Abstract
The cytochrome P450 sterol 14α-demethylase (CYP51) of Candida albicans is involved in an essential step of ergosterol biosynthesis and is the target for azole antifungal compounds. We have undertaken site-directed mutation of C. albicans CYP51 to produce a recombinant mutant protein with the amino acid substitution R467K corresponding to a mutation observed clinically. This alteration perturbed the heme environment causing an altered reduced-carbon monoxide difference spectrum with a maximum at 452 nm and reduced the affinity of the enzyme for fluconazole, as shown by ligand binding studies. The specific activity of CYP51(R467K) for the release of formic acid from 3β-[32-3H]hydroxylanost-7-en-32-ol was 70 pmol/nmol of P450/min for microsomal protein compared to 240 pmol/nmol of P450/min for microsomal fractions expressing wild-type CYP51. Furthermore, inhibition of activity by fluconazole revealed a 7.5-fold-greater azole resistance of the recombinant protein than that of the wild type. This study demonstrates that resistance observed clinically can result from the altered azole affinity of the fungal CYP51 enzyme.
Inhibition of the ergosterol biosynthetic pathway in fungi has attracted intense interest for the development and application of antifungal compounds (for a review, see reference 4). Although there are a number of inhibitors of sterol biosynthesis, the azole antifungal compounds represent the most widespread and are used in both medicine and agriculture. Azole antifungal compounds are known to bind and inhibit cytochrome P450 sterol 14α-demethylase (other names, CYP51 and P45014DM), which catalyzes the oxidative removal of the sterol C-14 methyl group. Inhibition of this step in sterol biosynthesis results in the accumulation of 14-methylated sterols, particularly 14α-methylergosta-8,24(28)-dien-3β-ol, which ultimately results in cell growth arrest in Candida albicans (7, 8). Recent work has indicated that the substrate of the 14α-demethylase reaction, lanosterol, can support growth if it is not modified by other sterol biosynthetic enzymes (2).
Fungal infection rates have increased due to organ transplantation, diseases and chemotherapies affecting the immune system, and, coupled to increased azole antifungal use, coupled to fungal resistance has emerged as a significant problem in the clinic, e.g., in >10% of AIDS patients (10). We have been interested in elucidating the molecular mechanisms of azole antifungal resistance in C. albicans and other fungi. Previously, our work has shown that alteration of the ergosterol biosynthetic pathway by a defect in sterol Δ5,6-desaturase (responsible for the conversion of ergosta-7,22-dienol to ergosterol) results in resistance to azole antifungals in Saccharomyces cerevisiae, as indicated by Mendelian genetic segregation. Defects in sterol Δ5,6-desaturation were also found in azole-resistant C. albicans isolated from the clinic and in Ustilago maydis (3, 19; S. L. Kelly, D. C. Lamb, D. E. Kelly, J. Loeffler, and H. Einsele, Letter, Lancet 348:1523–1524). A defect in this enzyme prevents the formation of the toxic sterol 14α-methylergosta-8,24(28)-dien-6β,3α-diol when the fungal cells are treated with azole and thus results in the formation of 14α-methylfecosterol, which allows cell growth in the presence of azole (5). Additionally, we reported, on the basis of laboratory studies, that alteration in the target enzyme CYP51 by a single amino acid, in this case a change of Thr315 to Ala by a single base substitution, produced an altered protein exhibiting reduced catalytic activity and reduced affinity for fluconazole and ketoconazole (9; S. L. Kelly, Abstr. 3rd Int. Symp. Cytochrome P450 Biodiversity, abstr. 28, 1995). Subsequently, a number of reports showing that differing genetic polymorphisms in the CYP51 gene from clinical C. albicans isolates are responsible for and/or associated with azole antifungal resistance have emerged, but no biochemical data on the isolated CYP51 protein have been available (11, 14, 20).
In the present study, we have undertaken site-directed mutagenesis of the C. albicans CYP51, heterologously expressed in S. cerevisiae, to assess the contribution of the mutation R467K to fluconazole resistance. This alteration is within the heme-binding domain and has previously been associated with the emergence of resistance by genetic means in clinical isolates of C. albicans (14, 20). We demonstrate that the alteration causes reduced affinity for the drug and alters the heme-binding environment in the protein active site.
