Validation | Training | Validation | Training | Validation | |
| Fold 1 | 0.007 | 0.017 | 0.980 | 0.860 | 0.119 | 0.228 |
| Fold 2 | 0.008 | 0.019 | 0.985 | 0.840 | 0.051 | 0.185 |
| Fold 3 | 0.010 | 0.021 | 0.988 | 0.824 | 0.099 | 0.190 |
| Fold 4 | 0.012 | 0.022 | 0.989 | 0.820 | 0.091 | 0.188 |
| Fold 5 | 0.010 | 0.016 | 0.994 | 0.873 | 0.076 | 0.204 |
| Average | 0.009 | 0.019 | 0.987 | 0.843 | 0.087 | 0.199 |
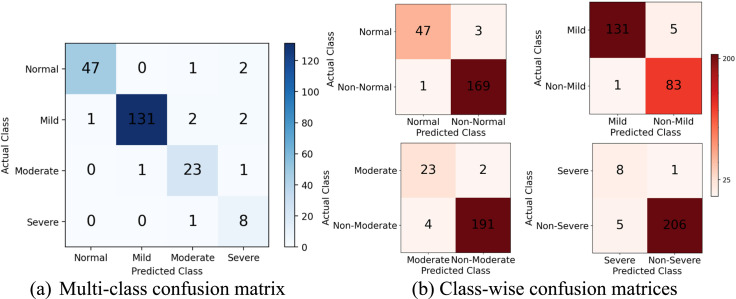
To evaluate the classifiable nature of the regression scores, the values are mapped to severity classes based on the scoring intervals. As described in the dataset section, the values closer to 0.00, 0.25, 0.50, and 0.75 get marked as Normal, Mild, Moderate, and Severe respectively. From Fig. 8, the model achieves excellent distinguishability between the categories. The recall metric is registered at over 94%, 96%, 92%, 88% for the individual classes respectively, which reinforces confidence in detecting the positive cases. There are only a few false positives resulting from misclassification.
Fig. 8.
Mapping the regression scores to severity classes to count true and false predictions under each category. Shown are the values recorded on the validation set averaged across 5 folds.
The trendline variation in the R2 score and decay of mean square error are visualized in Table 3. The values are tabulated in Table 4. The R2 curve shows an increasing trend that saturates in about 14 epochs. In all folds, the model converged close to 20 epochs. The minimal gap observed between training and validation curves suggests that the CNN generalized well for the data distribution. The decay in mean squared error follows a decreasing trend, where the validation loss initially staggers but stabilizes in the later phase. Fact that the mean difference in MSE across the folds is only 0.003, proves the model’s robustness to perform alike on different splits of unseen data. An optimal mean dice loss of 0.199 at the attention module suggests that it has significantly complemented the regression learning, by guiding the model’s focus towards infection
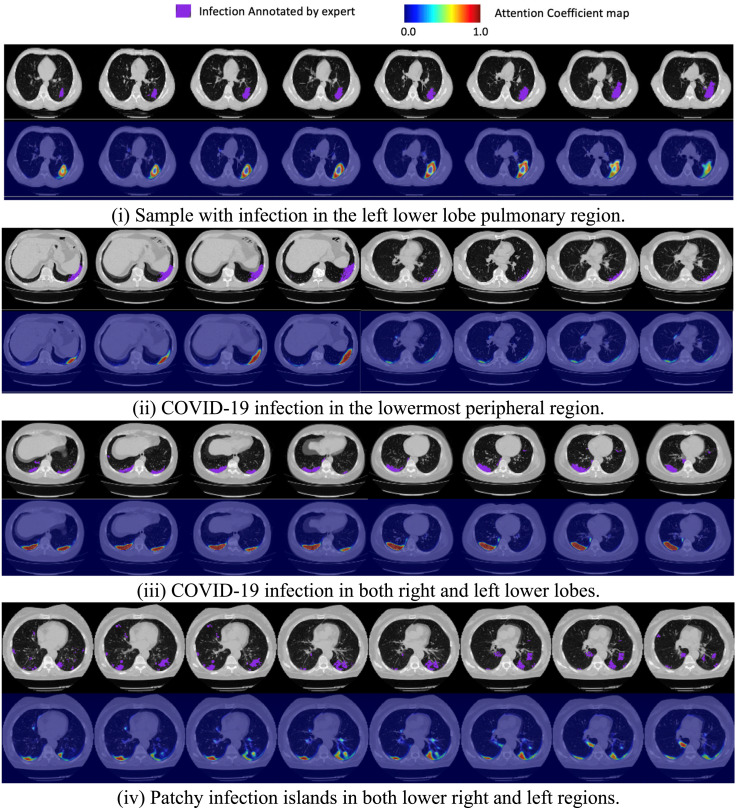
5.3. Infection localization
A major finding from this work is the ability to guide an attention model to learn focus areas with weak supervision. The severe regions map derived as in the non-local attention layer (refer Fig. 5) is visualized in Fig. 9. In all the four CT instances drawn from the test set, the CNN’s attention heatmap has roughly coincided with the ground truth infection label. This proves that explicitly guiding the attention learning even with a limited number of annotated samples leads to better interpretation of infected regions. It also assures applicability of this modeling to real-world data.
