Abstract
The process of protein ubiquitination and deubiquitination plays an important role in maintaining protein stability and regulating signal pathways, and protein homeostasis perturbations may induce a variety of diseases. The deubiquitination process removes ubiquitin molecules from the protein, which requires the participation of deubiquitinating enzymes (DUBs). Ubiquitin-specific protease 15 (USP15) is a DUB that participates in many biological cell processes and regulates tumorigenesis. A dislocation catalytic triplet was observed in the USP15 structure, a conformation not observed in other USPs, except USP7, which makes USP15 appear to be unique. USP15 has been reported to be involved in the regulation of various cancers and diseases, and the reported substrate functions of USP15 are conflicting, suggesting that USP15 may act as both an oncogene and a tumor suppressor in different contexts. The importance and complexity of USP15 in the pathological processes remains unclear. Therefore, we reviewed the diverse biological functions of USP15 in cancers and other diseases, suggesting the potential of USP15 as an attractive therapeutic target.
Keywords: USP15, deubiquitinatase, ubiquitin, target, cancer
1. Introduction
Protein degradation plays an important role in the physiological activities of cells. Disorders in protein degradation can cause tumors and some neurodegenerative diseases [1]. The main pathways of protein degradation in eukaryotic cells are ubiquitin-proteasome and autophagy. Autophagy is the process of transporting components to be degraded, which includes not only proteins, but also dysfunctional or redundant organelles, as well as lysosomes for catabolism. Ubiquitin can also participate in the signaling of the lysosomal degradation pathway [2]. Ubiquitination degrades proteins through the ubiquitin–proteasome system and through selective autophagy [3]. The proteasome is a catalytically active and ubiquitous protease, andits most common function is to degrade proteins through the 26s ubiquitin–proteasome system [4]. The 26s proteasome is composed of 20s core particles and 19s regulatory particles. Recent studies have found that the 20s proteasome degrades oxidatively damaged proteins, and unfolds independently of adenosine triphosphate (ATP)/ubiquitin [5,6].
Ubiquitin is a highly conserved peptide molecule, composed of 76 amino acids that post-translationally mark proteins for degradation. Ubiquitin-dependent proteolysis involves the sequential activation of three enzymes, as well as the attachment to target proteins. In the event of ATP supply, the ubiquitin-activating enzyme E1 delivers the ubiquitin molecule to the ubiquitin-conjugating enzyme E2; subsequently, the ubiquitin-ligase enzyme E3 attaches the ubiquitin molecules conjugated at E2 to the target protein, and, finally, this ubiquitinated protein is specifically degraded by the proteasome [7,8,9,10]. Occasionally, multiubiquitylation requires the additional activity of certain ubiquitin-chain elongation factors, such as the E4 enzyme. For example, yeast UFD2 binds to oligo-ubiquitylated substrates (proteins modified by only a few ubiquitin molecules) and catalyzes the multiubiquitin chain assembly in collaboration with E1, E2, and E3 [11,12]. The E4 enzyme has also been shown to be required for highly sustained prolongation [13]. Single or multiple ubiquitin molecules can be conjugated to the substrate protein via their lysine residues (K6, K11, K27, K29, K33, K48, and K63) or N-terminal methionine residues (M1) [14,15], where K48-linked and K63-linked chains are the most well-studied for directing substrate proteins for proteasomal and lysosomal degradation, respectively [16]. K48-linked ubiquitination is usually associated with proteasome-mediated degradation, while K63-linked ubiquitination is involved in autophagy and targets pure proteasomes [17,18,19]. Ubiquitination often affects protein stability, localization, activity, and interactions, and is widely involved in important physiological processes, including cell apoptosis [20,21,22], the cell cycle [23,24], DNA damage repair [25,26], ribosome biogenesis [27], stress responses [28], the induction of inflammatory responses [29], and transcriptional regulation [30]. The ubiquitination process is reversible. The removal of ubiquitin molecules from the ubiquitinated protein, and stabilizing the substrate protein, is known as the deubiquitination process [31,32], which requires the involvement of deubiquitinating enzymes (DUBs). Most DUBs have a clear regulatory role in human cancers and diseases [33]. For example, the loss of ubiquitinC-terminalhydrolaseL1 (UCHL1) activates mitochondrial autophagy in Parkinson’s disease [34], and the increased expression of cylindromatosis (CYLD) inhibits tumor inflammation and angiogenesis to prevent the development and progression of skin squamous cell tumors [35]. Ubiquitination and deubiquitination have been found to be dysregulated in different types of cancer, possibly due to mutations, deletions, or amplifications [36,37,38].
2. DUBs
DUBs are proteases which remove ubiquitin molecules from the ubiquitinated proteins. DUBs can regulate ubiquitination in four distinct mechanisms: (1) processing polyubiquitin molecules into single ubiquitin molecules, (2) recycling ubiquitin molecules, (3) preventing E3 ligase-mediated ubiquitin conjugation, and (4) cleaving polyubiquitin chains [39,40,41]. According to the structure of the catalytic domain, DUBs are divided into seven classes: ubiquitin-specific proteases (USPs), Josephin and JAB1/MPN+(MJP), JAB1/MPN/Mov34 metalloenzyme (JAMM), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), and the two recently discovered MIU-containing novel DUB (MINDY) and zinc finger-containing ubiquitin peptidase 1 (ZUP1) [41,42,43,44,45]. Except for ZUP1, which is a metalloprotease, the rest are cysteine proteases [46,47]. USPs are the most numerous family of DUBs [48]. USP15 is a member of the USPs family and is involved in various cellular processes. Here, we will provide a comprehensive overview of the role of USP15 in various cancers and other diseases, which will help us better understand the biological functions of USP15 and provide strategies for USP15 as a therapeutic target.
3. Structure and Characteristics of Ubiquitin-Specific Protease 15
The human USP15 gene is located on chromosome 12q14.1 and shares 60.5% sequence identity and 76% sequence similarity with the human homolog (UNP/Unph/USP4) of the mouse Unp proto-oncogene [49]. USP15 and USP4 have an overlap in their function and deubiquitination substrates. Fertility testing in mice showed that USP15 and USP4 have a sufficient functional redundancy to rescue inactivated mutations in a reciprocal manner [50]. The evolutionarily conserved skipping of exon 7 during the transcription of USP15 mRNA results in the deletion of a short element in the interregional link of USP15, thus producing two major isoforms, 1 and 2 [51], consisting of 981and 952 amino acid residues, respectively [52]. Kotani et al.found differences in the substrate preference between the two isoforms of USP15, that is, RNF213, a ubiquitin ligase associated with Moyamoya disease, is primarily recognized and deubiquitinated by isoform 1 [51], indicating the functional differences between the isoforms. In addition, the micronucleus phenotype can be rescued by USP15 isoforms when USP15 is depleted in U2OS cells [51]. Moreover, USP15 isoform 1 is preferentially upregulated in a set of non-small cell lung cancer cell lines, suggesting that heterodimer imbalance may contribute to genomic instability in cancer [53].
The domain structure of USP15 is composed of a catalytic region and a non-catalytic region. The catalytic region contains the typical catalytic triad, Cys-269, His-862, and Asp-879 [47], which participates in the formation of part of the active site of USP15. The mutation of Cys-269 to Ser suppresses the enzymatic activity of USP15 [54]. However, the catalytic triad of USP15 is in an inactive conformation, with the catalytic cysteine being far away from the catalytic histidine, which is atypical. There is a protease domain that harbors an approximately 300-amino acid insertion containing an ubiquitin-like (UBL) domain. The N-terminal’s non-catalytic domain includes the ubiquitin-specific protease domain (DUSP) that specifically binds to the substrate protein and the other UBL domain [55,56]. The USP15 UBL domain interacts with the DUSP domain through a UBL surface area that is rarely used as recognition site [57] (Figure 1a). A zinc (Zn) finger is discovered in the detailed analysis of USP15 and the related USPs sequences, which indicates that USP15, and the related USPs, require a functional Zn-finger to adopt a conformation that is needed for the binding and cleavage of is peptide-bond-linked polyubiquitin chains [58,59]. The structure of USP15, predicted by the Alpha Fold, is shown in Figure 1b.
Figure 1.
Schematic diagram of USP15 structure. (a) Schematic diagram of the domain composition of USP15 (isoform 2); (b) USP15 structure in front and top view directions. Available online: https://alphafold.ebi.ac.uk/entry/Q9Y4E8 (accessed on 26 October 2021).
