Abstract
Background
Testing for SARS-CoV-2, together with vaccination, is one of the most vital strategies in curbing the current COVID-19 pandemic. The pandemic has led to an unprecedented need for diagnostic testing and the rapid emergence of an abundance of commercial assays on the market. Due to the nature of the pandemic and in the interest of health protection, many of these assays received provisional authorisation for emergency use without thorough validation. To limit false negative and false positive results, it is key to define common criteria that SARS-CoV-2 assays need to fulfil. VALCOR or “VALidation of SARS-CORona Virus-2 assays” is a protocol designed to set up a framework for test validation of SARS-CoV-2 virus assays.
Objectives
VALCOR is a study protocol for the validation of assays used for confirmation of the presence of SARS-CoV-2 in patients with COVID-19 disease or the screening of carriers of SARS-CoV-2 virus by the identification of viral RNA in oropharyngeal and/or nasopharyngeal specimens or other specimens from the human respiratory tract.
Methods
The VALCOR panel of samples will contain clinical human specimens and standardised artificial specimens. The collection of clinical specimens will include nasopharyngeal or oropharyngeal specimens or other specimens from the respiratory tract obtained from COVID-19 patients and healthy carriers of SARS-CoV-2 as well as specimens from subjects not carrying SARS-CoV-2. Artificial specimens include calibrated amounts of viral RNA of SARS-CoV-2 sequences provided by established competent agencies that produce reference materials for the assessment of the limit of detection of each assay. The panel of samples are sent from a central reference laboratory (having access to biobanks of clinical specimens tested already for SARS-CoV-2 with a reference comparator assay) to participating laboratories for testing with a SARS-CoV-2 index assay that requires evaluation.
Discussion
VALCOR provides a harmonised and standard framework to benchmark the testing performance of SARS-CoV-2 assays that are rapidly evolving. As the pandemic incited an urgent need for testing capacity, there is a gap in the comprehensive validation of SARS-CoV-2 assays. This study will generate comprehensive validation data for assays used for the diagnosis of SARS-CoV-2 and may serve as a basis for other validation protocols.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13690-022-00869-4.
Keywords: SARS-CoV-2, Diagnostic test accuracy, COVID-19, Test validation, Quality control
Background
Coronavirus disease 2019 (COVID-19) is a potentially lethal respiratory disease, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). COVID-19 was initially reported in Wuhan (Hubei province, China) where it caused a large scale, yet localized epidemic near the end of 2019 [1]. Since then, SARS-CoV-2 has spread rapidly across all continents and has evolved into a global pandemic, with more than 450 million confirmed cases and over six million deaths, as of March 2022 [2]. The World Health Organization (WHO) and other international and national health authorities worldwide have underlined the urgent need to increase testing capacity as a vital strategy to reduce transmission of SARS-CoV-2 [3].
Assays that detect infection with SARS-CoV-2 are generally divided into two categories. The first include nucleic acid amplification tests (NAATs). These types of tests are used to detect targeted sequences of the viral genome. For SARS-CoV-2, NAATs identify viral ribonucleic acid (RNA) sequences contained in respiratory samples mainly collected with nasopharyngeal or oropharyngeal swabs. The most common and widely used assay is the reverse transcription-polymerase chain reaction (RT-PCR), which combines reverse transcriptase of RNA into deoxyribonucleic acid (DNA) (referred to as complementary DNA or cDNA) followed by PCR amplification of specific DNA targets. Other amplification methods to detect SARS-CoV-2 include isothermal amplification such as transcription-mediated amplification (TMA) and loop-mediated isothermal amplification (LAMP). Whereas RT-PCR requires cycling at different temperatures, isothermal amplification occurs at a constant temperature. Most NAATs are laboratory-based and require handling by trained personnel. Some NAATs are rapid and can be performed at the point of care, outside specialized laboratory settings; these may however not be as accurate as laboratory-based NAATs. The second category of SARS-CoV-2 tests detect viral antigens, i.e. proteins that surrounds the viral genome. Like the NAATs, antigen tests are used to detect current infection of SARS-CoV-2. Antigen tests differ from NAATs as they are easier to perform and provide rapid results. However, antigen tests have a lower sensitivity when compared to NAATs. Currently, the “gold standard” for the diagnosis of SARS-CoV-2 remains the laboratory-based NAATs, specifically RT-PCR assays [4–6].