Fig. 9.
Cross-section view of the CNN attention map against the expert annotated infection labeling for four different CT scan samples in the hold-out test set. The choice of samples is picked to show the model’s response to various heterogeneity in the lesion morphology.
The quantitative results summary of the attention learning is presented in Table 5. With a precision of 84.8%, the model has finely captured the infection morphology with fewer false positives. Given a 75.9% recall score, it has certainly identified the predominant regions of pulmonary involvement with a modest miss rate. The attention maps obtained against a heterogeneous distribution of lesion samples in Fig. 9, further validate these conclusions. Except for a few detached islands, majority of the infectious areas are well captured and accounted for in the severity quantification. In addition to the severity score, these estimated infected voxels can serve as additional radiological markers/insight for the clinician.
Table 5.
Fold-wise performance of the weakly supervised attention learning computed on the validation samples with infection labeling.
| Folds | Precision | Recall | DSC | IoU |
|---|---|---|---|---|
| Fold 1 | 0.824 | 0.726 | 0.772 | 0.623 |
| Fold 2 | 0.857 | 0.776 | 0.815 | 0.677 |
| Fold 3 | 0.876 | 0.753 | 0.810 | 0.695 |
| Fold 4 | 0.871 | 0.760 | 0.812 | 0.668 |
| Fold 5 | 0.811 | 0.781 | 0.796 | 0.672 |
| Average | 0.848 | 0.759 | 0.801 | 0.667 |
5.4. Ablation study
This section investigates the effectiveness of the three key building blocks of the proposed architecture - (1) Squeeze layer, (2) Channel-wise Attention layer and the (3) Non-local Attention layer. To quantify the usefulness of an individual component, the contribution of that component is measured as a percentage degrade in the CNN performance after its removal. The results of this evaluation are tabulated in Table 6.
Eliminating the squeeze layer increased the mean residual error to 0.192 and the R2 score dropped by 17.2%. This layer serves as the encoder. For this ablation experiment, in place of the squeeze encoder, the feature map was directly reduced to 32 channels and propagated downstream (refer Fig. 3). The inclusion of this layer primarily helps the CNN to process diverse receptive field information from different levels of the multi-scale feature pyramid. Low-level structures harnessed at the initial layers get streamlined into the CNN’s attention learning and serve as salient cues for infection localization.
The channel-wise attention layer weighs the relative significance of the channel information present in the densely fused contextual map from the squeeze layer (refer Fig. 4). In absence of this layer, this feature map is not channel-wise normalized, therefore patterns of random maximal activations emerge from the component maps and affect the rate of convergence. Thus the model saturated in more number of epochs, yet the performance quotient fell by 6.88% R2 score. This experiment confirms the usefulness of analyzing cross-channel relationships to determine salient features. Applying this step, especially in the deeper layers improves tuneability of the CNN features.
In the final ablation study, the non-local attention block is examined. This block performs decoding of the semantic features towards severity assessment. For the ablation experiment, this block is directly substituted with feedforward fully connected layers that learn regression. The fused keys matrix, is flattened and passed down these linear layers (refer Fig. 5). The outcome of removing this component has the highest impact on the results. The R2 score dropped by a huge factor of 28.9%. Thus, the non-local attention’s assessment of the significant encoded features plays a huge role in decoding the relevant details for evaluating severity.