4. Function of USP15 in Cancers and Other Diseases
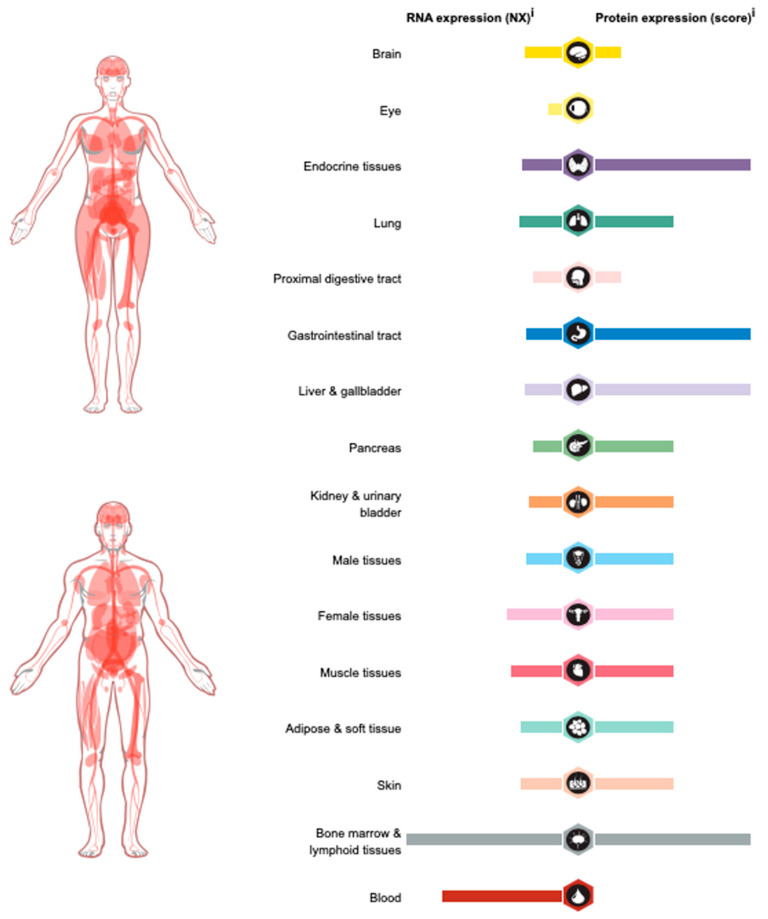
In humans, USP15 is highly expressed in endocrine tissues, the gastrointestinal tract, the liver, the gallbladder, bone marrow, lymphoid tissues, etc. (Figure 2).USP15 is upregulated in a variety of cancers, including glioblastoma [60], breast cancer [60], ovarian cancer [61], multiple myeloma [62], prostate cancer [63], gastric cancer [64,65], pancreatic ductal adenocarcinoma [66,67], and chronic myeloid leukemia [68]. However, the USP15 gene is deleted in 25.37% of pancreatic cancers and 10.9% of glioblastomas, implying a potential tumor suppressive function of USP15 in these cancers [69].USP15 is involved in various cellular processes, such as cell proliferation, cell invasion, apoptosis, autophagy, the cell cycle, genome integrity, transcription regulation, the immune response, and others. The complex and diverse biological functions indicate that USP15 is a critical protein that needs to be studied more comprehensively. The following sections will review, in detail, the important functions of USP15 in cancers and other diseases.
Figure 2.
The expression of USP15 RNA and proteins in various human tissues. Available online: https://www.proteinatlas.org/ENSG00000135655-USP15/tissue (accessed on 8 November 2021).
4.1. Proliferation, Migration, and Invasion
USP15 has been reported to promote or inhibit tumorigenesis and its progression in different cancers. Previous reports have proved that USP15 is highly expressed in breast cancer. Interleukin-1 receptor type 2 (IL1R2) binds with, and enhances, USP15 to deubiquitinate BMI1at the K81 residue, thereby promoting the breast cancer cell lines SUM159 and MB231, as well as their proliferation and invasion [70]. Moreover, Xia et al. revealed that USP15 promotes the proliferation of the breast cancer cell lines T47D and MCF-7 by deubiquitinating and stabilizing the estrogen receptor α (ERα) in vitro and in vivo, which is related to the cell cycle regulation, rather than cell apoptosis [71]. ERα can bind to RNA to regulate the growth of tumor cells and the efficacy of the anti-cancer drug, tamoxifen [72]. These findings indicate that USP15 may be a potential therapeutic target for breast cancer. Furthermore, Zhou et al. found that the expression of USP15is upregulated in the multiple myeloma cell lines, and USP15 overexpression enhances cell proliferation and inhibits cell apoptosis in multiple myeloma cell lines, such as RPMI 8226 and U266. Mechanistically, USP15 deubiquitinates and stabilizes NF-κB inhibitor α (IκBα), and USP15 is, in turn, positively regulated by NF-κB through increasing the activity of the USP15 promoter [62]. Most patients with hematological malignancies and multiple myeloma have developed resistance to immunomodulatory drugs. Cereblon (CRBN) is an E3 ubiquitin ligase and a direct protein target for the immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide, and USP15 antagonizes the ubiquitination of CRBN target proteins, such as glutamine synthetase, thereby preventing their degradation [73]. There are many reports that USPs participate in the development of gastric cancer and can be used as independent prognostic markers for gastric cancer patients. The silence of USP15 inhibits cell proliferation and invasion in the gastric cancer cell lines BGC-823 and MKN-45 in vivo and in vitro, and USP15 overexpression shows the opposite result. In terms of mechanisms, USP15 overexpression promotes the nuclear expression of β-catenin in gastric cancer cells, thereby activating the Wnt/β-caten in signaling pathway to promote the malignant progression of gastric cancer, suggesting that USP15 may be an oncogene of gastric cancer [65]. However, there has been another report stating that USP15 deubiquitinates IκBα to inhibit the NF-κB pathway, thereby blocking the proliferation and invasion of the gastric cancer cells lines SGC7901 and MKN45 [74].These data indicate that the functions of USP15 are complicated, and functions in the same types of cancer may be different, so continuous exploration is needed. Silencing USP15 also inhibits cell proliferation and invasion in glioblastoma cell lines U87-MG and U251-MG, indicating that USP15 may be a potentially effective treatment target for glioblastoma [75]. The knockdown of USP15 results in the decrease in the E3 ubiquitin ligase MDM2 protein in the melanoma cell line A375 and the colorectal cancer cell line HCT116, indicating that USP15 plays an important role in stabilizing MDM2. USP15 stabilizes MDM2 by the deubiquitination and regulation of p53 function, which, in turn, negatively regulates T-cell activation by targeting the degradation of the transcription factor, the nuclearfactorofactivatedT-cells2 (NFATc2), ultimately regulating both tumor growth and anti-tumor immunity [76]. Interestingly, the transforming growth factor β (TGFβ) enhances the production of USP15 by promoting the upregulation of USP15 translation, rather than transcription, let alone inhibiting USP15 degradation. USP15 interacts with, and stabilizes, p53 through deubiquitination, and it regulates the transcriptional activity of p53, thereby promoting the expression of p21 to inhibit human osteosarcoma cell line U2OS proliferation [77]. On the contrary, Padmanabhan et al. found that USP15 controls the protein expression of p53-R175H, but not p53 WT, through an ubiquitin-mediated lysosomal pathway in ovarian cancer cells [63]. Recent reports indicate that USP15 is highly expressed in liver cancer tissues compared with normal tissues, and the knockdown of USP15 inhibits cell proliferation and enhances cell apoptosis in the hepatocellular carcinoma cell lines SNU449 and Hep3B [78]. Furthermore, USP15 deubiquitinates and stabilizes kelch like ECH associated protein 1 (KEAP1) to degradate nuclear factor erythroid 2-related factor 2 (NRF2), which is enhanced by xanthine oxidoreductase, thereby leading to the accumulation of reactive oxygen species and changes in the oxidative environment, ultimately inducing cell apoptosis and suppressing the propagation of hepatocellular carcinoma [79]. Additionally, Chen et al. found that the expression of USP15 is upregulated in the rat model of epilepsy, and the suppression of USP15 activates the NRF2/HO-1 pathway, inhibiting the glutamate-induced oxidative damage in the mouse hippocampal neuron cell line HT22 [80]. USP15 also deubiquitinates and stabilizes NRF1 to promote NRF1-induced proteasome gene expression, thereby maintaining proteostasis [81]. USP15 can deubiquitinate the insulin receptor substrate 2 (IRS2), and IRS2 monoubiquitination increases insulin-like growth factor 1(IGF1) signaling in HEK293 cells [82]. However, IRS2 only binds to USP15 after it is connected to the ubiquitin molecule, thereby blocking IGF1-dependent IRS2 phosphorylation to weaken IGF-I signaling in prostate cancer PC-3 cells [83,84]. USP15 interacts with DEK and the kinesin family member 15 (KIF15) and stabilizes DEK to facilitate the proliferation of the leiomyosarcoma cell lines SK-UT-1 and SK-LMS-1 [85]. USP15 also deubiquitinates and stabilizes SMAD-specific E3 ubiquitin protein ligase 2 (SMURF2) at the K734 residue to affect the catalytic activity of SMURF2, which targets the TGFβ receptor complex for ubiquitin-mediated degradation, thereby regulating TGFβ-dependent cell migration [86]. Furthermore, USP15 deubiquitinates and stabilizesthe type I TGFβ receptor (TGFβRI) to enhance the TGFβ signaling pathway and promote glioblastoma multiforme cell proliferation and invasion [60,87]. In addition, USP15 deubiquitinates and stabilizes TGFβRI to enhance TGFβ/SMAD signaling and to promote the proliferation and invasion of hypertrophic scar-derived fibroblasts, suggesting that USP15 could be a potential target for the treatment of hypertrophic scars [88]. USP15 deubiquitinates and interacts with the extracellular-signal-regulated kinase 2 (ERK2), but it does not regulate the stability of ERK2, resulting in an increase in the pERK1/2 levels to further enhance the TGFβ/SMAD2 signaling pathway and suppress osteoarthritis progression. Moreover, overexpression of USP15 can repair the joint plane in the rat osteoarthritis model, showing that USP15 plays an important role in preventing cartilage damage in vivo and in vitro [89]. Additionally, USP15 deubiquitinates and interacts with the eukaryotic translation initiation factor 4A1 (EIF4A1) to promote the proliferation and migration of keratinocytes and to accelerate the re-epithelialization during wound healing in mice [90].