MATERIALS AND METHODS
Site-directed mutagenesis.
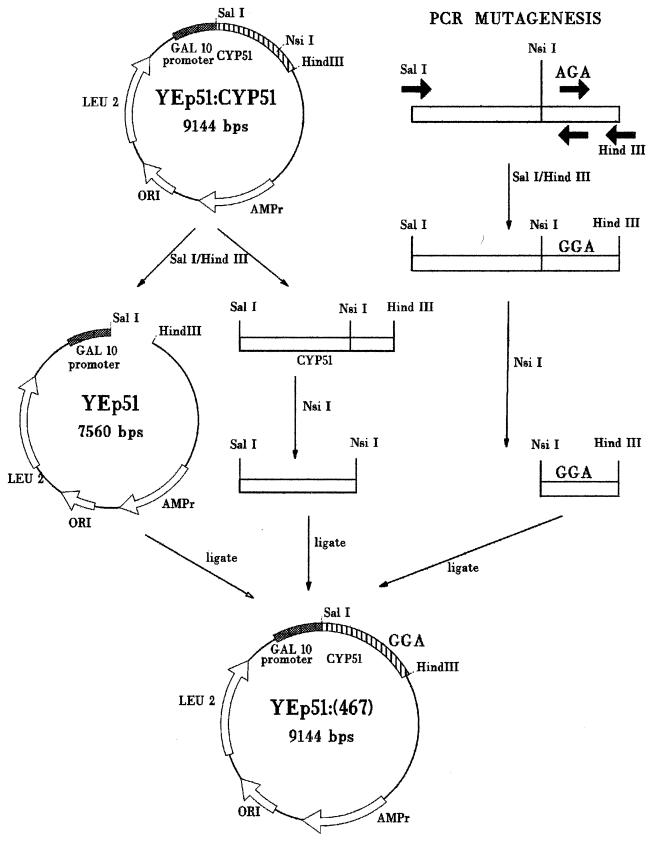
Our previous studies have employed a yeast expression system to express the C. albicans CYP51 by using the S. cerevisiae GAL 10 promoter in the vector YEp51 (17). Recombinant PCR was employed to replace codon 467 encoding arginine (AGA) with one encoding lysine (GGA). The following oligonucleotides were used as outside primers: 1, 5′-CGCGTCGACATAATGGCTATTGTTGAAACTGTC-3′, annealing to positions 1 to 21 of the C. albicans CYP51; 2, 5′-ACTAACGTTTTAAAACATACAAGTTTCTCT-3′, annealing to the 3′ end at positions 1569 to 1587 of CYP51. Inside primers used in the PCR mutagenesis were as follows: 1, 5′-GGTCGTGGTGGACATAGATGTATTGGG-3′; 2, 5′-CCCAATACATCTATGTCCACCACCACCAAA-3′. In a first step, two separate PCRs were carried out with outside primer 1 and inside primer 2 in one reaction and inside primer 1 and outside primer 2 in the other. The partially overlapping DNA fragments obtained were purified, mixed, and recombined in a later PCR step with outside primer 1 and outside primer 2. PCRs were performed on a Perkin-Elmer DNA thermal cycler with conditions as previously described. PCR was undertaken with Pfu polymerase (Stratagene, Amsterdam, The Netherlands). The resulting DNA fragment containing the R467K mutation was digested with NsiI/HindIII and ligated into the YEp51:CYP51 expression plasmid as shown in Fig. 1. Introduction of the mutation and maintenance of the authentic sequence were corroborated by DNA sequencing. All restriction enzymes and T4 DNA ligase were obtained from Promega (Southampton, United Kingdom), and the recommended conditions for use were applied.
FIG. 1.
Schematic representation of the strategy used for the generation of CYP51(R467K). Site-directed PCR mutagenesis was performed to change the triplet at position 467 from AGA to GGA as described in Materials and Methods. The NsiI-HindIII mutant fragment was ligated into the corresponding region of the wild-type gene in the yeast expression vector YEp51 allowing expression from the GAL10 promoter.
Heterologous expression of recombinant proteins.