Table 6.
Result analysis of ablation experiments.
| S. No. | Ablation experiment | MSE | RMSE | MAE | R score |
|---|---|---|---|---|---|
| 1 | Proposed model without squeeze layer | 0.038 | 0.194 | 0.192 | 0.698 |
| 2 | Proposed model without channel-wise attention layer | 0.027 | 0.164 | 0.142 | 0.785 |
| 3 | Proposed model without non-local attention layer | 0.050 | 0.223 | 0.218 | 0.599 |
| 4 | Proposed model with all components included | 0.019 | 0.139 | 0.133 | 0.843 |
5.5. Comparison of proposed architecture with the existing works
We present a performance analysis of the non-local attention CNN with existing CT-based works for COVID-19 diagnosis and severity prediction, as listed in Table 7. To ensure a fair comparison of the works, these compared methods were implemented as regression models on the same Mosmed dataset at hand and they were all evaluated on a common test partition.
Table 7.
Experimental performance validation of the proposed model with similar works for COVID-19 severity assessment from CT scans.
| S. No. | Source | Method | R score | MSE | RMSE | MAE |
|---|---|---|---|---|---|---|
| 1 | Aswathy et al. [35] | Transfer learning from combined ResNet50 and DenseNet201 | 0.557 | 0.493 | 0.243 | 0.238 |
| 2 | Yu et al. [34] | SVM classifier on DenseNet201 features | 0.568 | 0.484 | 0.235 | 0.233 |
| 3 | Goncharov et al. [46] | Residual U-Net | 0.605 | 0.470 | 0.221 | 0.215 |
| 4 | Qiblawey et al. [37] | Encoder–Decoder CNN | 0.636 | 0.457 | 0.209 | 0.206 |
| 5 | Naeem et al. [30] | CNN-LSTM autoencoder on SIFT, GIST features | 0.684 | 0.441 | 0.195 | 0.190 |
| 6 | Zhou et al. [41] | Spatial channel attention residual network | 0.691 | 0.437 | 0.191 | 0.188 |
| 7 | Mohammed et al. [42] | Spatial channel attention CNN-LSTM | 0.720 | 0.427 | 0.183 | 0.182 |
| 8 | Chatzitofis et al. [47] | Attention decoder CNN | 0.738 | 0.423 | 0.179 | 0.175 |
| 9 | Xiao et al. [48] | MIL on ResNet50 | 0.753 | 0.414 | 0.172 | 0.168 |
| 10 | He et al. [25] | Attention MIL | 0.771 | 0.407 | 0.166 | 0.160 |
| 11 | Li et al. [26] | Multi-view Dual-Siamese CNN | 0.802 | 0.392 | 0.154 | 0.147 |
| 12 | Ouyang et al. [49] | Dual sampling attention | 0.813 | 0.386 | 0.149 | 0.145 |
| 13 | Proposed work | Non-local squeeze attention CNN | 0.843 | 0.019 | 0.139 | 0.133 |
It is to be noted that though standard CNN architectures such as ResNet, DenseNet are accurate for most vision tasks, they still need additional customizations to recognize intricate CT structures contributing to severity. In a similar sense, the ML classifier on deep features also resulted in a lower R2 score of 56.8% [34]. To adapt U-Net for severity estimation, Goncharov et al. introduced a slice-wise correlation and achieved a better 6.51% R2 score and 0.215 MAE [46]. Similarly, the DenseNet161 encoder with U-Net quantified the infection severity accurately to a 63.6% R2 score [37]. Different from these encoder–decoder methods, the proposed CNN incorporates an attention fusion module that spatially orients the CNN focus to locations with high lesion likelihood.