4.2. Apoptosis
The occurrence of tumors is not only related to abnormal cell proliferation and differentiation, but also to abnormal apoptosis. USP15 has been reported to participate in various ways to promote cell apoptosis. USP15 deubiquitinates and stabilizes caspase-6 to increase cell apoptosis in the chronic myeloid leukemia cell K562, and miR-202-5p inhibits the expression of USP15 by directly targeting the 3’ untranslated region (3’UTR) of USP15 to suppress cell apoptosis [91]. USP15 also deubiquitinates and stabilizes procaspase-3 to promote paclitaxel-induced apoptosis in the human cervical cancer cell line HeLa [92]. Furthermore, Yu et al. analyzed the mRNA expression levels of nine DUB family members, including USP15, in eight normal tissues and 10 degenerative nucleus pulposus tissues, and found that only USP15 was highly expressed in the nucleus pulposus tissues, compared to the normal tissues. Subsequent findings have shown that USP15 deubiquitinates and stabilizes FK506 binding protein 5 (FKBP5), thereby promoting the apoptosis of nucleus pulposus cells by inhibiting the phosphatidylinositol-3-kinase (PI3K)/Akt pathway [93].Wang et al. also found that USP15 may dynamically target the endolysosomes on mitochondria and impair the mitochondrial outer membrane permeabilization functionality in the human breast cancer cell line MCF7, thereby participating in the process of apoptosis signaling [94].The role of USP15 in the regulation of apoptosis provides insights for the treatment of cancer and other diseases.
4.3. Autophagy
USP15 not only participates in the ubiquitin–proteasome system, but also plays a role in autophagy. Parkinson’s disease is a neurodegenerative disease which mainly occurs in the elderly, and its pathogenesis is mainly due to mitochondrial dysfunction. The E3 ligase PARKIN has been reported toplay a critical role in the autophagy clearance of defective mitochondria, which mediates the clearance of a variety of mitochondrial proteins after being activated [34,95]. USP15 can deubiquitinate PARKIN to impair mitochondrial autophagy, suggesting that USP15 inhibition could be a therapeutic strategy for Parkinson’s disease, caused by reduced PARKIN levels [96,97]. It has been reported that the expression of USP15 increased in the miR-26a knockdown mouse model, and miR-26adirectly targets USP15 to regulate cardiomyocyte autophagy, as well as playing a protective role in myocardial ischemic injury [98]. The above studies suggest that USP15 may be a potential target for the treatment of some diseases by participating in the regulation of autophagy.
4.4. Cell Cycle
The COP9 signalosome (CSN) affects the stability of microtubule filaments during the cell cycle [99]. USP15 has been reported to bind to CSN and protect CSN-related components [58]. Therefore, USP15 may be involved in cell cycle regulation. USP15 has been reported to accumulate on the newly synthesized RE1-silencing transcription factor (REST) in the early G1, playing an important role in the correct separation of the mitotic chromosome. Andrew et al. verified that USP15 deubiquitinates and stabilizes REST, but it does not antagonize the β-TrCP-mediated degradation of REST, which quickly replenishes REST when mitosis exits at the beginning of a new cell cycle [100,101,102]. Xiaet al. analyzed the cell cycle and the apoptosis-related proteins by flow cytometry after silencing USP15 in breast cancer cells, demonstrating that USP15 promotes breast cancer cell proliferation through the cell cycle [71]. Therefore, USP15 may regulate cancer progression by participating in the cell cycle.
4.5. Genome Integrity
It has been found that DNA is damaged in the USP15 knockout mice, and USP15 is phosphorylated by the ataxia telangiectasia mutated (ATM) kinase at the S678 residue, suggesting that USP15 plays a role in homologous recombination [54]. USP15 is enrolled by the mediator of DNA damage checkpoint 1 (MDC1) to the DNA double-strand break to deubiquitinate BRCA1 associated with RING domain 1 (BARD1), enhancing the BARD1-HP1γ interaction and the BRCA1/BARD1 retention at the DNA double-strand break, as well as reducing the PARP inhibitor sensitivity in breast and ovarian cancer cells [54]. There is a decrease in the survival rate and weight loss in the USP15 knockout mice, and USP15 interacts with and stabilizes the DNA repair factor FUS to promote the self-renewal of hematopoietic stem cells [68]. The squamous cell carcinoma antigen, recognized by T-cell 3 (SART3), shuttles USP15 to the nucleus through the nuclear localization sequence and promotes H2B deubiquitination in free histones by USP15, thereby affecting transcription, replication, and the DNA repair processes [103]. At the same time, SART3 also helps USP15 recruit free histones [104]. It has been reported that the knockdown of USP15 results in the expression decrease in the topoisomerase II α (TOP2A) protein, instead of mRNA, in the human non-small cell lung cancer cell line A549, indicating that USP15 regulates TOP2A to maintain genome integrity [53]. Moreover, USP15 cannot protect genome integrity and inhibit micronucleus formation by interfering with mitosis after being phosphorylated at the S229 residue [53]. These studies suggest that USP15 maintains genomic integrity by regulating DNA damage repair, replication, and transcription.
4.6. Transcriptional Regulation
USP15 acts in certain diseases through the transcriptional regulation of target genes. USP15 deubiquitinates and redistributes terminal uridylyl transferase 1 (TUT1) from the nucleolus to the nucleoplasm, resulting in the stabilization of U6 snRNA, thereby regulating the neurodegenerative phenotype [105]. USP15 also deubiquitinates and opposes R-SMAD monoubiquitylation to prevent R-SMADs recognizing the promoters involved in the differentiation of mesenchymal stem cells from osteoblasts [106]. In addition, USP15 has been reported to regulate dynamic protein-to-protein interactions in the spliceosome through the deubiquitination of the pre-mRNA-processingfactor31 (PRP31) and the DUSP-UBL domains of USP15 and PRP31, which only interact in the present of SART3 [107]. USP15 also deubiquitinatesand stabilizes the transcription factor, sulfurimitation1 (SLIM1), to regulate the hypertrophic responses of cardiac muscle in mice [108].