The plasmid YEp51:CYP51(R467K) was transformed into S. cerevisiae AH22 (MATa leu2-3,2-112 his3-11,3-15 canr). Yeast transformants were grown at 28°C and 250 rpm with 250-ml cultures in 500-ml flasks. The media used consisted of Difco (Becton Dickinson, Meylan, France) yeast nitrogen base without amino acids (1.34% [wt/vol]) supplemented with 100 mg of histidine/liter and 2% (wt/vol) glucose as the initial carbon source. Heterologous expression was induced when the glucose was exhausted at a cell density of approximately 108 cells/ml. The culture was left a further 4 h before galactose was added to a concentration of 3% (wt/vol). After 20 h of induction cells were harvested by centrifugation, resuspended in buffer containing 0.4 M sorbitol–50 mM Tris-HCl (pH 7.4), and broken with a disintegrator (Braun GmbH, Mesungen, Germany) with four bursts of 30 s together with cooling from liquid carbon monoxide. Cell debris was removed by centrifugation at 1,500 × g for 10 min with an MSE bench centrifuge. The resulting supernatant was centrifuged twice at 10,000 × g for 20 min to remove mitochondria and then at 100,000 × g for 90 min to yield the microsomal pellet. This was resuspended with a Potter-Elvehjeim glass homogenizer at about 10 mg of protein/ml in the same buffer described above. The P450 concentration was measured by reduced carbon monoxide difference spectroscopy (13), and protein concentrations were measured as described previously (12). NADPH-cytochrome P450 reductase levels were measured as described by Vermillion and Coon (18).
Spectrophotometric studies with recombinant proteins.
The interaction of fluconazole with C. albicans wild-type CYP51 and CYP51(R467K), isolated following heterologous expression in S. cerevisiae, was analyzed spectrophotometrically by using a Hitachi U3010 recording spectrophotometer. Microsomal wild-type CYP51 and CYP51(R467K) (0.2 nmol) were dissolved in 0.1 M potassium phosphate buffer, pH 7.2, containing 20% glycerol. The cytochrome was titrated with fluconazole. Further details are given in the figure legends.
Determination of microsomal sterol 14α-demethylase activity and inhibition by fluconazole.
Our previous studies have developed a novel method for studying the sterol 14α-demethylase activity of recombinant CYP51 following heterologous expression in yeast (17). Following the addition of the 32-tritiated CYP51 substrate 3β-[32-3H]hydroxylanost-7-en-32-ol (52 μg in 10 μl of dimethylformamide) to the microsomal protein, NADPH (final concentration, 1 mM) was added to the reaction mixture and the mixture was incubated at 37°C. After 20 min, the reaction was stopped by the addition of a mixture of dichloromethane (2 ml) and water (2 ml), and the mixture was immediately shaken and then centrifuged. The organic layer was discarded, further dichloromethane and water (2 ml each) were added, and the above procedure was repeated. To the resulting aqueous phase was added charcoal, and the suspension was shaken, left at 4°C for 1 h, and finally centrifuged to remove the charcoal. The radioactivity of the aqueous phase was measured by liquid scintillation counting. Sterol 14α-demethylase activities of the microsomal preparations of wild-type CYP51 and CYP51(R467K) were assayed as described above. Fluconazole was added to the reaction mixtures from 1,000-fold stock solutions. Double-reciprocal analysis and Lineweaver-Burk plots were used to calculate enzyme kinetic parameters based on mean values of duplicate experiments.
RESULTS
Site-directed mutagenesis and heterologous expression of CYP51 and CYP51(R467K).
