Abstract
MCPIP1 (Regnase-1, encoded by the ZC3H12A gene) regulates the mRNA stability of several inflammatory cytokines. Due to the critical role of this RNA endonuclease in the suppression of inflammation, Mcpip1 deficiency in mice leads to the development of postnatal multiorgan inflammation and premature death. Here, we generated mice with conditional deletion of Mcpip1 in the epidermis (Mcpip1EKO). Mcpip1 loss in keratinocytes resulted in the upregulated expression of transcripts encoding factors related to inflammation and keratinocyte differentiation, such as IL-36α/γ cytokines, S100a8/a9 antibacterial peptides, and Sprr2d/2h proteins. Upon aging, the Mcpip1EKO mice showed impaired skin integrity that led to the progressive development of spontaneous skin pathology and systemic inflammation. Furthermore, we found that the lack of epidermal Mcpip1 expression impaired the balance of keratinocyte proliferation and differentiation. Overall, we provide evidence that keratinocyte-specific Mcpip1 activity is crucial for the maintenance of skin integrity as well as for the prevention of excessive local and systemic inflammation.
Keywords: MCPIP1, Regnase-1, ZC3H12A, Skin inflammation
Introduction
The skin is the main interface between the body and the environment. It is essential for preventing water and electrolyte loss, as well as for protection against harmful substances and pathogens [1]. The impairment of skin barrier function can cause or aggravate skin disorders, including psoriasis, atopic dermatitis, and ichthyosis [2–5]. Although the epidermis is a highly organized stratified epithelium consisting of four distinct layers: stratum basale, stratum spinosum, stratum granulosum, and the uppermost stratum corneum, its barrier function is provided mainly by a number of factors in the stratum corneum (corneocytes), such as lipids, antibacterial peptides, proteases, transcription factors, and many others [6–8]. The unique crosstalk between epidermal layers, immune cells, and microbes is important for tissue repair and regeneration to maintain the skin barrier function [9].
Monocyte chemotactic protein-1-induced protein 1 (MCPIP1), also known as Regnase-1, is encoded by the ZC3H12A gene. MCPIP1 possesses a PIN domain that has RNase properties and selectively promotes the destabilization of mRNAs encoding certain inflammatory cytokines, signal transducers, and transcription factors, such as IL-6 and IL12p40 in macrophages and c-Rel, Ox40, and IL-2 in T cells [10–15]. ZC3H12A expression is induced by a number of proinflammatory factors, including IL-17A and IL-36 family members [16–18], and MCPIP1 is an essential regulator of inflammatory signaling activation and immune homeostasis [19–22].
The systemic role of MCPIP1 has been demonstrated in Mcpip1-knockout mice. Mice lacking functional Mcpip1 develop postnatal systemic inflammation, which manifests as splenomegaly, lymphadenopathy, abnormal responses of both innate and adaptive immune cells, enhanced cytokine production, and premature death. The lack of Mcpip1 in T cells leads to a similar phenotype, suggesting a key role of Mcpip1 in immune homeostasis [13, 21, 23, 24].
MCPIP1 RNase is upregulated at both the transcript and protein levels in the human psoriatic epidermis [16, 17, 25]. Recent studies have shown that Mcpip1 deficiency leads to exaggerated psoriasis-like skin inflammation in response to imiquimod in both heterozygous Mcpip1−/+ mice [16] and in mice with keratinocytespecific Mcpip1 depletion [17]. Importantly, Mcpip1−/+ mice do not show any signs of skin pathology at the basal level. Takaishi et al. further pointed out that IL-36 signaling plays a role in driving the development of psoriasiform inflammation in mice with ablated epidermal Mcpip1 function. The authors showed that the expression of some transcripts associated with skin inflammation, such as Il36a, S100a8, and Defb3, is upregulated specifically in Mcpip1-depleted keratinocytes, suggesting that Mcpip1 RNase may play a role in the regulation of keratinocyte biology [18]. However, no description of the in vivo effect of the loss of epidermal Mcpip1 function was provided; in particular, no baseline skin characteristics in the mice were reported [18], which is important to fully understand an impact of Mcpip1 on cutaneous pathophysiology.
To deepen our knowledge on the role of epidermal Mcpip1 in skin homeostasis, we generated keratinocyte-specific Mcpip1-knockout mice (Mcpip1EKO). Based on pathological, immunological, and transcriptional analyses, we found that loss of Mcpip1 function leads to an increase in epidermal thickness as a result of enhanced cell proliferation, skin lesion formation as a result of skin barrier impairment, and spontaneous skin inflammation development due to changes in the expression profile of some inflammatory mediators. Moreover, we found that conditional Mcpip1-knockout mice develop systemic inflammation.
Overall, we provide evidence that Mcpip1 is a novel factor that controls the transcriptional profile in keratinocytes, and is crucial for skin immunological functions and maintaining epidermal homeostasis via the regulation of the equilibrium between keratinocyte proliferation and differentiation. Moreover, we show that Mcpip1 expression positively correlates with the differentiation potential of keratinocytes and postulate that Mcpip1 is a novel factor in the squamous epidermis that is essential for the maintenance of skin integrity.
Materials and methods
Animals
The Cre-loxP system was used to generate Mcpip1EKO mice (C57BL/6NJ). The Mcpip1loxP/loxP (Mcpip1fl/fl) mice, herein control mice, have been described previously [26]. To generate Krt14CreMcpip1fl/fl mice, male Krt14Cre mice [27] were bred with female Mcpip1 loxP-flanked mice. The mice used in this study were sex- and age-matched littermates. All animal procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (Directive 2010/63/EU of the European Parliament) and carried out under a license from the Ethical Committee of Jagiellonian University.
Human primary keratinocyte differentiation
Normal human epidermal keratinocytes were obtained from Lonza Group Ltd. (Basel, Switzerland). The cells were cultured in a 75-cm2 cell-culture flask at 37 °C in a 5% CO2 atmosphere in serum-free Keratinocytes Growth Medium KGM-Gold (Lonza Group Ltd., Basel, Switzerland) supplemented with bovine pituitary extract, human endothelial growth factor, bovine insulin, hydrocortisone, gentamicinamphotericin B (GA-1000), epinephrine, and transferrin. To promote differentiation, primary keratinocytes were cultured in 6-well plates with serum-free keratinocyte growth medium KGM-Gold supplemented with 1.8 mM calcium.
Primary keratinocyte isolation
Mouse primary keratinocytes were isolated from newborn (0– 1 days) control (Mcpip1fl/fl) or Mcpip1-deficient (Krt14CreMcpip1fl/fl) pups. The newborn mice were killed by decapitation and incubated for 1 min in PBS (Lonza, MD, USA), 1 min in 70% EtOH, and 1 min in PBS with antibiotics (Lonza, MD, USA). To separate the epidermis from the dermis, the skin was cut into small pieces and incubated with 2 mL of dispase (StemCell Technologies, MA, USA) overnight at 4 °C. The next day, the skin was transferred into a 6-cm dish, and the epidermis was separated from the dermis with forceps. Epidermal sheets were incubated in 1 mL of 0.25% trypsin with EDTA for 15 min at room temperature (RT). Keratinocytes were washed out of the epidermal sheet using 5 mL of DMEM (Lonza, MD, USA) supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich). After centrifugation, keratinocytes were seeded in cell-culture plates in 10% FBS/DMEM. After 24 h, the medium was replaced with keratinocyte growth medium (keratinocyte cell basal medium supplemented with KGM-Gold™ SingleQuots™ (bovine pituitary extract, human endothelial growth factor, insulin (bovine), hydrocortisone, gentamicin-amphotericin B (GA-1000), epinephrine, and transferrin); Lonza, MD, USA). Keratinocytes were cultivated at 37 °C with 5% CO2, and the medium was refreshed every 2 days.
RNA isolation and quantitative real-time PCR
Skin samples or spleens were collected, placed in Eppendorf tubes, frozen in liquid nitrogen, and stored at − 80 °C. For RNA isolation, the samples were homogenized in Fenozol (A&A Biotechnology, Gdynia, Poland) using a tissue homogenizer (Miccra D-1, Germany). The quantity of ribosomal RNA and DNA contaminants was examined using electrophoresis in a 1% denaturing formaldehyde gel. The purity and concentration of total RNA were assessed using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription was performed with 1 μg of total RNA, oligo(dT) primer, and M-MLV reverse transcriptase (Promega, Madison, USA). The cDNA was diluted 5 times, and real-time PCR was performed using an Eco Real-Time PCR System (Illumina) with SYBR Green qPCR master mix (A&A Biotechnology). The relative abundance of transcripts was determined compared to the abundance of elongation factor-2 (EF2). The sequences of the primers (Sigma-Aldrich) are listed in Supplementary Table S1
RNA sequencing
RNA extraction
Total RNAwas extracted from mouse primary keratinocytes using a mirVana™ PARIS™ kit (Ambion) according to the manufacturer’s instructions. The concentration of each sample was measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). The total RNA quality was analyzed with a RNA 6000 Nano Kit on an Agilent 2100 bioanalyzer (Agilent). Only samples with an RNA integrity number (RIN) > 7 were considered for downstream analyses.
Mouse AmpliSeq transcriptome
An Ion AmpliSeq Transcriptome Mouse Gene Expression kit (Thermo Fisher Scientific) was used for library preparation from IMQ-treated samples according to the manufacturer’s protocol. Briefly, 100 ng of total RNA was reverse transcribed, and the cDNA was subjected to multiplex PCR to amplify fragments of the target transcripts. The amplicons were then subjected to partial digestion at the primer sequences followed by adaptor ligation to the amplicons and purification on magnetic beads. The generated library was quantified on a 2100 Bioanalyzer using a DNA 1000 kit (Agilent).
