Abstract
Antibodies and immune effectors (IE) are crucial for protecting humans from Gram-negative bacteria. Antibodies can bind outer membrane or cell surface (e.g. flagella) structures, thereby preventing adhesion, disrupting specific virulence functions, or targeting bacteria for phagocytosis. IE (antimicrobial peptides, cytokines and hormones) impinge on bacterial infections and regulate immune responses. A developing paradigm is that bacteria ‘recognize’ antibodies and IE, which alert them to challenging environments, promoting resistance phenotypes and increased virulence. A broader understanding of the interactions between bacteria and antibodies and IE will help define their relative contributions to pathogenesis, and perhaps indicate how we could use antibodies and IE to shape bacterial phenotypes that are easier for the immune system to control.
Bacterial responses to immune system effectors
The co-evolution of microbes and hosts has engendered mechanisms in each that provide the other with information as to their presence and perhaps intent. The vertebrate gut is illustrative of how bacteria and host communicate for mutual benefit [1]. Paneth cells secrete antimicrobial peptides that shape the endogenous flora to help fully vascularize the gut for nutrient adsorption [2]. The immune system can ‘sense’ products of pathogenic and non-pathogenic bacteria and in response constitute the immune system’s mucosal architecture. The mucosal antibody secretory (S)IgA is at high concentration in the gut lumen, where it interacts with pathogenic and non-pathogenic bacteria [3]. This learned response prevents aberrant inflammation and systemic antibody responses to commensal bacteria. Loss of SIgA leads to increased adherence of bacteria, resulting in hyperplasia and induction of systemic immune responses [4,5].
In contrast to the byplay between the cells of immune system and endogenous flora, invading bacterial pathogens are recognized by Toll-like receptors (TLR; see Glossary) that bind bacterial-derived molecules with recognizable ‘patterns’ [6]. Upon TLR engagement, the immune system mobilizes effectors of the innate and adaptive immune system. Neutrophils and macrophages respond to local and systemic immune effectors (IE) such as cytokines, chemokines and hormones, by migrating to sites of inflammation, becoming activated and secreting additional IE. Sustained infection and the presence of suitable immunogens induce the adaptive immune response, which includes T cells that can enhance phagocytic function via IE such as interferon (IFN)-γ. Immune effectors also activate antigen-specific B cells, which make antibodies targeted to soluble virulence factors and bacterial surface structures such as outer membrane proteins (OMP), lipopolysaccharide (LPS), flagella and pili.
The ability of immune systems to recognize bacterial products has developed over evolutionary time. The corollary to the host knowing that a pathogen is present is that bacteria can ‘sense’ the mobilized immune system. In this article, we review data suggesting that Gram-negative bacteria have an innate capacity to sense antibodies and/or IE and to modulate their phenotypes accordingly. There are at least four possible outcomes when bacteria engage antibodies or IE (Figure 1). We propose that the shaping of bacterial phenotypes by encounters with antibodies or IE is dynamic and, depending on the nature of the encounter, can either enhance bacterial virulence, or alternatively, can drive bacterial phenotypes to those that are less virulent.
Figure 1.
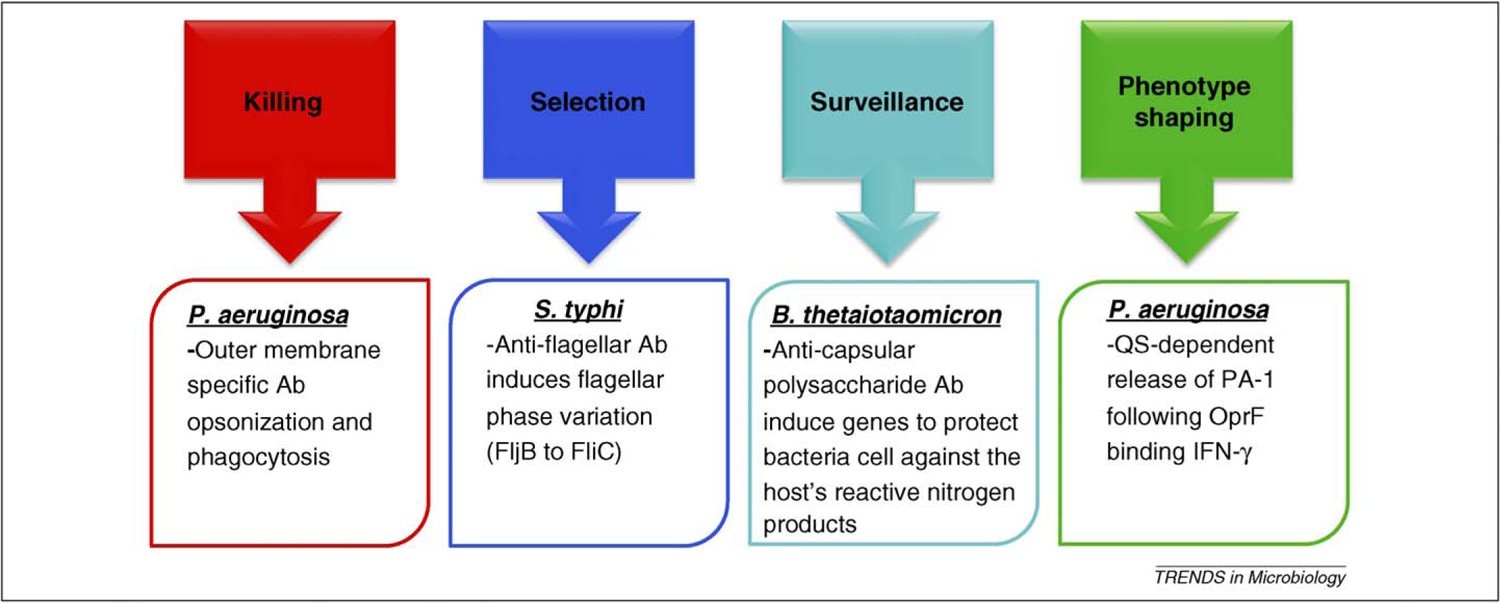
