Abstract
Among intra-cellular homeostasis mechanisms, ubiquitination plays a critical role in protein metabolism regulation by degrading proteins via activating a broad spectrum of ubiquitin chains. In fact, ubiquitination and sumoylation signaling pathways are characterized by increased complexity regarding the molecules and their interactions. The Ubiquitin-Proteasome System (Ub-PS) recognizes and targets a broad spectrum of protein substrates. Ubiquitin conjugation modifies each substrate protein determining its biochemical fate (degradation). A major functional activity of Ub-PS is autophagy mechanism regulation. Interestingly, Ub-PS promotes all stages of bulk autophagy (initiation, execution, and termination). Autophagy is a crucial catabolic process that provides protein degradation and for this reason the interaction with Ub-PS is crucial. Furthermore, ubiquitination controls and regulates specific types of protein targets. Ub-PS is also involved in oxidative cellular stress and DNA damage response. Additionally, the functional role of Ub-PS in ribosome machinery regulation seems to be crucial. Concerning carcinogenesis, Ub-PS is involved in malignant disease development and progression by negatively affecting the corresponding TGF-B-, MEEK/MAPK/ERK-JNK- dependent signaling pathways. In the current review article, we describe the role of Ub-PS biochemical modifications and alterations in oral squamous cell carcinoma (OSCC).
Keywords: Ubiquitination, carcinoma, oral, pathophysiology, protein degradation, review
Ubiquitin is a covalent protein modifier, with major role in protein modification. Ubiquitin pathways comprise a variety of intracellular protein modification systems, which regulate both the final form and the quantity of multiple proteins (1). A growing body of research has enriched our understating of the role of ubiquitination in a number of physiological processes (2). Existing data indicate that ubiquitination and sumoylation signaling pathways are characterized by increased complexity regarding the participating molecules and their interactions (3-5). The ubiquitin-proteasome system (Ub-PS) recognizes and targets a broad spectrum of protein substrates. Ubiquitin conjugation modifies each substrate protein determining its biochemical fate (degradation). A major functional activity of Ub-PS is autophagy mechanism regulation (6,7). Interestingly, Ub-PS promotes all stages of bulk autophagy (initiation, execution, and termination). Autophagy is a crucial catabolic process that provides final protein degradation and for this reason the interaction with Ub-PS is crucial (8). Furthermore, ubiquitination controls and regulates specific types of selective autophagy (9-11). Ub-PS is also involved in oxidative cell stress and DNA damage response (12). In conjunction, the functional role of Ub-PS in ribosome machinery regulation seems to be crucial. Ubiquitination is a significant and major post-translational mechanism for modifying ribosomal proteins. For this reason, the 40S ribosome subunit ubiquitination/ deubiquitination balance and the efficient translation eS7 are very important biochemical events (13-15).
Conversely to its physiological activity, abnormal Ub-PS modifications are implicated in non-malignant inflammatory and autoimmune phenotypes including arthritis, atherogenesis, psoriasis, uveitis, etc. (16-19). Furthermore, aberrant ubiquitination appears to be involved in malignant disease development and progression by negatively affecting the corresponding TGF-B-, MEEK/MAPK/ERK-, JNK-dependent signaling pathways (20,21). Therefore, in the current review article, we describe the role of Ub-PS modifications/alterations in the development of oral squamous cell carcinoma (OSCC) biochemical and molecular substrate.
Protein Ubiquitination: Molecules and Mechanisms
Ubiquitin discovery in the 1970s has eventually led to an improved understanding of the nature and interactions of post-translational modifications (22). Ubiquitin is a highly conserved 8.6-kDa (76 amino acids chain) protein, which includes seven lysine residues in its stereo-chemical structure. The ubiquitination procedure represents a major post-translational mechanism that drives protein substrates from synthesis and structural maturation to degradation/breakdown (23,24). Furthermore, it affects protein-protein interactions, protein localization, motivation, and kinase kinetics into the cell cycle (25,26). Besides these activities, ubiquitination regulates a variety of intracellular functions including endocytosis, DNA damage repair, and gene transcription and translation (27-29). Ubiquitination also affects inflammatory and autoimmune disease mechanisms by its involvement in the degradation of specific proteins (30).
