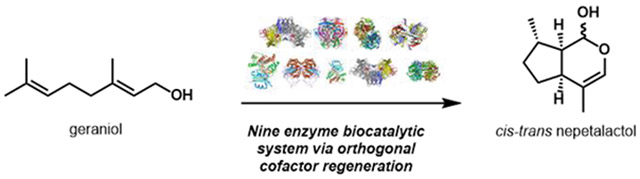
Abstract
Here we report the one-pot, cell-free enzymatic synthesis of the plant monoterpene nepetalactol starting from the readily available geraniol. A pair of orthogonal cofactor regeneration systems permitted NAD+-dependent geraniol oxidation followed by NADPH-dependent reductive cyclization without isolation of intermediates. The orthogonal cofactor regeneration system maintained a high ratio of NAD+ to NADH and a low ratio of NADP+ to NADPH. The overall reaction contains four biosynthetic enzymes, including a soluble P450; and five accessory and cofactor regeneration enzymes. Furthermore, addition of a NAD+-dependent dehydrogenase to the one-pot mixture led to ~1 g/L of nepetalactone, the active cat- attractant in catnip.
Keywords: synthetic biochemistry, natural products, biocatalysis, enzyme cascade, cofactor regeneration
Graphical Abstract
One-pot, cell-free synthesis of complex molecules using purified enzymes is a powerful technology to access natural products that are otherwise difficult to produce.1 This approach has been named total biosynthesis2 or synthetic biochemistry.3 Compared to synthetic chemistry approaches, “cell-free” biosynthesis exploits the precise regio- and stereoselectivities of enzymes to perform chemical transformations of unprotected substrates under mild reaction conditions. Cell-free biosynthesis platforms can also outperform microbial in vivo biosynthesis by eliminating competing metabolic pathways to overcome potential toxicity.4,5 Moreover, cell-free bioproduction benefits from modular and flexible pathway implementation, rapid design-build-test-learn cycles, and can approach theoretical conversion.6,7 Notwithstanding the increasing number of examples of cell-free biosynthesis, a number of challenges exist which limit the utility. In particular, efficient and orthogonal cofactor supply and regeneration is an ever-present obstacle, especially for more complex pathways in which multiple (redox) cofactors are involved. Numerous approaches have been developed to address this obstacle, including reengineering of enzyme cofactor specificity,8 use of chemically orthogonal unnatural cofactors9,10 and in an impressive demonstration, the use of a molecular rheostat and synthetic purge valve to manage excess cofactor buildup.11,12
Indeed, balancing cofactor usage is especially important in systems where both reductive and oxidative reactions are involved. When the biosynthetic pathway uses a single type of cofactor, such as NAD(H) or NADP(H), concomitant oxidation of reducing equivalents upon substrate reduction serves to regenerate oxidizing equivalents, and vice versa. During active metabolism, however, estimated ratios of NAD+:NADH range from 200:1 to 600:1,13 whereas estimated ratios of NADP+:NADPH range from 1:30 to 1:200.13 Thus, many biosynthetic pathway enzymes have evolved to use different types of cofactors. Indeed, natural product biosynthetic logic frequently employ both NAD+-dependent oxidation and NADPH-dependent reduction steps.14-17 Without an orthogonal cofactor regeneration system, combining all the enzymes in one pot will lead to futile redox cycles. Therefore, to achieve one-pot reconstitution of such pathways, it is essential to eliminate crosstalk when regenerating both cofactors. The situation is more complex when the thermodynamic equilibrium requires high concentrations of the correct cofactor which is true for many of the NAD(P)H-dependent oxidoreductases.18 Since the enzymes rely on a high ratio of the correct cofactor i.e., NAD+:NADH or NADPH:NADP+, maintaining an optimal ratio is key to drive the reaction to completion. In this study, we addressed the requirement of an orthogonal cofactor regeneration system to accomplish the one-pot cell-free synthesis of cis-trans nepetalactol (Figure 1).
Figure 1.
