Dear Editor,
Chromosome movement in mitosis is orchestrated by a microtubule-based protein machinery called the mitotic spindle (Hyman and Karsenti, 1996; Compton, 2000). The bipolar organization of the mitotic spindle is essential for the accurate segregation of chromosomes into daughter cells. Defects in bipolar spindle assembly can cause chromosome instability and aneuploidy, which are frequently observed in malignant tumors (Silkworth et al., 2009; McGranahan et al., 2012). The nuclear mitotic apparatus (NuMA) is a protein critical for bipolar spindle organization primarily due to its functions in the formation of spindle poles. Dysregulation of NuMA has been reported in a number of cancer types and is associated with cancer development (Bruning-Richardson et al., 2012). Therefore, it is of great significance to understand the molecular mechanisms by which NuMA contributes to spindle pole assembly.
NuMA contains an amino-terminal domain that mediates its interaction with the dynein motor complex, a central coiled-coil domain responsible for its self-assembly, and a carboxyl terminus that mediates its binding to microtubules and cortical proteins (Figure 1A; Compton and Cleveland, 1994; Cleveland, 1995). NuMA is localized in the nucleus in interphase cells and translocated to the spindle pole and cortex during mitosis (Lydersen and Pettijohn, 1980). Cortical NuMA is a fundamental component of the force-generating machinery that regulates spindle orientation (Okumura et al., 2018), whereas spindle pole-localized NuMA plays an important role in focusing microtubules at the poles (Chu et al., 2016). Despite the wealth of information about the consequences to spindles when NuMA function is perturbed, little is known about how NuMA organizes microtubule minus ends at spindle poles.
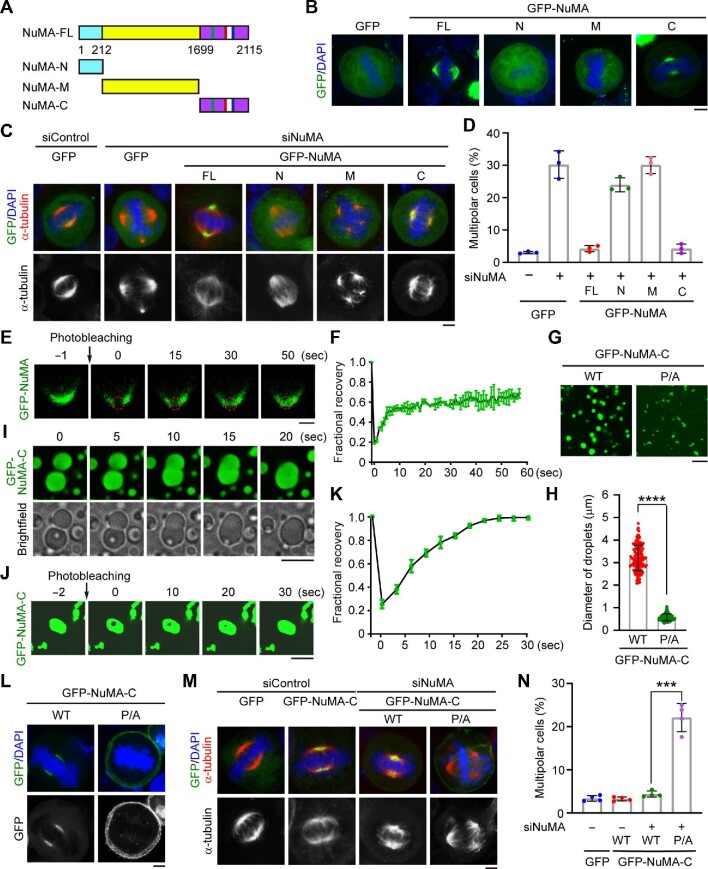
Figure 1.