CNN-LSTM based models generate an optimal level of fit with better results [30], [42]. Specifically, Naeem et al. used SIFT, GIST descriptors to encode the CT scan and reduced MAE to 0.190 [30]. Though this method analyzes global/local texture features, it adds an overhead for deriving these feature sets, also demands extensive hyperparameter fine-tuning. On the other hand, Mohammed et al.’s Spatial/Channel (SC) attention module generates slice-level severe regions prediction in a weakly supervised manner [42]. Nevertheless, it does not offer explicit guidance to aid in attention refinement or feature modulation, thus requiring more data and longer training to converge. The same model behavior was observed in the attention ResNet by Zhou et al. that registered a 69.1% R2 score [41]. Compared to these SC-attention modules, the proposed Squeeze-Channel attention layers form a fusion of various feature scales resulting in a global encoded representation. Furthermore, the non-local attention processing is modeled as a decoder function that exploits this context.
Extracting lesion-rich slices through attention decoding was achieved by Chatzitofis et al. which gave a 6.80% higher R2 score and 0.175 MAE [47]. While this approach considers only frontal slices to predict severity, accurate infection quantification should be congregated from different axial views of the scan.
Amongst weakly supervised techniques, MIL models converged faster and gave higher precision. He et al. projected severity score as a weighted linear combination of features from different 2D parts in the scan and attained a 77.1% R2 score, 0.160 MAE [25]. However compared to such dense pooling, the proposed non-local attention applies a rigorous cross-correlation of voluminal and channel-wise features to capture interdependencies across the 3D scan. Xiao et al.’s MIL pooling over ResNet features also does not consider the inclusion of multi-scale features, which is the key advantage in the proposed attention model [48].
The dual Siamese network proposed by Li et al. resulted in a high 81.3% R2 score and 0.145 MAE as it analyzed multiple views, yet it requires complementary information from other clinical markers to assess accurately [26]. Of the attention models, dual sampler attention induced size-aware sampling and attention refinement, therefore, has matched performance closer to the proposed work [49].
6. Conclusion
The attention framework presented in this paper was aimed at diagnosing the criticality of a patient’s medical condition from the CT scan. To the best of our knowledge, this is the first regression-based approach for COVID-19 severity scoring through a deep learning model. The CNN is customized as a multi-stage analyzer that encodes a range of cues into hierarchical attention layers, such as global contextual awareness, cross-channel interdependencies, and variably-sized receptive field information across different feature sizes. Specifically, multi-scale features are extracted and fused to construct a robust encoded representation for the decoder. Subsequently, we squeeze the structural and semantic details in the fused contextual map into a global reference encoding and apply cross-channel correlation. Finally, the processed feature set is matched against the base CT scan through a non-local attention module that decodes the lesion regions. Weakly supervising from a limited set of infection-labeled samples guides the CNN to converge towards infected areas. Designing explicit guidance to fine-tune the attention head is a notable highlight of this work.