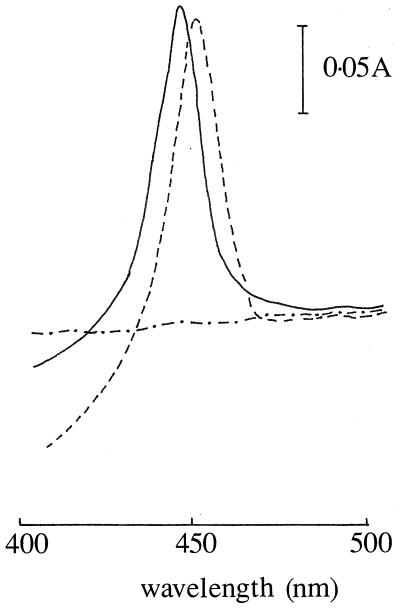
The expression system, which we have previously developed, had already included modifying the coding sequence at position 263 due to the presence of CTG, which encodes leucine in S. cerevisiae but serine in C. albicans (15). Introduction of TCT allowed the authentic amino acid (serine) to be included when CYP51 was expressed in S. cerevisiae. Further mutagenesis was undertaken to change Arg467 to Lys, a process involving a single base substitution changing AGA to GGA, as outlined in Fig. 1. S. cerevisiae AH22 transformed with CYP51(R467K) expressed levels of P450 comparable to those expressed by the wild-type enzyme, with up to 2.5 nmol of P450/mg of microsomal protein produced after expression from the GAL10 promoter of YEp51, as determined from the optical absorption spectrum of the carbon monoxide-bound form of reduced P450. NADPH-cytochrome P450 reductase levels in the microsomes were 0.85 ± 0.7 nmol/mg of protein for CYP51 and 0.94 ± 0.8 nmol/mg of protein for CYP51(R467K), indicating that differences in this electron donor did not contribute to different activities. Both CYP proteins exhibited stability under the conditions of the assays, as indicated by unchanged specific content measurements. The absorption maximum of the CO-bound form of wild-type CYP51 was located at 447 nm. However, the CO-bound form of CYP51(R467K) had the Soret absorption maximum at 452 nm (Fig. 2), which reflects pertubations in the heme environment of the protein.
FIG. 2.
Reduced-carbon monoxide difference spectrum of microsomal cytochrome CYP51(R467K). Shown are the reduced-carbon monoxide difference spectra of microsomal fractions, prepared as described in Materials and Methods, after induction of heterologous expression in yeast transformants containing YEp51 alone (— • —), CYP51 (——), and CYP51 (R467K) (–––).
Binding of fluconazole to CYP51 and CYP51(R467K).
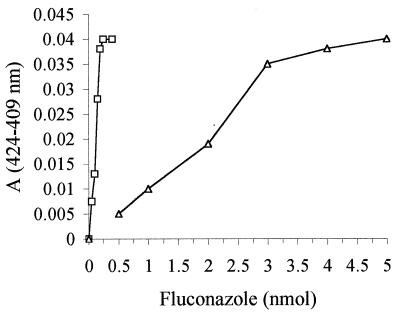
Fluconazole caused characteristic changes in the oxidized forms of both CYP51 and CYP51(R467K) due to a shift to a low-spin state for the heme on binding fluconazole. This interaction gave rise to the type II change, which is characterized by the displacement of the native sixth ligand of the heme iron (water molecule) by the nitrogen atom in the triazole ring (N-4) of fluconazole and which results in a spectral peak (420 to 427 nm) and a corresponding trough (390 to 410 nm) (21). The intensity of the resulting difference spectrum was found to be proportional to the amount of the azole-bound form of the cytochrome (22). Fluconazole caused the type IIa spectral change in both CYP51 and CYP51(R467K). The spectral peak was located at 424 nm and the trough was located at 409 nm for wild-type CYP51 and CYP51(R467K). The apparent affinity of fluconazole for CYP51(R467K) was greatly reduced compared to that for wild-type CYP51. Fluconazole bound stoichiometrically with wild-type CYP51, whereas the compound only showed saturation in binding with CYP51(R467K) at approximately 20-fold excess fluconazole over P450 (Fig. 3). Alteration in the affinity of fluconazole for CYP51(R467K) compared to that for wild-type CYP51 indicates a subtle alteration in the conformational environment around the P450 heme caused by this mutation, resulting in decreased fluconazole affinity.
FIG. 3.
Comparative analysis of type II binding spectra. The magnitudes of spectra obtained, as described in Materials and Methods, with different concentrations of fluconazole bound to 0.2 nmol of CYP51 (□) or CYP51(R467K) (▵) are shown. Data were reproducible in triplicate experiments.
Sterol 14α-demethylase activity of CYP51 and CYP51(R467K) and inhibition by fluconazole.