Phase separation of NuMA promotes spindle pole formation. (A) Schematic representations of full-length (FL) NuMA and truncated mutants. (B) Fluorescence images showing the localization of GFP-tagged full-length NuMA and truncated mutants in HeLa cells. Scale bar, 5 μm. (C and D) Immunofluorescence images of mitotic spindles (C) and quantification of the percentage of mitotic cells with multipolar spindles (D, n = 3 independent experiments) for HeLa cells transfected with the indicated siRNAs and plasmids. For each experiment, 60 mitotic cells were quantified. Scale bar, 5 μm. (E and F) FRAP analysis (E) and quantification (F, n = 2 independent experiments) of GFP-NuMA signals in HeLa cells. Scale bar, 2 μm. (G and H) Representative images (G) and size (H, n = 100 droplets from three independent experiments) of droplets formed by purified GFP-NuMA-C wild type (WT) or proline-to-alanine (P/A) mutant. Scale bar, 10 μm. (I) Time-lapse microscopy showing the fusion of GFP-NuMA-C droplets into a larger droplet. Scale bar, 10 μm. (J and K) FRAP analysis (J) and quantification (K, n = 3 independent experiments) of droplets formed by 10 μM purified GFP-NuMA-C. Scale bar, 5 μm. (L) Fluorescence images showing the localization of GFP-NuMA-C wild type and mutant in metaphase HeLa cells. Scale bar, 5 μm. (M and N) Immunofluorescence images of mitotic spindles (M) and quantification of the percentage of mitotic cells with multipolar spindles (N, n = 4 independent experiments) for HeLa cells transfected as indicated. For each experiment, 60 mitotic cells were quantified. Scale bar, 5 μm. Data are shown as mean ± SD. ***P < 0.001, ****P <0.0001.
To gain mechanistic insight into the role of NuMA in spindle pole formation, we examined its subcellular localization in HeLa cells. Consistent with the previous study (Chu et al., 2016), both endogenous NuMA and exogenous NuMA were found to be densely concentrated in the spindle poles of metaphase cells, and endogenous NuMA also showed robust localization at the cell cortex (Supplementary Figure S1A and B). Depletion of NuMA by specific siRNAs induced a high rate of multipolar spindles (Supplementary Figure S1C‒E). To identify the region responsible for the function of NuMA in spindle pole assembly, we constructed a series of truncated mutants (Figure 1A). Immunofluorescence staining revealed that only the carboxyl terminus of NuMA (NuMA-C) behaved similarly to the full-length protein and was able to rescue the spindle defects caused by NuMA depletion (Figure 1B‒D). Interestingly, time-lapse imaging showed that green fluorescent protein (GFP)-NuMA condensates were gradually fused into the spindle pole in metaphase cells (Supplementary Figure S1B), suggesting that this protein may undergo liquid‒liquid phase separation during mitosis. To examine the mobility of NuMA molecules, we performed fluorescence recovery after photobleaching (FRAP) experiments. We found that the fluorescence signal of GFP-NuMA in the spindle pole was largely recovered at 50 sec after bleaching, indicating that these droplets have liquid-like properties (Figure 1E and F). Taken together, these results suggest that NuMA has a phase-separation property during mitosis, which may be critical for the organization of spindle microtubules.
We then sought to identify the domain of NuMA that mediates its phase separation. Phase separation is typically driven by intrinsically disordered regions (low-complexity regions) and/or multivalent weak interactions, such as electrostatic and hydrophobic interactions (Dunker et al., 2001; Mitrea and Kriwacki, 2016). Therefore, we analyzed the electrical charge, hydrophobicity, and secondary structure of NuMA. We noticed that NuMA-C is intrinsically disordered and contains a number of charged and hydrophobic segments (Supplementary Figure S2A), indicating that this region may mediate its phase separation. To test this possibility, we purified GFP-tagged truncated mutants of NuMA-C and analyzed the phase-separation capacity in a buffer containing 10% polyethylene glycol (PEG)-8000. Fluorescence microscopy revealed that purified GFP-NuMA-C, but not GFP-NuMA-N or GFP-NuMA-M, formed droplets (Figure 1G‒I; Supplementary Figure S2B) and that the droplets exhibited concentration-dependent liquid‒liquid phase separation (Supplementary Figure S3A‒C). In addition, GFP-NuMA-C formed droplets in a PEG-8000 concentration-dependent manner (Supplementary Figure S3D‒F). FRAP analysis further revealed that the fluorescence signal of GFP-NuMA-C was quickly recovered after
bleaching (Figure 1J and K). Collectively, these data demonstrate that NuMA-C undergoes phase separation in vitro.