Ub-PS comprises of three main enzymes: E1 ubiquitin activating enzyme, E2 ubiquitin conjugating enzyme, and E3 ubiquitin ligase enzyme (31). The conjugation of ubiquitin to the lysine residue of other proteins is the result of their sequential action catalyzed by specific deubiquitinating enzymes (32). Biochemically, ubiquitination is based initially on the ubiquitin C-terminal glycine residue activation demanding ATP consumption. Progressively it finally leads to ubiquitin conjugation to the corresponding protein substrates (33). Mono- or multiple ubiquitin chains interact with Lys residues of target proteins. Alternatively, different ubiquitin moieties create new forms of chains by binding to N-terminal methionine residues or to the previously referred lysine residues. Furthermore, other biochemical reactions including acetylation, sumoylation, and phosphorylation provide complexity to the ubiquitin-based modifications. Another important regulatory action of ubiquitin is referred to E2F1 transcription factor regulation (34). Specific linked chains including K11/K48 lead to E2F1 degradation whereas K6, K27, K33, K63 demonstrate a non-degradative influence (35).
Besides, ubiquitination, autophagy plays also major role in intracellular protein degradation. Although these two mechanisms demonstrate different, independent functional pathways, it seems that there is some cross talk and interactions between them. Autophagy represents a degradation mechanism provided by lysosome activation (36). In fact, it acts as a critical catabolic process, as a response to intracellular stress. The formation of specific intracellular domains (autophagosomes) is followed by lysosome-dependent fusion process and molecule degradation (37). Hypoxia, cell organelle dysfunction and damage, oxidative stress and lack of critical ATP-based energy levels are intracellular signals for inducing autophagy motivation (38). At the molecular level, ULK1 serine/threonine kinase represents a major gene activated by stress signals leading to a cataract of molecules phosphorylation (39). The PI3K complex (lipid kinase VPS34, Beclin-1, VPS15, and ATG14) is activated by ULK1 and PI3P/ATG9/ATG2/WIPI interactions leading to autophagosome formation. Interestingly, ATG9 -as a transmembrane protein- is responsible for providing membrane substrates to autophagosomes (40). Concerning the influence of ubiquitination in autophagy regulation, it seems that there is a positive feedback loop. ULK1 and Beclin-1 molecules are targets for ubiquitination under the pressure of E3 ligases and especially the TRAF6, which is activated by the AMBRA1 gene, a PI3K component (41). This gene supports normal function ofULK1 by inducing K63-dependent ubiquitination (42). Similarly, TRAF6 enhances Beclin-1K63-dependent ubiquitination (43). In fact, the two genes Beclin-1 and AMBRA1 regulate not only initiation, but also termination of the autophagy phenomenon. Interestingly, Beclin-1subsequent degradation is mediated by the influence of HECT-type E3 ligase that provides Lys-11-linked ubiquitination of the gene (44). Additionally, Beclin-1is indirectly regulated by the E3 ligase PARKIN. Furthermore, AMBRA1 is negatively affected by the expression of phosphorylated mTORC1. Besides the main autophagy procedure, selective autophagy variants are also exposed to the influence of ubiquitination. E3 ligase is involved in aggrephagy, mitophagy, lysophagy, and xenophagy and all of them are stimulated by Lys-63 poly-Ub chains (45).