Sequencing on an Ion Proton System
Each library was diluted to ~ 80 pM prior to template preparation. Eight barcoded libraries were mixed at equal volumes and used for automatic template preparation on an Ion Chef (Thermo Fisher Scientific) instrument using reagents from an Ion PI Hi-Q 200 Kit (Thermo Fisher Scientific) and Ion PI v3 Proton Chip. The samples were sequenced on the Ion Proton System (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Gene expression analysis
Gene abundance was quantified with htseq-count (HTSeq framework version 0.6) [28] using Ensembl Gene gtf files from UCSC as a reference. Differential gene expression was analyzed with the R package DESeq2, version 1.10.1. The sequencing data (as mapped bam files) are available in the European Nucleotide Archive under accession number PRJEB31970.
Gene Ontology category analysis
Gene function annotation was performed using the Database for Annotation, Visualization, and Integrated Discovery v.6.8 (DAVID v.6.8). Gene Ontology Biological Processes (GO_BP) analyses were used to achieve functional annotation-based clustering of genes with upregulated and downregulated expression [29, 30].
Protein isolation and Western blotting
For protein isolation, skin or spleen samples were homogenized in RIPA buffer supplemented with a complete protease inhibitor cocktail (Roche, Basely, Switzerland) and a PhosSTOP phosphatase inhibitor cocktail (Roche) using a tissue homogenizer (Miccra D-1, Germany). Cultured keratinocytes were washed with PBS and lysed in RIPA buffer supplemented with protease and phosphatase inhibitors. The protein concentrations in the cell lysates were measured with the bicinchoninic acid assay. Tissue and cell lysates were separated by SDS/PAGE on 10% polyacrylamide gels and electrotransferred to PVDF membranes (Millipore, Billerica, MA, USA), which were blocked in 3% milk dissolved in Trisbuffered saline containing 0.05% Tween 20 (BioShop, Burlington, Canada). The membranes were incubated with primary antibodies overnight at 4 °C. Then, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies. The signal was detected using Immobilon Western Chemiluminescent HRP Substrate (Millipore) in a MicroChemi chemiluminescence detector (DNR Bio-Imaging Systems, Jerusalem, Israel). The antibodies used in this study are listed in Supplementary Table S2.
Histology and immunofluorescence staining
Skin tissue specimens were fixed in 4% formaldehyde for 2 h and then incubated overnight at 4 °C in 30% sucrose. The next day, the tissue samples were embedded in Tissue-Tek O.C.T. Compound (Fisher Scientific, Pittsburgh, PA, USA). Subsequently, 6–8-μm cryosections were cut and stained with hematoxylin and eosin (H&E). Antigen retrieval was performed in 10 mM citrate buffer (pH 6.0) for 30 min at 95 °C. For immunohistology, skin samples were stained using an EnVision G|2 System/AP, Rabbit/Mouse (Permanent Red) kit (Dako, Glostrup, Denmark) according to the manufacturer’s protocol. After staining, the skin samples were counterstained with Mayer’s hematoxylin, mounted in glycerol mounting medium (Dako), and examined with a Leica CTR6 LED. For immunofluorescence, nonspecific antibody binding was prevented by blocking with 5% horse/goat serum, 1% BSA, and 0.005% Tween (Sigma-Aldrich) in PBS for 1 h. Primary antibodies were incubated overnight at 4 °C in blocking buffer. The sections were rinsed in PBS and incubated with secondary antibodies for 1 h at room temperature. Nuclei were stained with Hoechst 33258 (Sigma-Aldrich). Samples were mounted with fluorescent mounting medium (Dako) and then examined with a Leica CTR6 LED (Leica Microsystems, Wetzlar, Germany) equipped with Lecia Application Suite X software. The antibodies utilized for staining are listed in Supplementary Table S2. Figures were prepared using ImageJ and Adobe Illustrator CC.
Barrier function assay
An in situ skin permeability assay using toluidine blue was performed as previously described [31]. Briefly, newborn mice were sacrificed and incubated in methanol/PBS (25, 50, 75, and 100%) for 1 min and then thoroughly washed with PBS. Pups were subsequently immersed in 0.0125% toluidine blue/PBS for 1 min. Destaining was performed with PBS washing before photographs were captured. Transepidermal water loss (TEWL) measurements were performed on adult mice (3 months old (mo)) using a Tewameter TM300 (Courage + Khazaka Electronic) according to the manufacturer’s operating instructions. Adult mice were dorsally shaved using a professional hair clipper. Twenty-four hours later, mice were anesthetized and after 10 min the measurements were taken. Data were expressed in g/m2h.
Flow cytometry
Blood samples were collected by retro-orbital bleeding into tubes containing 10 mM EDTA. Bone marrow cells were isolated from one femur by flushing with RPMI1640 medium (Biowest) supplemented with 2% FBS (Gibco). Then, 0.5cm2 back skin samples were cut into small pieces and incubated with 2.5 mg/mL Collagenase D (Roche Diagnostics) solution at 37 °C with continuous shaking at 1400 rpm for 45 min. Single-cell suspensions from the skin, spleen, and bone marrow were obtained by mashing the organs through 40-μm cell strainers in RPMI1640 medium (Biowest) supplemented with 2% FBS (Gibco). After the removal of red blood cells by treatment with lysis buffer (155 mM NH4Cl, 10 mM NaHCO3, 0.1 mM EDTA), the cells were washed in PBS and then stained for viability assessment (Zombie Aqua Fixable Viability Kit; BioLegend). After being washed, the cells were blocked with anti-CD16/CD32 antibodies (Fc block; eBioscience) for 10 min on ice. Then, the cells were stained with the appropriate directly conjugated antibodies listed in Supplementary Table S2 and then washed with PBS containing 1% BSA. Data were acquired on a BD LSRII (BD Biosciences). Singlets were selected based on FCS-A vs FCS-H. Dead cells were routinely excluded from the analysis. “Fluorescence minus one” (FMO) controls were routinely used to verify correct compensation and to set the thresholds for positive/negative events. Analyses were performed with FCS Express (De Novo Software).
Quantification and statistics
All analyses were performed with 3–11 independent biological replicates. GraphPad Prism 7 (GraphPad Software Inc., La Jolla, CA) was used for all analyses of numerical data and the generation of graphs and statistical tests, including one-way analysis of variance (ANOVA) and Student’s t test. Error bars represent the standard error of the mean.
Results
Mcpip1 RNase regulates the transcript level of immune response and terminal differentiation genes in keratinocytes
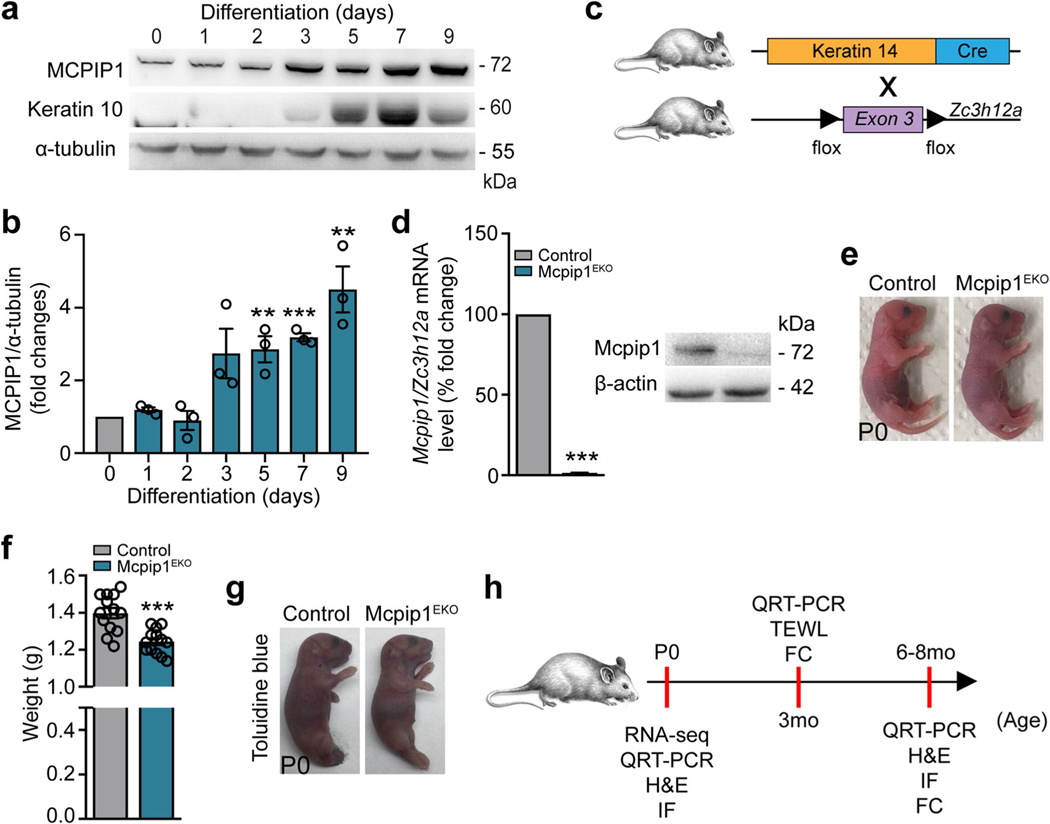
Epidermal keratinocytes undergo a multistep differentiation process that requires a tight balance between their proliferation and differentiation. To investigate the involvement of the MCPIP1 protein in this process, we characterized its expression in normal human keratinocytes and found that its expression is upregulated during Ca2+-induced differentiation in vitro, concurrently with that of keratin 10, a marker of epithelial differentiation (Fig. 1a, b). This finding is consistent with the observation that MCPIP1 protein localizes predominantly in the differentiated suprabasal layers of the normal human epidermis [17].
Fig. 1.
