Abstract
Three-dimensional (3D) bioprinting has emerged as an enabling tool for various biomedical applications, such as tissue regeneration and tissue model engineering. To this end, the development of bioinks with multiple functions plays a crucial role in the applications of 3D bioprinting technologies. In this study, we propose a new bioink based on two immiscible aqueous phases of gelatin methacryloyl (GelMA) and dextran, further endowed with anti-bacterial and anti-inflammatory properties. This micropore-forming GelMA-dextran (PGelDex) bioink exhibited excellent printability with vat-polymerization, extrusion, and handheld bioprinting methods. The porous structure was confirmed after bioprinting, which promoted the spreading of the encapsulated cells, exhibiting the exceptional cytocompatibility of this bioink formulation. To extend the applications of such a micropore-forming bioink, interleukin-4 (IL-4)-loaded silver-coated gold-nanorods (AgGNRs) and human mesenchymal stem cells (MSCs) were simultaneously incorporated, to display synergistic anti-infection behavior and immunomodulatory function. The results revealed the anti-bacterial properties of the AgGNR-loaded PGelDex bioink for both Gram-negative and Gram-positive bacteria. The data also indicated that the presence of IL-4 and MSCs facilitated macrophage M2-phenotype differentiation, suggesting the potential anti-inflammatory feature of the bioink. Overall, this unique anti-bacterial and immunomodulatory micropore-forming bioink offers an effective strategy for the inhibition of bacterial-induced infections as well as the ability of immune-regulation, which is a promising candidate for broadened tissue bioprinting applications.
Keywords: biofabrication, bioprinting, micropore-forming bioink, aqueous two-phase emulsion, anti-bacterial, immunomodulation
1. Introduction
As a form of additive manufacturing, three-dimensional (3D) printing has been used for various applications, such as the production of medical devices and implants, and as an intraoperative tool for wound-healing purposes [1–3]. Nevertheless, bacterial infection and inflammation around printed implants or dressing, remain challenging problems and are difficult avoid. There is no denying that bacterial adhesion and biofilm formation result in severe infection, followed by inflammation, leading to the failure of implant functions or wound closure [4]. A number of high-failure-profile cases have been reported where bacterial-related infection in orthopedics, including 3D-printed implants, is more than 3% [5, 6]. More importantly, excessive inflammation induced by infection delays the transition of the regeneration process and could lead to rejection of the implanted devices and scaffolding constructs [7, 8]. Biomaterial development featuring dual-functional anti-bacterial and anti-inflammatory performances can be a potential option to overcome this outstanding obstacle.
Nanoparticles have been used as an effective platform for both antimicrobial purposes and as bioagent carriers, due to their unique physical and chemical properties, including high specific surface areas and the ability of sustained release [9]. Silver nanoparticles (AgNPs) have received widespread attention for potential use as an antimicrobial agent [10]. The mechanism of the inhibitory characteristic of AgNPs against microbes has been extensively investigated [11]. The silver ion (Ag+) plays a crucial role in its antimicrobial activity by direct interaction with cellular components, including protein, DNA, and membranes. Moreover, AgNPs also can destroy the cell membranes with the association of “pit” formation in the cell walls of bacteria, leading to cell death. Much work so far has been done to immobilize AgNPs on implant surfaces or conjugated with injectable hydrogels for anti-bacterial applications [12–14]. Our previous study demonstrated a type of novel silver-coated gold-nanorods (AgGNRs), where the silver antimicrobial effect was augmented by its dumbbell-shape because of the further increased specific surface ratio and the sharp edges for weakening membrane integrity, comparing to conventional nanoparticle structures [15]. The unique shape of our AgGNRs also made them accessible from all dimensions, leading to large contact areas when interacting with bacterial membranes.
Inflammation is one of the responses of the immune system to bacterial infections, where macrophages are involved to defense against pathogens [16]. It is well-acknowledged that macrophages are classified into classically activated (M1) and alternatively activated (M2) populations based on surface receptors and inflammatory factor secretions [7, 8]. The M1 macrophages have potent anti-microbial activities, while the M2 macrophages pave the way for the resolution of the inflammation by phagocytosis, scavenging debris and apoptotic cells, and promoting tissue repair. The two types of macrophages can functionally reversibly change responding to cytokine environment and appropriate stimuli. This fact has been demonstrated in several studies where interleukin-4 (IL-4), an effective type-2 cytokine, could mediate the transformation of macrophages from the pro-inflammatory M1 phenotype to the anti-inflammatory M2 phenotype to achieve immunomodulation [17, 18]. Therapy using IL-4-loaded scaffolds was suggested to relieve the negative influence of inflammation by M1 macrophage on murine chondrocytes, which further exhibited enhanced regeneration of both subchondral bone and cartilage compared with scaffolds without IL-4 [19]. Other studies have alternatively suggested mesenchymal stem cells (MSCs) as an immunomodulator to treat inflammatory and immune disorders [20]. The influence of MSCs on both innate and adaptive immunity was shown to be through suppressing the activation and proliferation of immune cells, such as monocytes and macrophages [21]. The paracrine effects of released bioactive molecules, especially interleukins, are considered as a primary mechanism of MSCs immunomodulation [22].
On the other hand, the 3D bioprinting technology has emerged as a versatile tool to obtain reproducible tissue-mimicking functional 3D architectures through automated operations [23, 24]. Recent developments of a variety of bioprinting methods, such as extrusion bioprinting, inkjet bioprinting, and digital light processing (DLP)-based bioprinting, have attracted increasing attention [25, 26]. Particular emphasis is placed on engineering functional bioinks that consist of cells and bioprintable materials used in controllable tissue fabrication [27]. Therefore, the ideal bioink formulation should satisfy both printability and cytocompatibility requirements. Key features promoting cytocompatibility of a bioink include porosity for the diffusion of nutrients and oxygen, as well as correct biochemical cues of the biomaterial components to facilitate cell survival, adhesion, proliferation, and functions [28, 29]. Our previous studies demonstrated a unique micropore-forming bioink for enhanced bioprinting applications based on using an aqueous two-phase emulsion of gelatin methacryloyl (GelMA) as the continuous phase and poly(ethylene oxide) (PEO) droplets as the porogen [30–32]. We have shown that our micropore-forming GelMA-PEO bioink was superior in terms of cellular behaviors when compared with those in the absence of micropores. We hence hypothesized that this class of micropore-forming bioink serves as a powerful platform for cell-based bioprinting and is broadly applicable to a variety of bioprinting methods.
Leveraging this prior knowledge, the current study attempts to develop a dual-functional GelMA-dextran aqueous two-phase emulsion bioink, simultaneously formulated with IL-4-loaded AgGNRs and MSCs, for improved biomedical utilities (Figure 1). The printability performance of the micropore-forming GelMA-dextran (PGelDex) bioink was assessed in three bioprinting modalities, i.e., DLP, extrusion, and handheld bioprinting. The antimicrobial properties of the AgGNR-embedded micropore-forming bioink were further evaluated on both Gram-positive and Gram-negative bacterial species. We finally showed the possibility to redefine the anti-inflammatory microenvironment through both MSCs and IL-4 release to direct macrophage-polarization, ultimately allowing for an advanced micropore-forming bioink platform towards anti-infection and immunomodulation, enabling a broad range of relevant applications.
Figure 1.
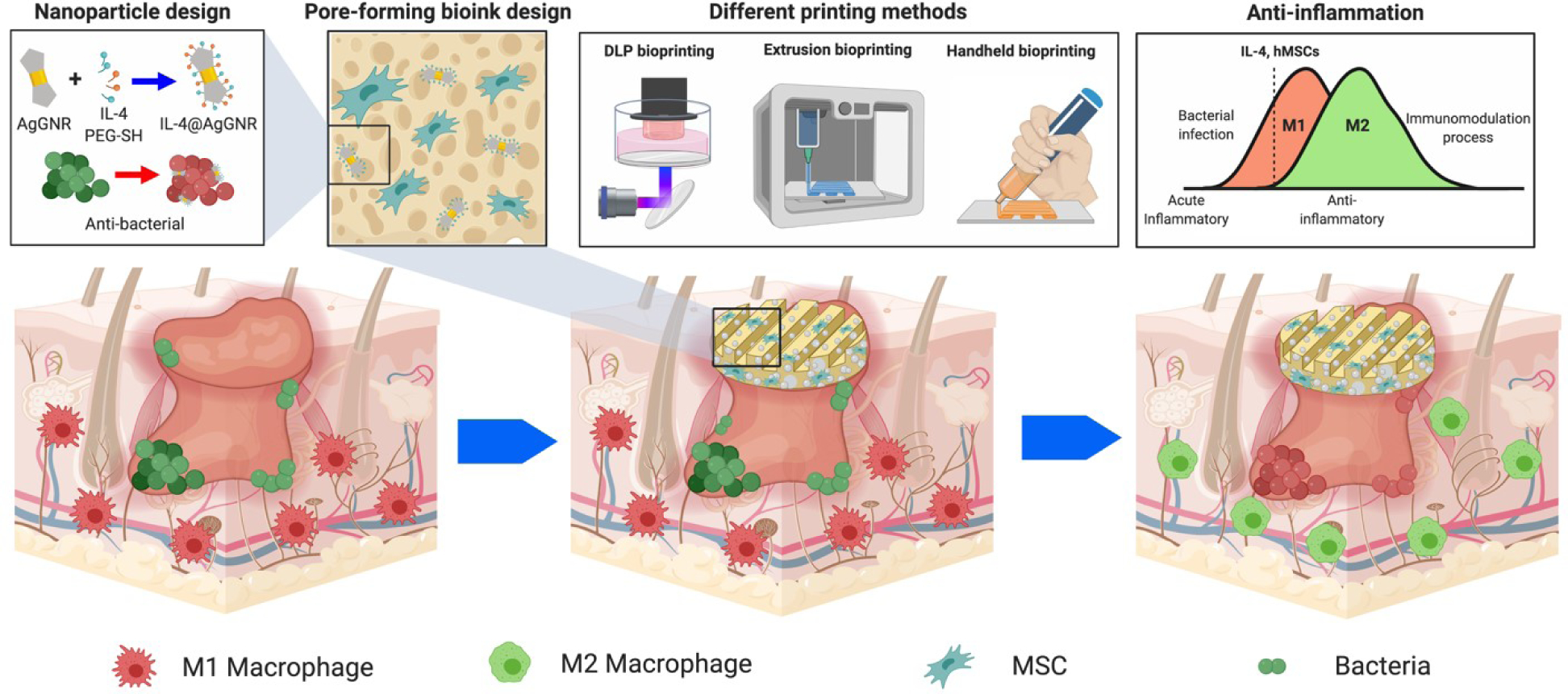
Schematics showing the formulation of the multi-functional two-phase aqueous emulsion PGelDex bioinks, the bioprinting processes, as well as their anti-bacterial and anti-inflammatory applications. Figure drawn with BioRender.