Ubiquitination Modifications in Oral Carcinoma
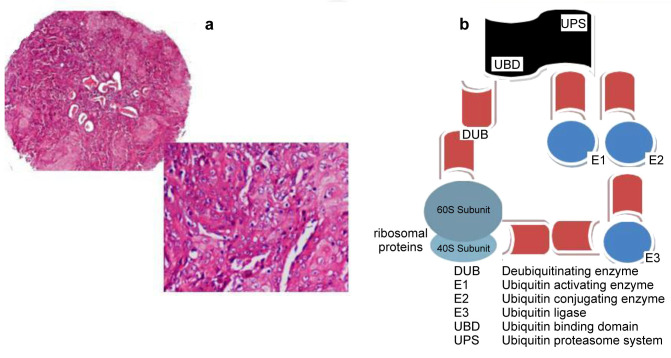
Oral squamous cell carcinoma (OSCC) represents a major malignancy in Head and Neck Squamous Cell Carcinoma (HNSCC) super-family. OSCCs frequently present an aggressive phenotype, with an increased tendency to present local and distant metastatic lymph nodes, as a result of severe genetic alterations (46). Etiopathogenetic factors that lead to OSCC development and progression include tobacco, alcohol chronic consumption and also viral-mediated deregulation (47). Concerning viral oncogenic activity, persistent Human Papilloma Virus (HPV) infection is responsible for malignant transformation of the affected oral/oropharyngeal epithelia modifying the host cell genome (48). Concerning ubiquitination modifications in OSCC, a variety of molecules are implicated in them (Figure 1). Ubiquitin D appears to play a pivotal role in malignancy. A study group evaluated its expression by implementing a combined immunohistochemistry (IHC), quantitative real-time polymerase chain (qRT-PCR) reaction and a western blot protocol, in a series of OSCCs (49). They reported that a significant over expression of the molecule correlated with induced proliferation, migration and invasion. They also suggested that ubiquitin D should be potentially considered a prognostic biomarker for monitoring OSCC patients.
Figure 1. Ideogram of ubiquitination signalling pathway in oral squamous cell carcinoma (OSCC). (a) A tissue microarray spot of an OSCC case (haematoxylin & eosin staining, original magnification 40×, 100×) (b) The Ubiquitin-Proteasome System (Ub-PS) recognizes and targets a broad spectrum of protein substrates. The ubiquitin-depended intracellular cascade includes a series of interactions between molecules such as E1/E2/E3 enzymes/ligases targeting ribosome protein domains (40S/60S subunits). The ubiquitination-deubiquitination procedure secures protein potential in a perfect balance promoting normal cell metabolism.
Another molecule involved in OSCC progression is the ubiquitin-specific protease 14 (USP14). Implementing experimental colony formation analysis in OSCC cell cultures and tissue specimens, a study group observed a significant over expression of the marker (50). Interestingly, USP14 up-regulation was associated with poor prognosis. Furthermore, USP14 over activation reduced apoptosis and autophagy leading to elevated radio-resistance in them. For these reasons, the authors suggested USP14 as a molecule essential for targeted therapeutic strategies. Similarly, another study focused on the aberrant expression of STUB1 based ubiquitin E3 ligase (51). Implementing a qRT-PCR assay on OSCCs they reported reduced STUB1 expression, associated to significant recurrence rates. Additionally, STUB1 progressive inactivation was correlated with TGM2 up-regulation in these cases, a protein related with malignancy aggressiveness. Another study group evaluated the impact of ubiquitin-specific processing proteases 17 (DUB3) expression on OSCC (52). Applying a qRT-PCR, IHC, western blot and flow cytometry combined assay they observed that DUB3 over expression led to increased cancerous cell proliferation and viability, combined with low apoptotic levels. They also suggested that DUB3 should be a potential target for OSCC therapeutic strategies.
Besides DUB3 over activation, increased expression of ubiquitin-specific protease 22 (USP22)-acting as deubiquitinating hydrolase- is involved in the development and progression of OSSC. A study group co-analyzed USP22, survivin and aurora-B molecules reported high expression levels for all of them. Interestingly, RING finger protein 139 (RNF139) E3 ubiquitin ligase activation reduced cancerous cells proliferation in a molecular study (53). The study group analyzed its expression in SCC9 and SCC25 cell cultures suggesting that the molecule is involved in aggressive phenotypes regarding OSSC (tongue carcinoma).