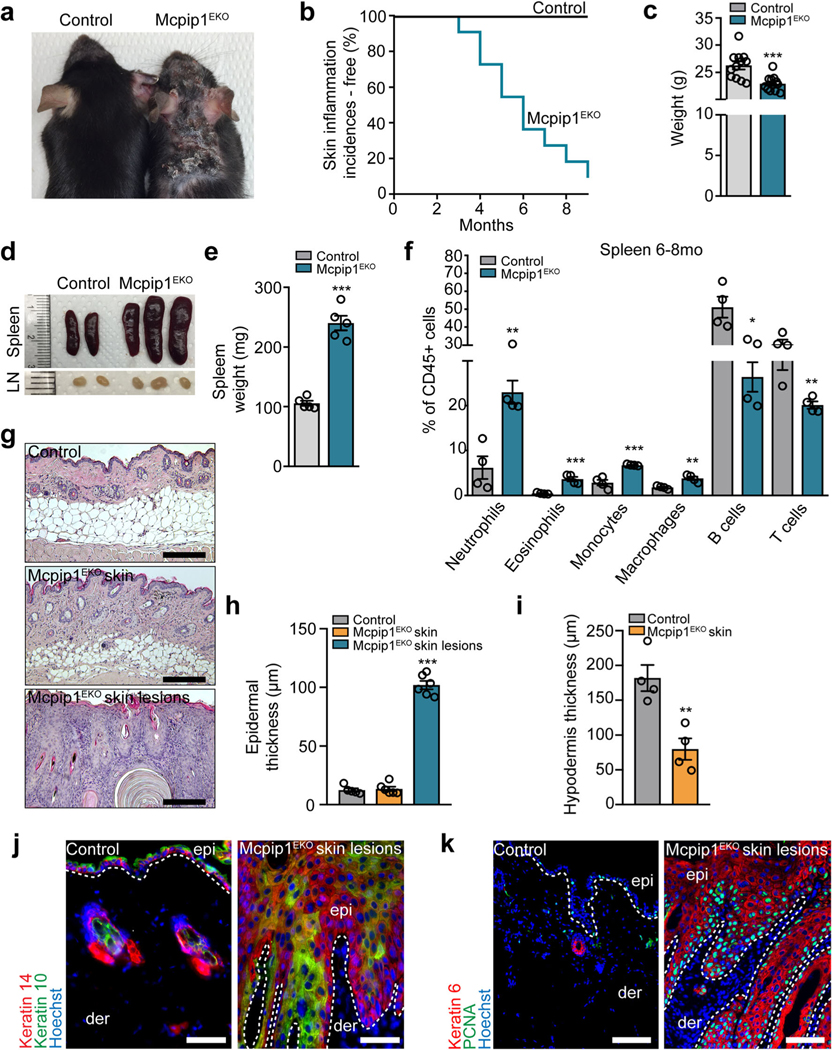
Conditional targeting of Mcpip1 in the epidermis. a Representative Western blot of three independent experiments for MCPIP1, keratin 10, and α-tubulin in primary human keratinocytes during Ca2+-induced differentiation. b Densitometric quantification of MCPIP1 levels (n = 3). c The generation of conditional Mcpip1EKO mice [26, 27]. d Left: QRT-PCR analysis of Mcpip1/Zc3h12a transcript levels (n = 7); right: Western blot for Mcpip1 and β-actin in isolated mouse keratinocytes. e Macroscopic appearance of newborn (P0) pups. f Body weights of newborn (P0) pups (n = 13). g Toluidine blue dye penetration assay at P0. h Schematic diagram representing the time points for RNA-Seq, QRT-PCR, H&E, immunofluorescence (IF), TEWL, and FC analyses. Data represent the mean ± SEM. **P < 0.01, ***P < 0.001 by unpaired t test
To determine the role of Mcpip1 in epidermal physiology in vivo, we generated Krt14CreMcpip1fl/fl (Mcpip1EKO) mice, in which Mcpip1 is specifically deleted in epidermal keratinocytes (Fig. 1c). Cre-mediated recombination resulted in the complete elimination of Mcpip1 mRNA and protein expression in primary keratinocytes (Fig. 1d and Supplementary Fig. S1). The Mcpip1EKO pups were born viable with a consistent body weight reduction of approximately 10% (Fig. 1e, f) but did not exhibit any obvious phenotypic abnormalities, including an intact outside-in stratum corneum barrier (Fig. 1g).
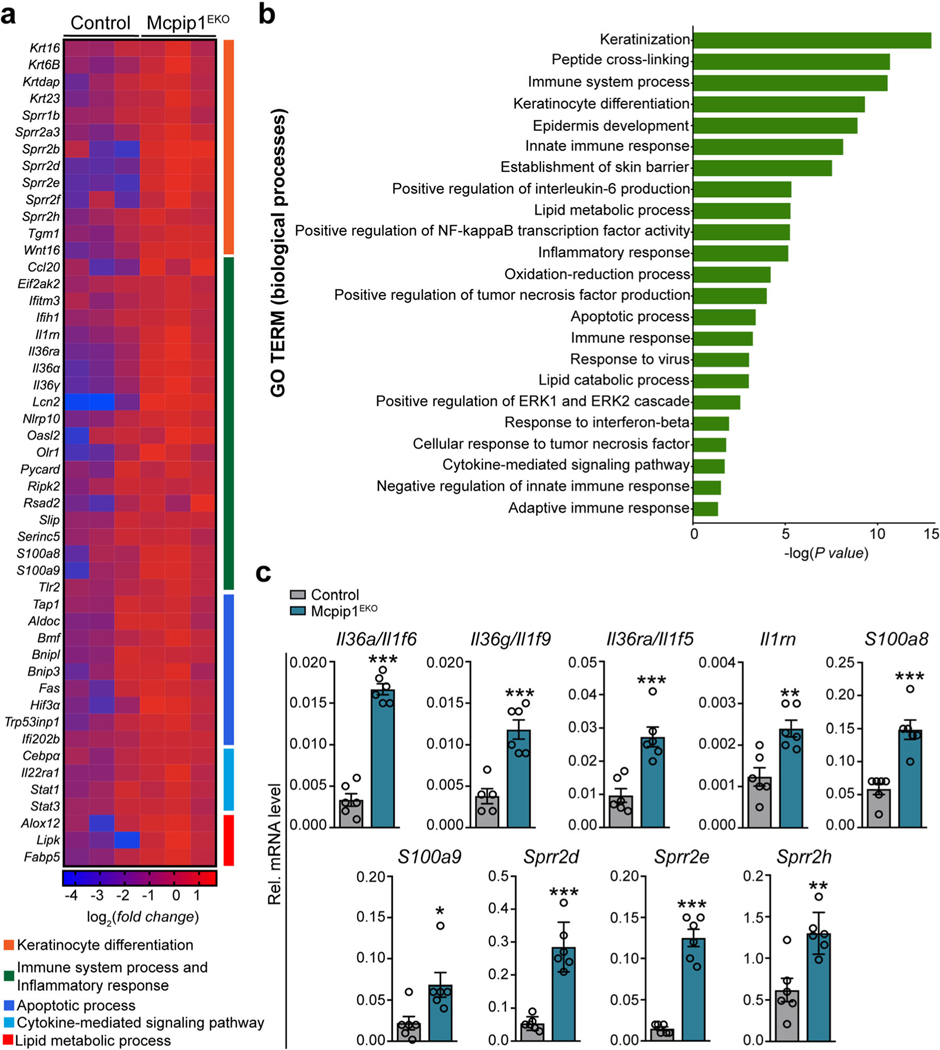
In the subsequent experiments, we performed a thorough biochemical and/or immunological characterization of Mcpip1EKO mice at three developmental stages: newborn (P0), adult (3 mo), and old (6–8 mo; Fig. 1h). We began the analysis by comparing the transcriptomes of keratinocytes isolated and cultured from the newborn control and Mcpip1EKO mice by RNA sequencing (RNA-Seq). The pairwise gene comparison indicated that the expression of 392 and 207 transcripts was significantly up- or downregulated (adj. P value < 0.05 and fold change > 1.5), respectively, in Mcpip1EKO cells. Functional analysis by Gene Ontology (GO) enrichment annotation revealed that the genes with upregulated expression in Mcpip1EKO mice were mainly assigned to groups related to keratinocyte differentiation and inflammatory responses (Fig. 2a, b). The expression of keratin (Krt6b, Krt16, and Krt23) and small proline-rich protein 2 (Sprr2d/e/h) family genes associated with epidermal growth, inflammation, and differentiation was enhanced in Mcpip1-deficient cells. The upregulated expression of interleukin-36α (IL-36α; Il36a/Il1f6) and IL-36γ (Il36g/Il1f9) cytokines, S100a8/a9 antibacterial peptides, and lipocalin-2 (Lcn2) indicated persistent proinflammatory signaling in Mcpip1EKO cells. Our RNASeq data are consistent with the known functions of MCPIP1 RNase as a negative regulator of immune responses. However, we also noticed the elevated expression of certain negative regulators of inflammation, such as Il1rn and Il36ra/Il1f5 transcripts that encode IL-1 and IL-36 receptor antagonists (Fig. 2a, c). Other pathways found to be elevated in Mcpip1EKO keratinocytes were related to lipid metabolism, oxidation-reduction processes, and apoptosis (Fig. 2a, b). Among the genes with upregulated expression that are important regulators of lipid metabolism processes are arachidonate 12-lipoxygenase, 12S type (Alox12), lipase, family member K (Lipk4), and fatty acid-binding protein 5 (Fabp5). Examples of transcripts encoding positive regulators of cell death shortlisted in our RNA-Seq data include TNF receptor superfamily member 6 (Fas), bone morphogenetic protein (Bmp4), and hypoxia-inducible factor 3-alpha (Hif3a).
Fig. 2.