2. Materials and Methods
2.1. Synthesis of GelMA
The synthesis of GelMA was performed based on our previously reported method [33, 34].First, 10.0 g of porcine gelatin (Sigma-Aldrich, USA) was dissolved in 100 mL of phosphate-buffered saline (PBS, Sigma-Aldrich) solution with heated stirring (50 °C), and then 5 mL of methacrylic anhydride (Sigma-Aldrich) was added dropwise with an extra 2 hours (h) of reaction. This reaction was stopped by the addition of 100 mL of warm PBS for 10 minutes (min). The final solution was dialyzed for up to 5 days using 12–14-kDa dialysis tubes (Spectrum Laboratories, USA) at 40 °C, followed by lyophilization with a freeze-dryer (Labconco, USA). The GelMA derived from porcine was termed pGelMA. By contrast, the GelMA derived from gelatin of cold-water fish skin (Sigma-Aldrich), termed as fGelMA, was synthesized using the same process, except that the addition amount of methacrylic anhydride was 8 mL [35].
2.2. Preparation of micropore-forming bioink
The formation of the aqueous two-phase emulsion bioink was conducted following the modified method as we previously described [30–32]. The 20 wt.% fGelMA or pGelMA solution was obtained by dissolving lyophilized fGelMA or pGelMA in PBS under constant stirring at room temperature or 37 °C, respectively. 10 wt.% dextran (molecular weight, Mw = 2,000 kDa, Aladdin, China) solution was also prepared in PBS. Then, the micropore-forming bioink was formulated by mixing the 10 wt.% dextran solution with pGelMA or fGelMA to reach the final concentration at 0.5%, 1.5%, or 3.0 wt.% through vigorous vortexing for 10 seconds (s). The size distribution of dextran droplets in GelMA solution was quantified immediately after emulsion-formation by the ImageJ software (National Institutes of Health, USA). The pGelMA-based micropore-forming bioink was designed for extrusion and handheld bioprinting, while the fGelMA-based one was intended for DLP bioprinting. Scanning electron microscopy (SEM, JSM-5600LV, JEOL, Japan) was adopted to visualize the porous microstructures of the micropore-forming bioinks. The bioinks formulated by different concentrations of dextran were lyophilized and further coated with 8-nm gold layer before SEM imaging.
2.3. Bioprinting
Bioinks used for DLP bioprinting were prepared with fish-derived micropore-forming GelMA-dextran (PGelDex) bioinks; 15 wt.% fGelMA, different concentrations of dextran solution, photoinitiator tris(2,2-bipyridyl)dichlororuthenium (II) hexahydrate/sodium persulfate (Ru/SPS, 2 mM/20 mM, Advanced Biomatrix, USA), and photoabsorber Ponceau 4R (2 wt.%, Sigma-Aldrich) were mixed and vortexed to achieve the final formulations. An in-house-built DLP bioprinter was used to perform bioprinting with micropore-forming bioinks [36–38]. For planar constructs, an oxygen plasma-cleaned glass slide with a layer of the micropore-forming bioink was exposed under visible light for 15 s. For 3D structures, cube, pyramid, and gyroid samples were bioprinted with a 300-μm thickness of each layer under 15-s exposure. After bioprinting, the samples were washed with PBS to remove the uncrosslinked bioinks.
A commercial extrusion bioprinter (Allevi 2, 3D Systems, USA) was used to conduct extrusion bioprinting. The prepared pGelMA-based PGelDex bioink was first loaded into the syringe and cooled down at 4°C to achieve proper viscosity for extrusion. All constructs were bioprinted at 300 mm min−1 and 38 pounds per square inch (psi), and then post-crosslinked via ultraviolet (UV) exposure (10 mW cm−2, 360–480 nm, Omini S2000, USA) for 30 s.
Handheld bioprinting was performed using an in-house-built handheld bioprinter (Figure S1), as we previously reported [30], where the open-source hardware and software packages are available and accessible. The speed of extrusion was adjustable to suit the hand-moving speed. In addition, external UV light was employed to enable post-crosslinking of the extruded patterns. The bioink formulation was the same as that for extrusion bioprinting.
The bioinks were mixed with a color dye (Createx, USA) to aid visualization when necessary. All these three bioprinting processes were proven to minimally influence cell viability when operated with proper parameters, as we previously showed [39–41].
2.4. AgGNR synthesis and characterizations
The synthesis of AgGNRs was conducted in three steps using the method that was previously reported by us [15]. Briefly, the seed solution was obtained by mixing 5 mL of 0.2-M cetyltrimethylammonium bromide (CTAB, Sigma-Aldrich) aqueous solution and 5 mL of 0.0005-M chloroauric acid (HAuCl4, Sigma-Aldrich) aqueous solution, following by the addition of 0.6 mL of 0.01-M sodium borohydride (NaBH4, Sigma-Aldrich) with stirring for 2 min. The growth solution was prepared by mixing 5 mL of 0.2-M CTAB, 0.3 mL of 0.004-M silver nitrate (AgNO3, Sigma-Aldrich) aqueous solution, 5 mL of 0.001-M HAuCl4 aqueous solution, and 70 μL of 0.0788-M ascorbic acid (Sigma-Aldrich). Then, 12 μL of the seed solution prepared in the first step was added to this growth solution. After reaction for 15 min, the solution was further centrifuged to remove unreacted reagents and resuspended. The third step was conducted by adding 1 mL of 0.001-M HAuCl4 aqueous solution, 0.75 mL of 0.004-M AgNO3 aqueous solution, and 25.38 μL of 0.0788-M ascorbic acid into the redispersed growth solution generated from the second step. The mixture solution was gently vortexed and kept at 37 °C for 30 min. The obtained solution was then centrifuged to remove unreacted reagents and redispersed in PBS. Further characterizations, including those on the shape and aspect ratios (the ratio of the length to the width), were carried out by transmission electron microscopy (TEM, JEOL) and energy-dispersive X-ray spectroscopy (EDX, JEOL) at 200 kV.
2.5. Surface-modification and IL-4-encapsulation
We next loaded recombinant human IL-4 (Peprotech, USA) to polyethylene glycol (PEG)-stabilized AgGNRs with previously described methods [42, 43]. To prevent aggregation and also load IL-4 to AgGNRs, a layer of PEG was conjugated to the AgGNR surfaces. We dissolved HS-PEG-COOH (Mw = 3 kDa, Sigma-Aldrich) was dissolved into PBS at the concentration of 1 mg mL−1, and IL-4 was dissolved at the concentration of 10 ng mL−1. Then, 50 μL of HS-PEG-COOH solution and 10 μL of IL-4 solution were added into 1-mL AgGNR suspension attained from the previous step. The mixture solution was left under stirring for 4 h under dark to ensure the conjugation of both PEG layer and IL-4 to the AgGNRs through thiol-gold (Au) and thiol-silver (Ag) bonds as well as electrostatic interactions. The obtained suspension was centrifuged to remove unconjugated reagents and redispersed into PBS. The surface charge of the obtained solution was analyzed using dynamic light scattering (Malvern Zetasizer, UK). The silver concentration, as the only antimicrobial functional ion, was determined by inductively coupled plasma mass spectrometry (PerkinElmer NexION, USA). The concentrations of AgGNRs used for the anti-bacterial and anti-inflammation studies were degreed into three levels, high (10 μg mL−1, termed as AgGNR-H), medium (5 μg mL−1, AgGNR-M), and low (2 μg mL−1, AgGNR-L).
2.6. Bioprinting with particle-loaded bioinks
The porous microstructures of AgGNR-loaded fish-derived PGelDex bioink and porcine-derived PGelDex bioink were visually evidenced by a fluorescence microscope (Eclipse, Nikon, Japan). Briefly, a droplet of the bioink was placed on a glass slide capped by a cover glass and observed at room temperature. The distribution of dextran droplet diameters was quantified by the ImageJ software. Then, DLP and extrusion bioprinting were used to analyze the printability of the AgGNR-loaded micropore-forming bioinks. Two-dimensional (2D) constructs were bioprinted with micropore-forming bioinks with and without AgGNRs using the same methods as described in Section 2.3. All bioprinted constructs were visualized under a microscope to confirm the presence of porous microstructures after bioprinting.
2.7. In vitro evaluations of anti-bacterial activities
The anti-bacterial properties of the micropore-forming bioinks with different AgGNR concentrations were evaluated by Gram-positive Staphylococcus aureus (S. aureus) and Gram-negative Escherichia coli (E. coli). Both bacteria were cultured in tryptic soy broth (TSB, Sigma-Aldrich) or on a TSB agar plate. The bioprinted samples formulated by GelMA and AgGNR-encapsulated PGelDex bioinks with different concentrations. To determine the attachment of the bacteria on the proposed bioinks, the proposed samples were incubated with 2 mL of bacteria suspension at the concentration of 1 × 108 colony-forming units (CFUs) mL−1. All samples in bacterial suspension-containing media were placed at 37 °C for 24 h. At the end of the incubation time, all bioprinted samples were gently rinsed with PBS to remove unattached bacteria. Each sample was transferred to 1-mL PBS and bacteria were detached with ultrasonic vibration (10 W, Branson Ultrasonics, USA) for 5 min. The bacterial solution was then serially diluted by 10-fold steps with sterile physiological saline. 10 μL of the diluted bacteria suspension was taken out and inoculated on the agar plates. After incubation for 24 h under 37 °C, bacterial colonies were counted and photographed by a digital single-lens reflex camera (Canon, Japan).