OM structure-specific antibodies and IE mediate diverse outcomes following interaction with bacteria. Killing: multiple OM structures (LPS, porins, capsule) can be bound by antibodies that interact with Fc receptors on phagocytes for effective killing of bacteria. Selection: antibodies to S. typhi flagella [24] result in selection of variants or induction of phase variation to change antibody targets. Surveillance: commensal bacteria can survey the lumen and respond to anti-LPS antibodies by expression of genes associated with protection from the host’s innate immune defenses [32]. Phenotype shaping: new evidence and extension of other observations suggest that antibodies and IE can regulate bacterial phenotypes. This could be via direct receptor–IE interaction (IFN-γ [18]) or indirectly by antibodies, for example, by preventing function of OM proteins (Opr86) and thus the cell adjusting gene expression for the perceived loss of general functions [48].
Bacteria can sense the immune system to their own advantage
Adaptability is crucial to bacterial survival in acutely changing environments; as such, bacteria know how to interrogate their world. It appears bacteria use several means to sense antibodies or IE as part of an early warning system to alert them to an impending immunologic attack. An early warning would allow the bacterium to implement strategies that avoid or resist the immune response.
Initial host recognition of bacterial infection results in production of proinflammatory cytokines (IFN-γ, tumor necrosis factor (TNF)-α, interleukin (IL)-1β, IL-6) that enhance recruitment and function of phagocytes at the infection site. IFN-γ is required for optimizing the phagocytic and bactericidal capacity of phagocytes. Several studies have shown that many bacteria have moderate to high affinity receptors for these cytokines, which signal enhanced growth or changes in virulence [7–14]. Patients with acute respiratory distress syndrome (ARDS) have high bacterial loads and elevated levels of cytokines in their lungs [13], and clinical isolates (Staphylococcus aureus, Pseudomonas aeruginosa and Acinetobacter spp.) from patients with ARDS cultured in vitro grow better in the presence of inflammatory cytokines at concentrations found in ARDS lungs.
Catecholamines (stress response hormones that regulate immune system effectors) can also regulate aspects of bacterial virulence. Norepinephrine affects production of the K99 pilus used for adhesion of enterotoxigenic Escherichia coli [15], E. coli O157:H7 responds to norepinephrine with Shiga toxin production [16], and Salmonella enterica serovar Typhimurium increases transcription of regulatory and signal transduction genes in response to epinephrine [17].
If regulation of bacterial phenotypes by antibodies or IE is a typical occurrence, criteria should be established to verify the relevance and pervasiveness of these responses, and to identify the mechanisms that regulate bacterial phenotypes in response to antibodies and IE. We believe there are at least five criteria (Box 1) that could serve as a framework to demonstrate the significance of antibodies and IE and the extent to which they influence bacterial pathogenesis.
Box 1. Criteria to verify the relevance of bacterial responses to antibodies and IE.
A specific receptor or receptors that bind IE or antibodies.
A signal transduction pathway to communicate antibody or IE binding to coordinate the bacterial response.
Induction of a specific response (i.e. altered transcription, or changes in protein activity, location, or function) by antibody or IE binding.
Response to antibody or IE binding, resulting in alteration of pathogenic potential, or the ability to evade or resist the immune response.
Development of novel therapeutic strategies wherein targeting with antibodies or IE decreases the ability of the microbe to access or maintain critical pathogenic phenotypes.
Criterion 1: does a specific receptor(s) bind antibodies or IE?