The proposed model achieved an average R2 score of 84.3% and a mean squared error of 0.133 on the MosMed data. Precise alignment of the infected hotspots with the clinician’s markings confirms the effectiveness of the model and its explainability of predictions. Results demonstrate that the approach has significant potential to augment known methodologies, as it outperformed recent works by a good margin. As future work, the research offers rich scope to expand into other prognostic tasks such as predicting severity conversion time, progression stages, and readmission rate. By including more clinical features and biochemical variables into attention learning it can cater to such diverse applications.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Feng Z., Yu Q., Yao S., Luo L., Zhou W., Mao X., Li J., Duan J., Yan Z., Yang M., Tan H., Ma M., Li T., Yi D., Mi Z., Zhao H., Jiang Y., He Z., Li H., … Wang W. Early prediction of disease progression in COVID-19 pneumonia patients with chest CT and clinical characteristics. Nature Commun. 2020;11(1) doi: 10.1038/s41467-020-18786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lassau N., Ammari S., Chouzenoux E., Gortais H., Herent P., Devilder M., Soliman S., Meyrignac O., Talabard M.-P., Lamarque J.-P., Dubois R., Loiseau N., Trichelair P., Bendjebbar E., Garcia G., Balleyguier C., Merad M., Stoclin A., Jegou S., … Blum M.G.B. Integrating deep learning CT-scan model, biological and clinical variables to predict severity of COVID-19 patients. Nature Commun. 2021;12(1) doi: 10.1038/s41467-020-20657-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn J.H., Choi E.Y. Expanded A-DROP score: A new scoring system for the prediction of mortality in hospitalized patients with community-acquired pneumonia. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-32750-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huespe I., Carboni Bisso I., Di Stefano S., Terrasa S., Gemelli N.A., Las Heras M. COVID-19 Severity Index: A predictive score for hospitalized patients. Med. Intensiv. 2020 doi: 10.1016/j.medin.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin G.D., Ryerson C.J., Haramati L.B., Sverzellati N., Kanne J.P., Raoof S., Schluger N.W., Volpi A., Yim J.-J., Martin I.B.K., Anderson D.J., Kong C., Altes T., Bush A., Desai S.R., Goldin J., Goo J.M., Humbert M., Inoue Y., … Leung A.N. The role of chest imaging in patient management during the COVID-19 pandemic. Chest. 2020;158(1):106–116. doi: 10.1016/j.chest.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu G., Yang P., Xie Y., Woodruff H.C., Rao X., Guiot J., Frix A.-N., Louis R., Moutschen M., Li J., Li J., Yan C., Du D., Zhao S., Ding Y., Liu B., Sun W., Albarello F., D’Abramo A., … Lambin P. Development of a clinical decision support system for severity risk prediction and triage of COVID-19 patients at hospital admission: an international multicenter study. Eur. Respir. J. 2020 doi: 10.1183/13993003.01104-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lieveld A.W.E., Azijli K., Teunissen B.P., van Haaften R.M., Kootte R.S., van den Berk I.A.H., van der Horst S.F.B., de Gans C., van de Ven P.M., Nanayakkara P.W.B. Chest CT in COVID-19 at the ED: Validation of the COVID-19 reporting and data system (CO-RADS) and CT severity score. Chest. 2021;159(3):1126–1135. doi: 10.1016/j.chest.2020.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zieleskiewicz L., Markarian T., Lopez A., Taguet C., Mohammedi N., Boucekine M., Baumstarck K., Besch G., Mathon G., Duclos G., Bouvet L., Michelet P., Allaouchiche B., Chaumoître K., Di Bisceglie M., Leone M. Comparative study of lung ultrasound and chest computed tomography scan in the assessment of severity of confirmed COVID-19 pneumonia. Intensiv. Care Med. 2020;46(9):1707–1713. doi: 10.1007/s00134-020-06186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang R., Li X., Liu H., Zhen Y., Zhang X., Xiong Q., Luo Y., Gao C., Zeng W. Chest CT severity score: An imaging tool for assessing severe COVID-19. Radiol.: Cardiothorac. Imaging. 2020;2(2) doi: 10.1148/ryct.2020200047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun D., Li X., Guo D., Wu L., Chen T., Fang Z., Chen L., Zeng W., Yang R. CT quantitative analysis and its relationship with clinical features for assessing the severity of patients with COVID-19. Korean J. Radiol. 2020;21(7):859. doi: 10.3348/kjr.2020.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazar Neto F., Marino L.O., Torres A., Cilloniz C., Meirelles Marchini J.F., Garcia de Alencar J.C., Palomeque A., Albacar N., Brandão Neto R.A., Souza H.P., Ranzani O.T., Bortolotto A.L., Müller Veiga A.D., Bellintani A.P., Fantinatti B.L., Nicolao B.R., Caldeira B.T., Umehara Juck C.E., Bueno C.G., … Gomez Gomez L.M. Community-acquired pneumonia severity assessment tools in patients hospitalized with COVID-19: a validation and clinical applicability study. Clin. Microbiol. Infect. 