2.8. Evaluating the cytocompatibility with encapsulated MSCs and human umbilical vein endothelial cells (HUVECs)
Human MSCs (Lonza, USA) were cultured in the MSC growth medium supplemented with BulletKit (Lonza). The GFP-labeled HUVECs (Angio-proteomie, USA) were cultured in human endothelial cell growth medium supplemented with BulletKit EGM-2 (Lonza). The cells were trypsinized and mixed with micropore-forming bioinks at the density of 3 × 106 cell mL−1 for subsequence bioprinting. Cytocompatibility analyses were performed by bioprinting MSCs or GFP-labeled HUVECs with micropore-forming bioinks of different formulations. All bioprinting processes were carried out under an aseptic condition with the DLP bioprinting method as described in Section 2.3. The bioprinted samples were washed with PBS to remove uncrosslinked bioink, and then cultured in a 5 vol.% CO2 incubator (Forma, USA) at 37 °C. After 1, 3, and 5 days or 1 day of culturing MSCs or GFP-labeled HUVECs, respectively, the viability values of the cells were measured by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS, Promega, USA) assay. All the bioprinted samples were incubated with the mixed assay solution in each well of a 48-well plate for 4 h under dark in the incubator. The absorbance at 490 nm was determined with a spectrophotometer (Molecular Devices, USA).
Additionally, the MSC samples were collected on day 1, 3, and 5 days after bioprinting and were stained with Alexa Fluor-488 phalloidin (Thermo Fisher, USA) overnight at 4 °C for F-actin and 4’,6-diamidino-2-phenylindole (DAPI, Vector Laboratories, USA) for nucleus observation. The samples were then visualized using confocal laser scanning microscopy (LSM880, Zeiss, Germany). The morphologies of the bioprinted GFP-labeled HUVECs were directly observed at 1 day after culture with a fluorescence microscope.
2.9. In vitro assessments of anti-inflammation performances
THP-1 monocyte-like cells (American Type Culture Collection, USA) were cultured in RPMI 1640 (Sigma-Aldrich) supplemented with 10 wt.% fetal bovine serum (ThermoFisher), 1% penicillin/streptomycin (ThermoFisher). 24-transwell plates (Sigma-Aldrich) and cell culture inserts (Sigma-Aldrich) were used to evaluate the immunomodulation of MSC-encapsulated and IL-4-loaded bioinks on macrophages. In this case, 1 × 106 cell mL−1 of THP-1 cells were cultured in the lower chamber, and co-cultured with 50 ng mL−1 of phorbol-12-myristate-13-acetate (PMA, Sigma-Aldrich) in a complete medium for 24 h [44–46]. M0-differentiated THP-1 cells were washed twice with PBS. Then, cells were treated with 10 ng mL−1 of lipopolysaccharides (LPS, Sigma-Aldrich) and 20 ng mL−1 of interferon-γ (IFN-γ, Sigma-Aldrich) for the following 24 h to obtain M1 macrophages. Scaffolds used for this study were prepared with fGelMA-derived PGelDex bioinks loaded with 1 × 106 cell mL−1 of MSCs and AgGNR-M. The bioinks consisted of 15 wt.% fGelMA, different concentrations of dextran solution, photoinitiator Ru/SPS (2 mM/20 mM), and photoabsorber Ponceau 4R (2 wt.%). The samples were bioprinted with 300-μm thickness of each layer under 15-s exposure. After bioprinting, the samples were washed with PBS to remove uncrosslinked bioinks and transferred into the upper chambers of the wells. After co-culturing for 12 h, the total mRNA of THP-1 cells was collected and used for the following analyses.
2.10. Quantitative real-time polymerase chain reaction (qRT-PCR) analyses
Total RNA of each sample was isolated using the TRIzol reagent (Invitrogen, USA) and purified following the manufacturer’s instruction. The concentration and purity were determined using NanoDrop 1000 (ThermoFisher). Reverse-transcription was carried out using a SuperScript VILO cDNA synthesis kit (ThermoFisher). We performed qRT-PCR with SYBR Green Master Mix kit (ThermoFisher) by QuantStudio 5 Real-Time PCR instrument (Applied Biosystems, USA). The human ribosomal protein L13a (RPL13A) and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes were used as the housekeeping genes for internal control to normalize the quantities of the target genes of MSCs and THP-1, respectively. GAPDH was selected as another optimal reference gene for qRT-PCR studies on human MSCs [47], while β-actin (ACTB) was utilized for THP-1 [48]. A series of targeted genes correlated with immunomodulatory functions of MSCs, including oxygenase (HMOX), cyclooxygenase-2 (COX-2), transforming growth factor-β (TGF-β), indolamine-2,3-dioxygenase (IDO), hepatocyte growth factor (HGF), TNF-stimulated gene-6 (TSG-6), C–C motif chemokine ligand-2 (CCL-2), and colony stimulating factor-1 (CSF-1), were evaluated. The sequences for both forward primers and reverse primers are listed in Table S1. In addition, the anti-inflammatory-related genes of THP-1, including tumor necrosis factor-α (TNF-α), IL-6 (IL-6), IL-1β (IL-1β), inducible nitric oxide synthase (iNOS), IL-10 (IL-10), mannose receptor (CD206), C-C motif chemokine ligand-22 (CCL22), and arginase 1(Arg-1), were also analyzed with the same method. Sequences of these forward primers and reverse primers are listed in Table S2.
2.11. Evaluating the hemocompatibility of functional porous bioinks
Hemocompatibility of the bioprinted constructs of differently formulated bioinks was evaluated by red blood cells (RBCs, Research Blood Components, USA) as described before [49]. The bioprinted constructs were co-cultured with 5 vol.% RBC suspensions for 1 h under standard incubation. The RBCs in PBS and 0.1 vol.% Triton X-100 solution (Sigma-Aldrich) were used as the negative and positive controls, respectively. After incubation, the bioprinted constructs with RBC suspension were centrifuged at 1,000 rpm for 10 min, and then 100 μL of the supernatant from each sample was transferred to the well of a 96-well plate. The solution was quantified by measuring absorbance at 540 nm. The hemolysis rate could be calculated via the following equation:
where Ap is the absorbance value in the experimental group, At is the absorbance of the Triton group, and Ab is the absorbance of the PBS group.
2.11. Statistical analyses
All the data are presented as means ± standard deviations (SDs) of the measurements in each group. The statistical analyses were performed using the GraphPad Prism 6.0 software (USA). Comparisons among the groups were analyzed by using a one-way analysis of variance (ANOVA) with Tukey’s multiple comparison method.
3. Results
3.1. Micropore-forming bioink formulations and characterizations
The fish-derived PGelDex bioinks were prepared by mixing 15 wt.% fGelMA solution with different concentrations of dextran. The micropore morphology of bioinks with different dextran concentrations was observed via optical microscopy, where the size of dextran droplets in the PGelDex emulsion was larger when the concentration of dextran was increased (Figure 2A). A similar pattern of results was obtained in porcine-derived PGelDex emulsion bioinks. Together, the findings confirmed that using dextran as porogen successfully generated the microporous microstructures within the GelMA-based bioinks. The size distribution of dextran droplets in the GelMA solution was quantified immediately after emulsion-formation. A narrow distribution of 7.5 ± 2.8 μm was observed at the concentration of 0.5 wt.% dextran (Figure 2B). The average sizes raised to 18.4 ± 5.9 μm and 39.0 ± 9.4 μm, respectively, and the size distributions of droplets became broader when the concentration of dextran was increased to 1.5 wt.% and 3.0 wt.%. For the porcine-derived PGelDex emulsion bioinks, the droplet sizes of 0.5, 1.5, and 3.0 wt.% dextran concentrations were increased from 5.4 ± 1.1 μm to 15.3 ± 5.8 μm and 21.5 ± 7.0 μm, respectively. As shown in Figure 2C, the typical microporous structures of fish-derived PGelDex emulsion bioinks were characterized by SEM, indicating good agreement with the observed pore morphologies from optical images. In addition, such an interconnected microporous structure was further confirmed by the confocal fluorescence image using 3.0 wt.% dextran in 15 wt.% fGelMA (Figure 2D).
Figure 2.
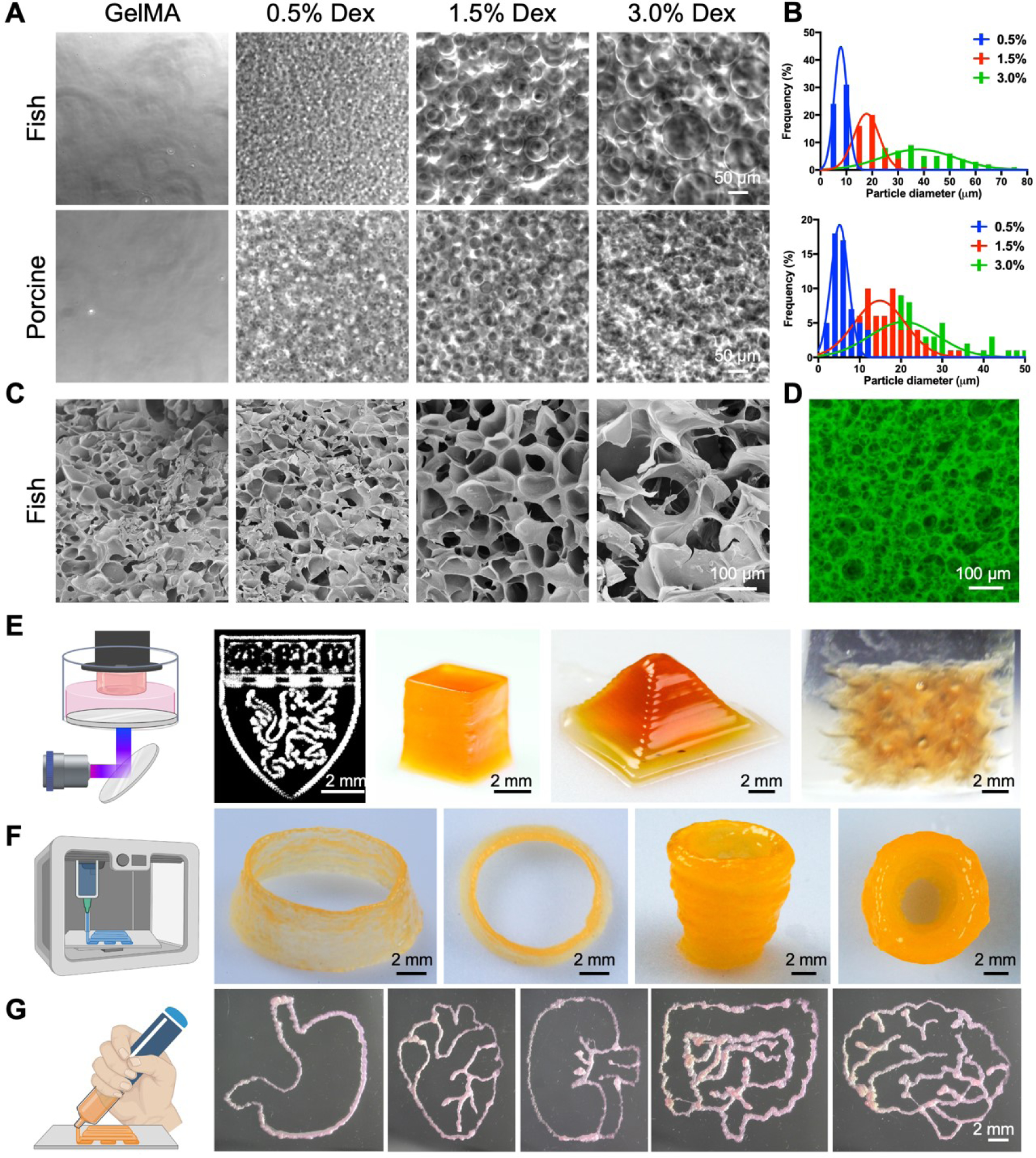
Characterizations and bioprinting of the PGelDex bioinks. (A) Optical micrographs showing the fish- and porcine-derived PGelDex bioinks containing different dextran concentrations (0.5, 1.5, and 3.0 wt.%). (B) Quantification data showing the size distributions of the dextran emulsion droplets of the PGelDex bioinks at different dextran concentrations (0.5, 1.5, and 3.0 wt.%). (C) SEM images showing the interconnected porous structures at dextran concentrations of 0.5, 1.5, and 3.0 wt.%. (D) Confocal fluorescence micrograph showing the interconnected porous structures in the construct with 3.0 wt.% of dextran. (E-G) 2D and 3D constructs fabricated with (E) DLP bioprinting, (F) extrusion bioprinting, and (G) handheld bioprinting using the PGelDex bioinks. Part of the figure drawn with BioRender.