The role of another molecule – the ubiquitin-specific protease 9 (USP9X) acting as deubiquitinase – in OSSC onset and progression is under investigation. A study group analyzed the co-expression of USP9X and MCL-1, an anti-apoptotic protein, by IHC immunofluorescence, and flow cytometry. In fact, USP9X deubiquitinates MCL-1 protein. They also reported high expression levels of both molecules. Interestingly, USP9X/MCL-1co-over expression was associated to aggressive phenotypes in OSCC combined with poor prognosis. USP9X plays also a significant role by deubiquitinating the critical immune checkpoint protein programmed cell death ligand 1 (PD-L1) in OSSC (54). USP9X stabilizes PD-L1 over expression enhancing also cancerous cell proliferation.
In conclusion, ubiquitination plays a critical role in protein metabolism regulation by degrading proteins, and by also providing intra-cellular homeostasis. Ubiquitination and sumoylation signaling pathways comprise a significant number of molecules. The Ub-PS recognizes and targets a broad spectrum of protein substrates. Ubiquitin conjugation modifies each substrate protein determining its biochemical fate (degradation). A major functional activity of Ub-PS is autophagy mechanism regulation. Interestingly, Ub-PS promotes all the stages of bulk autophagy (initiation, execution, and termination). Autophagy interacts with Ub-PS in the cell micro-environment. Furthermore, ubiquitination controls and regulates specific types of selective. Ub-PS is also involved in oxidative cellular stress and DNA damage response. Interestingly, the functional role of Ub-PS in ribosome machinery regulation seems to be crucial. Concerning carcinogenesis, it is involved in malignant diseases development and progression by negatively affecting the corresponding TGF-B-, MEEK/MAPK/ERK-, JNK-dependent signaling pathways. Continually enriched molecular data reveal new interactions involved in the ubiquitination process that drive cancerous cells to differences and imbalances regarding their metabolic potential providing new knowledge on this topic. For example, specific mutations in signalling pathways -such as NOTCH1C1133Y mutation- strongly interact with E3 ubiquitin ligase (FBXW7) by degrading it (55). Therefore, exploring and understanding the role of ubiquitination modifications in OSCC and neoplasia in general, is a crucial step for identifying patients with specific molecular signatures, potentially eligible for targeted therapeutic strategies.
Conflicts of Interest
The Authors declare no conflicts of interest associated with this article.
Authors’ Contributions
ET, EK, VR, VP, AC researched the literature and drafted the article, with ET a major contributor in writing the article. NM, LM, DR, PP, ODcollected the data provided by the corresponding references. All Authors read and approved the final article.
References
- 1.Akutsu M, Dikic I, Bremm A. Ubiquitin chain diversity at a glance. J Cell Sci. 2016;129(5):875–880. doi: 10.1242/jcs.183954. [DOI] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458(7237):422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18(6):579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 4.Chen RH, Chen YH, Huang TY. Ubiquitin-mediated regulation of autophagy. J Biomed Sci. 2019;26(1):80. doi: 10.1186/s12929-019-0569-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu C, Feng K, Zhao X, Huang S, Cheng Y, Qian L, Wang Y, Sun H, Jin M, Chuang TH, Zhang Y. Regulation of autophagy by E3 ubiquitin ligase RNF216 through BECN1 ubiquitination. Autophagy. 2014;10(12):2239–2250. doi: 10.4161/15548627.2014.981792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dikic I. Proteasomal and autophagic degradation systems. Annu Rev Biochem. 2017;86:193–224. doi: 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- 7.Khaminets A, Behl C, Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 2016;26(1):6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Zaffagnini G, Martens S. Mechanisms of selective autophagy. J Mol Biol. 2016;428(9 Pt A):1714–1724. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura N. Ubiquitin system. Int J Mol Sci. 2018;19(4):1080. doi: 10.3390/ijms19041080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K, Ingolia NT, Beckmann R, Inada T. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat Commun. 