4.7. Immune Response
USP15 participates in the regulation of multiple signal pathways and plays a key role in cellular immunity and the inflammatory response. USP15 deubiquitinatestetmethylcytosinedioxygenase2 (TET2) at the K1299 residue to inhibit the TET2 activity and to increase the expression of chemokines induced by interferon γ (IFNγ), resulting in a longer lifespan of mice [109]. There is a reduced Listeria load in the liver, a more enlarged spleen, and a higher level of serum IFNβ in the USP15 knockout mice [76]. Moreover, the TRAF-interacting protein with a forkhead-associated domain B (TIFAB) binds to the catalytic domain of USP15 to enhance USP15-deubiquitinating MDM2 at the K48 residue, thereby decreasing the expression of p53 [110]. USP15deubiquitinates and stabilizes the E6 protein of HPV to inhibit the E6-mediated p53 degradation in HPV-positive HeLa cells [111]. In addition, the E6 protein can block the deubiquitination of the tripartitemotifcontaining25 (TRIM25) by USP15, which ultimately leads to the degradation of TRIM25, thereby inhibiting the retinoic acid-inducible gene 1 (RIG1)-mediated antiviral signal transduction in the cervical cancer cell line C33A [112,113]. USP15 negatively regulates the activation of naive CD4+ T-cells and the generation of IFNγ, as well as the redundancy of IFNγ in USP15-deficient mice, which increases the expression of programmed death ligand 1 (PD-L1) and the C-X-C motif chemokine ligand 12 (CXCL12) to promote the immunosuppressive tumor microenvironment processed by immunosuppressive tumors induced by methylcholantrene [114]. USP15 deubiquitinates and stabilizes TANK-binding kinase 1(TBK1) at the K63 residue to inhibit type I IFN production in vitro and in vivo, which is mediated by the ubiquitin-conjugating enzyme UBE2S, thereby increasing virus replication [115]. However, USP15 also deubiquitinates and interacts with TRIM25 and prevents the linear ubiquitin chain assembly complex (LUBAC) and the E3 ligase-dependent degradation of TRIM25. IFNβ promoter activation caused by Sendai virus (SeV) infection is decreased after the knockdown of USP15 in HEK293T cells. The deubiquitination of TRIM25 by USP15 enhances the production of type I IFN and inhibits the replication of RNA viruses, resulting in an antiviral response [116,117]. Therefore, USP15 is not only recruited by UBE2S to suppress type I IFN production by deubiquitinating TBK1, but it also stabilizes TRIM25 to promote TRIM25 and RIG1-dependent type I IFN production, which is due to USP15 targeting different proteins and participating in different signaling pathways. Although USP15 does not interact with the hepatitis C virus (HCV) proteins and does not participate in innate immune responses, knockout of USP15 inhibits the translation of HCV RNA and the accumulation of lipid droplets in the hepatocellular carcinoma cell line Huh7, as well as the addition of palmitic acids, which partially recovers the production of infectious viral particles, suggesting that USP15 regulates HCV propagation by affecting viral RNA translation and lipid metabolism [118]. The above function of USP15 in the immune response suggests that USP15 may be a potential therapeutic target for cancer immunotherapy.
4.8. Others
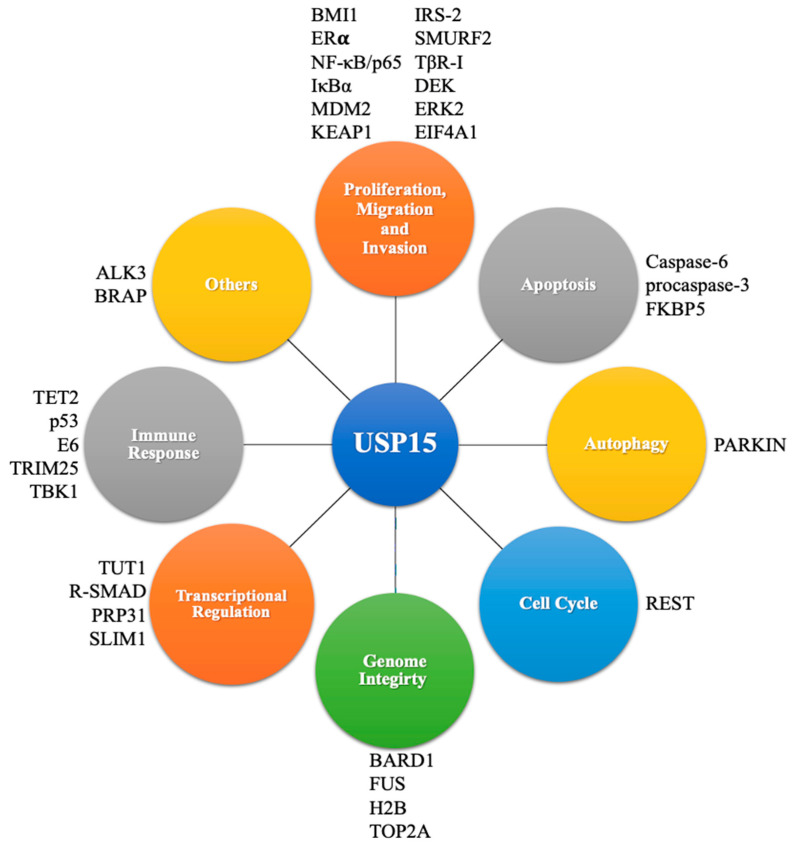
USP15 interacts with TGFβ-activated kinase 1 (TAK1)-binding protein (TAB) under the stimulation of the tumor necrosis factor α(TNFα), thereby inhibiting the degradation of TAB2/3 through the deubiquitination pathway, as well as inhibiting lysosomal degradation, or inhibiting the selective autophagy pathway. Therefore, USP15 can positively regulate the TNFα- and interleukin-1β-induced NF-κB activation [119,120]. USP15 deubiquitinates and stabilizes the hepatitis B virus X (HBx) protein in the human liver cancer cells Huh7, HepG2, and Hep3B [121]. Additionally, USP15 regulates the degradation of the human immunodeficiency virus (HIV) Nef and Gag proteins, thereby blocking the replication of HIV, indicating that USP15 can eliminate viral proteins by mediating protein degradation [122]. USP15 also deubiquitinates and stabilizes the bone morphogenetic protein (BMP) type I receptor ALK3 to promote BMP signal transduction [123]. Additionally, USP15 deubiquitinates and stabilizes E3 ubiquitin ligase BRAP, and BRAP, in turn, enhances the ubiquitination of USP15 to regulate MAPK signal transduction [124]. It has been reported that the endoplasmic reticulum-located E3 ubiquitin ligase Hrd1 can also ubiquitinate and interact with USP15 at the K21 residue in HEK293T cells, which does not cause USP15 to be degraded, but it loses its activity for IκBα deubiquitination during bacterial infection, leading to the excessive activation of NF-κB. Moreover, the knockout of Hrd1 in macrophages protects mice against septic shock induced by lipopolysaccharide, and the silence of USO15 in Hrd1-deficiency macrophages recovers the decreased production of interleukin-6 [125]. USP15 is also involved in the vesicle transport process. The interaction of the E3 ubiquitin ligase ring finger protein 26 (RNF26) and USP15 was detected in HEK293T cells. The overexpression of USP15 can deubiquitinate the substrate, sequestosome1 (SQSTM1) of RNF26; that is, USP15 and RNF26 are functionally competitive. Furthermore, RNF26 inhibits vesicle transport through ubiquitination, while USP15 releases trapped vesicles through deubiquitination, thereby achieving the cyclic regeneration of vesicle transport [126]. These studies provide sufficient information to enrich the biological functions of USP15 and to develop USP15-targeted therapeutic systems. The function, expression and target of USP15 in various cancers and other diseases were summarized in Table 1 and Figure 3.
Table 1.
The function, expression, and target of USP15 in various cancers and other diseases.
| Function | Cancer/Disease | Expression of USP15 | Target |
|---|---|---|---|
| Proliferation, migration, and invasion |
Breast cancer | Up [60] | BMI1 [70], ER𝛂 [71] |
| Multiple myeloma | Up [62] | NF-κB/p65 [62] | |
| Gastric cancer | Up [65] | IκBα [74] | |
| Colon cancer | Up [76] | MDM2 [76], p53 [77] | |
| Hepatocellular carcinoma | Up [78] | KEAP1 [79] | |
| Prostate cancer | Up [63] | IRS-2 [82,83] | |
| Glioblastoma multiforme | Up [60] | SMURF2 [86], TβR-I [60,87] | |
| Leiomyosarcoma | DEK [85] | ||
| Hypertrophic scar | TβR-I [88] | ||
| Osteoarthritis | ERK2 [89] | ||
| Wound healing | EIF4A1 [90] | ||
| Apoptosis | Chronic myeloid leukemia | Up [68] | Caspase-6 [91] |
| Procaspase-3 [92] | |||
| FKBP5 [93] | |||
| Autophagy | Parkinson’s disease | Up [96] | PARKIN [96,97] |
| Cell cycle | REST [100,101,102] | ||
| Genome integrity | Ovarian cancer | Up [61] | BARD1 [54] |
| Leukemia | Up [68] | FUS [68] | |
| H2B [104] | |||
| TOP2A [53] | |||
| Transcriptional regulation | Neurodegenerative diseases | TUT1 [105] | |
| R-SMAD [106] | |||
| PRP31 [107] | |||
| SLIM1 [108] | |||
| Immune response | Melanoma | Up [76] | TET2 [109] |
| Human papillomavirus | E6 [111], TRIM25 [116,117] | ||
| TBK1 [115] | |||
| Others | ALK3 [123] | ||
| BRAP [124] |
Figure 3.
Cellular processes and substrates involved in USP15.