Several NAATs and antigen tests have been developed in response to the pandemic. Many health agencies have set up frameworks for rapid test validation and provisional authorization [7, 8]. Sufficient availability of validated tests is required not only for diagnosis and correct management of hospitalized COVID-19 patients but is crucial for the identification of infected individuals, contact tracing, surveillance of the spread of infection and may be pivotal in decision making regarding the estimation of population-based contagious levels. This in turn informs and guides authorities on non-pharmaceutical interventions (NPIs) such as school and workplace closures, stay at home orders, national and international travel restrictions as well as quarantine measures in place [9].
Given the urgency and need for large scale testing, many regulatory agencies have issued provisional authorisations applying validation criteria based on the limit of detection, non-comparative analytical performance without claims for target benchmarks, in silico genome sequence and cross-contamination. As many new assays have arrived on the market, health authorities require lists of well-validated assays and more comprehensive validation criteria are needed [10]. Additionally, with the prospect of SARS-CoV-2 circulating in multiple waves and concerns over new variants of concern, massive testing capacity will have to be allocated globally for years to come [11]. As the majority of assays currently on the market are approved by emergency measures if anything, we propose a structured, open-source approach to the validation of SARS-CoV-2 diagnostic assays, specifically NAATs. Hereinafter, the word “assay(s)” is used to refer to NAATs used for the detection of SARS-CoV-2 unless otherwise specified.
We therefore establish a validation framework for the systematic performance evaluation of SARS-CoV-2 assays. The VALCOR (acronym for VALidation of severe acute respiratory CORona virus 2 assays) protocol is inspired on VALGENT (VALidation of HPVGENotyping Tests) which is a successful forum for comparison and validation of human papillomavirus tests usable for cervical cancer screening [12, 13]. In this paper, we describe the VALCOR study protocol in detail.
Methods
Study design
This is a multi-centre cross-sectional diagnostic test accuracy study to assess the diagnostic parameters of SARS-CoV-2 assays for the detection of SARS-CoV-2. The VALCOR consortium entails a collaboration between Sciensano (the National Scientific Institute of Public Health and currently responsible for the coordination of the VALGENT network) in Belgium, together with virology laboratories (hereinafter referred to as provider laboratories), preferentially with the role of national reference centres for SARS-CoV-2 control in Europe. Provider laboratories will compile a VALCOR panel composed of clinical and artificial samples which will be tested with an established reference SARS-CoV-2 test at the provider laboratory. Well-defined aliquots prepared from the VALCOR samples will be sent to client laboratories for testing with an index SARS-CoV-2 test. Virological sensitivity and specificity of index tests to detect the presence of SARS-CoV-2 as defined by the reference test will be the main outcome. Limit of detection will be assessed on a series of dilutions of clinical and artificial specimens.
Participating provider laboratories
Thus far, two VALCOR studies are in progress, a Belgian VALCOR and an Italian VALCOR. The Belgian VALCOR study has been approved by the Ethics Committee Research (EC Research) of University Hospitals Leuven (UZ Leuven) under the registration reference number S64233. The Italian VALCOR study has been approved by the Ethics Committee of the University of Milano – Bicocca under the registration reference number 0044362/20. Samples for the Belgian VALCOR are provided by the UZ Leuven, national reference laboratory for respiratory pathogens (Leuven, Belgium) and those for Italian VALCOR have been made available by the Clinical Microbiology and Virology Laboratory of the University of Milano-Bicocca (Monza, Italy).
Study population
The protocol foresees including clinical samples and artificial samples. Clinical samples (nasopharyngeal, oropharyngeal or other specimens from the respiratory tract) are obtained from COVID-19 patients or healthy carriers of SARS-CoV-2 as well as subjects not carrying SARS-CoV-2. Artificial samples with calibrated amounts of viral RNA are included as external quality control for the analytical validation of assays. Details of included samples are described below;
Composition of the VALCOR panel
A VALCOR panel will contain 220 clinical specimens (180 non-diluted and 40 diluted) as well as an additional number of dilutions of artificial standard reference viral RNA material.
-
Artificial specimens containing specified SARS-CoV-2 RNA sequences prepared by institutions specialised in the production of standard reference materials from microbiological agents (for instance Joint Research Centre of the European Commission [JRC] [14], National Institute of Standards and Technology [NIST] [15], American Type Culture Collection [ATCC] [16], BEI Resources [17] and others).