3.2. 3D bioprinting
The printing performances of the PGelDex bioinks were evaluated via DLP, extrusion, and handheld bioprinting methods. To investigate 3D bioprinting of the micropore-forming bioinks, an in-house-built DLP bioprinter was first introduced to print with a layer-by-layer photocrosslinking mechanism [36–38]. fGelMA-based bioink was used for DLP bioprinting due to its lower gelation temperature and thus the liquid state under room temperature, which is more suitable for vat-polymerization [50]. 15 wt.% fGelMA, 3.0 wt.% dextran, and 2-mM/20-mM Ru/SPS were used to produce the PGelDex bioink for DLP to fabricate 2D and 3D structures (Figure 2E). A planar pattern of the logo of Harvard University was achieved with a 15-s photocrosslinking. Moreover, we also successfully produced geometrically complex 3D constructs, such as a cube, a pyramid, and a gyroid.
The porcine-derived PGelDex bioink (15 wt.% pGelMA, 3.0 wt.% dextran, 0.2 wt.% lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP)) was further investigated using extrusion bioprinting. The micropore-forming bioink was prepared in the same way as described in Section 2.3, followed by cooling at 4°C and extruded at room temperature with post-crosslinking under UV exposure. As shown in Figure 2F, a multi-layered cylindrical tubular construct with a height of 4.5 mm and a diameter of 12 mm exhibited excellent structural integrity. A cylindrical cup presented a higher height and similar structural integrity.
We subsequently assessed the printability of our micropore-forming bioink using a handheld bioprinter. The handheld bioprinter used in this study was reported previously by us, which was built in-house with an ergonomic and portable design, providing convenient operational freedom using a single hand [30]. Upon loading the same porcine-derived PGelDex bioink as in extrusion bioprinting, we successfully hand-plotted a series of arbitrary patterns, including those emulating the stomach, the heart, the kidney, the intestine, and the brain, with simultaneous in situ UV-crosslinking (Figure 2G). The uniformity of those patterns displayed the consistency of bioprinting obtained by this handheld bioprinter. Overall, the developed micropore-forming bioinks were confirmed for their versatility in applications in multiple bioprinting modalities.
3.3. Characterizations of AgGNRs and bioprinting with AgGNR-loaded bioinks
The stability of the PEGylated, IL-4-conjugated Au particles has been previously assessed [42]. The conjugation between IL-4 and AgGNRs likely occurred through thiol-Au and thiol-Ag bonds as well as electrostatic interactions. The morphologies of the as-synthesized AgGNRs and AgGNRs surface-modified with IL-4 (IL-4@AgGNRs) were characterized using TEM. As shown in Figure 3A, both AgGNRs and IL-4@AgGNRs could be dispersed well. In addition, the morphological analyses of AgGNRs and IL-4@AgGNRs confirmed their dumbbell-like shape with two sharp tips on both ends. Statistical analyses of the size distributions were conducted via measuring the lengths, widths, and aspect ratios of randomly selected AgGNRs and IL-4@AgGNRs from TEM images. The results indicated that the coating of AgGNRs did not significantly influence their aspect ratios. We further applied EDX on the AgGNRs for elemental analyses, which described that both gold and silver elements were observed on the surfaces of the AgGNRs (Figure S2), suggesting the successful deposition of silver elements onto the gold nanorods. As revealed by Figure S3, compared to the positive-charged AgGNRs, the zeta potential of the IL-4@AgGNRs was close to neutral, which was due to the successful conjugation of PEG and IL-4 on the AgGNR surfaces.
Figure 3.
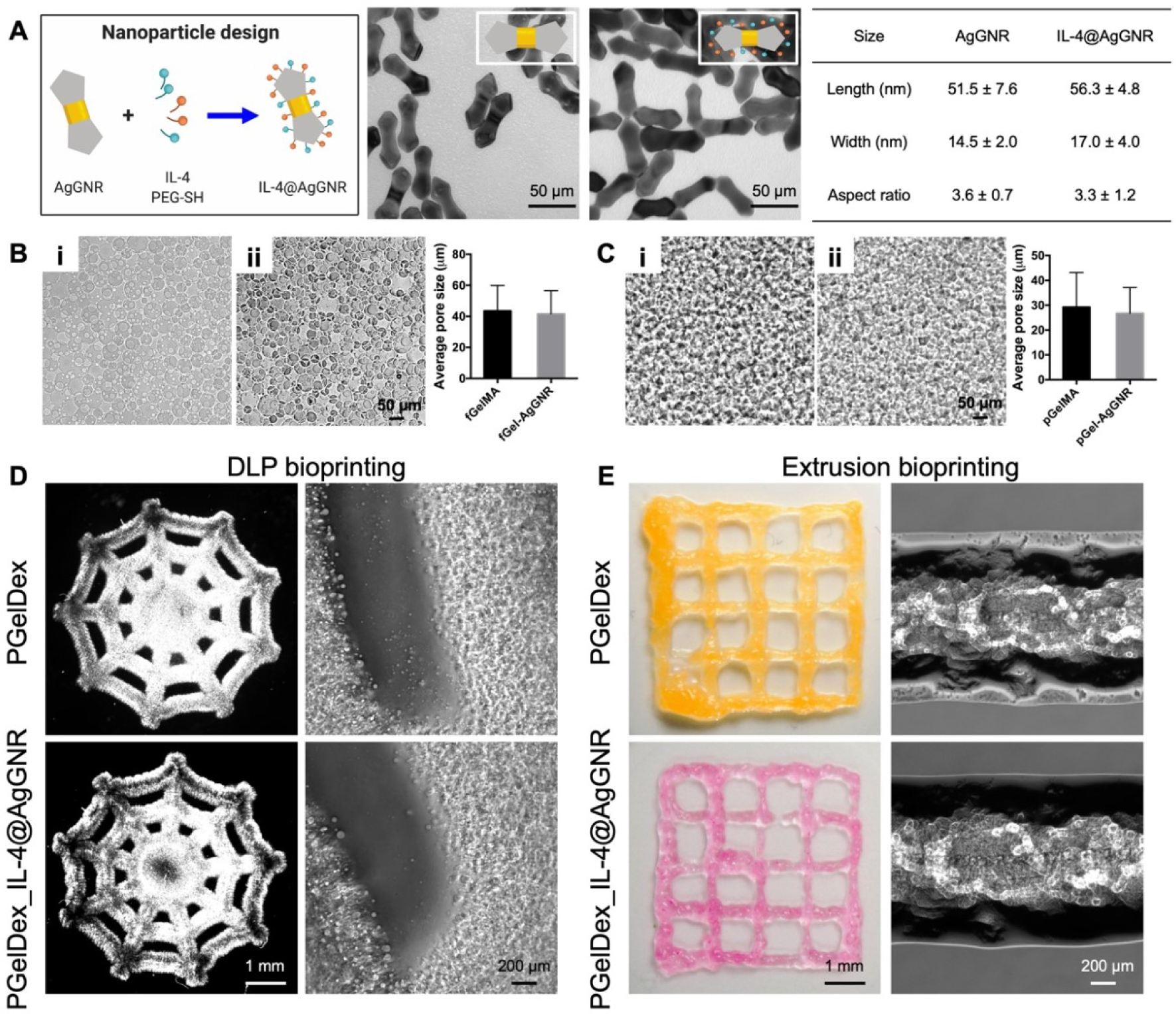
Characterizations of the IL-4@AgGNR-incorporated PGelDex bioinks. (A) TEM images and the size distributions (n = 100) of AgGNRs and IL-4@AgGNRs. (B) Optical micrographs showing fish-derived (i) PGelDex bioink and (ii) IL-4@AgGNR-incorporated PGelDex bioink. n = 3. (C) Optical micrographs showing porcine-derived (i) PGelDex bioink and (ii) IL-4@AgGNR-incorporated PGelDex bioink. (D) Optical micrographs showing DLP-bioprinted PGelDex and IL-4@AgGNR-incorporated PGelDex constructs. (E) Optical micrographs showing extrusion-bioprinted PGelDex and IL-4@AgGNR-incorporated PGelDex constructs.
The influence of encapsulating AgGNRs on the micropore-forming capacity and printability of the bioinks was subsequently examined. As shown in Figure 3B–C, the existence of AgGNRs had no noticeable impact on the formation of the emulsion for both pGelMA- and fGelMA-based micropore-forming bioinks. The average micropore size of the fish-derived PGelDex bioink at 3.0 wt.% dextran was 43.7 ± 7.5 μm, which was very close to the micropore size achieved with the same emulsion bioink after AgGNR-loading. A similar observation was confirmed in both optical micrographs and quantification results of porcine-derived PGelDex bioinks. The printability of AgGNR-encapsulated bioinks was further assessed with both DLP and extrusion bioprinting. Top views of the same structures produced with DLP bioprinting and fGelMA-derived pGelMA bioink are illustrated in Figure 3D, where a single-layered cobweb pattern could be readily bioprinted with the micropore-forming bioink in the absence or presence of AgGNRs. Further verification from optical micrographs of bioprinted samples elucidated the porous microstructures, suggesting the good agreement with the observation from the bioink before bioprinting. Similarly, the pGelMA-based micropore-forming bioink enabled extrusion bioprinting with a good shape fidelity and a stable micropore size distribution after adding AgGNRs (Figure 3E). We hence proved the consistency of the micropore-forming bioinks, either derived from fGelMA or pGelMA, without and with AgGNRs.