5. Conclusions
This review combines recent USP15-related research to illustrate the multi-faceted role of USP15 in cancer and other diseases. It is clear from the above discussion that USP15 is upregulated and plays an important role in the pathogenesis of cancer and other diseases. The involvement of USP15 in intracellular processes, such as apoptosis, autophagy, and the immune response has led to USP15 being an attractive therapeutic target. The comprehensive proteomic strategies accelerate the identification of USP15 substrate proteins. Evidence suggests that USP15 stabilizes oncoproteins by deubiquitination, thereby promoting proliferation in most cancers, including breast cancer, multiple myeloma, gastric cancer, glioblastoma, etc. However, some studies also point to USP15 acting as a negative regulator in certain tumor growth, which complicates the role of USP15 in tumors, suggesting that USP15 is a critical and promising target for tumor therapy. Therefore, the more comprehensive study of USP15 is needed to refine its biological function. Additionally, there is an urgent need to develop USP15-specific inhibitors to investigate its role in disease. Recently, a preclinical small-molecule inhibitor of USP15, USP15-Inh, selectively damages leukemia progenitor cells through re-engaging homeostatic redox responses [127]. Mitoxantrone has also been reported to inhibit the activity of USP15 to a certain extent [55]. The development of USP15 inhibitors undoubtedly provides a bright future for USP15-targeted therapy.
Author Contributions
Y.-C.L. and S.-W.C. wrote the manuscript and produced the figures; Y.-C.L., S.-W.C., Y.-B.S. and M.-W.C. reviewed and edited the manuscript. Z.S. supervised this work. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China No. 2017YFA0505104 (ZS), the National Natural Science Foundation of China Nos. 81772540 (ZS) and 51922111 (MWC), the Science and Technology Program of Guangdong No. 2019A050510023 (ZS) and the Science and Technology Development Fund, Macau SAR No. 0124/2019/A3 (MWC).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data herein presented are available in this article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kundu M., Thompson C.B. Autophagy: Basic principles and relevance to disease. Annu. Rev. Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 2.Chen R.H., Chen Y.H., Huang T.Y. Ubiquitin-mediated regulation of autophagy. J. Biomed. Sci. 2019;26:80. doi: 10.1186/s12929-019-0569-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaid S., Brandts C.H., Serve H., Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Nissan G., Sharon M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules. 2014;4:862–884. doi: 10.3390/biom4030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pacifici R.E., Kono Y., Davies K.J. Hydrophobicity as the signal for selective degradation of hydroxyl radical-modified hemoglobin by the multicatalytic proteinase complex, proteasome. J. Biol Chem. 1993;268:15405–15411. doi: 10.1016/S0021-9258(18)82272-4. [DOI] [PubMed] [Google Scholar]
- 6.Reinheckel T., Sitte N., Ullrich O., Kuckelkorn U., Davies K.J., Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Pt 3Biochem. J. 1998;335:637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hershko A., Heller H., Elias S., Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol Chem. 1983;258:8206–8214. doi: 10.1016/S0021-9258(20)82050-X. [DOI] [PubMed] [Google Scholar]
- 8.Hershko A., Eytan E., Ciechanover A., Haas A.L. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. Relationship to the breakdown of abnormal proteins. J. Biol. Chem. 1982;257:13964–13970. doi: 10.1016/S0021-9258(19)45327-1. [DOI] [PubMed] [Google Scholar]
- 9.Wilkinson K.D. Ubiquitin-dependent signaling: The role of ubiquitination in the response of cells to their environment. J. Nutr. 1999;129:1933–1936. doi: 10.1093/jn/129.11.1933. [DOI] [PubMed] [Google Scholar]
- 10.Ciechanover A., Elias S., Heller H., Hershko A. “Covalent affinity” purification of ubiquitin-activating enzyme. J. Biol. Chem. 1982;257:2537–2542. doi: 10.1016/S0021-9258(18)34957-3. [DOI] [PubMed] [Google Scholar]
- 11.Hoppe T. Multiubiquitylation by E4 enzymes: ‘one size’ doesn’t fit all. Trends Biochem. Sci. 2005;30:183–187. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Helliwell S.B., Losko S., Kaiser C.A. Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J. Cell Biol. 2001;153:649–662. doi: 10.1083/jcb.153.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koegl M., Hoppe T., Schlenker S., Ulrich H.D., Mayer T.U., Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/S0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 14.Akutsu M., Dikic I., Bremm A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016;129:875–880. doi: 10.1242/jcs.183954. [DOI] [PubMed] [Google Scholar]
- 15.Swatek K.N., Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schauer N.J., Magin R.S., Liu X., Doherty L.M., Buhrlage S.J. Advances in Discovering Deubiquitinating Enzyme (DUB) Inhibitors. J. Med. Chem. 2020;63:2731–2750. doi: 10.1021/acs.jmedchem.9b01138. [DOI] [PubMed] [Google Scholar]
- 17.Clague M.J., Heride C., Urbé S. The demographics of the ubiquitin system. Trends Cell Biol. 2015;25:417–426. doi: 10.1016/j.tcb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Kirkin V., McEwan D.G., Novak I., Dikic I. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 19.Grice G.L., Nathan J.A. The recognition of ubiquitinated proteins by the proteasome. Cell. Mol. Life Sci. 2016;73:3497–3506. doi: 10.1007/s00018-016-2255-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labbé K., McIntire C.R., Doiron K., Leblanc P.M., Saleh M. Cellular inhibitors of apoptosis proteins cIAP1 and cIAP2 are required for efficient caspase-1 activation by the inflammasome. Immunity. 2011;35:897–907. doi: 10.1016/j.immuni.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 21.Galan J.M., Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou P., Howley P.M. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell. 1998;2:571–580. doi: 10.1016/S1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- 23.Koepp D.M., Harper J.W., Elledge S.J. How the cyclin became a cyclin: Regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/S0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama K.I., Nakayama K. Ubiquitin ligases: Cell-cycle control and cancer. Nat. Rev. Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 25.Spence J., Sadis S., Haas A.L., Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell Biol. 1995;15:1265–1273. doi: 10.1128/MCB.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofmann R.M., Pickart C.M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/S0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 27.Finley D., Bartel B., Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- 28.Finley D., Ozkaynak E., Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh S., May M.J., Kopp E.B. NF-kappa B and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser P., Flick K., Wittenberg C., Reed S.I. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell. 2000;102:303–314. doi: 10.1016/S0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 31.Clague M.J., Barsukov I., Coulson J.M., Liu H., Rigden D.J., Urbé S. Deubiquitylases from genes to organism. Physiol. Rev. 2013;93:1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 32.Mevissen T.E.T., Komander D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 33.Hussain S., Zhang Y., Galardy P.J. DUBs and cancer: The role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle. 2009;8:1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 34.Ham S.J., Lee D., Xu W.J., Cho E., Choi S., Min S., Park S., Chung J. Loss of UCHL1 rescues the defects related to Parkinson’s disease by suppressing glycolysis. Sci. Adv. 2021;7:eabg4574. doi: 10.1126/sciadv.abg4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alameda J.P., García-García V.A., López S., Hernando A., Page A., Navarro M., Moreno-Maldonado R., Paramio J.M., Ramírez Á., García-Fernández R.A., et al. CYLD Inhibits the Development of Skin Squamous Cell Tumors in Immunocompetent Mice. Int. J. Mol. Sci. 2021;22:6736. doi: 10.3390/ijms22136736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love I.M., Shi D., Grossman S.R. p53 Ubiquitination and proteasomal degradation. Methods Mol. Biol. 2013;962:63–73. doi: 10.1007/978-1-62703-236-0_5. [DOI] [PubMed] [Google Scholar]