A series of dilutions will be prepared from which the limits of detection and/or ranges of detectability will be assessed. The origin, definition and preparation of dilutions are described in the supplementary material (p2–4).
Clinical specimens: residual original rough material or extracted RNA after SARS-CoV-2 testing stored in biobanks of the VALCOR provider laboratories (reference laboratories for SARS-CoV-2 or other laboratories mandated by these reference laboratories). The panel of clinical specimens will be composed as described below;
40 samples derived from hospitalized, SARS-CoV-2 confirmed cases
50 samples derived from non-hospitalized, SARS-CoV-2 confirmed cases
90 samples derived from SARS-CoV-2 negative cases
40 diluted samples from 2b (4 dilutions [1:2, 1:10, 1:20,1:50] of 10 samples randomly selected from the 50 non-hospitalized patients).
Each fresh sample of the VALCOR panel will be divided into aliquots of the original material and stored at − 80 °C. Aliquots upon request will be sent to the client’s laboratories in order to validate their assays Data on the pre-analytical phase of the testing such as the sample retrieval process, transport and storage conditions and storage media type will be recorded in as much detail as possible as these steps can impact test performance.
Testing procedures in VALCOR
One Belgian (VALCOR-BE1) and three Italian VALCOR panels have been compiled so far. The samples of VALCOR-BE1 were suspended in Universal Transport Medium (UTM) and Phosphate Buffered Saline (PBS). VALCOR-IT1 and VALCOR-IT2 were suspended in Universal Transport Medium (UTM) whereas the samples of VALCOR-IT3 were suspended in eNAT™ medium. Each sample of the VALCOR panels has to be tested with an established reference test in the provider laboratory and subsequently with index tests to be evaluated in client laboratories.
Transport of material and transmission of data
Transport of specimens
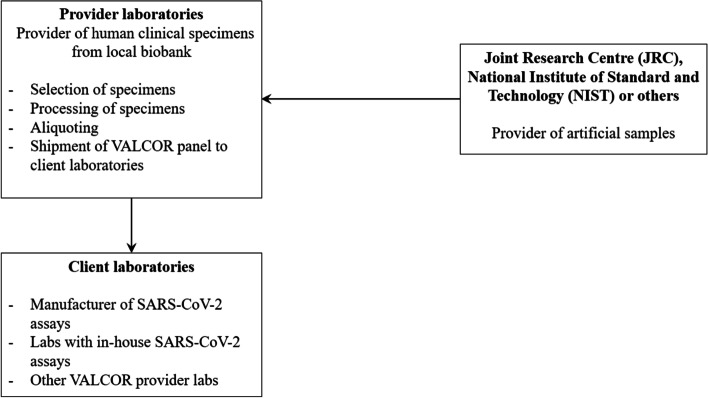
Transport of the VALCOR panel from the provider laboratory to the client laboratories will be regulated through the Material Transfer Agreement (Fig. 1).
Fig. 1.
Overview of the flow of specimens within the VALCOR study
Transmission of data
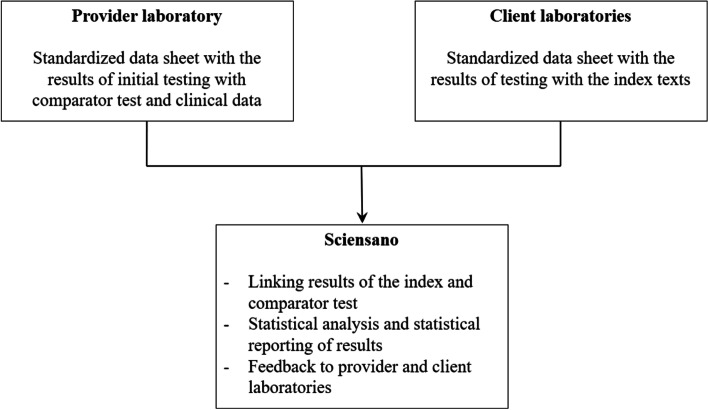
The provider laboratory will transmit data on clinical information and the results of the initial SARS-CoV-2 testing with the comparator assay described above in testing procedures to Sciensano (Fig. 2). The content of the data file is available in the supplementary material (p 5). The client laboratory will complete a standardised data sheet with the results of the SARS-CoV-2 testing with their assay and transmit this to Sciensano.
Fig. 2.