Epidermal Mcpip1 regulates the expression of genes involved in keratinocyte differentiation and the positive regulation of the immune response. RNA-Seq was performed on keratinocytes isolated from newborn Mcpip1EKO or control mice (n = 3). a Heatmap expression plot of select genes. b GO biological process terms enriched among genes with upregulated expression in Mcpip1EKO (adj. P value < 0.05 and fold change > 1.5). c QRT-PCR analysis of Il36a/Il1f6, Il36g/Il1f9, Il36ra/ Il1f5, Il1rn, S100a8, S100a9, Sprr2d, Sprr2e, and Sprr2h expression (n = 6). Data represent the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired t test
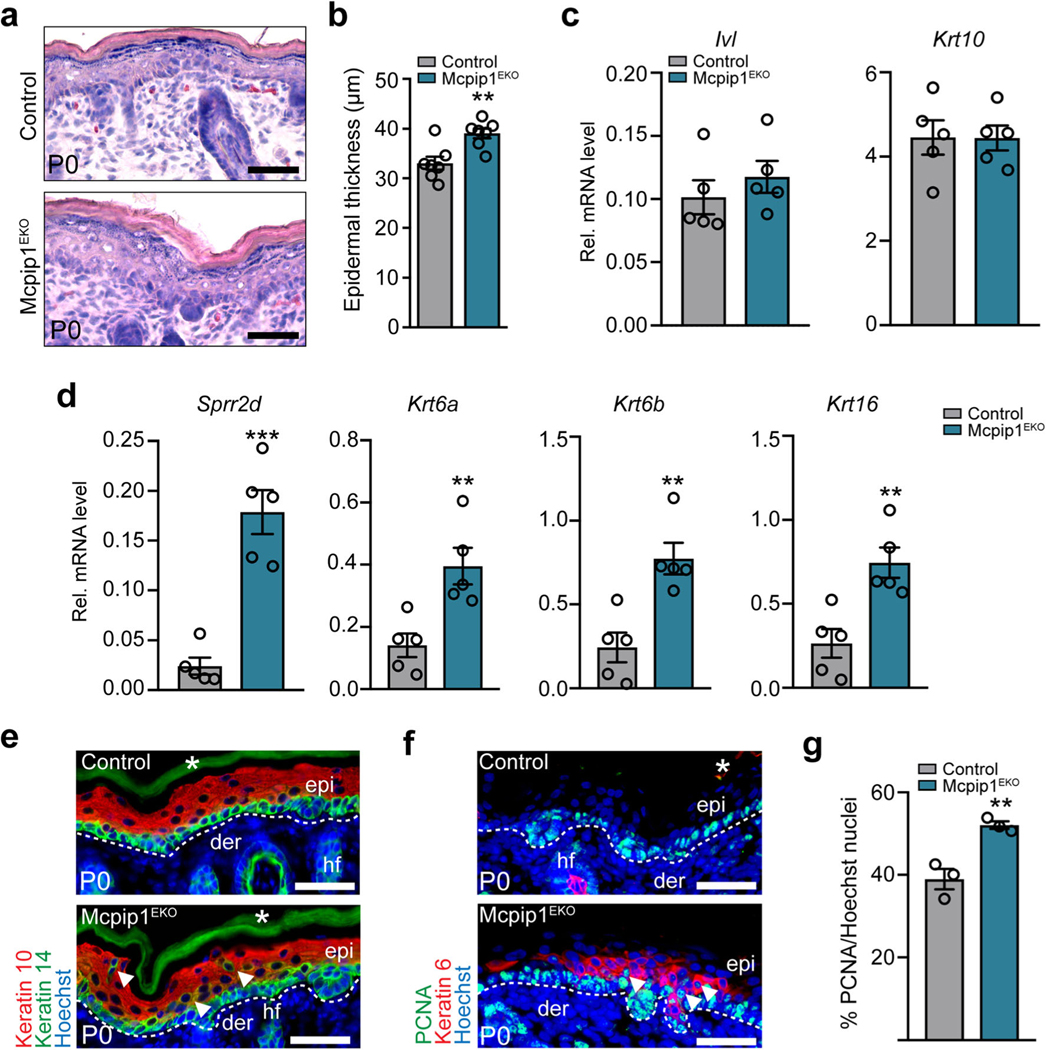
Newborn Mcpip1EKO mice exhibit disturbances in the distribution of the epidermal keratins Krt10/Krt14, Krt6, and PCNA
We next investigated the in vivo effects caused by the loss of keratinocyte Mcpip1 function. Histological analyses showed that the epidermis of newborn Mcpip1EKO mice was 1.2-fold thicker than those of control mice (Fig. 3a, b), suggesting alterations in epidermal proliferation and/or differentiation. QRT-PCR analyses of keratinocyte differentiation-associated markers indicated that the basal expression of involucrin (Ivl) and keratin 10 (Krt10) in the Mcpip1EKO neonatal epidermis was unchanged (Fig. 3c). In contrast, Mcpip1 loss resulted in the marked increase in Sprr2d, Krt6a, Krt6b, and Krt16 transcripts (Fig. 3d), suggesting an abnormal keratinocyte differentiation program in the Mcpip1EKO mouse epidermis. Subsequently, we analyzed the expression of several differentiation- and proliferation-specific markers by immunofluorescence. We found that there were occasionally keratin 10/keratin 14 (Krt10/Krt14) double-positive cells in suprabasal layers of Mcpip1-deficient epidermis, indicating the coexpression of basal and spinous keratins (Fig. 3e). The suprabasal expansion of Krt14 may be the result of a mild increase in inflammatory signaling within the Mcpip1EKO mouse epidermis. We further analyzed the protein expression of keratin 6 (Krt6). Under homeostatic conditions, in normal neonatal epidermis, Krt6 expression is restricted to the hair follicles. In response to biochemical or mechanical stress, under hyperproliferative conditions or due to keratinocyte differentiation defects, Krt6 expression is induced in the interfollicular epidermis [32–36]. Interestingly, we observed intermittent expression of Krt6 in the interfollicular epidermis of the Mcpip1EKO neonates, whereas in the control neonates, its expression was restricted to the hair follicles. This observation is consistent with our QRT-PCR data, which indicated increased levels of Krt6 mRNA in transgenic neonatal skin lysates. The induction of Krt6 expression in the suprabasal epidermis is a further indication of altered epidermal differentiation in Mcpip1EKO mice. In addition to the abnormalities in keratin expression, we observed a 1.3-fold increase in the abundance of epidermal proliferating cell nuclear antigen (PCNA)-positive cells, indicating the increased proliferation of Mcpip1EKO basal keratinocytes in vivo (Fig. 3f, g).
Fig. 3.
Newborn Mcpip1EKO mice exhibit disturbances in the expression of epidermal proliferation and differentiation markers. a H&E-stained back skin at P0. b Quantification of the epidermal thickness at P0 (n = 7). c QRT-PCR analysis of Ivl and Krt10 expression at P0 (n = 5). d QRT-PCR analysis of Sprr2d, Krt6a, Krt6b, and Krt16 expression at P0(n = 5). e Keratin 10 (Krt10) and keratin 14 (Krt14); f PCNA and keratin 6 immunofluorescence staining at P0. g Quantification of the epidermal PCNA-positive cells (n = 3). Arrowheads indicate Krt10/Krt14 double-positive suprabasal cells. Data represent the mean ± SEM. epi, epidermis; der, dermis; hf, hair follicles; *, nonspecific signal. The dashed line indicates the basal membrane. Scale bar, 100 μm. **P < 0.01, ***P < 0.001 by unpaired t test
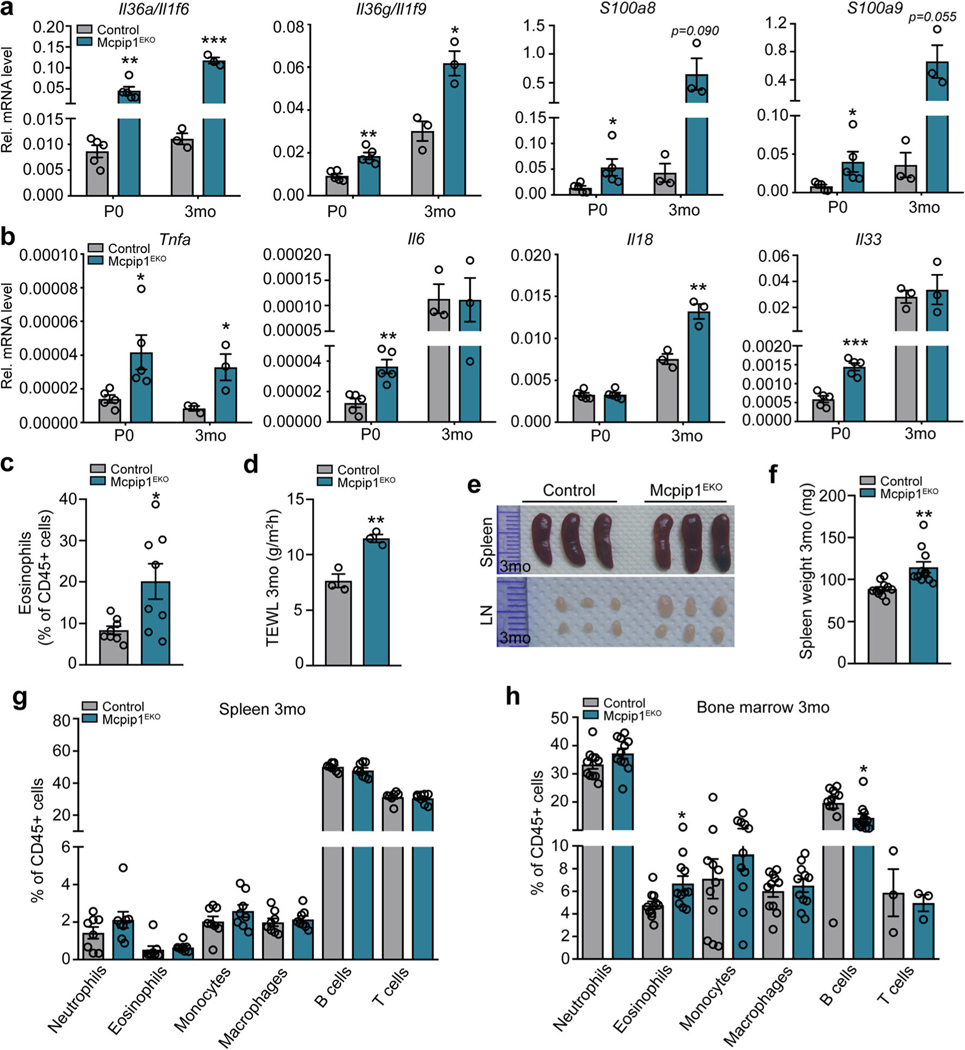
Increased expression of proinflammatory factors in newborn and young Mcpip1EKO mouse skin coincides with an impairment of skin integrity and the development of signs of systemic inflammation
We next carried out QRT-PCR expression analyses of the most significantly altered inflammation-related genes detected preliminarily in the Mcpip1EKO keratinocytes in the whole skin of both newborn (P0) and young mice (3 mo). We found that the levels of Il36a/g and S100a8/a9 were significantly elevated within Mcpip1EKO skin (Fig. 4a), consistent with our in vitro data (Fig. 2c). For instance, Il36a expression was increased in the P0 and 3-month-old Mcpip1EKO skin by ~ 5- and ~ 10-fold, respectively (Fig. 4a). The lack of epidermal Mcpip1 led to increased levels of other proinflammatory mediators in the skin. Notably, the mRNA expression of Tnfa, a cytokine associated with Th1 response, was increased in both newborn (P0) and young (3 mo) Mcpip1EKO skin (Fig. 4b). The expression of Il6 was ~ 3-fold elevated in Mcpip1EKO compared to the control (P0) pups, but its levels were not significantly altered in the skin of young (3 mo) mice (Fig. 4b). In contrast, the expression of Il18 mRNA was unchanged in the newborn (P0) mice and increased slightly in the young (3 mo) mice (Fig. 4b).