3.4. Evaluations of anti-bacterial performances of bioprinted constructs
The anti-bacterial performances of the bioprinted samples produced from the AgGNR-loaded bioinks were examined by measuring their abilities to inhibit S. aureus and E. coli growth on agar plates. After 24 h of incubation, colonies on agar plates indicated the bacteria attached to the bioprinted constructs. As indicated by the representative images shown in Figure 4A, the constructs bioprinted with AgGNR-encapsulated bioinks exhibited exceptional anti-bacterial properties compared to that of the plain bioinks. Moreover, higher bacterial reduction rates were observed in samples formed by AgGNR-loaded PGelDex bioinks when compared to those of the corresponding non-porous GelMA controls also containing AgGNRs. As a result of the porous structure, AgGNRs in the samples bioprinted with the micropore-forming bioinks were likely released faster through interconnected micropores than those made with the standard GelMA bioink. In addition, since AgGNRs were the primary anti-bacterial component in the bioink, when we further evaluated the influence of AgGNR concentration on bacteria-elimination, it was clear that such reduction rates of the bioprinted porous constructs loaded with high, medium, and low AgGNR concentrations (10, 5, and 2.5 μg mL−1, respectively) on E. coli were >65%, 75%, and 90%, respectively (Figure 4B). Overall, the AgGNR-loaded constructs bioprinted with micropore-forming bioinks could potentially be applied as a platform that exhibits favorable anti-bacterial properties.
Figure 4.
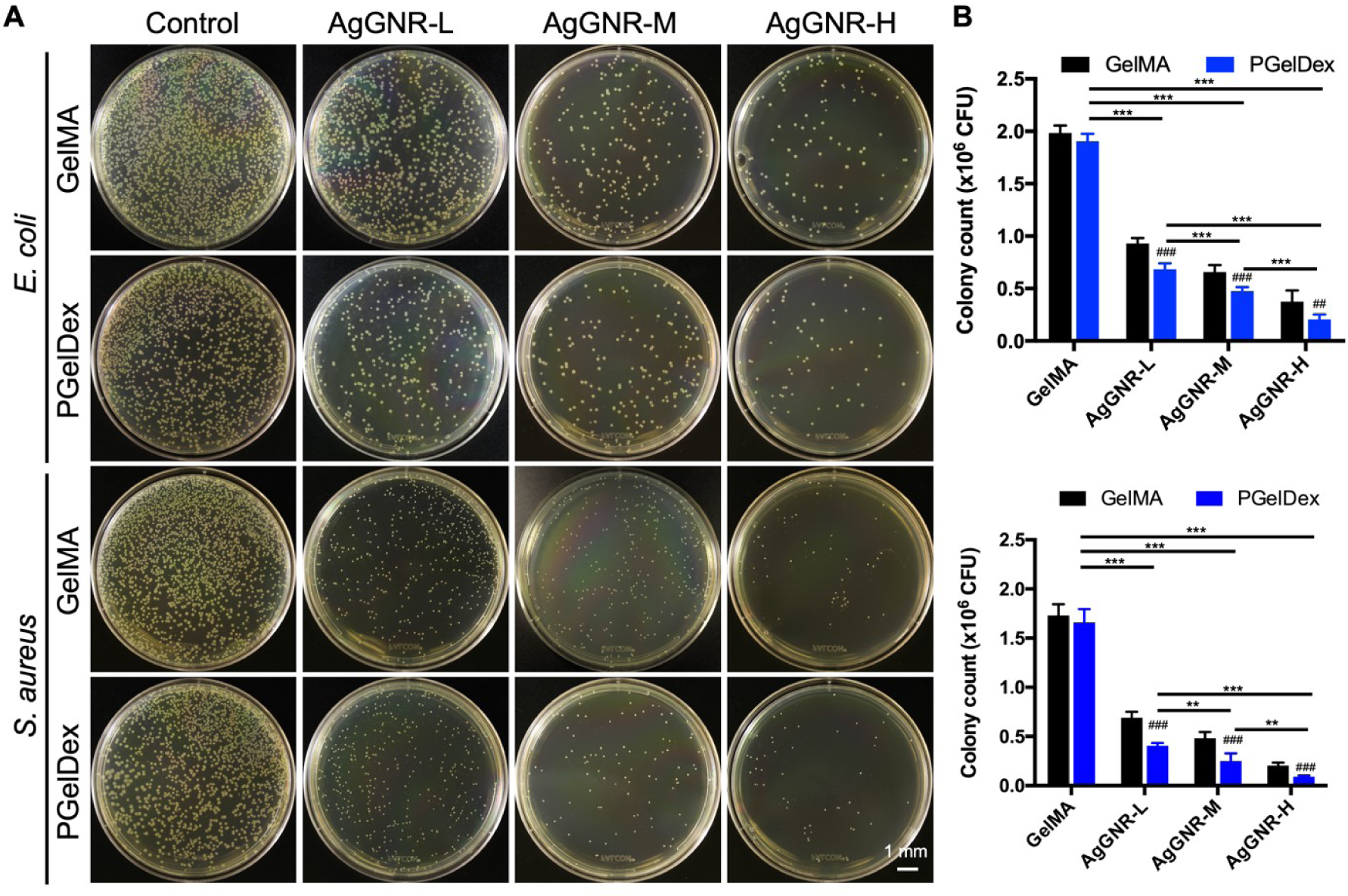
In vitro anti-bacterial performances of GelMA, PGelDex, and bioinks with different AgGNR concentrations (low, medium, and high) against E. coli and S. aureus. (A) Photographs of agar plates and (B) corresponding quantification results of colony numbers against E. coli (top graph) and S. aureus (bottom graph). *P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA; ## ###P < 0.01, P < 0.001; one-way ANOVA (compared with the GelMA control group); n = 3.
3.5. Evaluations of the bioink anti-inflammation performances
MSCs have been intensively investigated as an effective cell therapy due to their favorable immunomodulatory and regenerative properties [51–53]. The MSC-encapsulated micropore-forming bioinks were utilized for anti-inflammatory studies through a transwell assay. As a cell-laden bioink, the viability and activities of the encapsulated cells play a vital role in realizing their therapeutic efficacies, and accordingly the metabolic activities of encapsulated MSCs were assessed through the MTS assay. As shown in Figure 5A, the relative cell metabolic activities in all bioprinted constructs with PGelDex and PGelDex containing different concentrations of IL-4@AgGNRs were higher compared to GelMA control after 1 day of culture, suggesting the enhanced cytocompatibility. Among the samples bioprinted with IL-4@AgGNR-encapsulated PGelDex bioink, the cell metabolic activity was slightly decreased at higher concentrations of IL-4@AgGNRs after 5 days of culture. It is generally acknowledged that Ag products for medicinal or other purposes have activated Ag+, which might have a direct effect on biological systems by inducing cytotoxicity [54]. Therefore, it would be a major improvement for the use of Ag materials if we can reduce their concentrations while keeping their anti-bacterial properties. Our previous study had demonstrated that this type of AgGNRs could obtain decent anti-bacterial effects with lower Ag concentrations when compared to conventional AgNPs [15], which is an effective way to reduce related side effects. Accordingly, we selected PGelDex loaded with MSCs and medium concentration of AgGNR (PGelDex-IL-4@AgGNRs-M) as the bioink formulation for the subsequent anti-inflammation evaluation, which presented acceptable performances in anti-bacterial as well as cytocompatibility studies.
Figure 5.