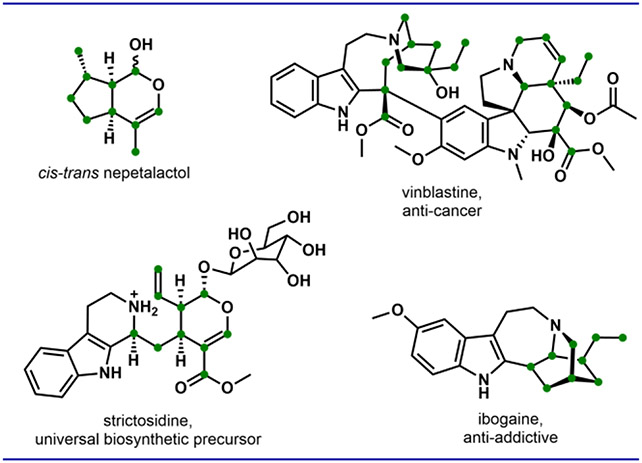
Cis-trans nepetalactol serves as the ten-carbon terpene core of strictosidine, the biosynthetic precursor to vinblastine, ibogaine, and ~3,000 additional monoterpene indole alkaloids.
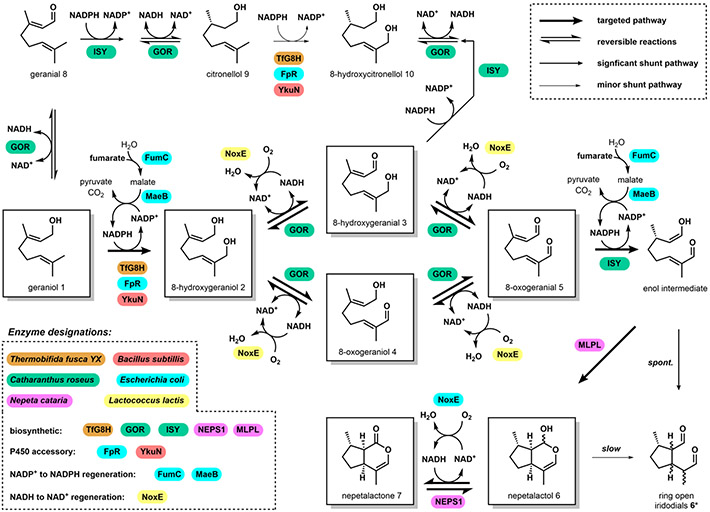
Biosynthesis of cis-trans nepetalactol 6 (also referred to hereafter as “nepetalactol”) from geraniol 1 requires NAD+-dependent oxidation and NADPH-dependent reductive cyclization (Figure 2). Geranyl pyrophosphate (GPP) is hydrolyzed by geraniol synthase to give geraniol 1.19 Regiospecific hydroxylation of one of the terminal methyl groups by the P450 geraniol-8-hydroxylase (G8H) provides 8-hydroxygeraniol 2.20 Next, tandem and reversible NAD+-dependent oxidation of 2 by geraniol oxidoreductase (GOR) generates the dialdehyde 8-oxogeranial 5 (via either 8-hydroxygeranial 3 or 8-oxogeraniol 4).21 Stereoselective reduction of 5 using NADPH to an enol intermediate by iridoid synthase (ISY), followed by enzyme-assisted cyclization by a major latex protein–like enzyme (MLPL) result in 6.22,23 5 can spontaneously form the ring-opened iridodials 6*, which can also be derived through the ring opening of 6, albeit at a slow rate.24 Dehydrogenation of 6 by nepetalactol-related short-chain reductase/dehydrogenase 1 (NEPS1) forms nepetalactone 7, which is a potent insect repellent and the active cat-attractant in catnip.25 Nepetalactol 6 can be further modified into strictosidine, precursor to >3,000 members of the monoterpene indole alkaloid family (Figure 1). Establishing cost-effective routes to nepetalactol and its derivatives is therefore an objective for both synthetic chemists and synthetic biologists.
Figure 2.
Biosynthesis of nepetalactol and nepetalactone along with possible shunt products. On pathway intermediates are boxed. Cofactor regeneration enzymes are only shown for main pathway reactions.