2021;27(7):1037.e1–1037.e8. doi: 10.1016/j.cmi.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C., Qin L., Li K., Wang Q., Zhao Y., Xu B., Liang L., Dai Y., Feng Y., Sun J., Li X., Hu Z., Xiang H., Dong T., Jin R., Zhang Y. A novel scoring system for prediction of disease severity in COVID-19. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao Y., Cao J., Wang Q., Shi Q., Liu K., Luo Z., Chen X., Chen S., Yu K., Huang Z., Hu B. D-dimer as a biomarker for disease severity and mortality in COVID-19 patients: a case control study. J. Intensive Care. 2020;8(1) doi: 10.1186/s40560-020-00466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durhan G., Ardali Duzgun S., Basaran Demirkazik F., Irmak I., Idilman I., Akpinar M.G., Akpinar E., Ocal S., Telli G., Topeli A., … Ariyurek O.M. Visual and software-based quantitative chest CT assessment of COVID-19: correlation with clinical findings. Diagn. Interv. Radiol. 2020;26(6):557–564. doi: 10.5152/dir.2020.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebrahimian S., Homayounieh F., Rockenbach M.A.B.C., Putha P., Raj T., Dayan I., Bizzo B.C., Buch V., Wu D., Kim K., Li Q., Digumarthy S.R., Kalra M.K. Artificial intelligence matches subjective severity assessment of pneumonia for prediction of patient outcome and need for mechanical ventilation: a cohort study. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-020-79470-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye B., Yuan X., Cai Z., Lan T. Severity assessment of COVID-19 based on feature extraction and V-Descriptors. IEEE Trans. Ind. Inf. 2021;17(11):7456–7467. doi: 10.1109/tii.2021.3056386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K., Ye L., Gao M., Zhou Z., Li L., Wang J., Yang Z., Cai H., Xu J., Yang L., … Wang G. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433.e11. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai W., Liu T., Xue X., Luo G., Wang X., Shen Y., Fang Q., Sheng J., Chen F., Liang T. CT quantification and machine-learning models for assessment of disease severity and prognosis of COVID-19 patients. Acad. Radiol. 2020;27(12):1665–1678. doi: 10.1016/j.acra.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schöning V., Liakoni E., Baumgartner C., Exadaktylos A.K., Hautz W.E., Atkinson A., Hammann F. Development and validation of a prognostic COVID-19 severity assessment (COSA) score and machine learning models for patient triage at a tertiary hospital. J. Transl. Med. 2021;19(1) doi: 10.1186/s12967-021-02720-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bats M.-L., Rucheton B., Fleur T., Orieux A., Chemin C., Rubin S., Colombies B., Desclaux A., Rivoisy C., Mériglier E., Rivière E., Boyer A., Gruson D., Pellegrin I., Trimoulet P., Garrigue I., Alkouri R., Dupin C., Moreau-Gaudry F., … Dabernat S. Covichem: A biochemical severity risk score of COVID-19 upon hospital admission. PLOS ONE. 2021;16(5) doi: 10.1371/journal.pone.0250956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cobre A. de F., Stremel D.P., Noleto G.R., Fachi M.M., Surek M., Wiens A., Tonin F.S., Pontarolo R. Diagnosis and prediction of COVID-19 severity: can biochemical tests and machine learning be used as prognostic indicators? Comput. Biol. Med. 2021;134 doi: 10.1016/j.compbiomed.2021.104531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quiroz J.C., Feng Y.-Z., Cheng Z.-Y., Rezazadegan D., Chen P.-K., Lin Q.-T., Qian L., Liu X.-F., Berkovsky S., Coiera E., Song L., Qiu X., Liu S., Cai X.-R. Development and validation of a machine learning approach for automated severity assessment of COVID-19 based on clinical and imaging data: Retrospective study. JMIR Med. Inform. 2021;9(2) doi: 10.2196/24572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Z., Zhao W., Xie X., Zhong Z., Shi F., Ma T., Liu J., Shen D. Severity assessment of COVID-19 using CT image features and laboratory indices. Phys. Med. Biol. 2021;66(3) doi: 10.1088/1361-6560/abbf9e. [DOI] [PubMed] [Google Scholar]