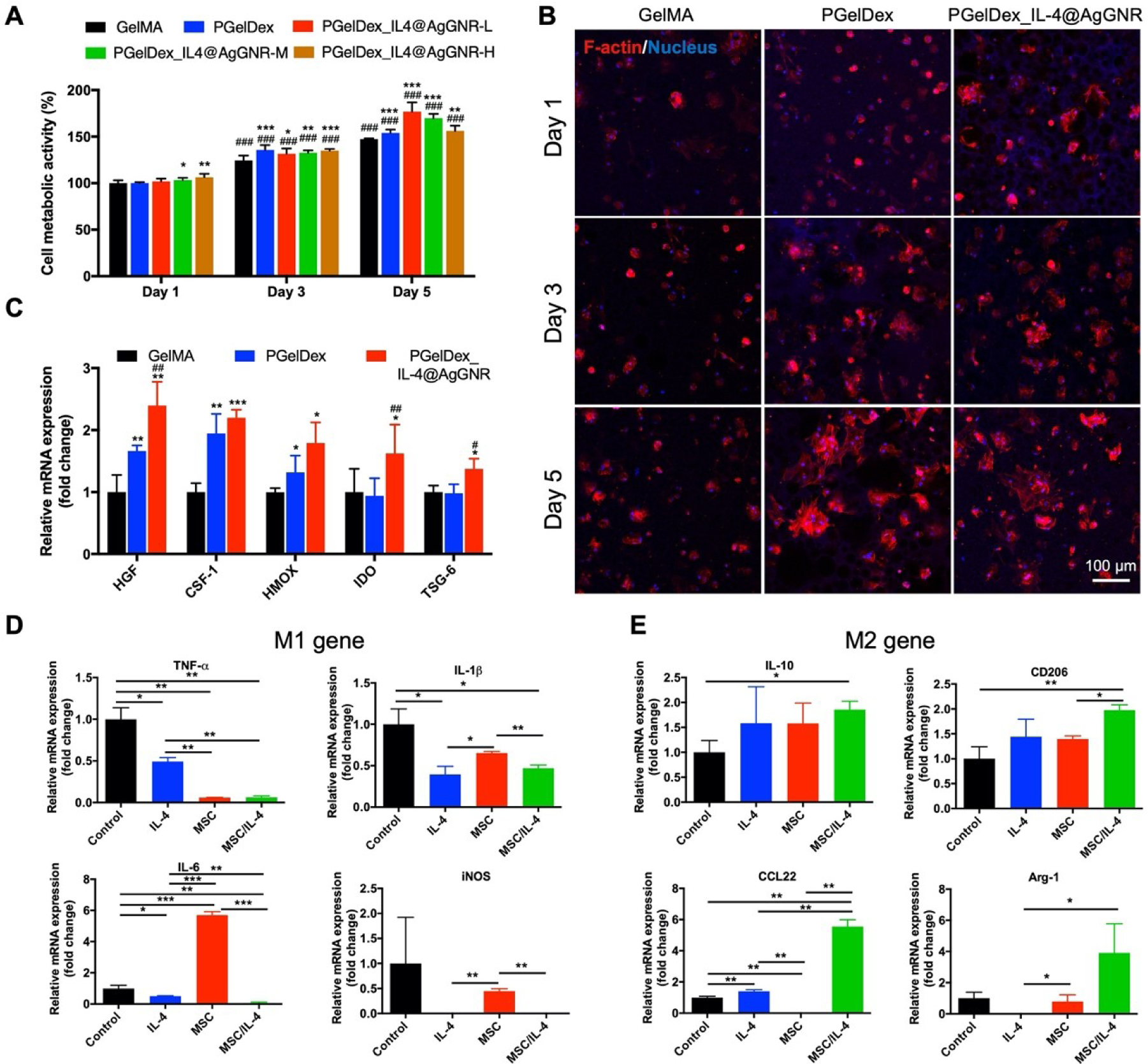
MSC- and IL-4@AgGNR-incorporated PGelDex micropore-forming bioink polarized macrophages into an anti-inflammatory phenotype. (A) Quantified cytocompatibility evaluations of MSCs cultured in the constructs bioprinted with GelMA, PGelDex, and PGelDex with different IL-4@AgGNR concentrations. (B) Fluorescence micrographs showing bioprinted MSCs in GelMA, PGelDex, and IL-4@AgGNR-incorporated PGelDex samples on days 1, 3, and 5 of culture. (C) Gene expressions of MSCs encapsulated in GelMA, PGelDex, and IL-4@AgGNR-incorporated PGelDex constructs normalized to reference gene RPL13A in GelMA. (D, E) Representative macrophage phenotype markers of THP-1 cells cultured in PGelDex, IL-4@AgGNR-incorporated PGelDex, MSC-encapsulated PGelDex, and MSC-IL-4@AgGNR-incorporated PGelDex constructs through a transwell assay. Relative mRNA expressions were normalized to reference gene GAPDH in GelMA. *P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA (A and C, compared with the GelMA control group; D and E, compared with the PGelDex control group); ###P < 0.001; one-way ANOVA (A, compared with the corresponding groups on day 1; C, compared with the PGelDex group); n = 3.
Interestingly, the MSCs were observed to spread in the bioprinted constructs made of both PGelDex and PGelDex-IL-4@AgGNRs-M bioinks under fluorescence microscopy. When the culture was continued for up to 5 days, MSC clusters were formed by a large portion of cells, implying the applicability of our micropore-forming bioink as an enabling platform to encapsulate MSCs (Figure 5B). By contrast, bioprinted MSCs in the conventional, non-porous GelMA samples exhibited less cell spreading and cluster-formation over 5 days of culture, which was in good consistency with our previous observations [30–32]. This promoted cytocompatibility of the bioprinted constructs may be ascribed to the porous microenvironment provided by the micropore-forming bioink, which might be attributed to the fact that the porous structure enabled improved diffusion of oxygen and nutrients than the non-porous controls, allowing for better encapsulated cell adhesion, spreading, and functions.
We further verified whether MSC encapsulation in our micropore-forming bioink with or without AgGNRs could still maintain immunomodulatory performances. A series of genes (HMOX, COX-2, TGF-β, IDO, HGF, TSG-6, CCL-2, and CSF-1) related to immunomodulation associated with MSCs were investigated via qRT-PCR. The immunomodulation of MSCs is determined by these genes, which is realized by the regulation of immune responses through secretomes and immune cell recruitment [55]. As can be seen from Figure 5C, the following immunosuppressive genes were upregulated in MSCs loaded within the micropore-forming bioink compared with MSCs in the non-microporous GelMA control: HGF, CSF-1, and HMOX; illustrating an enhanced immunomodulatory function. For MSCs encapsulated in the IL-4@AgGNR-loaded micropore-forming bioink, CSF-1 and HMOX, as well as anti-inflammatory genes, IDO and TSG-6 were further increased. By contrast, COX-2, TGF-β, CCL-2, and IL-6 were not identified to be significantly changed (Figure S4). Using another reference gene (GAPDH) for normalizations of gene expressions, the consistent results of upregulation of immunosuppressive genes were verified in the micropore-forming and IL-4@AgGNR-loaded micropore-forming bioink groups (Figure S5).
Finally, we investigated the immunomodulatory efficacy of MSC-encapsulated and IL-4@AgGNR-loaded micropore-forming bioink under LPS-induced inflammatory conditions (Figure 5D). M1 macrophages were induced from M0 macrophage under IFN-γ and LPS treatment, and then were co-cultured with bioprinted constructs encapsulating MSCs and IL-4@AgGNRs. After 24 h of incubation, qRT-PCR analysis revealed that the M1 phenotype markers, including TNF-α, IL-6, IL-1β, and iNOS were significantly decreased in IL-4 and IL-4+MSC groups when compared with the control group in the absence of both. These results identified that the anti-inflammatory bioink loaded with IL-4 and MSCs successfully polarized macrophages into the M2 phenotype. However, the M2 macrophage markers, such as IL-10, CD206, CCL22, and Arg-1, exhibited the opposite tendency with remarkably enhanced levels in IL-4 and IL-4+MSC groups. Furthermore, significant upregulation of M2 macrophage markers confirmed the synergistic anti-inflammatory effects of IL-4 and MSCs. Similar conclusions were obtained in evaluations where the ACTB was applied as the reference gene (Figure S6).
Previous studies have investigated the immunomodulatory effects of both IL-4 and MSCs. In a recent study He et al. reported that the release of IL-4 from high-stiffness transglutaminase-crosslinked gelatins promoted the polarization of macrophages to M2 phenotype [56]. Mooney and colleagues revealed that hydrogel encapsulated-MSC had anti-inflammatory functions and could promote polarization of M1 macrophages to M2 [21, 57]. Here, we revealed that IL-4-conjugated AgGNRs and MSCs in our micropore-forming bioink were able to shift the macrophages away from the M1 phenotype towards the M2 state in vitro following LPS-induced inflammation.
3.6. Additional evaluations of cytocompatibility and hemocompatibility
Biocompatibility plays an essential role in the various biomedical applications of a bioink. To preliminarily assess the biocompatibility of our formulations, the cytocompatibility and hemocompatibility of the micropore-forming bioink and IL-4@AgGNR-loaded micropore-forming bioinks at different concentrations were carried out by bioprinting in the presence of GFP-labeled HUVECs. The fluorescence images of HUVECs shown in Figure 6A exhibited notable cytocompatible as a large number of HUVECs were visualized within all the bioprinted samples after 24 h of culture. In addition, the metabolic activities of HUVECs after 1 day of incubation was also examined by MTS assay. As shown in Figure 6B, no significant cytotoxicity was observed in response to the bioinks, even with a high concentration of embedded IL-4@AgGNRs. Those results suggested the promising applicability of IL-4@AgGNR-loaded bioinks in biomedical applications. Longer-term cytotoxicity assessments after 6 days of culture further revealed that both the micropore-forming bioink and the IL-4@AgGNR-loaded micropore-forming bioink were cytocompatible showing better-spread HUVECs within the pores than the cells in the non-porous GelMA construct (Figure S7).
Figure 6.
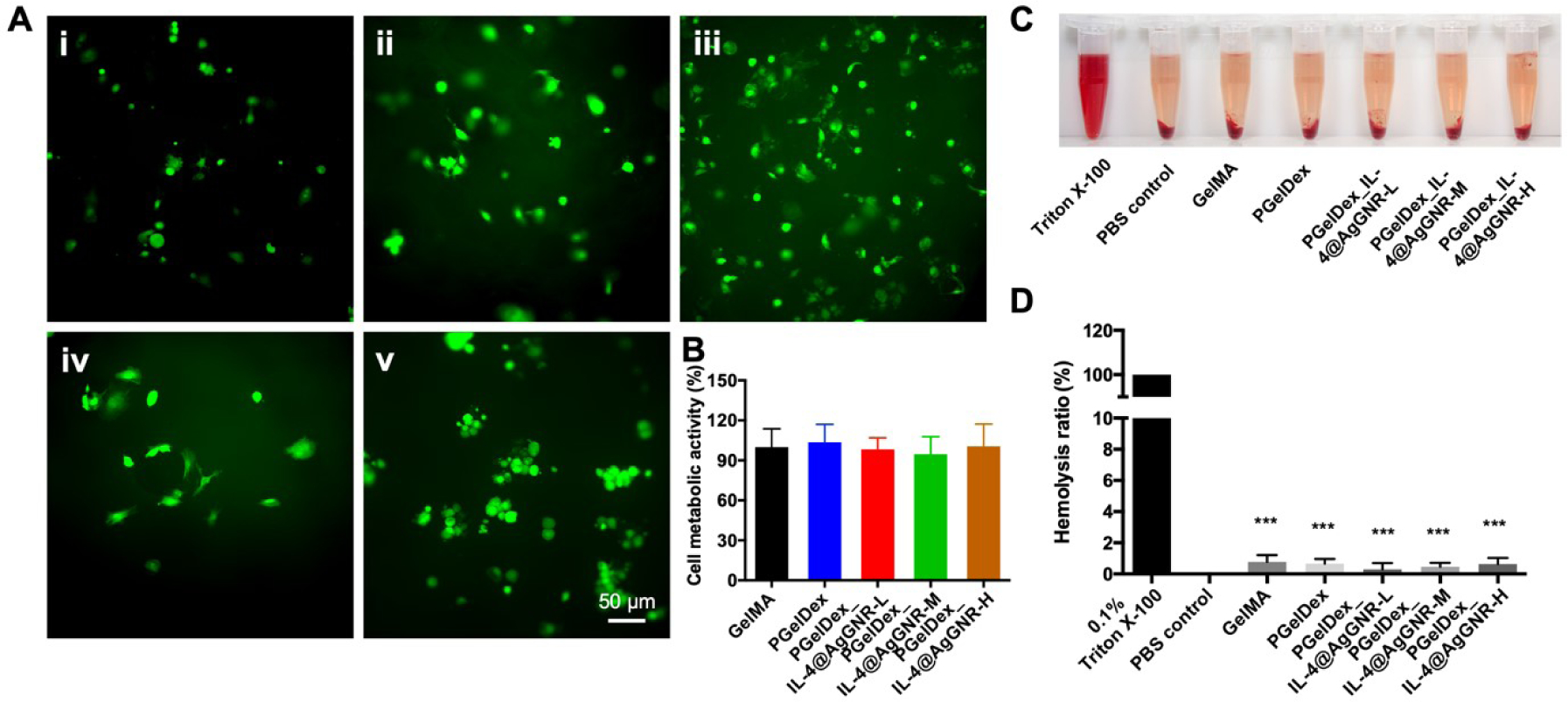
(A) Morphology of HUVECs after 1 day of culturing in constructs bioprinted from (i) GelMA, (ii) PGelDex, (iii) PGelDex-IL-4@AgGNR-H, (iv) PGelDex-IL-4@AgGNR-M, and (v) PGelDex-IL-4@AgGNR-L for 24 h. (B) Quantitative cytocompatibility evaluations by the MTS assay of the GelMA, PGelDex, and PGelDex with different IL-4@AgGNR concentrations by contact-culture with HUVECs. (C) Photograph showing hemolytic activities of the constructs made of GelMA, PGelDex, and PGelDex with different IL-4@AgGNR concentrations. (D) Quantified hemolytic percentages of the constructs made of GelMA, PGelDex, and PGelDex with different IL-4@AgGNR concentrations. ***P < 0.001; one-way ANOVA (D, compared with the Triton X-100 control group); n = 8.