Overview of the flow of data within the VALCOR study
Standard data record sheets will be used by all VALCOR partners. Epidemiologists from the department of Public Health and Surveillance of the Sciensano institute (Brussels) will receive the virological results of index test(s) from each participating client laboratory and will link with the virological data of the reference test performed in the provider laboratory.
Outcome measures and data analysis
The statistical analysis will subsequently be performed using Stata 16.0 (College Station, TX, USA).
Outcomes will be virological sensitivity and specificity of the index tests vs the comparator test. If output data from index and comparator tests include a quantitative metric (signal strength) related to viral load, scatterplots will be made which may be explored to assess possible cut-off optimisations.
Network meta-analyses using methods to pool multiple testing accuracy data will be applied as soon as results from different VALCOR panels will become available [18, 19]. The matrices of VALCOR test data will be enriched with data extracted from studies included in a systematic review and meta-analysis of published reports (peer-reviewed and grey literature).
After a few runs of VALCOR, one or more standard comparator tests might be chosen based upon consensus to be reached with the comity of collaborating VALCOR partners. Benchmark values for validation will be defined as well taking into account the use of the test (clinical diagnosis, tracking of contacts, screening in the population or defined communities, point-of-care testing). The focus will be on virological sensitivity.
Discussion
The VALCOR protocol builds further on the experience of the VALGENT concept which is a successful forum for comparison and validation of human papillomavirus (HPV) tests usable for cervical cancer screening [12, 13]. The rationale and validation panel design principles of VALGENT is converted into a system to validate assays that detect SARS-CoV-2. VALCOR is the first validation study that will facilitate a standardised validation framework for systematic performance evaluation of SARS-CoV-2 assays.
Testing for COVID-19 is ultimately one of the key approaches to tackle and confine the SARS-CoV-2 pandemic. In particular, for both screening and contact tracing of communities, SARS-CoV-2 assays with a high sensitivity and negative predictive value (depending on the background prevalence) are needed. Specificity is important as well however good positive predictive values will be guaranteed in clinical settings given the clinical pre-triage and suggestive symptoms. Assay’s sensitivity and specificity also need to take into account the different sample types which are used for SARS-CoV-2 detection, such as nasopharyngeal, oropharyngeal, nasal swabs or less invasive saliva samples. Different suspension liquid or media (such as saline or PBS solution, UTM, viral transport medium (VTM), eNAT, etc.) may influence the stability of viral nucleic acid of the sample over time. Additionally, the pre-analytical procedures such as retrieval and transport, rapid or traditional nucleic acid extraction methods, the use of appropriate assays’ internal controls (to evaluate sample adequacy) may impact test performance [20, 21].
Since the emergence of variants of SARS-Cov-2, surveillance testing is now routinely carried out to monitor and track the emergence of new variants of concerns. As many countries now require positive samples to be further tested for whole-genome sequencing, there is a limited volume of residual material that is stored in reference laboratories. This may pose a limitation as there could be a lack of available clinical specimens required for validation studies such as VALCOR. Besides, specimens are usually stored in biobanks and retested. The stability of viral RNA over time could affect the assay’s results as degradation of virus material in frozen samples is expected.
As a result of the rapid spread of COVID-19, many regulatory bodies were forced to allow emergency authorisations for SARS-CoV-2 assays to fulfil the unprecedented need for testing requirements worldwide. The lack of vigorous validation procedures resulted in the subpar performance of many SARS-CoV-2 assays in the real-world setting. Our VALCOR protocol provides a basis for a standardised validation framework.
Extensions of the VALCOR protocol
Nasopharyngeal samples are considered the reference specimen for the detection of SARS-CoV-2. However, alternate sampling methods such as saliva and oral swabs which are less invasive are a viable option for the diagnosis of SARS-CoV-2 as well. Hence, the validation of diagnostic tests using different samples is necessary to evaluate virological sensitivity and specificity of SARS-COV-2 assays on different types of specimens. VALCOR-like protocols may be adapted to include validation of other samples besides nasopharyngeal samples. Additionally, the VALCOR protocol may be adapted to accommodate other methods of testing besides NAAT such as antigen testing to detect SARS-CoV-2 proteins under the condition that viral proteins are conserved in the appropriate transport medium.