- 24.Karthik R., Menaka R., Hariharan M., Kathiresan G.S. AI for COVID-19 detection from radiographs: Incisive analysis of state of the art techniques, key challenges and future directions. IRBM. 2021 doi: 10.1016/j.irbm.2021.07.002. Elsevier BV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irmak E. COVID-19 disease severity assessment using CNN model. IET Image Process. 2021;15(8):1814–1824. doi: 10.1049/ipr2.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z., Zhao S., Chen Y., Luo F., Kang Z., Cai S., Zhao W., Liu J., Zhao D., Li Y. A deep-learning-based framework for severity assessment of COVID-19 with CT images. Expert Syst. Appl. 2021;185 doi: 10.1016/j.eswa.2021.115616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karthik R., Menaka R., M H. Learning distinctive filters for COVID-19 detection from chest X-ray using shuffled residual CNN. Appl. Soft Comput. 2021;99:106744. doi: 10.1016/j.asoc.2020.106744. Elsevier BV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karthik R., Menaka R., H M., Won D. Contour-enhanced attention CNN for CT-based COVID-19 segmentation. Pattern Recognit. 2022;125 doi: 10.1016/j.patcog.2022.108538. Elsevier BV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samala R.K., Hadjiiski L., Chan H.-P., Zhou C., Stojanovska J., Agarwal P., Fung C. In: Medical Imaging 2021: Computer-Aided Diagnosis. Drukker K., Mazurowski M.A., editors. SPIE; 2021. Severity assessment of COVID-19 using imaging descriptors: a deep-learning transfer learning approach from non-COVID-19 pneumonia. [DOI] [Google Scholar]
- 30.Naeem H., Bin-Salem A.A. A CNN-LSTM network with multi-level feature extraction-based approach for automated detection of coronavirus from CT scan and X-ray images. Appl. Soft Comput. 2021 doi: 10.1016/j.asoc.2021.107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aboutalebi H., Pavlova M., Shafiee M.J., Sabri A., Alaref A., Wong A. Research Square Platform LLC; 2021. COVID-Net CXR-S: Deep Convolutional Neural Network for Severity Assessment of COVID-19 Cases from Chest X-Ray Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L., Zhen H., Fang X., Wan S., Ding W., Guo Y. A unified two-parallel-branch deep neural network for joint gland contour and segmentation learning. Future Gener. Comput. Syst. 2019;100:316–324. doi: 10.1016/j.future.2019.05.035. Elsevier BV. [DOI] [Google Scholar]
- 33.Zhao Y., Li H., Wan S., Sekuboyina A., Hu X., Tetteh G., Piraud M., Menze B. Knowledge-aided convolutional neural network for small organ segmentation. IEEE J. Biomed. Health Inform. 2019;23(4):1363–1373. doi: 10.1109/jbhi.2019.2891526. Institute of Electrical and Electronics Engineers (IEEE) [DOI] [PubMed] [Google Scholar]
- 34.Yu Z., Li X., Sun H., Wang J., Zhao T., Chen H., Ma Y., Zhu S., Xie Z. Rapid identification of COVID-19 severity in CT scans through classification of deep features. BioMed. Eng. OnLine. 2020;19(1) doi: 10.1186/s12938-020-00807-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aswathy A.L., Hareendran A., SS V.C. COVID-19 diagnosis and severity detection from CT-images using transfer learning and back propagation neural network. J. Infect. Public Health. 2021;14(10):1435–1445. doi: 10.1016/j.jiph.2021.07.015. Elsevier BV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Y.-Z., Liu S., Cheng Z.-Y., Quiroz J.C., Rezazadegan D., Chen P.-K., Lin Q.-T., Qian L., Liu X.-F., Berkovsky S., Coiera E., Song L., Qiu X.-M., Cai X.-R. Cold Spring Harbor Laboratory; 2020. Severity Assessment and Progression Prediction of COVID-19 Patients based on the LesionEncoder Framework and Chest CT. [DOI] [Google Scholar]
- 37.Qiblawey Y., Tahir A., Chowdhury M.E.H., Khandakar A., Kiranyaz S., Rahman T., Ibtehaz N., Mahmud S., Maadeed S.A., Musharavati F., Ayari M.A. Detection and severity classification of COVID-19 in CT images using deep learning. Diagnostics. 2021;11(5):893. doi: 10.3390/diagnostics11050893. MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lessmann N., Sánchez C.I., Beenen L., Boulogne L.H., Brink M., Calli E., Charbonnier J.-P., Dofferhoff T., van Everdingen W.M., Gerke P.K., Geurts B., Gietema H.A., Groeneveld M., van Harten L., Hendrix N., Hendrix W., Huisman H.J., Išgum I., Jacobs C., … van Ginneken B. Automated assessment of COVID-19 reporting and data system and chest CT severity scores in patients suspected of having COVID-19 using artificial intelligence. Radiology. 