A specific response of a bacterium to antibodies or IE would predicate the existence of specific receptors with physiologically relevant binding constants. Several recent studies support the existence of such specific receptors.
IFN-γ is required for optimizing phagocyte function. IFN-γ specifically binds to Pseudomonas aeruginosa via the outer membrane (OM) porin OprF, which initiates quorum sensing (QS)-dependent release of PA-1, a lectin that increases the permeability of the host’s epithelial barrier [18]. Physiologic levels of IFN-γ are in the 5–50 ng/ml range but might reach higher concentrations at the site of infection [19]. The affinity of OprF–IFN-γ binding has not yet been determined, but the maximum expression of PA-1 was obtained at 10ng/ml of IFN-γ. Similarly, in enterohemorrhagic E. coli, adrenergic agonists (IE, hormones) bind to QseC, the sensor kinase of a two-component signal transduction system [20], resulting in enhanced transcription of type III secretion system genes, flagella and Shiga toxin (a toxin that can kill white blood cells).
There are numerous examples of antibodies that bind OM structures on Gram-negative bacteria. Of course, not all of these OM binding events induce classic signal transduction. Some OM structures such as siderophore receptors are linked to TonB-dependent signaling processes required for siderophore utilization [21]. Thus, interruption of siderophore acquisition by antibodies specific for the siderophore would prevent ligand transport, which normally induces TonB-dependent signaling via allosteric changes in the receptor. Antibodies that block iron entry and TonB-dependent signaling would have negative consequences for the cells.
In our opinion, a more pervasive type of signaling at the OM would be based on loss or reduction of function. For example, blocking the ability of porins to transport nutrients or other small molecules might be interpreted as a physiologic deficiency, requiring gene transcription to correct the deficit [22]. Other antibody receptors might include established virulence factors, such as flagella, for which blocking their function mimics changes in the environment that invoke a response involving gene activation [23,24]. The examples we note represent a fraction of the OM antibodies or IE interactions that probably occur. The question is: how many such interactions are important and can their effects be shaped?
Criterion 2: are specific signal transduction pathways used to communicate the presence of IE or antibodies?
The second criterion invokes the need for a specific, but not necessarily dedicated, signal transduction pathway to communicate the binding of antibodies and IE to the bacterial receptor. That is, this signal transduction event might respond to multiple inputs, including the engagement of components of the immune system. However, this signal transduction pathway should be clearly defined and fit the classic definition of such pathways (i.e. two-component systems, QS, and ligand-mediated transcriptional regulation).
Kappa opioid peptides (e.g. dynorphin) are released in response to stress or local hypoxia, both components of enteric infections [25,26]. Dendritic cells (antigen-presenting cells) can bind kappa agonists, and if LPS is also present, their T cell stimulatory capacity is increased [27]. Dynorphin intersects with the QS-related quinolone signaling system (PQS) and a transcriptional regulator, MvfR/PqsR, to increase pyocyanin production, which is toxic to macrophages [25]. As described above, binding of OprF by IFN-γ also results in signaling via QS, and adrenergic agonists signal via a two-component system. Thus, at least for IE, there are several documented examples of well-characterized signal transduction pathways required for bacterial response to these immunity factors.
Criterion 3: is there a specific set of responses in response to antibody or IE binding?
After detection of antibodies or IE and the subsequent transduction of that event, is there an explicit bacterial response? In many ways, this criterion goes hand-in-hand with criterion 2 in that one would expect a predictable response to activation of a particular signal transduction pathway.
An important class of IE comprises the antimicrobial peptides, such as polymyxin B and LL-37 [28], which can bind multiple macromolecules (e.g. DNA and LPS). Evidence from different two-component systems (PmrAB and PhoPQ) suggest that known signal transduction systems that bind antimicrobial peptides (polymxyin B) cause specific gene expression outputs, particularly, the induction of genes and their products known to modify LPS structure, which in turn reduces antimicrobial peptide binding [29].
Low concentrations of polymyxin B also inhibit P. aeruginosa biofilm formation [30]. Treatment of P. aeruginosa with sublethal LL-37 concentrations enhanced twitching motility and alters the expression of multiple Pseudomonas aeruginosa genes (311 up, 475 down) [31]. Although it is difficult to consider the differential expression of ~700 genes ‘specific’, a gene set required for type IV pili function was upregulated and thus might account for increased pili-mediated motility and reduced biofilm formation. Although the receptor and signal transduction pathway mediating response to LL-37 are unknown, the fact that other forms of motility were not affected and twitching motility is typically a tightly regulated process implies that LL-37 was mediating a specific change in bacterial behavior. Changes in LPS structure via LL-37 exposure are probably advantageous to Pseudomonas aeruginosa, but whether the loss of biofilm formation is beneficial to the microbe or to the host remains to be determined.