2017;8(1):159. doi: 10.1038/s41467-017-00188-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeuchi K, Tesina P, Matsuo Y, Sugiyama T, Cheng J, Saeki Y, Tanaka K, Becker T, Beckmann R, Inada T. Collided ribosomes form a unique structural interface to induce Hel2-driven quality controlpathways. EMBO J. 2019;38(5):e100276. doi: 10.15252/embj.2018100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takehara Y, Yashiroda H, Matsuo Y, Zhao X, Kamigaki A, Matsuzaki T, Kosako H, Inada T, Murata S. The ubiquitination-deubiquitination cycle on the ribosomal protein eS7A is crucial for efficient translation. iScience. 2021;24(3):102145. doi: 10.1016/j.isci.2021.102145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDowell GS, Philpott A. Non-canonical ubiquitylation: mechanisms and consequences. Int J Biochem Cell Biol. 2013;45(8):1833–1842. doi: 10.1016/j.biocel.2013.05.026. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol. 2006;3(4):255–261. [PubMed] [Google Scholar]
- 15.Wilck N, Fechner M, Dan C, Stangl V, Stangl K, Ludwig A. The effect of low-dose proteasome inhibition on pre-existing atherosclerosis in LDL receptor-deficient mice. Int J Mol Sci. 2017;18(4):781. doi: 10.3390/ijms18040781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi S, Gesualdo C, Maisto R, Trotta MC, Di Carluccio N, Brigida A, Di Iorio V, Testa F, Simonelli F, D’Amico M, Di Filippo C. High levels of serum ubiquitin and proteasome in a case of HLA-B27 uveitis. Int J Mol Sci. 2017;18(3):505. doi: 10.3390/ijms18030505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyengar PV. Regulation of ubiquitin enzymes in the TGF-β pathway. Int J Mol Sci. 2017;18(4):877. doi: 10.3390/ijms18040877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Chen Y, Gan X, Huang Z, Zou M, Fu W, Xing W, Xu D. SAK-HV decreases the self-ubiquitination of MEKK1 to promote macrophage proliferation via MAPK/ERK and JNK pathways. Int J Mol Sci. 2017;18(4):835. doi: 10.3390/ijms18040835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, Boyse EA. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci USA. 1975;72(1):11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12(5):295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258(13):8206–8214. [PubMed] [Google Scholar]
- 22.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhananjayan SC, Ismail A, Nawaz Z. Ubiquitin and control of transcription. Essays Biochem. 2005;41:69–80. doi: 10.1042/EB0410069. [DOI] [PubMed] [Google Scholar]
- 24.Hurst JH, Dohlman HG. Dynamic ubiquitination of the mitogen-activated protein kinase kinase (MAPKK) Ste7 determines mitogen-activated protein kinase (MAPK) specificity. J Biol Chem. 2013;288(26):18660–18671. doi: 10.1074/jbc.M113.475707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jentsch S, McGrath JP, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 26.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84(2):277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 27.Dougherty SE, Maduka AO, Inada T, Silva GM. Expanding role of ubiquitin in translational control. Int J Mol Sci. 2020;21(3):1151. doi: 10.3390/ijms21031151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 29.Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18(5):520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- 30.Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, Yang XW, Martinez-Martin N, Matsumoto ML, Dixit VM, Rape M. Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell. 2017;171(4):918–933.e20. doi: 10.1016/j.cell.2017.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbé S. Deubiquitylases from genes to organism. Physiol Rev. 2013;93(3):1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 32.Grumati P, Dikic I. Ubiquitin signaling and autophagy. J Biol Chem. 2018;293(15):5404–5413. doi: 10.1074/jbc.TM117.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer. 2017;17(3):184–197. doi: 10.1038/nrc.2016.143. [DOI] [PubMed] [Google Scholar]
- 34.Dubrez L. Regulation of E2F1 transcription factor by ubiquitin conjugation. Int J Mol Sci. 2017;18(10):2188. doi: 10.3390/ijms18102188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13(8):508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 36.Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. 2018;20(5):521–527. doi: 10.1038/s41556-018-0092-5. [DOI] [PubMed] [Google Scholar]