The catalytic activities of purified CYP51 and CYP51(R467K) were assayed by measuring the release of [3H]formic acid during the conversion of 3β-[32-3H]hydroxylanost-7-en-32-ol to its C-14 demethylated product, 4,4-dimethyl-5α-cholesta-8,14,24-trien-3β-ol. Heterologously expressed microsomal CYP51(R467K) showed a reduction in its ability to metabolize the alcohol derivative substrate compared to the wild-type CYP51 (60% reduction in the catalytic activity of CYP51(R467K) compared to CYP51). As expected, the demethylation activity was below the baseline of detectability in microsomes from the host strain harboring the parent vector, YEp51 (data not shown). Wild-type CYP51 and CYP51(R467K) were shown to have similar Km values (20 μM). However, the maximal enzymatic rates for both proteins differed [240 pmol of product formed/min/nmol of CYP51 compared to 70 pmol of product formed/min/nmol of CYP51(R467K)], showing the reduced catalytic activity of the mutant enzyme compared to that of the wild-type form.
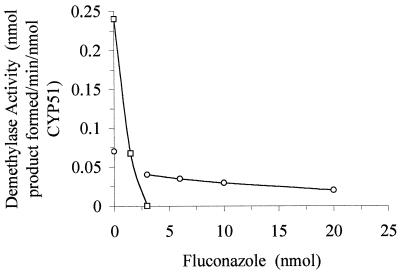
Figure 4 shows the results of experiments in which microsomal CYP51 and CYP(R467K) (3 nmol of P450) activities were inhibited with increasing amounts of fluconazole. For CYP51, the inhibition of enzymatic activity was dependent on azole concentration, and total inhibition of enzyme activity occurred at a concentration equimolar to that of CYP51 (3 nmol of CYP51 required 3 nmol of fluconazole for complete inhibition and 1.5 nmol for 50% inhibition. However, for CYP51(R467K) 38 nmol of fluconazole was needed for complete inhibition (by extrapolation) and 11.3 nmol for 50% inhibition. For comparison of 50% inhibition points this was a 7.5-fold excess of fluconazole needed for CYP51(R467K) inhibition compared to CYP51.
FIG. 4.
Inhibition of sterol 14α-demethylase activity. Comparative plot of sterol demethylase activity in the presence of various amounts of fluconazole for CYP51 and CYP51(R467K). P450 was used to examine the inhibitory effects of fluconazole for CYP51 (□) and CYP51(R467K) (○).
DISCUSSION
Resistance to azole antifungal compounds has been shown to be attributed to one or a combination of the following mechanisms: (i) reduced accumulation of the compound within the fungal cell, (ii) alteration of the ergosterol biosynthetic pathway to prevent formation of fungistatic sterols, and (iii) alteration in the target enzyme resulting in reduced affinity for the azole antifungal compound (6). In the present study we have proved at the biochemical level that the resistance mechanism resulting from the substitution R467K in CYP51 is due to reduced affinity for the drug. This demonstration shows that alterations in CYP51 can produce resistance, and diagnostic tests resulting in rapid adjustment of treatment regimens can be envisaged.
Fluconazole inhibits CYP51 through coordination of N-4 in the triazole ring with the heme iron of the cytochrome. In addition, hydrophobic interactions between the N-1 substituent and the apoprotein of the active site have also been proposed as being important in determining the affinity of fluconazole for the enzyme. In particular molecular modeling, based on the known structure of P450CAM, has suggested an aromatic interaction between the fluorophenyl moiety and either Phe233 or Phe235 in the apoprotein (1; D. C. Lamb, P. Boscott, B. C. Baldwin, D. E. Kelly, and S. L. Kelly, Abstr. Proc. Int. Symp. Cytochrome P450 Microorganisms Plants, abstr. P-32, 1993). The observation that a 20-fold increase in fluconazole was required to saturate CYP51(R467K) compared to that necessary to saturate wild-type CYP51 clearly reveals reduced affinity of fluconazole for CYP51(R467K). Since fluconazole can elicit a type II spectral change, the low affinity of this compound cannot be explained by steric hindrance or exclusion from the active site. Therefore, the mutation has altered the stereo- and regiospecific interactions of the N-1 substituent with the apoprotein near the heme involved in binding fluconazole. Reduced affinity of fluconazole for CYP51(R467K) was substantiated when a 7.5-fold-excess concentration of compound was required to inhibit sterol 14α-demethylase activity, as identified by total inhibition of activity. This reduction in fluconazole affinity has arisen by replacement of the arginine for lysine at position 467, altering the heme environment in CYP51. This was also reflected in the altered reduced-CO difference spectrum, where the Soret maximum moved from 447 nm in the wild type to 452 nm in the mutant. One possible explanation for reduced fluconazole affinity may be that the R467K mutation results in a slight tilting of the heme or some other alteration in the position of the heme so that the interaction of N-1 substituent groups of fluconazole with CYP51 apoprotein is decreased and hence reduced affinity results. The residue under investigation forms part of the conserved heme-binding domain observed in all cytochrome P450 molecules (Fig. 5) (16). In all the known CYP51 genes isolated to date the same triplet encodes arginine, but other members of the superfamily have lysine at the same position where it was found in the mutant protein (16). This may explain the retention of catalytic activity by the mutant protein altered in this important region of all cytochrome P450 molecules, i.e., this alteration is compatible with a functional heme-binding environment.