Next, we investigated whether NuMA-C promotes spindle pole formation through its phase-separation property. We assessed the amino acid composition of NuMA and found that NuMA-C contains a strikingly high percentage of prolines (Supplementary Figure S4A), which are conserved among different species (Supplementary Figure S4B). To explore whether these prolines are required for NuMA-C to undergo phase separation, we constructed two NuMA-C mutants, each containing a set of proline-to-alanine mutations; in the GFP-NuMA-C-P/A-1 mutant, the first 20 prolines were mutated, while in the GFP-NuMA-C-P/A-2 mutant, the last 18 prolines were mutated. In vitro phase-separation assays with purified proteins showed that both mutants reduced the size of GFP-NuMA-C droplets (Supplementary Figure S5A), indicating that both sets of mutations attenuated the ability of NuMA-C to undergo phase separation. To further confirm the role of prolines in regulating the NuMA phase-separation property and function, we constructed another mutant (GFP-NuMA-C-P/A), in which all the 38 prolines in the carboxyl terminus were mutated to alanines. Microtubule co-sedimentation assays showed that the P/A mutations did not disrupt the interaction between GFP-NuMA-C and microtubules (Supplementary Figure S5B). However, the P/A mutant exhibited a significantly weakened phase-separation property (Figure 1G and H). In addition, this mutant was localized at the cell cortex instead of spindle poles (Figure 1L; Supplementary Figure S5C and D). Given that NuMA is recruited to the cell cortex via its interaction with cortical proteins (Zhu et al., 2011; Okumura et al., 2018; Takayanagi et al., 2019), it will be interesting to investigate whether the phase-separation property of NuMA controls its interaction with cortical proteins to promote its localization at the cell cortex. Furthermore, we found that the P/A mutant could not rescue the spindle pole defects caused by NuMA depletion (Figure 1M and N). These results therefore suggest that the conserved prolines in NuMA-C are essential for its phase-separation property and its role in promoting spindle pole formation.
NuMA is known to stimulate the assembly of spindle poles and microtubule asters in dividing cells (Harborth et al., 1999; Nachury et al., 2001; Wiese et al., 2001). However, the underlying molecular mechanisms remain elusive. In this study, our findings suggest that the carboxyl terminus of NuMA triggers its phase separation to form condensates, which in turn promote spindle pole assembly (Supplementary Figure S6). In this scenario, the phase-separation property of NuMA may act as an efficient mechanism to regulate its localization at spindle poles and subsequently accumulate diverse proteins at spindle poles for organizing microtubules. Certainly, alternative mechanisms may exist to regulate the localization and action of NuMA to promote spindle pole assembly. In the future, it will be important to examine whether the dysregulation of NuMA in tumor tissues disrupts its phase-separation property to cause spindle defects. In addition, further studies are warranted to elucidate how NuMA phase separation-mediated spindle pole assembly controls cell division in various physiological processes and cancer development.
We would like to thank Drs Jingyan Fu (China Agricultural University), Haiyang Yu, and Xueliang Zhu (Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences) for critical discussion. This work was supported by grants from the National Natural Science Foundation of China (31730050, 31871347, 32000481, and 32070708) and the National Key R&D Program of China (2017YFA0503502). J.Z. supervised the project. H.M. and J.Z. designed the experiments. H.M., F.Q., L.J., S.X., J.R., and M.L. performed the experiments. H.M., F.Q., and J.G. analyzed data. H.M., F.Q., and J.Z. wrote the manuscript.]
Supplementary Material
Contributor Information
Huixian Ma, Shandong Provincial Key Laboratory of Animal Resistance Biology, Collaborative Innovation Center of Cell Biology in Universities of Shandong, Institute of Biomedical Sciences, College of Life Sciences, Shandong Normal University, Jinan 250014, China.
Feifei Qi, Shandong Provincial Key Laboratory of Animal Resistance Biology, Collaborative Innovation Center of Cell Biology in Universities of Shandong, Institute of Biomedical Sciences, College of Life Sciences, Shandong Normal University, Jinan 250014, China.
Li Ji, Shandong Provincial Key Laboratory of Animal Resistance Biology, Collaborative Innovation Center of Cell Biology in Universities of Shandong, Institute of Biomedical Sciences, College of Life Sciences, Shandong Normal University, Jinan 250014, China.
Songbo Xie, Shandong Provincial Key Laboratory of Animal Resistance Biology, Collaborative Innovation Center of Cell Biology in Universities of Shandong, Institute of Biomedical Sciences, College of Life Sciences, Shandong Normal University, Jinan 250014, China.