Criterion 4: do antibodies or IE regulate the pathogenic potential or the ability of the microbe to deal with hostile host environments?
Criteria 1–3 focus on documenting the mechanistic underpinning of responsiveness to antibodies or IE engagement to investigate whether microbes have evolved specific means of sensing and responding to these host factors, rather than employing a general ‘stress response’ strategy. In regards to clinical implications of bacteria countering antibody or IE attack, it is important to establish the functional significance of such a response; two such examples are presented below.
Salmonella express flagella that can sense the environment [32,33]. First, binding of Salmonella enterica serovar Typhi z66 with α-flagella antisera causes increased transcription of 187 genes, particularly, the genes for phase variation of FilB to FilC9 [24]. Phase variation is a known mechanism of immune system avoidance by bacteria. Second, the anti-Bacteroides thetaiotaomicron capsular polysaccharide (SIgA) of mono-associated mice resulted in upregulation of three bacterial genes used for defense against the products of inducible nitric oxide synthase, a phagocyte enzyme that generates bactericidal reactive nitrogen factors [34]. For both these examples, bacteria respond to antibodies by actively counteracting known host defense systems. The next significant challenge is to design experimental systems to answer similar questions in vivo; that is, can we conclusively demonstrate that a specific response to antibodies or IE enhances pathology?
Criterion 5: is the ability of antibodies and IE to shape bacterial phenotypes sufficiently robust to exploit for novel therapeutic strategies?
The last criterion addresses the issue of whether what we learn by satisfying the first four criteria will be useful for improving human health. Several therapeutic and basic science advances should follow if a new paradigm of antibody- or IE-induced staging of bacterial phenotypes is established.
Targeting IE such as proinflammatory cytokines or hormones for therapy is not practical because of their general utility to host defense, thus we believe that focusing on bacterial OM structures is a more practical strategy. OM structures are numerous and accessible, and serve as excellent targets for therapies that can shape bacterial phenotypes. Antibodies to explore phenotype shaping will include those already existing and known to target OM, and those yet to be identified and isolated from recombinant antibody library strategies (outlined in Figure 2). The use of recombinant libraries with established diversity (preassembled specificities) obviates the issues associated with producing antibodies by conventional immunization, which may result in structures that might not be highly immunogenic. Antibody libraries could be readily screened for antibodies specificities that alter virulence factor elaboration and thus alter the microbe’s phenotype to one inconsistent with colonization or virulence. Such antibodies might be effective antimicrobial agents alone or in combination with current routine antibiotics. To identify such ‘antimicrobial’ or ‘anti-infective’ antibodies, high-throughput screens must be configured, and using microarray technology would be an appropriate means to discover the global effect of antibody–OM binding events on virulence and microbial biology.