Fig. 4.
Newborn and young Mcpip1EKO mice have elevated levels of inflammatory factors in their skin and develop mild systemic inflammation. QRT-PCR analysis of selected transcript levels in the control and Mcpip1EKO newborn (P0, n = 5) and young (3 mo, n = 3) mice. a Il36a/Il1f6, Il36g/Il1f9, S100a8, S100a9 transcript levels. b Tnfa, Il6, Il18, and Il33 transcript levels. c Flow cytometric analysis of 3-monthold skin eosinophils (n = 8). d Quantification of TEWL in 3-month-old mice (n = 3). e Spleenandlymph node images of 3-month-old controland Mcpip1EKO mice. f Spleen weights of 3-month-old control and Mcpip1EKO mice (n = 10). g Flow cytometric analysis of 3-month-old mouse splenic CD45+ cells (n = 8). h Flow cytometric analysis of 3-month-old mouse bone marrow CD45+ cells (n = 3–11). Data represent the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired t test
In newborn mice (P0), we also noticed the differential expression of transcripts encoding the cytokine IL-33 that exerts both pro- and anti-inflammatory effects [37]. In the Mcpip1EKO skin (P0), the Il33 mRNA levels were increased ~ 2.5-fold (Fig. 4b). We next assayed whether elevated inflammatory signaling within the Mcpip1EKO mouse skin is reflected in the phenotype of skin immune cells. Flow cytometry analyses of 3-month-old mouse back skin homogenates indicated excessive presence of eosinophils in the Mcpip1EKO dermis (Fig. 4c). There were no significant changes in the relative frequencies of other leukocytes, including neutrophils, monocytes, macrophages, and T lymphocytes (Supplementary Fig. S2a). Cutaneous B cells were barely detected in both the Mcpip1EKO and control mice (data not shown).
To determine whether elevated levels of inflammatory mediators are associated with changes in skin barrier integrity in Mcpip1EKO mice, we measured TEWL. The TEWL was analyzed in the 3-month-old mice showing no macroscopic signs of skin inflammation. The Mcpip1EKO mice demonstrated significantly (1.5-fold) higher TEWL rates than control mice, suggesting reduced skin barrier function (Fig. 4d).
Despite molecular, immunological, and skin barrier function defects, the skin of 3-month-old Mcpip1EKO mice did not show any phenotypic signs of skin pathology. We also did not observe alterations in body weight (Supplementary Fig. S2b). However, young Mcpip1EKO mice showed a mild systemic effect, as evidenced by the enlarged lymph nodes and spleens (Fig. 4e), which were 1.4-fold heavier than those of the control (Fig. 4f). The flow cytometry analysis of splenic and blood CD45+ cells did not yet show any significant differences between the control and Mcpip1EKO mice (Fig. 4g and Supplementary Fig. 2c). However, in the bone marrow, statistically significant increase and decrease in population of eosinophils and B cells, respectively, was observed (Fig. 4h).
Old Mcpip1EKO mice have aggravated skin and systemic inflammation
Although newborn and young (3 mo) Mcpip1EKO mice did not develop a skin inflammation phenotype, aging Mcpip1EKO mice progressively developed skin inflammation. At approximately 4 months of age, the mice started to develop chronic wounds around their cheeks, ears, necks, and trunks, which were accompanied by hair loss within the affected areas (Fig. 5a, b). Adult (6 mo) Mcpip1EKO mice exhibited 13% reduced body weight (Fig. 5c), splenomegaly, and enlarged lymph nodes (Fig. 5d). The spleens of old Mcpip1EKO mice were 2.2-fold heavier than those of control mice (Fig. 5e). Flow cytometric analyses showed a significant increase in neutrophil (~ 4-fold), eosinophil (~ 8-fold), monocyte (2.4-fold), and macrophage (2.1-fold) numbers in Mcpip1EKO old mice compared to control mice, but a decreased presence of the B and T lymphocytes among splenocytes (Fig. 5f). Together, these data indicate substantial systemic changes in immune compartment of the adult Mcpip1EKO mice. At the histological level, the skin lesions exhibited hyperkeratosis with dysplastic keratinocytes populating the invaginations, which led to a profound increase in the epidermal thickness within the Mcpip1EKO skin lesions (Fig. 5g, h). In addition, compared to the control mice, the aging (6 mo) Mcpip1EKO mice exhibited a profound 2.3-fold reduction in the thickness of the hypodermis within the unaffected skin (Fig. 5i), possibly as a result of the elevated inflammation in Mcpip1EKO mice.
Fig. 5.
Development of skin lesions and systemic inflammation in old Mcpip1EKO mice. a Macroscopic appearance of 6-month-old mice. b Kaplan-Meier plots for skin inflammation incidences (n = 22). c Body weights of 6- to 8-month-old mice (n = 12). d Spleen and lymph node images of 6-month-old mice. e Spleen weights of 6- to 8-month-old mice (n = 5). f Flow cytometric analysis of 6- to 8-month-old mouse splenic CD45+ cells (n = 4). g H&E staining of control and unaffected Mcpip1EKO back skin and Mcpip1EKO skin lesions. h Quantification of the epidermal thickness (n = 6). Mcpip1EKO lesional skin was compared to control skin. i Quantification of hypodermis in 6-month-old mice (n = 4). j Keratin 14 and keratin 10; k PCNA and keratin 6 immunofluorescence staining. Data represent the mean ± SEM; epi, epidermis; der, dermis. The dashed line indicates the basal membrane. Scale bar, 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired t test or one-way ANOVA
We next characterized the skin pathologies that developed in old mice upon Mcpip1 deletion in detail and found that significant thickening of the Mcpip1EKO lesional epidermis corresponded with altered Krt14/Krt10 expression. We noticed the suprabasal expansion of basal Krt14 expression and reduction in the early differentiation marker Krt10 (Fig. 5j). The hyperproliferative phenotype was further confirmed by staining for the proliferation markers Krt6 and PCNA, which revealed an increase in the number of actively proliferating cells in both basal and suprabasal compartments (Fig. 5k).