Initial efforts to recapitulate nepetalactol production using cell-free biosynthesis were prompted by the expensive cost from commercial vendors, the low titers observed in microbial hosts,26-29 and difficulties in implementing synthetic routes.30-35 Reported syntheses suffer from low yields and enantioselectivities; or rely on the costly synthon (−)-citronellol as an enantiopure starting material.30-35 To perform cell-free biosynthesis, geraniol 1 was selected as the starting material because of its high abundance as an essential oil and low cost. Since the plant homologues of G8H are membrane-bound and not suitable for in vitro biocatalysis, a functionally equivalent, soluble bacterial P450 was used. CYP154E1 (TfG8H) from Thermobifida fusca YX was reported to perform the same hydroxylation as G8H and can be reductively regenerated by the NADPH-dependent cytochrome P450 flavodoxin/ferredoxin reductase (FpR from Escherichia coli) and flavodoxin (YkuN) from Bacillus subtillis.36,37 Using TfG8H, FpR and YkuN, we were able to observe hydroxylation of 1 to 8-hydroxygeraniol 2 (Figure S2). We next confirmed the oxidation of 2 to 8-hydroxygeranial 3, 8-oxogeraniol 4 and 8-oxogeranial 5 when combined with GOR and excess NAD+ (Figure S3). The reaction was observed to reach an equilibrium between these four compounds due to depletion of cofactor and reaction reversibility. Upon incubation of 5 (330 mg/L) together with ISY and MLPL, nepetalactol 6 was almost exclusively formed using excess NADPH, with a small amount of ring-opened iridodials 6* being detected. ISY could utilize both NADPH and NADH as reducing cofactors, albeit showing strong preference for NADPH.
Despite demonstration of competent in vitro activities of the individual enzymes, one-pot synthesis of 6 from 1 as shown in Figure 2 using sub-stoichiometric amounts of cofactors is challenging. First, an orthogonal cofactor regeneration system is required to regenerate NAD+ from NADH (for the GOR oxidation step), while not oxidizing NADPH, which is required for TfG8H and ISY turnover. Similarly, the cofactor regeneration system must also regenerate NADPH from NADP+, while not reducing NAD+ to NADH. Second, the enzyme activities of the regeneration systems must be carefully tuned to match the differential reactivities TfG8H, GOR and ISY. The oxidation of 2 to 5 is stepwise and readily reversible, and thus can accumulate mono-aldehyde intermediates 3 and 4 (Figure 2). With 3, ene-reduction catalyzed by ISY follow by aldehyde reduction catalyzed by GOR can give irrecoverable shunt products such as 8-hydroxycitronellol 10. Hence, the ratio of NAD+ to NADH available for GOR oxidation must be well-controlled.
For the oxidizing enzyme that can selectively oxidize NADH instead of NADPH, we use the NADH-oxidase (NoxE) from Lactococcus lactis (Figure S5B).38 When coupled, 10 μM GOR and 5 μM NoxE were able to fully convert 340 mg/L 2 to 5 in the presence of limiting 100 μM NAD+ (Figure S6) within 1 hour (Figure S7). In contrast, a cost-effective NADPH regeneration system that does not reduce NAD+ was not readily available. The conventional glucose-6-phosphate (G6P) dehydrogenase or glyceraldehyde-3-phosphate (GAP) dehydrogenase, which convert G6P to 6-phospho-D-glucono-1,5-lactone and GAP to 1,3-bisphosphoglycerate, respectively,39,40 use the expensive substrates G6P and GAP. Other NADPH-regeneration enzymes such as glucose-1-dehydrogenase and isocitrate dehydrogenase were also not suitable due to their non-specific cofactor usage or high substrate cost. Combining the requirements of cofactor orthogonality, ease of enzyme expression and cost effectiveness, we chose a two-enzyme system which consists of fumarate hydratase (FumC) and NADP+-dependent malic enzyme (MaeB) from Escherichia coli. MaeB catalyzes the decarboxylation of (S)-malate to generate pyruvate in a strictly NADP+-dependent manner.41 While (S)-malate acid is relatively expensive, it can be readily generated from the hydration of the inexpensive fumarate by FumC. In addition to its cofactor selectivity, the decarboxylation reaction catalyzed by MaeB is irreversible and thus drives the coupled reaction forward. Cloning and characterization of MaeB and FumC confirmed that the two-enzyme system displayed excellent selectivity towards NADP+ over NAD+ in the presence of fumarate (Figure S5C). To test this system in catalysis, we performed the coupled reaction of TfG8H/FpR/YkuN and FumC/MaeB with limiting concentrations of NADPH. Full conversion of 310 mg/L (2 mM) geraniol 1 to 8-hydroxygeraniol 2 with 5 μM TfG8H, 10 μM FpR, 10 μM YkuN, 1 μM FumC, 10 μM MaeB and 100 μM NADPH (Figure S8) was observed within 1.5 hours (Figure S9). This full conversion establishes a robust in vitro P450 biocatalytic reaction, which typically requires excess NADPH due to suboptimal electron transfer between the P450 and its partner enzymes.42 The FumC/MaeB regeneration system was also fully compatible with ISY and MLPL and supported the full conversion of 330 mg/L (2 mM) 5 to nepetalactol 6 in the presence of 0.5 μM ISY, 5 μM MLPL, 1 μM FumC, 10 μM MaeB and 100 μM NADPH (Figure S11). Neither TfG8H- nor ISY-catalyzed reaction was inhibited by the accumulated pyruvate (Figures S10 and S12).