FIG. 5.
Amino acid sequences of P450s in the heme-binding region. Shown are an alignment of CYP51 amino acid sequences from humans, rats, S. cerevisiae (S.C.), Sorghum bicolor (S.B.), Mycobacterium tuberculosis (M.T.), and C. albicans (C.A.) and a comparison to the same region in CYP51(R467K) and various mammalian CYP isoforms involved in foreign compound metabolism. The conserved arginine and lysine residues are in boldface.
The alteration in the heme environment was also reflected when the catalytic activity of the mutant enzyme was measured. The catalytic turnover was reduced by the mutation, with no significant effect on the substrate binding constant (Km) indicating that the affinity of the enzyme for sterol was not altered. An alteration in the location of molecular oxygen vis-à-vis the C-14 methyl group of the sterol substrate during catalysis may account for this observation.
Here we demonstrate altered fluconazole affinity for mutant CYP51(R467K), which has been observed in separate fluconazole-resistant strains of C. albicans from AIDS patients (14, 20). It can be anticipated to be a common mechanism of resistance, although an epidemiological study of the types of resistant strains is not yet completed. Other molecular changes in these resistant strains have also been observed, and the contribution of the CYP51(R467K) mutation to resistance in cells is under further evaluation by reintroduction into C. albicans. Single-base-pair changes in CYP51 can be detected by rapid PCR-based methods (11) and this may allow adjustment of therapy where the sensitivity of such strains to alternative antifungal agents has been previously characterized.
ACKNOWLEDGMENT
We are grateful for support from the University of Wales Aberystwyth Research Fund.
REFERENCES
- 1.Boscott P E, Grant G H. Modeling cytochrome P450 14 alpha demethylase (Candida albicans) from P450cam. J Mol Graph. 1994;12:185–92. doi: 10.1016/0263-7855(94)80086-3. [DOI] [PubMed] [Google Scholar]
- 2.Gachotte D, Pierson C A, Lees N D, Barbuch R, Koegel C, Bard M. A yeast sterol auxotroph (erg25) is rescued by addition of azole antifungals and reduced levels of heme. Proc Natl Acad Sci USA. 1997;94:11173–11178. doi: 10.1073/pnas.94.21.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joseph-Horne T, Manning N J, Hollomon D, Kelly S L. Defective sterol delta 5(6)-desaturase as a cause of azole resistance in Ustilago maydis. FEMS Microbiol Lett. 1995;127:29–34. doi: 10.1111/j.1574-6968.1995.tb07445.x. [DOI] [PubMed] [Google Scholar]
- 4.Kelly S L, Arnoldi A, Kelly D E. Molecular genetic analysis of azole antifungal mode of action. Biochem Soc Trans. 1993;21:1034–1038. doi: 10.1042/bst0211034. [DOI] [PubMed] [Google Scholar]
- 5.Kelly S L, Lamb D C, Corran A J, Baldwin B C, Kelly D E. Mode of action and resistance to azole antifungals associated with the formation of 14 α-methylergosta-8,24(28)-dien-3 β,6 α-diol. Biochem Biophys Res Commun. 1995;207:910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 6.Kelly S L, Lamb D C, Kelly D E. Inhibitors of CYP51 as antifungal agents and resistance to azole antifungals, 157–172. In: Arinc E, Schenkman J B, Hodgson E, editors. Molecular and applied aspects of oxidative drug metabolizing enzymes. New York, N.Y: Plenum; 1998. [Google Scholar]