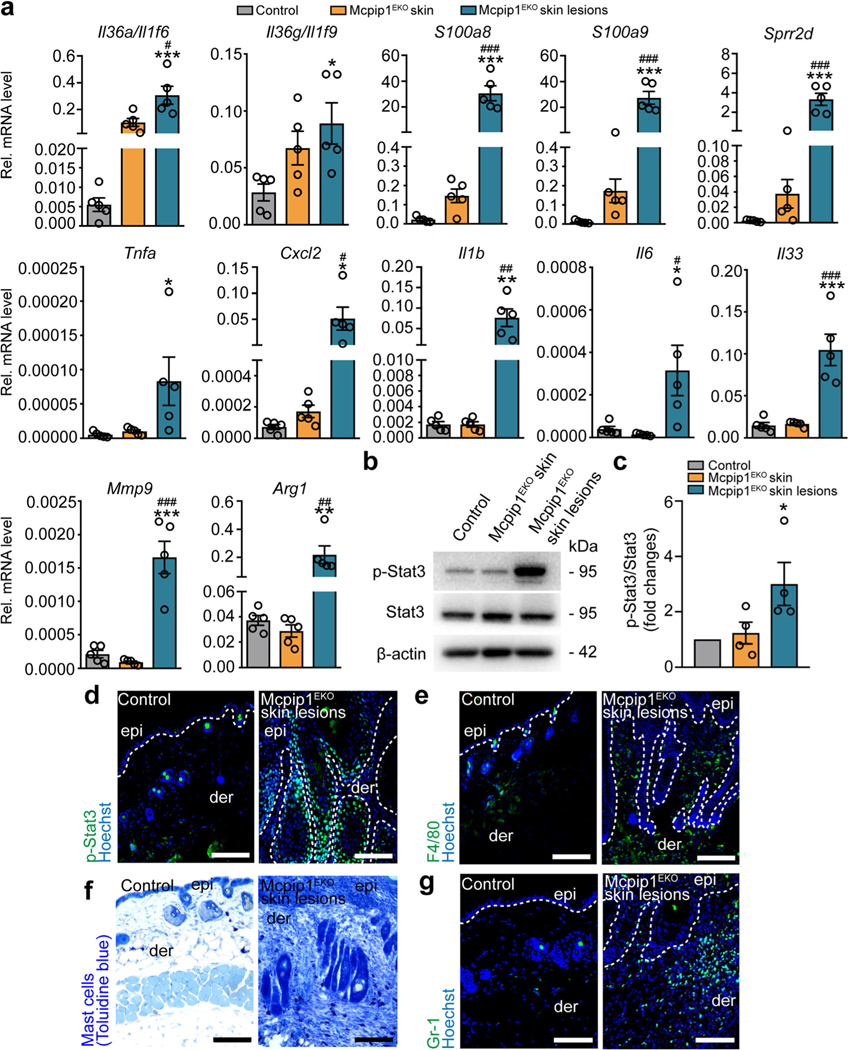
Cutaneous inflammatory phenotype of adult Mcpip1EKO mice also manifested in the enhanced production of various proinflammatory factors. In particular, the transcript levels of Il36a/g, Tnfa, Il1b, and Il6 cytokines; Cxcl2 chemokine; S100a8/a9 antibacterial peptides; and Sprr2d increased profoundly within lesional Mcpip1EKO skin (Fig. 6a). We also noticed a significant increase in the Il33 transcript level, as well as elevated matrix metallopeptidase 9 (Mmp9) and arginase-1 (Arg1; Fig. 6a). The activation of proinflammatory factors within the Mcpip1EKO skin lesion was positively correlated with the activation of Stat3 (Fig. 6b–d). In addition, the infiltration of the Mcpip1EKO lesional dermis by macrophages (Fig. 6e), mast cells (Fig. 6f), and neutrophils (Fig. 6g) was observed, suggestive of skin inflammatory phenotype acquired by the Mcpip1EKO mice upon aging.
Fig. 6.
Old Mcpip1EKO mice spontaneously develop skin inflammation. a QRT-PCR analysis of Il36a/Il1f6, Il36g/Il1f9, S100a8, S100a9, Sprr2d, Tnfa, Cxcl2, Il1b, Il6, Il33, Mmp9, and Arg1 transcript levels in the healthy skin of the control and Mcpip1EKO mice (6–8 mo) and in the skin lesions of Mcpip1EKO mice (n = 5). b Representative Western blot for p-Stat3, Stat3, and β-actin in the control and Mcpip1EKO mice (6–8 mo) from four independent experiments. c Densitometric quantification of p-Stat3/Stat3 levels (n = 4). d P-Stat3 immunofluorescence staining of the skin sections. e F4/80 immunostaining. f Toluidine blue immunostaining. g Gr-1 immunostaining. Scale bar, 100 μm. Data represent the mean ± SEM; epi, epidermis; der, dermis. The dashed line indicates the basal membrane. *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA. * refers to the comparison of control and Mcpip1EKO skin lesions; # refers to the comparison of Mcpip1EKO skin and Mcpip1EKO skin lesions
Discussion
In response to pathogens or other environmental factors, keratinocytes release proinflammatory mediators that attract and stimulate immune cells, initiating an inflammatory response. Therefore, keratinocytes serve as a physical barrier and provide tight control of inflammatory processes to avoid overreaction and tissue damage. MCPIP1 is an important regulator of inflammatory responses; however, there is also evidence that it is a regulator of proliferation, differentiation, angiogenesis, and cell metabolism [38–41]. To explore the role of MCPIP1 in skin homeostasis, we generated keratinocyte - specific Mcpip1-knockout mice (Mcpip1EKO).
Although newborn and young Mcpip1EKO mice did not exhibit any obvious macroscopic abnormalities, histological analyses indicated epidermal thickening with the presence of Krt14/Krt10 double-positive cells in the suprabasal layers that correlated with the increased proliferation of basal and suprabasal keratinocytes. The perturbed Krt14/Krt10 distribution was reflected in our Mcpip1-depleted keratinocyte RNA-Seq results, in which the levels of transcripts associated with keratinocyte differentiation were increased compared to those in control keratinocytes. For example, the expression of “stress keratins,” Krt6b and Krt16, which are associated with hyperproliferation and inflammation of the epidermis [42], was enhanced. This observation was confirmed by QRT-PCR analyses of whole skin lysates of newborn Mcpip1EKO pups. Furthermore, immunostaining showed abnormal Krt6 expression in the interfollicular epidermis of Mcpip1EKO pups, most likely due to the elevated inflammatory signaling present in their skin. We also noticed a prominent increase in the transcriptional expression of the terminal differentiation marker Sprr2d in both isolated keratinocytes and newborn mouse skin. Together, these data strongly suggest that there is an imbalance in keratin reprogramming within the Mcpip1EKO epidermis. Significant differences in the profiles of transcripts encoding proinflammatory mediators, such as the IL-36α/γ cytokines, were observed in Mcpip1EKO keratinocytes or in the skin of young mice. Nevertheless, this increase in the levels of inflammatory mediators did not lead to macroscopically visible skin inflammation in young mice, possibly due to the sufficient levels of negative regulators of inflammation, the transcript levels of which were also elevated in Mcpip1EKO keratinocytes. These negative regulators include IL-36/IL-1 receptor antagonists and interleukin-1 receptor-associated kinase (Irak3). The increase in the mRNA levels of IL-36α and of the antimicrobial proteins S100a8, S100a9, and Lcn2 in Mcpip1-depleted keratinocytes is consistent with the results of previous reports [17, 18, 43].
Our comprehensive QRT-PCR analysis of the transcriptional expression of several inflammatory factors in the whole skin of newborn and young (3 mo) mice revealed that the levels of Il36a, Il36g, S100a8, and S100a9 detected preliminarily in our RNA-Seq analysis were also elevated in the whole skin of newborn and young Mcpip1EKO mice. In addition to the elevated levels of these factors, we noticed elevated transcript levels of Il6, Tnfa, and Il33 in the newborn mice and of Tnfa and Il18 in the young (3 mo) mice. The elevated expression of a plethora of inflammatory mediators in the Mcpip1EKO skin that are released into the circulation is likely responsible for triggering the development of mild systemic inflammation in young (3 mo) mice, which manifests as the enlargement of the spleen and lymph nodes. To ensure the specificity of our conditional knockout model, we validated that the transcriptional and translational expression of Mcpip1 in the spleens of 3-month-old mice was unaltered (Supplementary Fig. S3a and S3b).
Despite the disturbances observed at the molecular level (elevated expression of inflammatory factors and abnormal epidermal proliferation/differentiation patterns), the skin of newborn and young (3 mo) Mcpip1EKO mice did not show any overall macroscopic changes. The skin barrier of newborn Mcpip1EKO mice was intact (indicated by the toluidine blue dye penetration assay), whereas the young (3 mo) mice showed impairment of skin barrier function, as indicated by the significantly elevated TEWL level, which most likely was the cause of the progressive development of local and possibly mild systemic inflammatory responses. However, upon aging, Mcpip1EKO mice gradually developed local skin pathologies. As a result of systemic inflammation and impairment of the skin barrier function, 4-month-old mice began to exhibit chronic skin lesions, with noticeable dermal presence of mast cells, neutrophils, and macrophages. The appearance of keratin pearls, which are characteristic of squamous cell carcinoma but can also occur in benign hyperproliferative tissue, indicates an abnormal keratinization pattern in the Mcpip1EKO adult skin and is most likely the result of excessive proliferation. The abnormally high levels of transcripts encoding Sprr2d protein as well as the altered expression of Krt14/ Krt10, Krt6, and PCNA correlated with the perturbed differentiation and proliferation within the Mcpip1EKO skin lesions. It was reported that enhanced levels of the cornified envelope protein Sprr2d lead to corneocyte fragility, resulting in a barrier defect, mild inflammation, and keratinocyte hyperproliferation [44]. The upregulation of Krt6 expression manifested within Mcpip1EKO skin lesions in old mice was associated with the downregulation of Krt10 expression and upregulation of the transcriptional expression of inflammatory factors (e.g., Tnfα, IL-1β, and IL-6) that are generally involved in the activation of stress keratins [45–47]. In addition to their mechanical properties, these keratins have specialized functions upon barrier breach and play important roles in various pathologies. Generally, the induction of Krt6 and Krt16 expression occurs at the expense of the Krt1/Krt10 pair in the postmitotic layers of the interfollicular epidermis under conditions of environmental stress (e.g., tissue injury, UV exposure, and viral infection) and in some diseases (e.g., psoriasis and carcinoma) [32–36]. In addition, the elevated mRNA expression of a plethora of proinflammatory mediators and chemokines, such as IL-1β, IL-6, IL-36α, S100a8/a9, and Cxcl2, within Mcpip1EKO skin lesion, and activation of Stat3, is associated with the enhanced proliferation rate and the exaggeration of the inflammatory response in the skin. The transcriptional expression of Il18, which was transiently activated in young Mcpip1EKO mice, was not altered in the old mouse skin (Supplementary Fig. S4a). Similarly, no differences in the expression of Il23a were found, suggesting that the Th17 response is not involved in the development of Mcpip1EKO skin lesions (Supplementary Fig. S4a). However, we noticed a prominent increase in the Il33 transcript level in skin lesions. This pleiotropic cytokine acts as an “alarmin” in response to external stimuli or tissue damage. The IL-33/ST2 pathway regulates the balance between extensive inflammation and tissue remodeling [37, 48, 49]. In skin lesions of Mcpip1EKO mice, we also noticed the transcriptional activation of Mmp9 metallopeptidase expression, most likely enhanced by IL-33 [49, 50]; and of arginase-1, a marker of alternatively activated macrophages, the expression of which increases upon tissue injury and repair [51, 52]. The extracellular matrix proteins of the basement membrane zone (BMZ) are important components of the intrafollicular epidermis (IFE) stem cell niche and functionally connect the dermis and epidermis. Type XVII collagen is expressed in basal keratinocytes and is involved in keratinocyte proliferation and migration [53, 54]. Here, we did not observe significant differences in collagen XVII expression between skin lysates from old Mcpip1EKO and control mice healthy skin. However, a small reduction in the ectodomain in the Mcpip1EKO skin lesions was observed (Supplementary Fig. S4b and S4c). Collagen XVII undergoes posttranslational modifications, such as ectodomain shedding and degradation, in physiological and pathological settings through the action of several proteases, such as disintegrin and metalloproteinases ADAM9/10/17, MMP9, neutrophil elastase, and other serine proteases [55, 56]. In the Mcpip1EKO mouse skin lesions, we observed the upregulation of Mmp9 expression and neutrophil infiltration. Our results suggest that ectodomain shedding of collagen XVII may be involved in Mcpip1EKO skin lesion development; however, this observation requires further analysis. In conclusion, we identified the activation of both proand anti-inflammatory signaling pathways in Mcpip1EKO mouse skin lesions; however, the levels of anti-inflammatory factors were not sufficient to resolve the chronic inflammation and promote skin remodeling.