The hemocompatibility of the constructs bioprinted with different formulations of the bioinks was then confirmed by an in vitro hemolysis assay. The photograph in Figure 6C exhibited the apparent difference in color between the negative control, the positive control, and the constructs produced with GelMA, PGelDex, and PGelDex with different IL-4@AgGNR concentrations. Compared to the bright red color of the Triton X-100 group, which indicated a complete RBC hemolysis, all the other groups displayed near-transparent colors. The quantitative results revealed that the addition of IL-4@AgGNRs could slightly increase RBC hemolysis, yet the absorbance was still close to that of the negative control (Figure 6D). On the basis of these outcomes, we concluded that IL-4@AgGNR-loaded PGelDex bioink had excellent cytocompatibility and hemocompatibility, thus enabling a promising potential for a number of 3D bioprinting applications.
4. Discussion
Bacteria-induced infection and the following inflammation are drivers of disease progression for many prevalent conditions ranging from wound healing to sepsis [58]. As is known, traditional treatment strategies often involve systemic antibiotic administration. Nevertheless, antibiotic-resistance following long-term exposure to antibiotics remains a critical issue in the treatment of infections, and therefore, compounds that can replace the anti-bacterial effect of antibiotics are urgently needed [59]. AgNPs were found to be an effective treatment option for a broad spectrum of extracellular bacteria, as their usage benefits from avoiding antibiotic-resistance [60]. In addition to AgNPs, gold nanorods (AuNRs) have received considerable attention in the field of nanomedicine and drug delivery because of their hyperthermal properties under near-infrared (NIR) excitation, and the fact that their surfaces can be conveniently functionalized with diverse ligands [61]. This photothermal effect is responsible for the broadened therapeutic applications of AuNRs, such as photothermal ablation of cancer cells and eradication of several biofilms [62]. Therefore, the rational combination of AgNPs and AuNRs would leverage the anti-bacterial and photothermal potentials for the ablation of biofilms. We demonstrated in our study that the synthesized sharp-edged dumbbell AgGNRs exhibited superior anti-bacterial activity due to the disruption of cellular structures, which may result in cytotoxicity, genotoxicity, and ultimately cell death. Compared with conventional AgNPs, AgGNRs presented more effective anti-bacterial properties. The addition of AgGNRs in the bioink showed no adverse effects on printability in both DLP and extrusion bioprinting.
Early bacterial-induced inflammation (1–3 days) is triggered by bacterial debris and the infiltration of macrophages and neutrophils into the infected area, which mediate tissue debridement by phagocytosis [7]. While the M1 macrophages should polarize to an M2 phenotype for the resolution of inflammation by secreting anti-inflammatory factors that promote bone-formation [8, 63], failure to progress to an M2 phenotype can lead to poor tissue regeneration [64, 65]. One way to overcome this limitation is through immunomodulation. Previous studies have shown that IL-4-mediated macrophage-enabled immunomodulation can significantly facilitate tissue regeneration both in vitro and in vivo. However, it remains challenging to directly use soluble IL-4, since it was reported to lose its immunomodulatory performance to improve muscle functions after acute injury in healthy mice [42, 43]. Further studies carried out by Mooney and co-workers displayed that gold nanoparticle (AuNP)-conjugated IL-4 had a sustained release profile and retained its bioactivity and ability to polarize M2 macrophage phenotype in vitro [43, 66]. Compared with AuNPs, AgGNRs not only can be used as a drug delivery platform but also possess anti-bacterial properties. Our study demonstrated that partial PEGylation of AgGNRs, then IL-4 conjugation, yielded stable and monodispersed nanoparticles for bioactive IL-4-delivery. It was found that AgGNR-conjugated IL-4 retained its biological activity and the ability to direct macrophage phenotype in vitro.
The results of these studies indicated an enabling micropore-forming bioink for multiple bioprinting methods (extrusion and DLP bioprinting) with cell- and biomolecule-loading efficiencies. More importantly, we illustrated that IL-4-conjugated AgGNRs loaded in the micropore-forming bioink had an anti-bacterial effect, and promoted immunomodulation along with MSCs, a condition representative of clinically relevant bacteria-induced inflammation. The presence of dextran as the porogen within GelMA provided the successful formation of the microporous microstructures. Also importantly, our micropore-forming bioinks exhibited good printability through the evaluations of DLP, extrusion, and handheld bioprinting methods.
5. Conclusions
In this study, an anti-bacterial and anti-inflammatory bioink was developed based on our micropore-forming bioink formulation enabled by aqueous two-phase emulsion of GelMA and dextran, in which two functional agents, IL-4@AgGNRs and MSCs, were loaded to eliminate bacterial-induced infection and direct macrophage polarization into the M2 phenotype. Our results revealed that this functional micropore-forming bioink embedded with AgGNRs could successfully suppress both Gram-positive and Gram-negative bacterial growth. Compared with AgGNR-loaded GelMA bioink, PGelDex containing AgGNRs exhibited a more notable suppression of bacterial growth possibly due to the fast release of AgGNRs from the microporously structured constructs. Furthermore, the PGelDex bioinks exhibited excellent printability in DLP, extrusion, and handheld bioprinting methods. Of interest, the MSC-encapsulated bioink displayed favorable spreading for the cells within the pore areas, suggesting the excellent cytocompatibility of our formulations. In addition, immunomodulatory properties of the PGelDex bioink containing IL-4 and MSCs were evaluated in vitro via a transwell study. Our data illustrated that the presence of IL-4 and MSCs could synergistically induce macrophage polarization towards an anti-inflammatory M2 phenotype. Thus, with the aid of AgGNRs as an IL-4-carrier, our MSC-laden micropore-forming bioink is multi-functional, providing an exciting platform for potentially widespread applications where anti-bacterial, anti-inflammatory, and cell-instructive properties are simultaneously required.
It should be noted that in our study however, the THP-1 cell line was used as an alternative source to primary monocyte-derived macrophages, to overcome the problems of limited lifespan and inter-individual variability of primary monocyte-derived human macrophages. THP-1 derived from the peripheral blood of a childhood case of acute monocytic leukemia, is recognized as an immortalized monocyte-like cell line [67]. It has been extensively applied as an alternative model to primary human monocytes in vitro to study the functions, mechanisms, and responses of monocytes or macrophages [68]. However, the THP-1 cell line presents some limitations, such as poor response to LPS compared with the primary monocytes [68]. It has been extensively reported that phorbol-12-myristate-13-acetate (PMA) is the most effective differentiation agent to produce THP-1 monocyte-derived macrophages [69]. Following the differentiation, the induced cells could be confirmed to present functional characteristics of mature macrophages, such as adherence to culture plates, alteration in the morphology into flat and amoeboid shapes, as well as the well-developed Golgi apparatuses, rough endoplasmic reticula, and a number of ribosomes in the cytoplasm. Despite so, additional characterizations of our bioink formulations using primary monocytes and macrophages may still be needed to fully understand their immunomodulative potential.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institutes of Health (R21EB026175, R21EB025270, R00CA201603, R01EB028143, R01GM134036), the National Science Foundation (CBET-EBMS-123859), and the Brigham Research Institute. AMG would like to acknowledge funding from the European Union’s Horizon2020 research and innovation programme under grant agreement 760921 (PANBioRA). We also thank the NeuroTechnology Studio at Brigham and Women’s Hospital for providing confocal microscopy access and consultation on data acquisition and data analysis.