- 37.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19(6):349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 38.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8(11):931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 39.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wirth M, Joachim J, Tooze SA. Autophagosome formation—the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013;23(5):301–309. doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 41.Antonioli M, Albiero F, Nazio F, Vescovo T, Perdomo AB, Corazzari M, Marsella C, Piselli P, Gretzmeier C, Dengjel J, Cecconi F, Piacentini M, Fimia GM. AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev Cell. 2014;31(6):734–746. doi: 10.1016/j.devcel.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 42.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, Cecconi F. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15(4):406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 43.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3(123):ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Platta HW, Abrahamsen H, Thoresen SB, Stenmark H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem J. 2012;441(1):399–406. doi: 10.1042/BJ20111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali J, Sabiha B, Jan HU, Haider SA, Khan AA, Ali SS. Genetic etiology of oral cancer. Oral Oncol. 2017;70:23–28. doi: 10.1016/j.oraloncology.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 46.Grégoire V, Lefebvre JL, Licitra L, Felip E, EHNS-ESMO-ESTRO Guidelines Working Group Squamous cell carcinoma of the head and neck: EHNS-ESMO-ESTRO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21(Suppl 5):v184–v186. doi: 10.1093/annonc/mdq185. [DOI] [PubMed] [Google Scholar]
- 47.Reder H, Wagner S, Gamerdinger U, Sandmann S, Wuerdemann N, Braeuninger A, Dugas M, Gattenloehner S, Klussmann JP, Wittekindt C. Genetic alterations in human papillomavirus-associated oropharyngeal squamous cell carcinoma of patients with treatment failure. Oral Oncol. 2019;93:59–65. doi: 10.1016/j.oraloncology.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 48.Song A, Wang Y, Jiang F, Yan E, Zhou J, Ye J, Zhang H, Ding X, Li G, Wu Y, Zheng Y, Song X. Ubiquitin D promotes progression of oral squamous cell carcinoma via NF-Kappa B signaling. Mol Cells. 2021;44(7):468–480. doi: 10.14348/molcells.2021.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie W, Xu L. Ubiquitin-specific protease 14 promotes radio-resistance and suppresses autophagy in oral squamous cell carcinoma. Exp Cell Res. 2021;398(2):112385. doi: 10.1016/j.yexcr.2020.112385. [DOI] [PubMed] [Google Scholar]
- 50.Liu CM, Yu CC, Lin T, Liao YW, Hsieh PL, Yu CH, Lee SS. E3 ligase STUB1 attenuates stemness and tumorigenicity of oral carcinoma cells via transglutaminase 2 regulation. J Formos Med Assoc. 2020;119(10):1532–1538. doi: 10.1016/j.jfma.2020.06.004. [DOI] [PubMed] [Google Scholar]
- 51.Luo F, Zhou Z, Cai J, Du W. DUB3 facilitates growth and inhibits apoptosis through enhancing expression of EZH2 in oral squamous cell carcinoma. Onco Targets Ther. 2020;13:1447–1460. doi: 10.2147/OTT.S230577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu T, Liu J, Chen Q, Jin S, Mi S, Shao W, Kudo Y, Zeng S, Qi G. Expression of USP22 and the chromosomal passenger complex is an indicator of malignant progression in oral squamous cell carcinoma. Oncol Lett. 2019;17(2):2040–2046. doi: 10.3892/ol.2018.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Yin W, Shi C. E3 ubiquitin ligase, RNF139, inhibits the progression of tongue cancer. BMC Cancer. 2017;17(1):452. doi: 10.1186/s12885-017-3438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jingjing W, Wenzheng G, Donghua W, Guangyu H, Aiping Z, Wenjuan W. Deubiquitination and stabilization of programmed cell death ligand 1 by ubiquitin-specific peptidase 9, X-linked in oral squamous cell carcinoma. Cancer Med. 2018;7(8):4004–4011. doi: 10.1002/cam4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng Y, Song A, Wang C, Zhang W, Liang D, Ding X, Li G, Zhang H, Zhang W, Du Y, Zhou J, Wu H, Wu Y, Song X. Isoform specific FBXW7 mediates NOTCH1 Abruptex mutation C1133Y deregulation in oral squamous cell carcinoma. Cell Death Dis. 2020;11(8):615. doi: 10.1038/s41419-020-02873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]