Systematically, as a result of the enhanced skin inflammation, a vast range of proinflammatory factors that were increased in the old (6–8 mo) Mcpip1EKO mouse skin were further released into the circulation and likely exacerbated the systemic inflammation, which phenotypically manifested as enlarged spleens and reduced body weight. We hypothesize that the transcriptional activation of IL-33 observed within the skin lesions of old Mcpip1EKO mice contributes to the activation of the immune system via the ST2 receptor and exaggerates neutrophil- and eosinophil-dominated systemic inflammation [37, 57, 58]. IL-33-overexpressing mice are born with reduced body weight and progressively develop systemic inflammation with neutrophilia and increased myelopoiesis [58]. In a recent study, Peng and co-authors showed that in mice Mcpip1 loss contributes to the development of Th2-associated allergic airway inflammation. It was proposed that Mcpip1 is a negative regulator of Th2 function through the Notch/Gata3 pathway [59]. It would be interesting to evaluate the mechanism that contributes to the inflammatory phenotype of the Mcpip1EKO mice. The systemic, myeloid celldominated inflammatory phenotype observed in old, 6–8 mo, Mcpip1EKO mice suggests that myelopoiesis may be enhanced in Mcpip1EKO mice. This hypothesis is supported by the increased frequencies of myeloid cell populations, including neutrophils, eosinophils, monocytes, and macrophages, and reduced lymphocyte (B and T cells) populations in the spleens of Mcpip1EKO mice. In addition, myeloid cells tend to be more prevalent, whereas lymphocytes less prevalent among leukocytes in the bone marrow of young Mcpip1EKO mice, with significantly increased and decreased frequencies of eosinophils and B cells, respectively, supporting our hypothesis.
The prevalent factor triggering “spontaneous” skin inflammation in Mcpip1EKO mice remains elusive. It would be interesting to investigate whether these mice develop skin lesions in germ-free conditions because it has been shown that antibiotic treatment can improve inflammation and survival in completely Mcpip1-deficient mice [24]. Therefore, Mcpip1EKO mice may respond to the otherwise harmless skin microbiota. Alternatively, these skin lesions could be the consequence of elevated levels of proinflammatory factors in the skin, mild allergic reactions, and progressive systemic inflammation that contribute to itching and the development of mild scratching-induced wounds, the healing of which is impaired by the absence of Mcpip1. The progressive loss of skin integrity can be caused by several factors, including skin dehydration, thinning of the subcutaneous lipid layer, and systemic inflammation.
In conclusion, we postulate that epidermal Mcpip1 is essential for proper keratinocyte differentiation and epidermal functioning. Our study demonstrates a previously undescribed function of Mcpip1, which is required in keratinocytes for their proper differentiation and the maintenance of skin integrity. Our data also indicate that the normal functioning of Mcpip1 in keratinocytes is essential for the maintenance of systemic homeostasis, as the deficiency of this protein in aging mouse keratinocytes results in an inflammatory response that impacts multiple tissues and organs, including adipose tissue and the spleen.
Supplementary Material
Key messages.
Loss of murine epidermal Mcpip1 upregulates transcripts related to inflammation and keratinocyte differentiation.
Keratinocyte Mcpip1 function is essential to maintain the integrity of skin in adult mice.
Ablation of Mcpip1 in mouse epidermis leads to the development of local and systemic inflammation.
Acknowledgments
For the Krt14Cre mice, we are very thankful to Prof. Carien Niessen (Germany). We are grateful to the staff of the animal facility of the Faculty of Biochemistry, Biophysics and Biotechnology for help with animal breeding.
Funding information This research was supported by grants from the National Science Centre: PRELUDIUM 2014/13/N/NZ3/00729 (to P.K.) and OPUS 2016/23/B/NZ3/00792 (to J.J.). The Faculty of Biochemistry, Biophysics and Biotechnology of Jagiellonian University is a partner of the Leading National Research Centre (KNOW) supported by the Ministry of Science and Higher Education. W.D. was supported by the Flanders Institute for Biotechnology (VIB), a UGent grant (GOA-01G01914) and by Methusalem grant (BO16/MET_V/007, Ghent University).
Footnotes
Compliance with ethical standards
Conflict of interests The authors state no conflict of interests.
Declaration of ethics approval These experiments are performed according to law of EU.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00109-019-01853-2) contains supplementary material, which is available to authorized users.
References
- 1.Nestle FO, Di Meglio P, Qin J-Z, Nickoloff BJ (2009) Skin immune sentinels in health and disease. Nat Rev Immunol 9:679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elias PM, Williams ML, Holleran WM, Jiang YJ, Schmuth M (2008) Pathogenesis of permeability barrier abnormalities in the ichthyoses: inherited disorders of lipid metabolism. J Lipid Res 49:697–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guttman-Yassky E, Nograles KE, Krueger JG (2011) Contrasting pathogenesis of atopic dermatitis and psoriasis—part I: clinical and pathologic concepts. J Allergy Clin Immunol 127:1110–1118 [DOI] [PubMed] [Google Scholar]
- 4.Segre JA (2006) Epidermal barrier formation and recovery in skin disorders. J Clin Invest 116:1150–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Smeden J, Bouwstra JA (2016) Stratum corneum lipids: their role for the skin barrier function in healthy subjects and atopic dermatitis patients. Curr Probl Dermatol 49:8–26 [DOI] [PubMed] [Google Scholar]
- 6.Elias PM, Gruber R, Crumrine D, Menon G, Williams ML, Wakefield JS, Holleran WM,Uchida Y (1841) Formation and functions of the corneocyte lipid envelope (CLE). Biochim Biophys Acta 2014:314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Natsuga K (2014) Epidermal barriers. Cold Spring Harb Perspect Med 4:a018218–a018218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsuruta D, Green KJ, Getsios S, Jones JCR (2002) The barrier function of skin: how to keep a tight lid on water loss. Trends Cell Biol 12:355–357 [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi T, Naik S, Nagao K (2019) Choreographing immunity in the skin epithelial barrier. Immunity 50:552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu M, Blackshear PJ (2017) RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol 17:130–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uehata T, Akira S (1829) mRNA degradation by the endoribonuclease Regnase-1/ZC3H12a/MCPIP-1. Biochim Biophys Acta 2013:708–713 [DOI] [PubMed] [Google Scholar]
- 12.Iwasaki H, Takeuchi O, Teraguchi S, Matsushita K, Uehata T, Kuniyoshi K, Satoh T, Saitoh T, Matsushita M, Standley DM et al. (2011) The IkappaB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat Immunol 12:1167–1175 [DOI] [PubMed] [Google Scholar]
- 13.Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, Satoh T, Mino T, Suzuki Y, Standley DM et al. (2013) Malt1-induced cleavage of Regnase-1 in CD4(+) helper T cells regulates immune activation. Cell. 153:1036–1049 [DOI] [PubMed] [Google Scholar]
- 14.Li M, Cao W, Liu H, Zhang W, Liu X, Cai Z, Guo J, Wang X, Hui Z, Zhang H et al. (2012) MCPIP1 down-regulates IL-2 expression through an ARE-independent pathway. PLoS One 7:e49841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizgalska D, Wegrzyn P, Murzyn K, Kasza A,Koj A,Jura J, Jarzab B, Jura J (2009) Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1beta mRNA. FEBS J 276:7386–7399 [DOI] [PubMed] [Google Scholar]
- 16.Monin L, Gudjonsson JE, Childs EE, Amatya N, Xing X, Verma AH, Coleman BM, Garg AV, Killeen M, Mathers A et al. (2017) MCPIP1/Regnase-1 restricts IL-17A- and IL-17C-dependent skin inflammation. J Immunol 198:767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz-Romeu E, Ferran M, Gimenez-Arnau A, Bugara B, Lipert B, Jura J, Florencia EF, Prens EP, Celada A, Pujol RM et al. (2016) MCPIP1 RNase is aberrantly distributed in psoriatic epidermis and rapidly induced by IL-17A. J Invest Dermatol 136:1599–1607 [DOI] [PubMed] [Google Scholar]
- 18.Takaishi M, Satoh T, Akira S, Sano S (2018) Regnase-1, an immunomodulator, limits the IL-36/IL-36R autostimulatory loop in keratinocytes to suppress skin inflammation. J Invest Dermatol 138:1439–1442 [DOI] [PubMed] [Google Scholar]
- 19.Jeltsch KM, Hu D, Brenner S, Zoller J, Heinz GA, Nagel D, Vogel KU, Rehage N, Warth SC, Edelmann SL et al. (2014) Cleavage of Roquin and Regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat Immunol 15:1079–1089 [DOI] [PubMed] [Google Scholar]