With an orthogonal cofactor regeneration system in hand, a one-pot enzymatic synthesis of nepetalactol 6 was attempted using all purified enzymes. As expected from the substrate promiscuity of ISY and GOR (Figure 2), incubation of all nine enzymes along with 310 mg/L 1 led to a number of shunt products. Although 5 was produced (15% yield), a considerable amount of citronellol 9 and 8-hydroxycitronellol 10 were formed (Figures 2 and S13). The alcohol 9 is formed when 1 is oxidized to geranial 8 by GOR, which then undergoes ene-reduction by ISY, followed by a GOR-catalyzed reduction (Figure 2). To reduce the formation of 9 which is an irrecoverable shunt product, we used a multi-step, one-pot approach in which ISY was added after G8H and GOR reactions were completed. A 200 μL-scale, two-step approach was successful in producing a higher amount of nepetalactol (65% yield) (Figure S14). However, the formation of shunt products remained and was particularly problematic in a larger scale reaction (10 mL) (Figure S15). We hypothesized formation of 9 is due to low G8H hydroxylation activity caused by oxygen transport deficiency in the reaction vessel, which led to the O2-independent GOR oxidation and subsequent conversion to 9.22,23,43 When the agitation rate was raised from 250 rpm to 300 rpm, aggregation and precipitation of enzymes were observed.
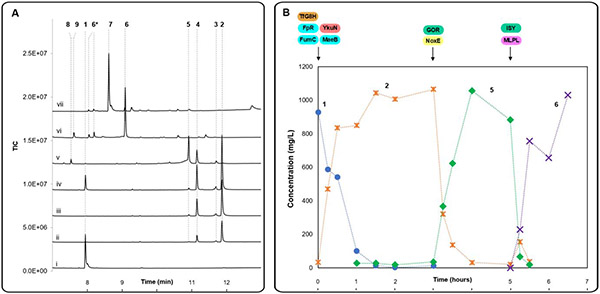
To eliminate the formation of 9 and 10 as major shunt products, a one-pot drop-in strategy was pursued, where the biosynthetic enzymes were added sequentially after the upstream reaction was completed. As an initial test, 310 mg/L 1 was incubated with TfG8H, FpR, YkuN, FumC, MaeB along with 100 μM NADPH and 6 mM fumarate for 2 hours. Upon full conversion of 1 to 2, GOR, NoxE and 100 μM NAD+ were added directly to the mixture, which was incubated for 2 additional hours. Finally, ISY, MLPL and 6 mM fumarate were added and reacted for 2 hours. This scheme fully converted 3.1 mg geraniol to nepetalactol in a 10 mL reaction mixture (Figure S17), forming >3 mg nepetalactol in the reaction (>95% conversion). We then increased the amount of substrate added to the reaction through the addition of multiple aliquots of 1 (Figure 3A). The G8H reaction was supplemented with an additional 310 mg/L geraniol and 6 mM fumarate every 1.5 hours, and substrate hydroxylation was monitored (Figure 3A, trace i-iv). Our NADPH-regeneration system supported hydroxylation of a combined 930 mg/L of 1 to 2 within 4.5 hours. Subsequent addition of GOR and reaction for two hours led to complete conversion of 2 to 3, 4, and 5, and only very minor amounts of 8 (Figure 3A, trace v). Finally, addition of ISY and MLPL resulted in the formation of 940 mg/L of nepetalactol 6 (93% yield) in two additional hours (Figure 3A, trace vi). Overall, this one-pot mixture operating at the 10 mL scale produced ~1 g/L of 6 after 8.5 hours (Figure 3A, trace vi).