- 37.Lu Z., Hunter T. Ubiquitylation and proteasomal degradation of the p21(Cip1), p27(Kip1) and p57(Kip2) CDK inhibitors. Cell Cycle. 2010;9:2342–2352. doi: 10.4161/cc.9.12.11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul I., Batth T.S., Iglesias-Gato D., Al-Araimi A., Al-Haddabi I., Alkharusi A., Norstedt G., Olsen J.V., Zadjali F., Flores-Morales A. The ubiquitin ligase Cullin5(SOCS2) regulates NDR1/STK38 stability and NF-κB transactivation. Sci. Rep. 2017;7:42800. doi: 10.1038/srep42800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Andrea A., Pellman D. Deubiquitinating enzymes: A new class of biological regulators. Crit. Rev. Biochem. Mol. Biol. 1998;33:337–352. doi: 10.1080/10409239891204251. [DOI] [PubMed] [Google Scholar]
- 40.Komander D., Clague M.J., Urbé S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 41.Poondla N., Chandrasekaran A.P., Kim K.S., Ramakrishna S. Deubiquitinating enzymes as cancer biomarkers: New therapeutic opportunities? BMB Rep. 2019;52:181–189. doi: 10.5483/BMBRep.2019.52.3.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdul Rehman S.A., Kristariyanto Y.A., Choi S.Y., Nkosi P.J., Weidlich S., Labib K., Hofmann K., Kulathu Y. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol. Cell. 2016;63:146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das T., Shin S.C., Song E.J., Kim E.E. Regulation of Deubiquitinating Enzymes by Post-Translational Modifications. Int. J. Mol. Sci. 2020;21:4028. doi: 10.3390/ijms21114028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mansour M.A. Ubiquitination: Friend and foe in cancer. Int.J. Biochem. Cell Biol. 2018;101:80–93. doi: 10.1016/j.biocel.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ambroggio X.I., Rees D.C., Deshaies R.J. JAMM: A metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amerik A.Y., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Das T., Song E.J., Kim E.E. The Multifaceted Roles of USP15 in Signal Transduction. Int. J. Mol. Sci. 2021;22:4728. doi: 10.3390/ijms22094728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker R.T., Wang X.W., Woollatt E., White J.A., Sutherland G.R. Identification, functional characterization, and chromosomal localization of USP15, a novel human ubiquitin-specific protease related to the UNP oncoprotein, and a systematic nomenclature for human ubiquitin-specific proteases. Genomics. 1999;59:264–274. doi: 10.1006/geno.1999.5879. [DOI] [PubMed] [Google Scholar]
- 50.Vlasschaert C., Xia X., Coulombe J., Gray D.A. Evolution of the highly networked deubiquitinating enzymes USP4, USP15, and USP11. BMC Evol. Biol. 2015;15:230. doi: 10.1186/s12862-015-0511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kotani Y., Morito D., Sakata K., Ainuki S., Sugihara M., Hatta T., Iemura S.I., Takashima S., Natsume T., Nagata K. Alternative exon skipping biases substrate preference of the deubiquitylase USP15 for mysterin/RNF213, the moyamoya disease susceptibility factor. Sci. Rep. 2017;7:44293. doi: 10.1038/srep44293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chou C.K., Chang Y.T., Korinek M., Chen Y.T., Yang Y.T., Leu S., Lin I.L., Tang C.J., Chiu C.C. The Regulations of Deubiquitinase USP15 and Its Pathophysiological Mechanisms in Diseases. Int. J. Mol. Sci. 2017;18:483. doi: 10.3390/ijms18030483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fielding A.B., Concannon M., Darling S., Rusilowicz-Jones E.V., Sacco J.J., Prior I.A., Clague M.J., Urbé S., Coulson J.M. The deubiquitylase USP15 regulates topoisomerase II alpha to maintain genome integrity. Oncogene. 2018;37:2326–2342. doi: 10.1038/s41388-017-0092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng Y., Liao Q., Tan W., Peng C., Hu Z., Chen Y., Li Z., Li J., Zhen B., Zhu W., et al. The deubiquitylating enzyme USP15 regulates homologous recombination repair and cancer cell response to PARP inhibitors. Nat. Commun. 2019;10:1224. doi: 10.1038/s41467-019-09232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward S.J., Gratton H.E., Indrayudha P., Michavila C., Mukhopadhyay R., Maurer S.K., Caulton S.G., Emsley J., Dreveny I. The structure of the deubiquitinase USP15 reveals a misaligned catalytic triad and an open ubiquitin-binding channel. J. Biol. Chem. 2018;293:17362–17374. doi: 10.1074/jbc.RA118.003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elliott P.R., Liu H., Pastok M.W., Grossmann G.J., Rigden D.J., Clague M.J., Urbé S., Barsukov I.L. Structural variability of the ubiquitin specific protease DUSP-UBL double domains. FEBS Lett. 2011;585:3385–3390. doi: 10.1016/j.febslet.2011.09.040. [DOI] [PubMed] [Google Scholar]
- 57.Winget J.M., Mayor T. The diversity of ubiquitin recognition: Hot spots and varied specificity. Mol. Cell. 2010;38:627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Hetfeld B.K., Helfrich A., Kapelari B., Scheel H., Hofmann K., Guterman A., Glickman M., Schade R., Kloetzel P.M., Dubiel W. The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr. Biol. 2005;15:1217–1221. doi: 10.1016/j.cub.2005.05.059. [DOI] [PubMed] [Google Scholar]
- 59.Harper S., Besong T.M., Emsley J., Scott D.J., Dreveny I. Structure of the USP15 N-terminal domains: A β-hairpin mediates close association between the DUSP and UBL domains. Biochemistry. 2011;50:7995–8004. doi: 10.1021/bi200726e. [DOI] [PubMed] [Google Scholar]
- 60.Eichhorn P.J., Rodón L., Gonzàlez-Juncà A., Dirac A., Gili M., Martínez-Sáez E., Aura C., Barba I., Peg V., Prat A., et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat. Med. 2012;18:429–435. doi: 10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- 61.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou L., Jiang H., Du J., Li L., Li R., Lu J., Fu W., Hou J. USP15 inhibits multiple myeloma cell apoptosis through activating a feedback loop with the transcription factor NF-κBp65. Exp. Mol. Med. 2018;50:1–12. doi: 10.1038/s12276-018-0180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Padmanabhan A., Candelaria N., Wong K.K., Nikolai B.C., Lonard D.M., O’Malley B.W., Richards J.S. USP15-dependent lysosomal pathway controls p53-R175H turnover in ovarian cancer cells. Nat. Commun. 2018;9:1270. doi: 10.1038/s41467-018-03599-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., Li B., Liu X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–w514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhong M., Zhou L., Fang Z., Yao Y.Y., Zou J.P., Xiong J.P., Xiang X.J., Deng J. Ubiquitin-specific protease 15 contributes to gastric cancer progression by regulating the Wnt/β-catenin signaling pathway. World J. Gastroenterol. 2021;27:4221–4235. doi: 10.3748/wjg.v27.i26.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang B., Zhou L., Lu J., Wang Y., Liu C., Liang Z., Zhou W., You L., Guo J. Clinicopathological and prognostic significance of ubiquitin-specific peptidase 15 and its relationship with transforming growth factor-β receptors in patients with pancreatic ductal adenocarcinoma. J. Gastroenterol. Hepatol. 2021;36:507–515. doi: 10.1111/jgh.15244. [DOI] [PubMed] [Google Scholar]
- 67.Srihari S., Ragan M.A. Systematic tracking of dysregulated modules identifies novel genes in cancer. Bioinformatics. 2013;29:1553–1561. doi: 10.1093/bioinformatics/btt191. [DOI] [PubMed] [Google Scholar]