Supplementary Information
Acknowledgements
We thank the European Commission, Joint Research Centre, Directorate F – Health, Consumers and Reference Materials, Geel, Belgium and the National Institute of Standards and Technology (NIST, USA) for providing the reference materials included in the Belgian and Italian VALCOR study. We thank Gerhard Buttinger, scientific and technical support officer at JRC who coordinated and measured the artificial samples. The authors wish to express their gratitude to Dr. Peter M. Vallone for assisting with the artificial materials from NIST. We would also like to thank all those colleagues who provided support for the setup of the Italian VALCOR panels: Marialuisa Lavitrano, Director of BBMRI.it, a national node of BBMRI-ERIC; Elena Pariani, Director of the Regional SARS-CoV-2 Reference Laboratory, University of Milan; Annalisa Cavallero, Sergio Malandrin and Gianluca Emanuele Calcagno, from the Clinical Microbiology Laboratory, San Gerardo Hospital Monza; Francesca Borgo and Annalisa Cianflone from the Microbiology Laboratory, PTP Science Park, Lodi.
Abbreviations
- COVID-19
Coronavirus disease 2019
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- WHO
World Health Organization
- NAATs
Nucleic acid amplification tests
- RNA
Ribonucleic acid
- RT-PCR
Reverse transcriptase-polymerase chain reaction
- DNA
Deoxyribonucleic acid
- TMA
Transcription mediated amplification
- LAMP
Loop-mediated isothermal amplification
- NPIs
Non-pharmaceutical interventions
- VALCOR
Validation of severe acute respiratory coronavirus 2 assays
- VALGENT
Validation of HPV genotyping tests
- JRC
Joint Research Centre
- NIST
National Institute of Standards and Technology
- ATCC
(American Type Culture Collection)
- LOD
Limit of detection
- IVT
In vitro transcripted
- ssRNA
Single-stranded ribonucleic acid
- EQA
External Quality Control
- RGTM
Research Grade Test Material
- UTM
Universal Transport Medium
- PBS
Phosphate Buffered Saline
- HPV
Human Papilloma Virus
Authors’ contributions
MA: Principal investigator, protocol development, statistical analysis; SKD: statistical analysis, protocol development, writing of the manuscript; LC, Jannes B, CC and MM: coordination of samples, compilation of clinical data; PC: coordination and advice regarding artificial samples; Jesper B, CC, PC: assistance in the development of study protocol. CS and SKD: practical coordination of the study. All authors reviewed and/or edited the manuscript. All co-authors approved the final manuscript and its submission to this journal. The author(s) read and approved the final manuscript.
Funding
The Belgian VALCOR is funded by emergency funding from the Federal Belgian Government (https://www.sciensano.be/en/projects/validation-sars-corona-virus-2-assays). The Italian VALCOR has not received specific funding for this project. VALCOR is a researcher induced study. Manufacturers of devices and assays can participate by offering devices or test kits and contribute funding for laboratory work and statistical analyses. No commercial influence is accepted regarding the publication of the study protocol and results.
Availability of data and materials
Final study datasets generated by VALCOR will be stored locally and securely at Sciensano. Anonymised will be made available by request to the corresponding author on a case-by-case basis pending approval from the information security coordinator at Sciensano.
Declarations
Ethics approval and consent to participate
This protocol has been reviewed by the Ethics Committee Research (EC Research) of University Hospitals Leuven (UZ Leuven) and the Ethics Committee of the University of Milano – Bicocca.
Should additional amendments arise, then these will be submitted to the aforementioned Medical Ethics Committee. Samples for Belgian VALCOR are provided by the UZ Leuven, national reference laboratory for respiratory pathogens (Leuven, Belgium) and those for Italian VALCOR by the Clinical Microbiology and Virology Laboratory of the University of Milano-Bicocca (Monza, Italy). As the data are pseudonymised and the data generated through VALCOR would not enable personal identification of patients from whom the samples were taken, informed consent was waived and authorisation from the Information Security Committee (https://www.ehealth.fgov.be/ehealthplatform/nl/informatieveiligheidscomite in Dutch Informatieveiligheidscomité [IVC]) was not required.
Consent for publication
This manuscript contains no individual person’s data.