Figure 3.
One pot enzymatic synthesis of nepetalactol and nepetalactone using an orthogonal cofactor regeneration system. (A) GC-MS chromatograms for 10 mL-scale one-pot conversion of 6 mM geraniol to nepetalactol and nepetalactone. Final reaction contained 925.5 mg/L geraniol 1, 5 μM TfG8H, 10 μM FpR, 10 μM YkuN, 10 μM GOR, 0.5 μM ISY, 5 μM MLPL, 1 μM FumC, 10 μM MaeB, 100 μM NADPH, 100 μM NAD+ and 18 mM fumarate in BTP buffer (pH 9.0) unless otherwise specified. (i) starting material, 2 mM 1, (ii) 1.5-hour reaction with TfG8H, (iii) additional 1.5-hour reaction with TfG8H and an additional aliquot of 2 mM 1 added, (iv) additional 1.5-hour reaction with TfG8H and an additional aliquot of 2 mM 1 added, (v) 2-hour reaction after GOR was added to (iv), (vi) 2-hour reaction after ISY and MLPL were added to (v), (vii) 2-hour reaction after ISY/MLPL and NEPS1 were added to (v). Peak identities were deduced from GC-MS and by comparison to authentic standards (see Supporting Information). (B) Substrate (1, blue circle) and products' (2, orange star; 5, green diamond; 6, purple cross) concentrations measured over time, with timing of added enzymes indicated with arrows atop. The reaction condition is as specified above with starting 1 concentration of 957 mg/L (6.2 mM).
Encouraged by this result in Figure 3A, we performed the 10 mL reaction with starting batch concentration of 1 at 957 mg/L (6.2 mM) (Figure 3B, and S17 for the GC/MS traces), with no additional aliquots. The concentrations of 1, 2, 5 and 6 were measured and plotted as a function of time in Figure 3B. GOR and NoxE was added after three hours when all of 1 were converted to 2; while ISY and MLPL were added after five hours when all of 2 were converted to 5. After two additional hours, all of the 5 were converted to 6 with a final concentration of ~ 1 g/L.
To probe the compatibility of the one-pot reaction with downstream biosynthetic enzymes that act on 6, the nepetalactol-related short-chain reductase/dehydrogenase (NEPS1) was introduced to form 7. Since NEPS1 utilizes NAD+ to convert nepetalactol to nepetalactone 7, it was added to the reaction mixture at the same time as ISY and MLPL without any additional cofactors or coenzymes. The near complete conversion of 1 to 7 (930 mg/L) was observed after 8.5 hours (Figure 3A, trace vii; and Figure S17). Production of nepetalactone by recycling sub-stoichiometric concentrations of each nicotinamide cofactor corresponds to 180- and 120-fold decreases in the required molar loading of NAD+ and NADPH, respectively.
In summary, we report the production of nearly 1 g/L nepetalactol 6 or nepetalactone 7 from geraniol through the use of a pair of orthogonal cofactor regeneration enzymes. The reaction requires up to five biosynthetic and five auxiliary enzymes and can be operated in a one-pot fashion. Our results highlight a major advantage permitted by cell-free systems – the precise temporal control of enzymatic action which is difficult to program via metabolic engineering.44-47 Future protein engineering efforts aimed at lowering ISY promiscuity, or implementing shunt metabolite rescue,48 could further simplify the reaction sequence. Our system produces nepetalactol at a titer ~130-fold greater than the highest reported in a microbial platform.28 Depending on the estimated cost of protein (Table S4), our total material cost ranges from 60 USD to 120 USD to generate 1 g of 6, which is significantly lower than current commercial sources. Our platform establishes a cost-effective method to produce 6, which is useful in the biosynthetic investigation and synthesis of the monoterpene indole alkaloid natural products.