- 68.van den Berk P., Lancini C., Company C., Serresi M., Sanchez-Bailon M.P., Hulsman D., Pritchard C., Song J.Y., Schmitt M.J., Tanger E., et al. USP15 Deubiquitinase Safeguards Hematopoiesis and Genome Integrity in Hematopoietic Stem Cells and Leukemia Cells. Cell Rep. 2020;33:108533. doi: 10.1016/j.celrep.2020.108533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oikonomaki M., Bady P., Hegi M.E. Ubiquitin Specific Peptidase 15 (USP15) suppresses glioblastoma cell growth via stabilization of HECTD1 E3 ligase attenuating WNT pathway activity. Oncotarget. 2017;8:110490–110502. doi: 10.18632/oncotarget.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L., Qiang J., Yang X., Wang D., Rehman A.U., He X., Chen W., Sheng D., Zhou L., Jiang Y.Z., et al. IL1R2 Blockade Suppresses Breast Tumorigenesis and Progression by Impairing USP15-Dependent BMI1 Stability. Adv. Sci. 2020;7:1901728. doi: 10.1002/advs.201901728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xia X., Huang C., Liao Y., Liu Y., He J., Shao Z., Hu T., Yu C., Jiang L., Liu J., et al. The deubiquitinating enzyme USP15 stabilizes ERα and promotes breast cancer progression. Cell Death Dis. 2021;12:329. doi: 10.1038/s41419-021-03607-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu Y., Huangyang P., Wang Y., Xue L., Devericks E., Nguyen H.G., Yu X., Oses-Prieto J.A., Burlingame A.L., Miglani S., et al. ERα is an RNA-binding protein sustaining tumor cell survival and drug resistance. Cell. 2021;184:5215–5229.e5217. doi: 10.1016/j.cell.2021.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nguyen T.V. USP15 antagonizes CRL4(CRBN)-mediated ubiquitylation of glutamine synthetase and neosubstrates. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2111391118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng J., Zhang H., Ma R., Liu H., Gao P. Long non-coding RNA KRT19P3 suppresses proliferation and metastasis through COPS7A-mediated NF-κB pathway in gastric cancer. Oncogene. 2019;38:7073–7088. doi: 10.1038/s41388-019-0934-z. [DOI] [PubMed] [Google Scholar]
- 75.Xu K., Pei H., Zhang Z., Wang H., Li L., Xia Q. Ubiquitin-specific protease 15 promotes tumor cell invasion and proliferation in glioblastoma. Oncol. Lett. 2018;15:3846–3851. doi: 10.3892/ol.2018.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zou Q., Jin J., Hu H., Li H.S., Romano S., Xiao Y., Nakaya M., Zhou X., Cheng X., Yang P., et al. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat. Immunol. 2014;15:562–570. doi: 10.1038/ni.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu W.T., Huang K.Y., Lu M.C., Huang H.L., Chen C.Y., Cheng Y.L., Yu H.C., Liu S.Q., Lai N.S., Huang H.B. TGF-β upregulates the translation of USP15 via the PI3K/AKT pathway to promote p53 stability. Oncogene. 2017;36:2715–2723. doi: 10.1038/onc.2016.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yao X.Q., Li L., Piao L.Z., Zhang G.J., Huang X.Z., Wang Y., Liang Z.L. Overexpression of Ubiquitin-Specific Protease15 (USP15) Promotes Tumor Growth and Inhibits Apoptosis and Correlated With Poor Disease-Free Survival in Hepatocellular Carcinoma. Technol. Cancer Res Treat. 2020;19:1533033820967455. doi: 10.1177/1533033820967455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun Q., Zhang Z., Lu Y., Liu Q., Xu X., Xu J., Liu Y., Yu H., Yu D., Sun B. Loss of Xanthine Oxidoreductase Potentiates Propagation of Hepatocellular Carcinoma Stem Cells. Hepatology. 2020;71:2033–2049. doi: 10.1002/hep.30978. [DOI] [PubMed] [Google Scholar]
- 80.Chen X., Bao G., Liu F. Inhibition of USP15 Prevent Glutamate-Induced Oxidative Damage by Activating Nrf2/HO-1 Signaling Pathway in HT22 Cells. Cell. Mol. Neurobiol. 2020;40:999–1010. doi: 10.1007/s10571-020-00789-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fukagai K., Waku T., Chowdhury A., Kubo K., Matsumoto M., Kato H., Natsume T., Tsuruta F., Chiba T., Taniguchi H., et al. USP15 stabilizes the transcription factor Nrf1 in the nucleus, promoting the proteasome gene expression. Biochem. Biophys. Res. Commun. 2016;478:363–370. doi: 10.1016/j.bbrc.2016.07.045. [DOI] [PubMed] [Google Scholar]
- 82.Fukushima T., Yoshihara H., Furuta H., Kamei H., Hakuno F., Luan J., Duan C., Saeki Y., Tanaka K., Iemura S., et al. Nedd4-induced monoubiquitination of IRS-2 enhances IGF signalling and mitogenic activity. Nat. Commun. 2015;6:6780. doi: 10.1038/ncomms7780. [DOI] [PubMed] [Google Scholar]
- 83.Fukushima T., Yoshihara H., Furuta H., Hakuno F., Iemura S.I., Natsume T., Nakatsu Y., Kamata H., Asano T., Komada M., et al. USP15 attenuates IGF-I signaling by antagonizing Nedd4-induced IRS-2 ubiquitination. Biochem. Biophys. Res. Commun. 2017;484:522–528. doi: 10.1016/j.bbrc.2017.01.101. [DOI] [PubMed] [Google Scholar]
- 84.White M.F. IRS proteins and the common path to diabetes. Am. J. Physiol. Endocrinol. Metab. 2002;283:E413–E422. doi: 10.1152/ajpendo.00514.2001. [DOI] [PubMed] [Google Scholar]
- 85.Ge W., Chen Y., Guo Y., Zhao D., Mu L., Zhang K., Zhuo W. KIF15 upregulation promotes leiomyosarcoma cell growth via promoting USP15-mediated DEK deubiquitylation. Biochem. Biophys. Res. Commun. 2021;570:117–124. doi: 10.1016/j.bbrc.2021.07.042. [DOI] [PubMed] [Google Scholar]
- 86.Iyengar P.V., Jaynes P., Rodon L., Lama D., Law K.P., Lim Y.P., Verma C., Seoane J., Eichhorn P.J. USP15 regulates SMURF2 kinetics through C-lobe mediated deubiquitination. Sci. Rep. 2015;5:14733. doi: 10.1038/srep14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang L., Zhou F., García de Vinuesa A., de Kruijf E.M., Mesker W.E., Hui L., Drabsch Y., Li Y., Bauer A., Rousseau A., et al. TRAF4 promotes TGF-β receptor signaling and drives breast cancer metastasis. Mol. Cell. 2013;51:559–572. doi: 10.1016/j.molcel.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 88.Tu L., Lin Z., Huang Q., Liu D. USP15 Enhances the Proliferation, Migration, and Collagen Deposition of Hypertrophic Scar-Derived Fibroblasts by Deubiquitinating TGF-βR1 In Vitro. Plast. Reconstr. Surg. 2021;148:1040–1051. doi: 10.1097/PRS.0000000000008488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang W., Zhu Y., Sun Z., Jin C., Wang X. Positive feedback regulation between USP15 and ERK2 inhibits osteoarthritis progression through TGF-β/SMAD2 signaling. Arthritis Res. Ther. 2021;23:84. doi: 10.1186/s13075-021-02456-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao Y., Huang X., Zhang Z., Zhang Y., Zhang G., Zan T., Li Q. USP15 Enhances Re-epithelialization Through Deubiquitinating EIF4A1 During Cutaneous Wound Repair. Front. Cell Dev. Biol. 2020;8:529. doi: 10.3389/fcell.2020.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nie Z.Y., Yao M., Yang Z., Yang L., Liu X.J., Yu J., Ma Y., Zhang N., Zhang X.Y., Liu M.H., et al. De-regulated STAT5A/miR-202-5p/USP15/Caspase-6 regulatory axis suppresses CML cell apoptosis and contributes to Imatinib resistance. J. Exp. Clin. Cancer Res. 2020;39:17. doi: 10.1186/s13046-019-1502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu M., Takanashi M., Oikawa K., Tanaka M., Nishi H., Isaka K., Kudo M., Kuroda M. USP15 plays an essential role for caspase-3 activation during Paclitaxel-induced apoptosis. Biochem. Biophys. Res. Commun. 2009;388:366–371. doi: 10.1016/j.bbrc.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 93.Yu B., Shen B., Ba Z., Liu Z., Yuan J., Zhao W., Wu D. USP15 promotes the apoptosis of degenerative nucleus pulposus cells by suppressing the PI3K/AKT signalling pathway. J. Cell Mol. Med. 2020;24:13813–13823. doi: 10.1111/jcmm.15971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang T.S., Coppens I., Saorin A., Brady N.R., Hamacher-Brady A. Endolysosomal Targeting of Mitochondria Is Integral to BAX-Mediated Mitochondrial Permeabilization during Apoptosis Signaling. Dev. Cell. 2020;53:627–645.e627. doi: 10.1016/j.devcel.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Durcan T.M., Fon E.A. The three ‘P’s of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev. 2015;29:989–999. doi: 10.1101/gad.262758.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cornelissen T., Haddad D., Wauters F., Van Humbeeck C., Mandemakers W., Koentjoro B., Sue C., Gevaert K., De Strooper B., Verstreken P., et al. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum. Mol. Genet. 2014;23:5227–5242. doi: 10.1093/hmg/ddu244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cornelissen T., Vilain S., Vints K., Gounko N., Verstreken P., Vandenberghe W. Deficiency of parkin and PINK1 impairs age-dependent mitophagy in Drosophila. Elife. 2018;7 doi: 10.7554/eLife.35878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liang H., Su X., Wu Q., Shan H., Lv L., Yu T., Zhao X., Sun J., Yang R., Zhang L., et al. LncRNA 2810403D21Rik/Mirf promotes ischemic myocardial injury by regulating autophagy through targeting Mir26a. Autophagy. 2020;16:1077–1091. doi: 10.1080/15548627.2019.1659610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peth A., Boettcher J.P., Dubiel W. Ubiquitin-dependent proteolysis of the microtubule end-binding protein 1, EB1, is controlled by the COP9 signalosome: Possible consequences for microtubule filament stability. J. Mol. Biol. 2007;368:550–563. doi: 10.1016/j.jmb.2007.02.052. [DOI] [PubMed] [Google Scholar]