Figure 2.
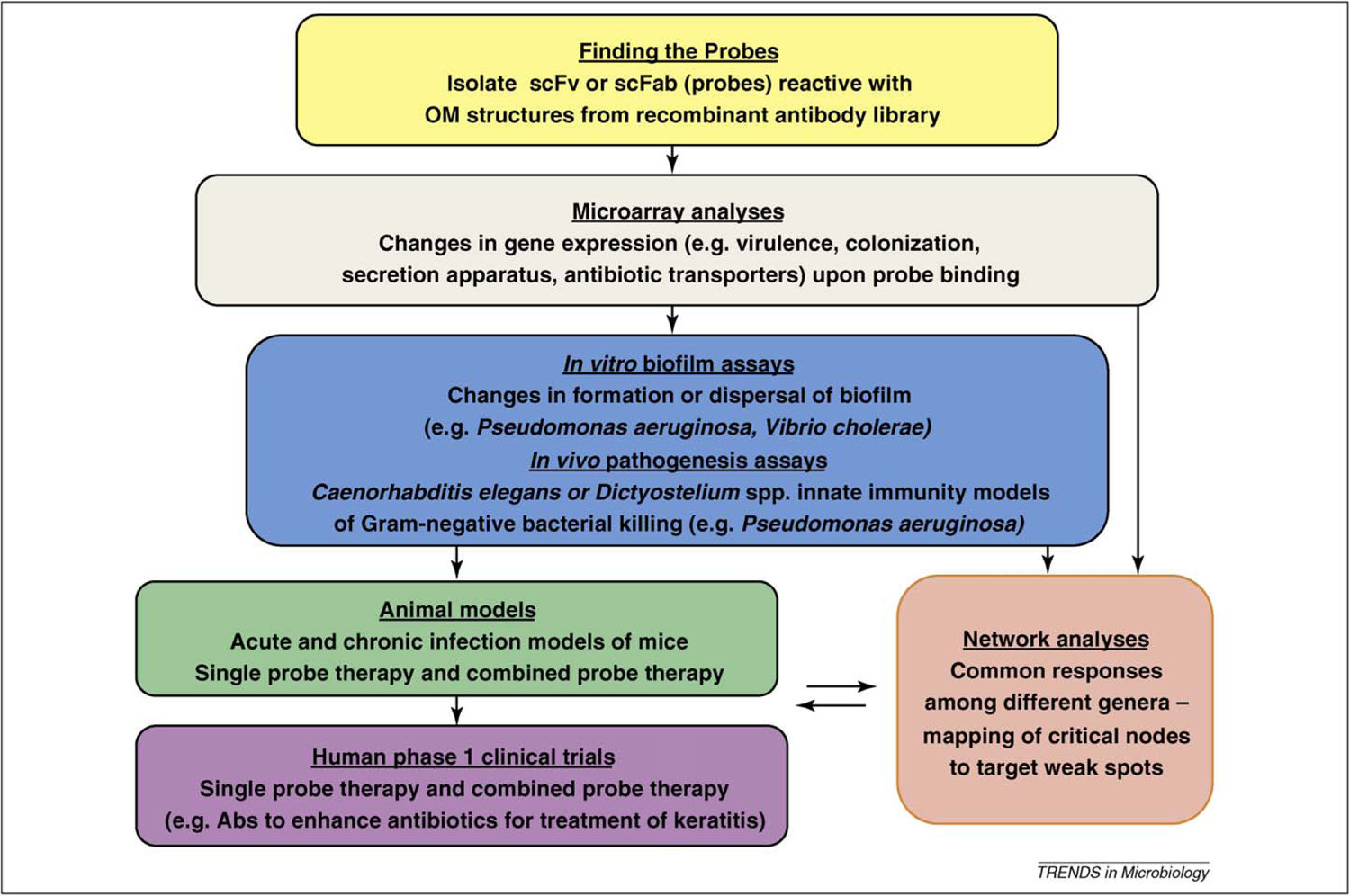
Flow chart of a reagent development path to discover new antibodies to enhance the efficacy of current therapeutics against Gram-negative bacteria. Multiple recombinant antibody libraries containing most of the expressed antibody repertoire (scFv and scFab) can be presented on the surface coat of phage, thus allowing their screening in vitro. Manipulation of bacterial growth, biofilm formation or other aspects of bacterial biology will provide suitable targets against which to adsorb the scFv and scFab libraries, and thus lead to the identification of a comprehensive set of OM probes for different Gram-negative pathogens. These antibodies, used singularly or in combination, or in combination with antibiotics, can be screened in high-throughput assays (for example, assessing altered virulence gene expression or effects on biofilm formation), followed by further assessment of candidates for their global effect on the microbes with microarray studies. A similar screen could be performed with invertebrates, exploiting the fact that pathogenic bacteria such as P. aeruginosa are known to kill nematodes and amoeba upon ingestion by the host. For example, antibodies can be bound to bacteria before feeding to the hosts, and the changes in the pathogenesis noted. The antibodies that have positive effects in the different assays should be tested in appropriate mouse models with successful candidates moving to safety trials. Contemporaneous with the microarray studies, antibody-specific and bacteria-specific networks should be generated to identify crucial nodes that might suggest the optimal pairing of probes. Furthermore, if known virulence pathways are regulated by antibodies and these are known to be required for virulence in any of the models, these candidates should be among the first investigated in clinical trials.
In vitro and in vivo assessment of antibody and IE effects on pathogenesis
What in vitro models systems might serve to help to identify those antibodies and IE that are able to provide a therapeutic benefit? In vitro biofilm formation is an accepted model of in vivo biofilm biology and such microbial communities are a component in many chronic bacterial infections [35]. Antibodies directed towards OM proteins could be used both for studying biofilm biology and for defining gene activation pathways that negatively regulate biofilm formation. Antibodies or IE that can disperse or weaken the biofilm structure might reduce the inherent resistance of the bacteria to antibiotics and phagocytes. In vitro biofilm formation or dispersal models are also excellent high-throughput systems to explore how multiple antibodies or IE shape bacterial phenotypes.
We also suggest that the utility of in vitro analysis of therapeutic antibodies should be complemented by invertebrate models of bacterial pathogenesis [36–40]. Antibodies that bind pathogenic bacterial OM structures should be investigated for their potential to prevent bacterial-induced death of Caenorhabditis elegans or Dictyostelium spp. [35,36], and such systems are also adaptive to high-throughput assays [41,42]. Later, a focused antibody or IE set should be tested in disease-specific small animal models before phase I safety trials in humans are contemplated.
Why might this concept be a major advance?