Supplementary Material
ACKNOWLEDGMENT
This work is support by NIH R01AT010001 to YT. The plasmid used for expression of NoxE was a gift from Prof. James Bowie.
Footnotes
Supporting Information
Experimental details, spectroscopic and computational data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Hayashi Y Pot Economy and One-Pot Synthesis. Chem. Sci 7, 866–880 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts AA; Ryan KS; Moore BS; Gulder TAM Total (Bio)Synthesis: Strategies of Nature and of Chemists. Top. Curr. Chem 297, 149–203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowie JU; Sherkhanov S; Korman TP; Valliere MA; Opgenorth PH; Liu H Synthetic Biochemistry: The Bio-Inspired Cell-Free Approach to Commodity Chemical Production. Trends Biotechnol. 38, 766–778 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Korman TP; Opgenorth PH; Bowie JU A Synthetic Biochemistry Platform for Cell Free Production of Monoterpenes from Glucose. Nat. Commun 8, 15526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherkhanov S; Korman TP; Chan S; Faham S; Liu H; Sawaya MR; Hsu W-T; Vikram E; Cheng T; Bowie JU Isobutanol Production Freed from Biological Limits Using Synthetic Biochemistry. Nat. Commun 11, 4292 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claassens NJ; Burgener S; Vögeli B; Erb TJ; Bar-Even A A Critical Comparison of Cellular and Cell-Free Bioproduction Systems. Curr. Opin. Biotechnol 60, 221–229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudley QM; Karim AS; Jewett MC Cell-Free Metabolic Engineering: Biomanufacturing beyond the Cell. Biotechnol. J 10, 69–82 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black WB; Zhang L; Mak WS; Maxel S; Cui Y; King E; Fong B; Sanchez Martinez A; Siegel JB; Li H Engineering a Nicotinamide Mononucleotide Redox Cofactor System for Biocatalysis. Nat. Chem. Biol 16, 87–94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King E; Maxel S; Li H Engineering Natural and Noncanonical Nicotinamide Cofactor-Dependent Enzymes: Design Principles and Technology Development. Curr. Opin. Biotechnol 66, 217–226 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson KN; Black WB; Li H Aldehyde Production in Crude Lysate- and Whole Cell-Based Biotransformation Using a Noncanonical Redox Cofactor System. ACS Catal. 10, 8898–8903 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Opgenorth PH; Korman TP; Iancu L; Bowie JU A Molecular Rheostat Maintains ATP Levels to Drive a Synthetic Biochemistry System. Nat. Chem. Biol 13, 938–942 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Opgenorth PH; Korman TP; Bowie JU A Synthetic Biochemistry Molecular Purge Valve Module That Maintains Redox Balance. Nat. Commun 5, 4113 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Walsh CT & Tang Y The Chemical Biology of Human Vitamins., RSC Publishing, United Kingdom. (2018). [Google Scholar]
- 14.Walsh CT & Tang Y Natural Product Biosynthesis. RSC Publishing, United Kingdom. (2017). [Google Scholar]
- 15.Tang M-C; Zou Y; Watanabe K; Walsh CT; Tang Y Oxidative Cyclization in Natural Product Biosynthesis. Chem. Rev 117, 5226–5333 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris JS; Caldo KMP; Liang S; Facchini PJ PR10/Bet v1-like Proteins as Novel Contributors to Plant Biochemical Diversity. ChemBioChem 22, 264–287 (2021). [DOI] [PubMed] [Google Scholar]