Jie Ran, Shandong Provincial Key Laboratory of Animal Resistance Biology, Collaborative Innovation Center of Cell Biology in Universities of Shandong, Institute of Biomedical Sciences, College of Life Sciences, Shandong Normal University, Jinan 250014, China.
Min Liu, Shandong Provincial Key Laboratory of Animal Resistance Biology, Collaborative Innovation Center of Cell Biology in Universities of Shandong, Institute of Biomedical Sciences, College of Life Sciences, Shandong Normal University, Jinan 250014, China.
Jinmin Gao, Shandong Provincial Key Laboratory of Animal Resistance Biology, Collaborative Innovation Center of Cell Biology in Universities of Shandong, Institute of Biomedical Sciences, College of Life Sciences, Shandong Normal University, Jinan 250014, China.
Jun Zhou, Shandong Provincial Key Laboratory of Animal Resistance Biology, Collaborative Innovation Center of Cell Biology in Universities of Shandong, Institute of Biomedical Sciences, College of Life Sciences, Shandong Normal University, Jinan 250014, China; State Key Laboratory of Medicinal Chemical Biology, Department of Genetics and Cell Biology, College of Life Sciences, Nankai University, Tianjin 300071, China.
References
- Bruning-Richardson A., Bond J., Alsiary R.et al. (2012). NuMA overexpression in epithelial ovarian cancer. PLoS One 7, e38945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X., Chen X., Wan Q.et al. (2016). Nuclear mitotic apparatus (NuMA) interacts with and regulates astrin at the mitotic spindle. J. Biol. Chem. 291, 20055–20067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W. (1995). NuMA: a protein involved in nuclear structure, spindle assembly, and nuclear re-formation. Trends Cell Biol. 5, 60–64. [DOI] [PubMed] [Google Scholar]
- Compton D.A. (2000). Spindle assembly in animal cells. Annu. Rev. Biochem. 69, 95–114. [DOI] [PubMed] [Google Scholar]
- Compton D.A., Cleveland D.W. (1994). NuMA, a nuclear protein involved in mitosis and nuclear reformation. Curr. Opin. Cell Biol. 6, 343–346. [DOI] [PubMed] [Google Scholar]
- Dunker A.K., Lawson J.D., Brown C.J.et al. (2001). Intrinsically disordered protein. J. Mol. Graph. Model. 19, 26–59. [DOI] [PubMed] [Google Scholar]
- Harborth J., Wang J., Gueth-Hallonet C.et al. (1999). Self assembly of NuMA: multiarm oligomers as structural units of a nuclear lattice. EMBO J. 18, 1689–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A.A., Karsenti E. (1996). Morphogenetic properties of microtubules and mitotic spindle assembly. Cell 84, 401–410. [DOI] [PubMed] [Google Scholar]
- Lydersen B.K., Pettijohn D.E. (1980). Human-specific nuclear protein that associates with the polar region of the mitotic apparatus: distribution in a human/hamster hybrid cell. Cell 22, 489–499. [DOI] [PubMed] [Google Scholar]
- McGranahan N., Burrell R.A., Endesfelder D.et al. (2012). Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 13, 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea D.M., Kriwacki R.W. (2016). Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 14, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M.V., Maresca T.J., Salmon W.C.et al. (2001). Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104, 95–106. [DOI] [PubMed] [Google Scholar]
- Okumura M., Natsume T., Kanemaki M.T.et al. (2018). Dynein‒dynactin‒NuMA clusters generate cortical spindle-pulling forces as a multi-arm ensemble. eLife 7, e36559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth W.T., Nardi I.K., Scholl L.M.et al. (2009). Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One 4, e6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi H., Hayase J., Kamakura S.et al. (2019). Intramolecular interaction in LGN, an adaptor protein that regulates mitotic spindle orientation. J. Biol. Chem. 294, 19655–19666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C., Wilde A., Moore M.S.et al. (2001). Role of importin-β in coupling Ran to downstream targets in microtubule assembly. Science 291, 653–656. [DOI] [PubMed] [Google Scholar]
- Zhu J., Wen W., Zheng Z.et al. (2011). LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and Gαi/LGN/NuMA pathways. Mol. Cell 43, 418–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.