- 100.Guardavaccaro D., Frescas D., Dorrello N.V., Peschiaroli A., Multani A.S., Cardozo T., Lasorella A., Iavarone A., Chang S., Hernando E., et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Faronato M., Patel V., Darling S., Dearden L., Clague M.J., Urbé S., Coulson J.M. The deubiquitylase USP15 stabilizes newly synthesized REST and rescues its expression at mitotic exit. Cell Cycle. 2013;12:1964–1977. doi: 10.4161/cc.25035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xirodimas D.P. DUBs “found in translation”: USP15 controls stability of newly synthesized REST. Cell Cycle. 2013;12:2536–2537. doi: 10.4161/cc.25844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Das T., Kim E.E., Song E.J. Phosphorylation of USP15 and USP4 Regulates Localization and Spliceosomal Deubiquitination. J. Mol. Biol. 2019;431:3900–3912. doi: 10.1016/j.jmb.2019.07.023. [DOI] [PubMed] [Google Scholar]
- 104.Long L., Thelen J.P., Furgason M., Haj-Yahya M., Brik A., Cheng D., Peng J., Yao T. The U4/U6 recycling factor SART3 has histone chaperone activity and associates with USP15 to regulate H2B deubiquitination. J. Biol. Chem. 2014;289:8916–8930. doi: 10.1074/jbc.M114.551754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim J., Nakamura J., Hamada C., Taketomi T., Yano S., Okajima T., Kashiwabara S.I., Baba T., Sato B., Chiba T., et al. USP15 Deubiquitinates TUT1 Associated with RNA Metabolism and Maintains Cerebellar Homeostasis. Mol. Cell. Biol. 2020;40:e00098-20. doi: 10.1128/MCB.00098-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Inui M., Manfrin A., Mamidi A., Martello G., Morsut L., Soligo S., Enzo E., Moro S., Polo S., Dupont S., et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat. Cell. Biol. 2011;13:1368–1375. doi: 10.1038/ncb2346. [DOI] [PubMed] [Google Scholar]
- 107.Das T., Park J.K., Park J., Kim E., Rape M., Kim E.E., Song E.J. USP15 regulates dynamic protein-protein interactions of the spliceosome through deubiquitination of PRP31. Nucleic Acids Res. 2017;45:4866–4880. doi: 10.1093/nar/gkw1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Isumi Y., Hirata T., Saitoh H., Miyakawa T., Murakami K., Kudoh G., Doi H., Ishibashi K., Nakajima H. Transgenic overexpression of USP15 in the heart induces cardiac remodeling in mice. Biochem. Biophys. Res. Commun. 2011;405:216–221. doi: 10.1016/j.bbrc.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 109.Chen L.L., Smith M.D., Lv L., Nakagawa T., Li Z., Sun S.C., Brown N.G., Xiong Y., Xu Y.P. USP15 suppresses tumor immunity via deubiquitylation and inactivation of TET2. Sci. Adv. 2020;6:eabc9730. doi: 10.1126/sciadv.abc9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Niederkorn M., Hueneman K., Choi K., Varney M.E., Romano L., Pujato M.A., Greis K.D., Inoue J.I., Meetei R., Starczynowski D.T. TIFAB Regulates USP15-Mediated p53 Signaling during Stressed and Malignant Hematopoiesis. Cell Rep. 2020;30:2776–2790.e2776. doi: 10.1016/j.celrep.2020.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li S., Hong X., Wei Z., Xie M., Li W., Liu G., Guo H., Yang J., Wei W., Zhang S. Ubiquitination of the HPV Oncoprotein E6 Is Critical for E6/E6AP-Mediated p53 Degradation. Front. Microbiol. 2019;10:2483. doi: 10.3389/fmicb.2019.02483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chiang C., Pauli E.K., Biryukov J., Feister K.F., Meng M., White E.A., Münger K., Howley P.M., Meyers C., Gack M.U. The Human Papillomavirus E6 Oncoprotein Targets USP15 and TRIM25 To Suppress RIG-I-Mediated Innate Immune Signaling. J. Virol. 2018;92:e01737-17. doi: 10.1128/JVI.01737-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Poirson J., Biquand E., Straub M.L., Cassonnet P., Nominé Y., Jones L., van der Werf S., Travé G., Zanier K., Jacob Y., et al. Mapping the interactome of HPV E6 and E7 oncoproteins with the ubiquitin-proteasome system. FEBS J. 2017;284:3171–3201. doi: 10.1111/febs.14193. [DOI] [PubMed] [Google Scholar]
- 114.Zou Q., Jin J., Xiao Y., Zhou X., Hu H., Cheng X., Kazimi N., Ullrich S.E., Sun S.C. T Cell Intrinsic USP15 Deficiency Promotes Excessive IFN-γ Production and an Immunosuppressive Tumor Microenvironment in MCA-Induced Fibrosarcoma. Cell Rep. 2015;13:2470–2479. doi: 10.1016/j.celrep.2015.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huang L., Liu H., Zhang K., Meng Q., Hu L., Zhang Y., Xiang Z., Li J., Yang Y., Chen Y., et al. Ubiquitin-Conjugating Enzyme 2S Enhances Viral Replication by Inhibiting Type I IFN Production through Recruiting USP15 to Deubiquitinate TBK1. Cell Rep. 2020;32:108044. doi: 10.1016/j.celrep.2020.108044. [DOI] [PubMed] [Google Scholar]
- 116.Pauli E.K., Chan Y.K., Davis M.E., Gableske S., Wang M.K., Feister K.F., Gack M.U. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci. Signal. 2014;7:ra3. doi: 10.1126/scisignal.2004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Torre S., Polyak M.J., Langlais D., Fodil N., Kennedy J.M., Radovanovic I., Berghout J., Leiva-Torres G.A., Krawczyk C.M., Ilangumaran S., et al. USP15 regulates type I interferon response and is required for pathogenesis of neuroinflammation. Nat. Immunol. 2017;18:54–63. doi: 10.1038/ni.3581. [DOI] [PubMed] [Google Scholar]
- 118.Kusakabe S., Suzuki T., Sugiyama Y., Haga S., Horike K., Tokunaga M., Hirano J., Zhang H., Chen D.V., Ishiga H., et al. USP15 Participates in Hepatitis C Virus Propagation through Regulation of Viral RNA Translation and Lipid Droplet Formation. J. Virol. 2019;93 doi: 10.1128/JVI.01708-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou Q., Cheng C., Wei Y., Yang J., Zhou W., Song Q., Ke M., Yan W., Zheng L., Zhang Y., et al. USP15 potentiates NF-κB activation by differentially stabilizing TAB2 and TAB3. FEBS J. 2020;287:3165–3183. doi: 10.1111/febs.15202. [DOI] [PubMed] [Google Scholar]
- 120.Braun H., Staal J. Stabilization of the TAK1 adaptor proteins TAB2 and TAB3 is critical for optimal NF-κB activation. FEBS J. 2020;287:3161–3164. doi: 10.1111/febs.15210. [DOI] [PubMed] [Google Scholar]
- 121.Su Z.J., Cao J.S., Wu Y.F., Chen W.N., Lin X., Wu Y.L., Lin X. Deubiquitylation of hepatitis B virus X protein (HBx) by ubiquitin-specific peptidase 15 (USP15) increases HBx stability and its transactivation activity. Sci. Rep. 2017;7:40246. doi: 10.1038/srep40246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pyeon D., Timani K.A., Gulraiz F., He J.J., Park I.W. Function of ubiquitin (Ub) specific protease 15 (USP15) in HIV-1 replication and viral protein degradation. Virus Res. 2016;223:161–169. doi: 10.1016/j.virusres.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Herhaus L., Al-Salihi M.A., Dingwell K.S., Cummins T.D., Wasmus L., Vogt J., Ewan R., Bruce D., Macartney T., Weidlich S., et al. USP15 targets ALK3/BMPR1A for deubiquitylation to enhance bone morphogenetic protein signalling. Open Biol. 2014;4:140065. doi: 10.1098/rsob.140065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hayes S.D., Liu H., MacDonald E., Sanderson C.M., Coulson J.M., Clague M.J., Urbé S. Direct and indirect control of mitogen-activated protein kinase pathway-associated components, BRAP/IMP E3 ubiquitin ligase and CRAF/RAF1 kinase, by the deubiquitylating enzyme USP15. J. Biol. Chem. 2012;287:43007–43018. doi: 10.1074/jbc.M112.386938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lu Y., Qiu Y., Chen P., Chang H., Guo L., Zhang F., Ma L., Zhang C., Zheng X., Xiao J., et al. ER-localized Hrd1 ubiquitinates and inactivates Usp15 to promote TLR4-induced inflammation during bacterial infection. Nat. Microbiol. 2019;4:2331–2346. doi: 10.1038/s41564-019-0542-2. [DOI] [PubMed] [Google Scholar]
- 126.Jongsma M.L., Berlin I., Wijdeven R.H., Janssen L., Janssen G.M., Garstka M.A., Janssen H., Mensink M., van Veelen P.A., Spaapen R.M., et al. An ER-Associated Pathway Defines Endosomal Architecture for Controlled Cargo Transport. Cell. 2016;166:152–166. doi: 10.1016/j.cell.2016.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Niederkorn M., Ishikawa C., K M.H., Bartram J., Stepanchick E., J R.B., A E.C.-C., Bolanos L.C., Uible E., Choi K., et al. The deubiquitinase USP15 modulates cellular redox and is a therapeutic target in acute myeloid leukemia. Leukemia. 2021:1–14. doi: 10.1038/s41375-021-01394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data herein presented are available in this article.