The blind spot
Disease pathogenesis is based on the conflict between pathogens and the host’s immune system. Over evolutionary time, bacteria have developed a stochastic response (heterogeneous gene activation or ‘on or off’) to environmental changes to ‘hedge their bets’ as to how the new environmental changes will affect their future [43]. The sooner the cells define their environment, the more quickly they can respond with energy-efficient survival strategies. If the cell population responds to a signal and their environmental assessment is wrong, then the population is at risk. If assessment of the environmental clues is not timely, then the population is also at risk. It follows that if OM binding ‘events’ can provide contradictory signals that result in maximum ‘bet hedging’, then the progression of cells as a group to virulence expression might be retarded or even prevented.
The end game
Why might these newly discovered roles of antibodies be opportune as antibiotic resistance spreads? We propose that many antibodies specific for OM structures are largely outside the evolutionary ‘memory’ of the typical host that bacteria encounter. This is because many OM structures are either not normally immunodominant immunogens (during infection) or are not bound in concert with other antibodies or IE. Thus, having a set of OM probes for specific Gram-negative pathogens and knowing how they interact to shape phenotypes might be a means to impinging on crucial network nodes that control function. It might be especially difficult for the bacterial cells to develop resistance against this type of noncidal attack. Antibodies that produce signals interpreted as deficiencies (i.e. loss of intracellular Fe3+ by blocking siderophore receptors) or too many unfolded proteins in the porin transport pathway (i.e. blocking Opr86) will globally alter gene and protein expression or activity necessary to cope with the perceived deficits in the bacterial cell. This global response to multiple immune assaults will, we predict, increase the ‘noise’ in regulatory systems, increase the molecular crowding of proteins in the transport pathways, activate stress responses and probably have additional deleterious effect on the microbe, which is already withstanding the onslaught of standard antimicrobials.
Concluding remarks
Antibodies have well documented roles in protecting hosts from bacterial infection, such as enhanced phagocytosis of opsonized bacteria. Other protective roles of antibodies are more unusual. N-3-oxo-dodecanoly homoserine lactone-specific antibodies can moderate QS and expression of pyocyanin, thereby protecting macrophages from pyocyanin-mediated toxicity [44–46]. At the OM, antibodies catalyze oxidation of water to generate reactive oxygen species [47]. Opr86-specific antibodies inhibit biofilm formation without killing the cells [48]. These diverse mechanisms suggest that antibodies have extensive protective capacities that are not fully appreciated. To fill in the blanks, we propose that a comprehensive effort based on recombinant antibody libraries should be undertaken to define the complete set of OM targets expressed by Gram-negative pathogens (Figure 2). Similar strategies could be employed with IE.
Systems biology attempts to integrate multiple, complex and seemingly unrelated parameters to produce a comprehensive network model of how ‘things’ work [49,50]. Robust biological networks (gene regulation in bacteria) can withstand a large number of random alterations in individual components and still maintain their proper function. Systems biologists use data sets from microarray (transcription network) and yeast two-hybrid interaction (protein–protein network) screens to define interactors in the network. Bottlenecks in the network are nodes that receive ‘information’ flow crucial to regulation [51–53]. Incorrect passage of information or interrupted (nonsynchronous) flow can induce noise in the system that is incompatible with efficient energy usage or phenotype transitions [54]. Bottleneck nodes predicted by networks are often virulence factors that have been verified by standard microbiological approaches [51,53]. Thus, a long-term goal is to understand the networks on which OM structures impinge, and how to regulate them to positive effect.
The five criteria we listed, if satisfied, will provide a solid foundation that bacteria do ‘sense’ and ‘respond’ to antibodies and IE that this regulation predictably affects pathogenesis. The challenge then becomes how to advance this perspective to develop therapeutic strategies. Currently, discovery of the protective roles of antibodies needs to be expanded from individual OM of isolated bacteria. The breadth of regulation by different OM-specific Abs needs to be uncovered so bacterial phenotypes can be shaped and exploited. The current trend to antibiotic resistance indicates a need for alternative therapies that might be filled by OM-specific antibodies and IE.