Competing interests
VALCOR is a researcher induced study protocol where manufacturers can participate under the condition of providing test kits and covering research costs. Researchers do not receive any financial advantage by collaborating in VALCOR. Jesper B is the PI of a study at AHH-Hvidovre Hospital on SARS-CoV-2 antigen test where BD Diagnostics delivered test kits without cost. CC is co-founder of Hiantis Srl and has ongoing research collaborations with Copan Italia, BD, Seegene, GeneFirst and Novosanis. All other authors declare no personal conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Marc Arbyn and Sharonjit K. Dhillon contributed equally to this work
References
- 1.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO). Weekly Operational Update on COVID-19. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. (Accessed 15 Mar 2022).
- 3.World Health Organization (WHO). Regulation and Prequalification. https://www.who.int/teams/regulation-prequalification/eul. (Accessed 14 Apr 2021).
- 4.Centers for Disease Control and Prevention (CDC). Overview of Testing for SARS-CoV-2 (COVID-19). https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html. (Accessed 25 Oct 2021).
- 5.Kevadiya BD, Machhi J, Herskovitz J, et al. Diagnostics for SARS-CoV-2 infections. Nat Mater. 2021;20(5):593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu X, Wang X, Han L, et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Europen Commission Joint Research Centre. COVID-19 In Vitro Diagnostic Devices and Test Methods Database. https://covid-19-diagnostics.jrc.ec.europa.eu/. (Accessed 09 July 2020).
- 8.FIND Diagnosis for all. SARS-CoV-2 Diagnosis Pipeline. https://www.finddx.org/covid-19/pipeline/ (Accessed 09 Oct 2021).
- 9.Ebrahim SH, Ahmed QA, Gozzer E, Schlagenhauf P, Memish ZA. Covid-19 and community mitigation strategies in a pandemic. BMJ. 2020;368:m1066. doi: 10.1136/bmj.m1066. [DOI] [PubMed] [Google Scholar]
- 10.Europen Commission (EC). Current performance of COVID-19 test methods and devices and proposed performance criteria. https://ec.europa.eu/docsroom/documents/40805. (Accessed 20 Oct 2020).
- 11.Cheng VC-C, Siu GK-H, Wong S-C, et al. Complementation of contact tracing by mass testing for successful containment of beta COVID-19 variant (SARS-CoV-2 VOC B.1.351) epidemic in Hong Kong. Lancet Reg Health West Pac. 2021;17:100281. doi: 10.1016/j.lanwpc.2021.100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arbyn M, Depuydt C, Benoy I, et al. VALGENT: a protocol for clinical validation of human papillomavirus assays. J Clin Virol. 2016;76(Suppl 1):S14–S21. doi: 10.1016/j.jcv.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Bonde J, Ejegod DM, Cuschieri K, et al. The Valgent4 protocol: robust analytical and clinical validation of 11 HPV assays with genotyping on cervical samples collected in SurePath medium. J Clin Virol. 2018;108:64–71. doi: 10.1016/j.jcv.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Joint Research Centre (JRC). EURM-019 single stranded RNA (ssRNA) fragments of SARS-CoV-2. https://crm.jrc.ec.europa.eu/p/EURM-019. (Accessed 10 Aug 2020).
- 15.National Institute of Standards and Technology (NIST). SARS-CoV-2 Research Grade Test Material. https://www.nist.gov/programs-projects/sars-cov-2-research-grade-test-material. (Accessed 10 Aug 2020).
- 16.American Type Culture Collection (ATCC). ATCC Resources representing FDA-recommended species for SARS-CoV-2 molecular diagnostic development https://www.atcc.org/. (Accessed 15 Apr 2021).
- 17.Resources B. Supporting Infectious Disease Research https://www.beiresources.org/Home.aspx. (Accessed 15 Apr 2021).
- 18.Nyaga VN, Aerts M, Arbyn M. ANOVA model for network meta-analysis of diagnostic test accuracy data. Stat Methods Med Res. 2018;27(6):1766–1784. doi: 10.1177/0962280216669182. [DOI] [PubMed] [Google Scholar]
- 19.Nyaga VN, Arbyn M, Aerts M. Beta-binomial analysis of variance model for network meta-analysis of diagnostic test accuracy data. Stat Methods Med Res. 2018;27(8):2554–2566. doi: 10.1177/0962280216682532. [DOI] [PubMed] [Google Scholar]
- 20.Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin Chem Lab Med. 2020;58(7):1070–1076. doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y-W, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58(6):e00512–e00520. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Final study datasets generated by VALCOR will be stored locally and securely at Sciensano. Anonymised will be made available by request to the corresponding author on a case-by-case basis pending approval from the information security coordinator at Sciensano.