References
- [1].Moroni L, Burdick JA, Highley C, Lee SJ, Morimoto Y, Takeuchi S, Yoo JJ, Nat Rev Mater 2018. 3(5) 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ma X, Liu J, Zhu W, Tang M, Lawrence N, Yu C, Gou M, Chen S, Adv Drug Deliv Rev 2018. 132 235–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Prendergast ME, Burdick JA, Adv Mater 2020. 32(13) e1902516. [DOI] [PubMed] [Google Scholar]
- [4].Mofazzal Jahromi MA, Sahandi Zangabad P, Moosavi Basri SM, Sahandi Zangabad K, Ghamarypour A, Aref AR, Karimi M, Hamblin MR, Adv Drug Deliv Rev 2018. 123 33–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Campoccia D, Montanaro L, Arciola CR, Biomaterials 2006. 27(11) 2331–9. [DOI] [PubMed] [Google Scholar]
- [6].Darouiche RO, New England Journal of Medicine 2004. 350(14) 1422–1429. [DOI] [PubMed] [Google Scholar]
- [7].Alvarez MM, Liu JC, Trujillo-de Santiago G, Cha BH, Vishwakarma A, Ghaemmaghami AM, Khademhosseini A, J Control Release 2016. 240 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Labonte AC, Tosello-Trampont AC, Hahn YS, Mol Cells 2014. 37(4) 275–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Makabenta JMV, Nabawy A, Li CH, Schmidt-Malan S, Patel R, Rotello VM, Nat Rev Microbiol 2021. 19(1) 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chaloupka K, Malam Y, Seifalian AM, Trends in biotechnology 2010. 28(11) 580–588. [DOI] [PubMed] [Google Scholar]
- [11].Kim JS, Kuk E, Yu KN, Kim J-H, Park SJ, Lee HJ, Kim SH, Park YK, Park YH, Hwang C-Y, Nanomedicine: Nanotechnology, biology and medicine 2007. 3(1) 95–101. [DOI] [PubMed] [Google Scholar]
- [12].Reithofer MR, Lakshmanan A, Ping AT, Chin JM, Hauser CA, Biomaterials 2014. 35(26) 7535–7542. [DOI] [PubMed] [Google Scholar]
- [13].Zhao L, Wang H, Huo K, Cui L, Zhang W, Ni H, Zhang Y, Wu Z, Chu PK, Biomaterials 2011. 32(24) 5706–16. [DOI] [PubMed] [Google Scholar]
- [14].Gunputh UF, Le H, Handy RD, Tredwin C, Materials Science and Engineering: C 2018. 91 638–644. [DOI] [PubMed] [Google Scholar]
- [15].Zhang J, Wang M, Webster TJ, International journal of nanomedicine 2018. 13 6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shi C, Pamer EG, Nat Rev Immunol 2011. 11(11) 762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chen J, Li M, Yang C, Yin X, Duan K, Wang J, Feng B, Colloids Surf B Biointerfaces 2018. 163 336–345. [DOI] [PubMed] [Google Scholar]
- [18].Spiller KL, Nassiri S, Witherel CE, Anfang RR, Ng J, Nakazawa KR, Yu T, Vunjak-Novakovic G, Biomaterials 2015. 37 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gong L, Li J, Zhang J, Pan Z, Liu Y, Zhou F, Hong Y, Hu Y, Gu Y, Ouyang H, Zou X, Zhang S, Acta Biomater 2020. 117 246–260. [DOI] [PubMed] [Google Scholar]
- [20].Nauta AJ, Fibbe WE, Blood, The Journal of the American Society of Hematology 2007. 110(10) 3499–3506. [DOI] [PubMed] [Google Scholar]
- [21].Gonzalez-Pujana A, Vining KH, Zhang DKY, Santos-Vizcaino E, Igartua M, Hernandez RM, Mooney DJ, Biomaterials 2020. 257 120266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Song N, Scholtemeijer M, Shah K, Trends Pharmacol Sci 2020. 41(9) 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang YS, Yue K, Aleman J, Moghaddam KM, Bakht SM, Yang J, Jia W, Dell’Erba V, Assawes P, Shin SR, Dokmeci MR, Oklu R, Khademhosseini A, Ann Biomed Eng 2017. 45(1) 148–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Murphy SV, De Coppi P, Atala A, Nat Biomed Eng 2020. 4(4) 370–380. [DOI] [PubMed] [Google Scholar]
- [25].Li W, Mille LS, Robledo JA, Uribe T, Huerta V, Zhang YS, Adv Healthc Mater 2020. 9(15) e2000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang M, Li W, Tang G, Garciamendez-Mijares CE, Zhang YS, Advanced Healthcare Materials 2021. 2100380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang YS, Khademhosseini A, Science 2017. 356(6337). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gungor-Ozkerim PS, Inci I, Zhang YS, Khademhosseini A, Dokmeci MR, Biomater Sci 2018. 6(5) 915–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wu T, Gao YY, Su J, Tang XN, Chen Q, Ma LW, Zhang JJ, Wu JM, Wang SX, Climacteric 2021. 1–9. [DOI] [PubMed] [Google Scholar]
- [30].Ying G, Manriquez J, Wu D, Zhang J, Jiang N, Maharjan S, Hernandez Medina DH, Zhang YS, Mater Today Bio 2020. 8 100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ying G, Jiang N, Parra C, Tang G, Zhang J, Wang H, Chen S, Huang NP, Xie J, Zhang YS, Adv Funct Mater 2020. 30(46). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ying GL, Jiang N, Maharjan S, Yin YX, Chai RR, Cao X, Yang JZ, Miri AK, Hassan S, Zhang YS, Adv Mater 2018. 30(50) e1805460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gong J, Schuurmans CCL, Genderen AMV, Cao X, Li W, Cheng F, He JJ, Lopez A, Huerta V, Manriquez J, Li R, Li H, Delavaux C, Sebastian S, Capendale PE, Wang H, Xie J, Yu M, Masereeuw R, Vermonden T, Zhang YS, Nat Commun 2020. 11(1) 1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Maharjan S, Alva J, Cámara C, Rubio AG, Hernández D, Delavaux C, Correa E, Romo MD, Bonilla D, Santiago ML, Matter 2021. 4(1) 217–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yoon HJ, Shin SR, Cha JM, Lee SH, Kim JH, Do JT, Song H, Bae H, PLoS One 2016. 11(10) e0163902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Miri AK, Nieto D, Iglesias L, Goodarzi Hosseinabadi H, Maharjan S, Ruiz-Esparza GU, Khoshakhlagh P, Manbachi A, Dokmeci MR, Chen S, Advanced Materials 2018. 30(27) 1800242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Li W, Wang M, Mille LS, Robledo Lara JA, Huerta V, Uribe Velazquez T, Cheng F, Li H, Gong J, Ching T, Murphy CA, Lesha A, Hassan S, Woodfield TBF, Lim KS, Zhang YS, Adv Mater 2021. 33(35) e2102153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang M, Li W, Mille LS, Ching T, Luo Z, Tang G, Garciamendez CE, Lesha A, Hashimoto M, Zhang YS, Adv Mater 2022. 34(1) e2107038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li H, Cheng F, Orgill DP, Yao J, Zhang YS, Essays in biochemistry 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Li H, Cheng F, Li W, Cao X, Wang Z, Wang M, Robledo-Lara JA, Liao J, Chavez-Madero C, Hassan S, Xie J, Trujillo-de Santiago G, Alvarez MM, He J, Zhang YS, Biofabrication 2020. 12(4) 045027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li W, Wang M, Mille LS, Robledo JA, Huerta V, Uribe T, Cheng F, Li H, Gong J, Ching T, Murphy CA, Lesha A, Hassan S, Woodfield T, Lim KS, Zhang YS, Advanced Materials 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Raimondo TM, Mooney DJ, Proc Natl Acad Sci U S A 2018. 115(42) 10648–10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Raimondo TM, Mooney DJ, Science Advances 2021. 7(26) eabh3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Starr T, Bauler TJ, Malik-Kale P, Steele-Mortimer O, PLoS One 2018. 13(3) e0193601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Baxter E, Graham A, Re N, Carr I, Robinson J, Mackie S, Morgan A, Journal of immunological methods 2020. 478 112721. [DOI] [PubMed] [Google Scholar]
- [46].Gatto F, Cagliani R, Catelani T, Guarnieri D, Moglianetti M, Pompa PP, Bardi G, Nanomaterials 2017. 7(10) 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li X, Yang Q, Bai J, Yang Y, Zhong L, Wang Y, Mol Med Rep 2015. 11(2) 1304–11. [DOI] [PubMed] [Google Scholar]
- [48].Maeß MB, Sendelbach S, Lorkowski S, BMC molecular biology 2010. 11(1) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Liang Y, Zhao X, Hu T, Chen B, Yin Z, Ma PX, Guo B, Small 2019. 15(12) e1900046. [DOI] [PubMed] [Google Scholar]
- [50].Zhu W, Ma X, Gou M, Mei D, Zhang K, Chen S, Curr Opin Biotechnol 2016. 40 103–112. [DOI] [PubMed] [Google Scholar]
- [51].Nasef A, Ashammakhi N, Fouillard L, Regen Med 2008. 3(4) 531–46. [DOI] [PubMed] [Google Scholar]
- [52].Wang Y, Chen X, Cao W, Shi Y, Nat Immunol 2014. 15(11) 1009–16. [DOI] [PubMed] [Google Scholar]
- [53].Gao F, Chiu SM, Motan DA, Zhang Z, Chen L, Ji HL, Tse HF, Fu QL, Lian Q, Cell Death Dis 2016. 7 e2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Aueviriyavit S, Phummiratch D, Maniratanachote R, Toxicology letters 2014. 224(1) 73–83. [DOI] [PubMed] [Google Scholar]
- [55].Lin L, Du L, Cell Immunol 2018. 326 24–32. [DOI] [PubMed] [Google Scholar]
- [56].He XT, Li X, Xia Y, Yin Y, Wu RX, Sun HH, Chen FM, Acta Biomater 2019. 88 162–180. [DOI] [PubMed] [Google Scholar]
- [57].Mao AS, Ozkale B, Shah NJ, Vining KH, Descombes T, Zhang L, Tringides CM, Wong SW, Shin JW, Scadden DT, Weitz DA, Mooney DJ, Proc Natl Acad Sci U S A 2019. 116(31) 15392–15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ma CY, Shi GY, Shi CS, Kao YC, Lin SW, Wu HL, J Immunol 2012. 188(12) 6328–37. [DOI] [PubMed] [Google Scholar]
- [59].Fraimow HS, Semin Plast Surg 2009. 23(2) 90–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhou Y, Kong Y, Kundu S, Cirillo JD, Liang H, J Nanobiotechnology 2012. 10 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Patra JK, Das G, Fraceto LF, Campos EVR, Rodriguez-Torres MDP, Acosta-Torres LS, Diaz-Torres LA, Grillo R, Swamy MK, Sharma S, Habtemariam S, Shin HS, J Nanobiotechnology 2018. 16(1) 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Al-Bakri AG, Mahmoud NN, Molecules 2019. 24(14) 2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wynn TA, Vannella KM, Immunity 2016. 44(3) 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Löffler J, Sass FA, Filter S, Rose A, Ellinghaus A, Duda GN, Dienelt A, Frontiers in immunology 2019. 2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lee J, Byun H, Madhurakkat Perikamana SK, Lee S, Shin H, Adv Healthc Mater 2019. 8(4) e1801106. [DOI] [PubMed] [Google Scholar]
- [66].Raimondo TM, Mooney DJ, Proceedings of the National Academy of Sciences 2018. 115(42) 10648–10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K, Int J Cancer 1980. 26(2) 171–6. [DOI] [PubMed] [Google Scholar]
- [68].Bosshart H, Heinzelmann M, Annals of translational medicine 2016. 4(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Daigneault M, Preston JA, Marriott HM, Whyte MK, Dockrell DH, PLoS One 2010. 5(1) e8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