Glossary
- Antibodies
soluble antigen-specific structures (e.g. SIgA) able to access portals of pathogenic bacterial entry
- IE
host-derived molecules such as cytokines, which are produced locally but act systemically, and with the ascribed role of activating and recruiting immune cells. These can include hormones such as dynorphin and catecholamines (e.g. epinephrine), which bind receptors on lymphocytes and activate them
- Proinflammatory cytokine
signaling molecules able to amplify the innate and adaptive immune response. Some are characteristic of cell-mediated immunity (e.g. IFN-γ)
- TLR
host receptors that detect microbial ‘patterns’ including flagella, pili, CpG-modified DNA and LPS
- Porins
proteins found in the outer membrane of Gram-negative bacteria that typically act as size selective pores (~600–800 mw) and that can be immunodominant protective antigens (e.g. OprF). Used to export antibiotics and regulate susceptibility to antimicrobial peptides
- Quorum sensing molecules
soluble, fatty acid-derived molecules secreted by bacteria, and used as a means for cell–cell communication and assessing bacterial population size. These molecules also have pro-inflammatory activity
- Recombinant antibodies libraries (scFv/scFab)
single chain (sc) antigen-specific proteins that represent the repertoire of antigen specificities possible in intact immunoglobulins. These scFv/scFab proteins can be expressed on the surface of bacterial phage, thus allowing selection for exact specificities
References
- 1.Cheesman SE and Guillemin K (2007) We know you are in there: conversing with the indigenous gut microbiota. Res. Microbiol 158, 2–9 [DOI] [PubMed] [Google Scholar]
- 2.Stappenbeck TS et al. (2002) Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. U. S. A 24, 15451–15455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki K et al. (2007) Intestinal IgA synthesis: a primitive form of adaptive immunity that regulates microbial communities in the gut. Semin. Immunol 2, 127–135 [DOI] [PubMed] [Google Scholar]
- 4.Fagarasan S et al. (2002) Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 298, 1424–1427 [DOI] [PubMed] [Google Scholar]
- 5.Suzuki K et al. (2004) Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. U. S. A 101, 1981–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar H (2009) Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun 388, 621–625 [DOI] [PubMed] [Google Scholar]
- 7.Denis M et al. (1991) Interleukin-2 and granulocyte-macrophage colony-stimulating factor stimulate growth of a virulent strain of Escherichia coli. Infect. Immun 59, 1853–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porat R et al. (1991) Enhancement of growth of virulent strains of Escherichia coli by interleukin-1. Science 254, 430–432 [DOI] [PubMed] [Google Scholar]
- 9.Hogan JS et al. (1993) Growth responses of coliform bacteria to recombinant bovine cytokines. J. Dairy Sci 76, 978–982 [DOI] [PubMed] [Google Scholar]
- 10.Luo G et al. (1993) Tumor necrosis factor alpha binding to bacteria: evidence for a high-affinity receptor and alteration of bacterial virulence properties. Infect. Immun 61, 830–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hultgren O (1998) Staphylococcus aureus-induced septic arthritis and septic death is decreased in IL-4-deficient mice: role of IL-4 as promoter for bacterial growth. J. Immunol 160, 5082–5087 [PubMed] [Google Scholar]
- 12.Kanangat S et al. (2001) Enhanced extracellular growth of Staphylococcus aureus in the presence of selected linear peptide fragments of human interleukin (IL)-1beta and IL-1 receptor antagonist. J. Infect. Dis 183, 65–69 [DOI] [PubMed] [Google Scholar]
- 13.Meduri GU (2002) Clinical review: a paradigm shift: the bidirectional effect of inflammation on bacterial growth. Clinical implications for patients with acute respiratory distress syndrome. Crit. Care 6, 24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zav’yalov VP et al. (1995) Specific high affinity binding of human interleukin 1 beta by Caf1A usher protein of Yersinia pestis. FEBS. Lett 371, 65–68 [DOI] [PubMed] [Google Scholar]
- 15.Lyte M (1997) Norepinephrine-induced expression of the K99 pilus adhesin of enterotoxigenic Escherichia coli. Biochem. Biophys. Res. Commun 232, 682–686 [DOI] [PubMed] [Google Scholar]
- 16.Lyte M et al. (1996) Production of Shiga-like toxins by Escherichia coli O157:H7 can be influenced by the neuroendocrine hormone norepinephrine. J. Lab. Clin. Med 128, 392–398 [DOI] [PubMed] [Google Scholar]
- 17.Karavolos MH et al. (2008) Epinephrine modulates the global transcriptional profile of Salmonella revealing a role in the antimicrobial peptide and oxidative stress resistance responses. BMC. Genomics 9, e458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L et al. (2005) Recognition of host immune activation by Pseudomonas aeruginosa. Science 309, 774–777 [DOI] [PubMed] [Google Scholar]
- 19.Sahiratmadja E et al. (2007) Dynamic changes in Pro- and anti-inflammatory cytokine profiles and gamma interferon receptor signaling integrity correlate with tuberculosis disease activity and response to curative treatment. Infect. Immun 75, 820–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke MB et al. (2006) The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U. S. A 103, 10420–10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koebnik R (2005) TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13, 343–347 [DOI] [PubMed] [Google Scholar]