2021;298(1):E18–E28. doi: 10.1148/radiol.2020202439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang L., Han R., Ai T., Yu P., Kang H., Tao Q., Xia L. Serial quantitative chest CT assessment of COVID-19: A deep learning approach. Radiol.: Cardiothorac. Imaging. 2020;2(2) doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pu J., Leader J.K., Bandos A., Ke S., Wang J., Shi J., Du P., Guo Y., Wenzel S.E., Fuhrman C.R., Wilson D.O., Sciurba F.C., Jin C. Automated quantification of COVID-19 severity and progression using chest CT images. Eur. Radiol. 2020;31(1):436–446. doi: 10.1007/s00330-020-07156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou J., Zhang X., Zhu Z., Lan X., Fu L., Wang H., Wen H. Cohesive multi-modality feature learning and fusion for COVID-19 patient severity prediction. IEEE Trans. Circuits Syst. Video Technol. 2021:1. doi: 10.1109/tcsvt.2021.3063952. Institute of Electrical and Electronics Engineers (IEEE) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohammed A., Wang C., Zhao M., Ullah M., Naseem R., Wang H., Pedersen M., Cheikh F.A. Weakly-supervised network for detection of COVID-19 in chest CT scans. IEEE Access. 2020;8:155987–156000. doi: 10.1109/access.2020.3018498. Institute of Electrical and Electronics Engineers (IEEE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z., Zhao W., Shi F., Qi L., Xie X., Wei Y., Ding Z., Gao Y., Wu S., Liu J., Shi Y., Shen D. A novel multiple instance learning framework for COVID-19 severity assessment via data augmentation and self-supervised learning. Med. Image Anal. 2021;69 doi: 10.1016/j.media.2021.101978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He K., Zhao W., Xie X., Ji W., Liu M., Tang Z., Shi Y., Shi F., Gao Y., Liu J., Zhang J., Shen D. Synergistic learning of lung lobe segmentation and hierarchical multi-instance classification for automated severity assessment of COVID-19 in CT images. Pattern Recognit. 2021;113 doi: 10.1016/j.patcog.2021.107828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue W., Cao C., Liu J., Duan Y., Cao H., Wang J., Tao X., Chen Z., Wu M., Zhang J., Sun H., Jin Y., Yang X., Huang R., Xiang F., Song Y., You M., Zhang W., Jiang L., … Xie M. Modality alignment contrastive learning for severity assessment of COVID-19 from lung ultrasound and clinical information. Med. Image Anal. 2021;69 doi: 10.1016/j.media.2021.101975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goncharov M., Pisov M., Shevtsov A., Shirokikh B., Kurmukov A., Blokhin I., Chernina V., Solovev A., Gombolevskiy V., Morozov S., Belyaev M. CT-based COVID-19 triage: Deep multitask learning improves joint identification and severity quantification. Med. Image Anal. 2021;71:102054. doi: 10.1016/j.media.2021.102054. Elsevier BV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatzitofis A., Cancian P., Gkitsas V., Carlucci A., Stalidis P., Albanis G., Karakottas A., Semertzidis T., Daras P., Giannitto C., Casiraghi E., Sposta F.M., Vatteroni G., Ammirabile A., Lofino L., Ragucci P., Laino M.E., Voza A., Desai A., … Savevski V. Volume-of-Interest aware deep neural networks for rapid chest CT-based COVID-19 patient risk assessment. Int. J. Environ. Res. Public Health. 2021;18(6):2842. doi: 10.3390/ijerph18062842. MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao L., Li P., Sun F., Zhang Y., Xu C., Zhu Hongbo, Cai F.-Q., He Y.-L., Zhang W.-F., Ma S.-C., Hu C., Gong M., Liu L., Shi W., Zhu Hong. Development and validation of a deep learning-based model using computed tomography imaging for predicting disease severity of coronavirus disease 2019. Front. Bioeng. Biotechnol. 2020 doi: 10.3389/fbioe.2020.00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ouyang X., Huo J., Xia L., Shan F., Liu J., Mo Z., Yan F., Ding Z., Yang Q., Song B., Shi F., Yuan H., Wei Y., Cao X., Gao Y., Wu D., Wang Q., Shen D. Dual-Sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging. 2020;39(8):2595–2605. doi: 10.1109/tmi.2020.2995508. Institute of Electrical and Electronics Engineers (IEEE. [DOI] [PubMed] [Google Scholar]