- 20.Jura J, Skalniak L, Koj A (1823) Monocyte chemotactic protein-1induced protein-1 (MCPIP1) is a novel multifunctional modulator of inflammatory reactions. Biochim Biophys Acta 2012:1905–1913 [DOI] [PubMed] [Google Scholar]
- 21.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H et al. (2009) Zc3h12a is anRNase essential for controlling immune responses by regulating mRNA decay. Nature. 458:1185–1190 [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z, Miao R, Huang S, Elder B, Quinn T, Papasian CJ, Zhang J, Fan D, Chen YE, Fu M (2013) MCPIP1 deficiency in mice results in severe anemia related to autoimmune mechanisms. PLoS One 8: e82542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, Kolattukudy PE, Fu M (2010) MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J Exp Med 207:2959–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao R, Huang S, Zhou Z, Quinn T, Van Treeck B, Nayyar T, Dim D, Jiang Z, Papasian CJ, Eugene Chen Y et al. (2013) Targeted disruption of MCPIP1/Zc3h12a results in fatal inflammatory disease. Immunol Cell Biol 91:368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swindell WR, Sarkar MK, Liang Y, Xing X, Gudjonsson JE (2016) Cross-disease transcriptomics: unique IL-17A signaling in psoriasis lesions and an autoimmune PBMC signature. J Invest Dermatol 136:1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Huang X, Huang S, He H, Lei T, Saaoud F, Yu XQ, Melnick A, Kumar A, Papasian CJ et al. (2017) Central role of myeloid MCPIP1 in protecting against LPS-induced inflammation and lung injury. Signal Transduct Target Ther 2:17066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hafner M, Wenk J, Nenci A, Pasparakis M, Scharffetter-Kochanek K,Smyth N, Peters T, Kess D,Holtkotter O, Shephard P et al. (2004) Keratin 14Cre transgenic miceauthenticate keratin14as an oocyteexpressed protein. Genesis 38:176–181 [DOI] [PubMed] [Google Scholar]
- 28.Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57 [DOI] [PubMed] [Google Scholar]
- 30.Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardman MJ, Sisi P, Banbury DN, Byrne C (1998) Patterned acquisitionofskinbarrier functionduring development. Development 125:1541–1552 [DOI] [PubMed] [Google Scholar]
- 32.Heyden A, Lutzow-Holm C, Clausen OP, Brandtzaeg P, Huitfeldt HS (1994) Expression of keratins K6 and K16 in regenerating mouse epidermis is less restricted by cell replication than the expression of K1 and K10. Epithelial Cell Biol 3:96–101 [PubMed] [Google Scholar]
- 33.Stoler A, Kopan R, Duvic M, Fuchs E (1988) Use of monospecific antisera and cRNA probes to localize the major changes in keratin expression during normal and abnormal epidermal differentiation. J Cell Biol 107:427–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mils V, Vincent C, Croute F, Serre G (1992) The expression of desmosomal and corneodesmosomal antigens shows specific variations during the terminal differentiation of epidermis and hair follicle epithelia. J Histochem Cytochem 40:1329–1337 [DOI] [PubMed] [Google Scholar]
- 35.Coulombe PA (1997) Towards a molecular definition of keratinocyte activation after acute injury to stratified epithelia. Biochem Biophys Res Commun 236:231–238 [DOI] [PubMed] [Google Scholar]
- 36.McGowan K, Coulombe PA (1998) The wound repair-associated keratins 6, 16, and 17. Insights into the role of intermediate filaments in specifying keratinocyte cytoarchitecture. Subcell Biochem 31:173–204 [PubMed] [Google Scholar]
- 37.Liew FY, Girard J-P, Turnquist HR (2016) Interleukin-33 in health and disease. Nat Rev Immunol 16:676–689 [DOI] [PubMed] [Google Scholar]
- 38.Vrotsos EG, Kolattukudy PE, Sugaya K (2009) MCP-1 involvement in glial differentiation of neuroprogenitor cells through APP signaling. Brain Res Bull 79:97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niu J, Azfer A, Zhelyabovska O, Fatma S, Kolattukudy PE (2008) Monocyte chemotactic protein (MCP)-1 promotes angiogenesis via a novel transcription factor, MCP-1-induced protein (MCPIP). J Biol Chem 283:14542–14551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao J, Dai X, Pena T, Doyle DA, Guenther TM, Carlson MA (2015) MCPIP1 regulates fibroblast migration in 3-D collagen matrices downstream of MAP kinases and NF-kappaB. J Invest Dermatol 135:2944–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipert B, Wegrzyn P, Sell H, Eckel J, Winiarski M, Budzynski A, Matlok M, Kotlinowski J, Ramage L, Malecki M et al. (1843) Monocyte chemoattractant protein-induced protein 1 impairs adipogenesis in 3T3-L1 cells. Biochim Biophys Acta 2014:780–788 [DOI] [PubMed] [Google Scholar]
- 42.Zhussupbekova S, Sinha R, Kuo P, Lambert PF, Frazer IH, Tuong ZK (2016) A mouse model of hyperproliferative human epithelium validated by keratin profiling shows an aberrant cytoskeletal response to injury. EBioMedicine. 9:314–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garg AV, Amatya N, Chen K, Cruz JA, Grover P, Whibley N, Conti HR, Hernandez Mir G, Sirakova T, Childs EC et al. (2015) MCPIP1 endoribonuclease activity negatively regulates interleukin-17-mediated signaling and inflammation. Immunity. 43:475–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schafer M, Farwanah H, Willrodt A-H, Huebner AJ, Sandhoff K, Roop D, Hohl D, Bloch W, Werner S (2012) Nrf2 links epidermal barrier function with antioxidant defense. EMBO Mol Med 4:364–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haines RL, Lane EB (2012) Keratins and disease at a glance. J Cell Sci 125:3923. [DOI] [PubMed] [Google Scholar]
- 46.Lessard JC, Piña-Paz S, Rotty JD, Hickerson RP, Kaspar RL, Balmain A, Coulombe PA (2013) Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc Natl Acad Sci U S A 110:19537–19542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helenius TO, Antman CA, Asghar MN, Nystrom JH, Toivola DM (2016) Keratins are altered in intestinal disease-related stress responses. Cells. 5. 10.3390/cells5030035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Salvo E, Ventura-Spagnolo E, Casciaro M, Navarra M, Gangemi S (2018) IL-33/IL-31 axis: a potential inflammatory pathway. Mediat Inflamm 2018:3858032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kotsiou OS, Gourgoulianis KI, Zarogiannis SG (2018) IL-33/ST2 axis in organ fibrosis. Front Immunol 9:2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinto SM, Subbannayya Y, Rex DAB, Raju R, Chatterjee O, Advani J, Radhakrishnan A, Keshava Prasad TS, Wani MR, Pandey A (2018) A network map of IL-33 signaling pathway. J Cell Commun Signal 12:615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Campbell L, Saville CR, Murray PJ, Cruickshank SM, Hardman MJ (2013) Local arginase 1 activity is required for cutaneous wound healing. J Invest Dermatol 133:2461–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wynn TA, Vannella KM (2016) Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 44:450–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe M, Natsuga K, Nishie W, Kobayashi Y, Donati G, Suzuki S, Fujimura Y, Tsukiyama T, Ujiie H, Shinkuma S et al. (2017) Type XVII collagen coordinates proliferation in the interfollicular epidermis. eLife. 6. 10.7554/eLife.26635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jackow J, Schlosser A, Sormunen R, Nystrom A, Sitaru C, Tasanen K, Bruckner-Tuderman L, Franzke C-W (2016) Generation of a functional non-shedding collagen XVII mouse model: relevance of collagen XVII shedding in wound healing. J Invest Dermatol 136:516–525 [DOI] [PubMed] [Google Scholar]
- 55.Hirako Y, Usukura J, Uematsu J, Hashimoto T, Kitajima Y, Owaribe K (1998) Cleavage of BP180, a 180-kDa bullous pemphigoid antigen, yields a 120-kDa collagenous extracellular polypeptide. J Biol Chem 273:9711–9717 [DOI] [PubMed] [Google Scholar]
- 56.Nishie W (2014) Update on the pathogenesis of bullous pemphigoid: an autoantibody-mediated blistering disease targeting collagen XVII. J Dermatol Sci 73:179–186 [DOI] [PubMed] [Google Scholar]
- 57.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X et al. (2005) IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 23:479–490 [DOI] [PubMed] [Google Scholar]
- 58.Talabot-Ayer D, Martin P, Vesin C, Seemayer CA, Vigne S, Gabay C, Palmer G (2015) Severe neutrophil-dominated inflammation and enhanced myelopoiesis in IL-33-overexpressingCMV/IL33 mice.J Immunol 194:750–760 [DOI] [PubMed] [Google Scholar]
- 59.Peng H, Ning H, Wang Q, Lu W, Chang Y, Wang TT, Lai J, Kolattukudy PE, Hou R, Hoft DF et al. (2018) Monocyte chemotactic protein-induced protein 1 controls allergic airway inflammation by suppressing IL-5-producing TH2 cells through the Notch/ Gata3 pathway. J Allergy Clin Immunol 142:582–594.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.