- 22.Hancock RE and Brinkman FS (2002) Function of pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol 56, 17–38 [DOI] [PubMed] [Google Scholar]
- 23.McCarter LL (2001) Polar flagellar motility of the Vibrionaceae. Microbiol. Mo. Boil. Rev 65, 445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang et al. (2009) Global transcriptional response of Salmonella enterica serovar Typhi to anit-z66 antiserum. FEMS Microbiol. Lett 298, 51–55 [DOI] [PubMed] [Google Scholar]
- 25.Zaborina O et al. (2007) Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PloS. Pathog 3, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel NJ et al. (2007) Recognition of intestinal epithelial HIF-1alpha activation by Pseudomonas aeruginosa. Am. J. Physiol. Gastrointest. Liver Physiol 292, G134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Messmer D et al. (2006) Morphine reciprocally regulates IL-10 and IL-12 produciton by monocyte-derived human dendritic cells and enhances T cell activation. Mol. Med 12, 284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan Z and Tam VH (2008) Polymyxin B: a new strategy for multidrug-resistant Gram-negative organisms. Expert Opin. Investig. Drugs 17, 661–668 [DOI] [PubMed] [Google Scholar]
- 29.Gunn JS (2006) the Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16, 284–290 [DOI] [PubMed] [Google Scholar]
- 30.Gooderham WJ et al. (2008) Induction by cationic antimicrobial peptides and involvement in intrinsic polymyxin and antimicrobial peptide resistance, biofilm formation, and swarming motility of PsrA in Pseudomonas aeruginosa. J. Bacteriol 190, 5624–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overhage J et al. (2008) Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun 76, 4176–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q et al. (2005) Sensing wetness: a new role for the bacterial flagellum. EMBO J. 24, 2034–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mashimo T et al. (2007) Temperature-hypersensitivi sites of the flagellar switch component FliG in Salmonella enteric serovar Typhimurium. J. Bacteriol 189, 5153–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson DA et al. (2007) IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2, 328–339 [DOI] [PubMed] [Google Scholar]
- 35.Hall-Stoodley L and Stoodley P (2009) Cell Microbiol. 11, 1034–1043 [DOI] [PubMed] [Google Scholar]
- 36.Hilbi H et al. (2007) Environmental predators as models for bacterial pathogenesis. Environ. Microbiol 9, 563–575 [DOI] [PubMed] [Google Scholar]
- 37.Cosson P and Soldati T (2008) Eat, kill or die: when amoeba meets bacteria. Curr. Opin. Microbiol 11, 271–276 [DOI] [PubMed] [Google Scholar]
- 38.Tan MW et al. (1999) Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U. S. A 96, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alibaud L et al. (2008) Pseudomonas aeruginosa virulence genes identified in a Dictyostelium model. Cell Microbiol. 10, 729–740 [DOI] [PubMed] [Google Scholar]
- 40.Matz C et al. (2008) Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J. 2, 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haskins KA (2008) Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Dev. Cell 15, 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carilla-Latorre S et al. (2008) Dictyostelium transcriptional responses to Pseudomonas aeruginosa: common and specific effects from PA01 and PA14 strains. BMC Microbiol. 8, 109–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veening JW et al. (2008) Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol 62, 193–210 [DOI] [PubMed] [Google Scholar]
- 44.Kaufmann GF et al. (2008) Bacterial quorum sensing: a new target for anti-infective immunotherapy. Expert Opin. Biol. Ther 8, 719–724 [DOI] [PubMed] [Google Scholar]
- 45.Kaufmann GF et al. (2006) Antibody interference with N-acyl homoserine lactone-mediated bacterial quorum sensing. J. Am. Chem. Soc 128, 2802–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaufmann GF et al. (2008) The quorum quenching antibody RS2–1G9 protects macrophages from the cytotoxic effects of the Pseudomonas aeruginosa quorum sensing signalling molecule N-3-oxo-dodecanoyl-homoserine lactone. Mol. Immunol 45, 2710–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nieva J et al. (2006) Immunoglobulins can utilize riboflavin (Vitamin B2) to activate the antibody-catalyzed water oxidation pathway. Immunol. Lett 103, 33–38 [DOI] [PubMed] [Google Scholar]
- 48.Tashiro Y et al. (2008) Opr86 is essential for viability and is a potential candidate for a protective antigen against biofilm formation by Pseudomonas aeruginosa. J. Bacteriol 190, 3969–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma’ayan A (2009) Network integration an graph analysis in mammalian molecular systems biology. IET. Syst. Biol 2, 206–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunlop MJ et al. (2008) Regulatory activity revealed by dynamic correlations in gene expression noise. Nat. Genet 40, 1493–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McDermott JE et al. (2009) Bottlenecks and hubs in inferred networks are important for virulence in Salmonella typhimurium. J. Comput. Biol 16, 169–180 [DOI] [PubMed] [Google Scholar]
- 52.Oltvai ZN and Barabási AL (2002) Systems biology. Life’s complexity pyramid. Science 298, 763–764 [DOI] [PubMed] [Google Scholar]
- 53.Yu H et al. (2007) The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PloS. Comput. Biol 3, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeong H et al. (2001) Lethality and centrality in protein networks. Nature 411, 41–42 [DOI] [PubMed] [Google Scholar]