- 7.Kelly S L, Lamb D C, Kelly D E, Loeffler J E, Einsele H. Resistance to fluconazole in Candida albicans from AIDS patients involving cross-resistance to amphotericin. Lancet. 1996;348:1523–1524. doi: 10.1016/S0140-6736(05)65949-1. [DOI] [PubMed] [Google Scholar]
- 8.Kelly S L, Lamb D C, Kelly D E, Manning N J, Loeffler J, Hebart H, Schumacher U, Einsele H. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta 5,6-desaturation. FEBS Lett. 1997;400:80–82. doi: 10.1016/s0014-5793(96)01360-9. [DOI] [PubMed] [Google Scholar]
- 9.Lamb D C, Kelly D E, Schunck W H, Shyadehi A Z, Akhtar M, Lowe D J, Baldwin B C, Kelly S L. The mutation T315A in Candida albicans sterol 14α-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 10.Law D, Moore C B, Wardle H M, Ganguli L A, Keaney M G, Denning D W. High prevalence of antifungal resistance in Candida spp. from patients with AIDS. J Antimicrob Chemother. 1994;34:659–668. doi: 10.1093/jac/34.5.659. [DOI] [PubMed] [Google Scholar]
- 11.Loffler J, Kelly S L, Hebart H, Schumacher U, Lass-Florl C, Einsele H. Molecular analysis of CYP51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol Lett. 1997;151:263–268. doi: 10.1111/j.1574-6968.1997.tb12580.x. [DOI] [PubMed] [Google Scholar]
- 12.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 13.Omura T, Sato R. The carbon monoxide binding pigment of liver microsomes. I. Evidence for its haemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 14.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α- demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santos A S, Keith G, Tuite M F. Non-standard translational events in Candida albicans mediated by an unusual seryl-tRNA with a 5′-CAG-3′ (leucine) anticodon. EMBO J. 1993;12:607–616. doi: 10.1002/j.1460-2075.1993.tb05693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu T, Hirano K, Takahashi M, Hatano M, Fujii-Kuriyama Y. Site directed mutagenesis of rat liver cytochrome P450d: axial ligand and heme incorporation. Biochemistry. 1988;27:4138–4141. doi: 10.1021/bi00411a035. [DOI] [PubMed] [Google Scholar]
- 17.Shyadehi A Z, Lamb D C, Kelly S L, Kelly D E, Schunck W H, Wright J N, Corina D, Akhtar M. The mechanism of the acyl-carbon bond cleavage reaction catalyzed by recombinant sterol 14α-demethylase of Candida albicans (other names are: lanosterol 14α-demethylase, P-45014DM, and CYP51) J Biol Chem. 1996;271:12445–12450. doi: 10.1074/jbc.271.21.12445. [DOI] [PubMed] [Google Scholar]
- 18.Vermillion J L, Coon M J. Purified liver microsomal NADPH-cytochrome P450 reductase: spectral characterization of oxidation-reduction states. J Biol Chem. 1978;253:2694–2704. [PubMed] [Google Scholar]
- 19.Watson P F, Rose M E, King D J, Ellis S W, England H, Kelly S L. Defective sterol C5-6 desaturation and azole resistance: a new hypothesis on the mode of action of azole antifungals. Biochem Biophys Res Commun. 1989;164:1170–1175. doi: 10.1016/0006-291x(89)91792-0. [DOI] [PubMed] [Google Scholar]
- 20.White T C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α-demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;41:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiggins T, Baldwin B C. Binding of azole fungicides related to diclobutrazol to cytochrome P450. Pesticide Sci. 1984;15:206–209. [Google Scholar]
- 22.Yoshida Y, Aoyama Y. Interaction of azole antifungal agents with cytochrome P45014DM purified from Saccharomyces cerevisiae microsomes. Biochem Pharmacol. 1987;36:229–235. doi: 10.1016/0006-2952(87)90694-0. [DOI] [PubMed] [Google Scholar]