- 17.R. Lichman B The Scaffold-Forming Steps of Plant Alkaloid Biosynthesis. Nat. Prod. Rep 38, 103–129 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Sellés Vidal L; Kelly CL; Mordaka PM; Heap JT Review of NAD(P)H-Dependent Oxidoreductases: Properties, Engineering and Application. Biochim. Biophys. Acta BBA - Proteins Proteomics 1866, 327–347 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Brown S; Clastre M; Courdavault V; O’Connor SE De Novo Production of the Plant-Derived Alkaloid Strictosidine in Yeast. Proc. Natl. Acad. Sci 112, 3205–3210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collu G; Unver N; Peltenburg-Looman AMG; van der Heijden R; Verpoorte R; Memelink J Geraniol 10-Hydroxylase11The Nucleotide Sequence Newly Reported in This Paper Has Been Deposited in the GenBank/EMBL Data Banks with the Accession Number AJ251269, a Cytochrome P450 Enzyme Involved in Terpenoid Indole Alkaloid Biosynthesis. FEBS Lett. 508, 215–220 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Miettinen K; Dong L; Navrot N; Schneider T; Burlat V; Pollier J; Woittiez L; van der Krol S; Lugan R; Ilc T; Verpoorte R; Oksman-Caldentey K-M; Martinoia E; Bouwmeester H; Goossens A; Memelink J; Werck-Reichhart D The Seco-Iridoid Pathway from Catharanthus Roseus. Nat. Commun 5, 1–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geu-Flores F; Sherden NH; Courdavault V; Burlat V; Glenn WS; Wu C; Nims E; Cui Y; O’Connor SE An Alternative Route to Cyclic Terpenes by Reductive Cyclization in Iridoid Biosynthesis. Nature 492, 138–142 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Lichman BR; Godden GT; Hamilton JP; Palmer L; Kamileen MO; Zhao D; Vaillancourt B; Wood JC; Sun M; Kinser TJ; Henry LK; Rodriguez-Lopez C; Dudareva N; Soltis DE; Soltis PS; Buell CR; O’Connor SE The Evolutionary Origins of the Cat Attractant Nepetalactone in Catnip. Sci. Adv 6, eaba0721 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindner S; Geu-Flores F; Bräse S; Sherden NH; O’Connor SE Conversion of Substrate Analogs Suggests a Michael Cyclization in Iridoid Biosynthesis. Chem. Biol 21, 1452–1456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lichman BR; Kamileen MO; Titchiner GR; Saalbach G; Stevenson CEM; Lawson DM; O’Connor SE Uncoupled Activation and Cyclization in Catmint Reductive Terpenoid Biosynthesis. Nat. Chem. Biol 15, 71 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell A; Bauchart P; Gold ND; Zhu Y; De Luca V; Martin VJJ Engineering of a Nepetalactol-Producing Platform Strain of Saccharomyces Cerevisiae for the Production of Plant Seco-Iridoids. ACS Synth. Biol 5, 405–414 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Billingsley JM; DeNicola AB; Barber JS; Tang M-C; Horecka J; Chu A; Garg NK; Tang Y Engineering the Biocatalytic Selectivity of Iridoid Production in Saccharomyces Cerevisiae. Metab. Eng 44, 117–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yee DA; DeNicola AB; Billingsley JM; Creso JG; Subrahmanyam V; Tang Y Engineered Mitochondrial Production of Monoterpenes in Saccharomyces Cerevisiae. Metab. Eng 55, 76–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan Y; Liu J; Du Y; Pei X; Li M Aspergillus Oryzae Biosynthetic Platform for de Novo Iridoid Production. J. Agric. Food Chem 69, 2501–2511 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Kouda R; Yakushiji F Recent Advances in Iridoid Chemistry: Biosynthesis and Chemical Synthesis. Chem. – Asian J 15, 3771–3783 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Lee S; Paek S-M; Yun H; Kim N-J; Suh Y-G Enantioselective Total Synthesis of a Natural Iridoid. Org. Lett 13, 3344–3347 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Sim J; Yoon I; Yun H; An H; Suh Y-G Divergent Synthetic Route to New Cyclopenta[c]Pyran Iridoids: Syntheses of Jatamanin A, F, G and J, Gastrolactone and Nepetalactone. Org. Biomol. Chem 14, 1244–1251 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Harnying W; Neudörfl J-M; Berkessel A Enantiospecific Synthesis of Nepetalactones by One-Step Oxidative NHC Catalysis. Org. Lett 22, 386–390 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Sakai K; Ishiguro Y; Funakoshi K; Ueno K; Suemune H A Novel Synthesis of Cis-3,4-Disubstituted Cyclopentanones. Tetrahedron Lett. 25, 961–964 (1984). [Google Scholar]
- 35.Suemune H; Oda K; Saeki S; Sakai K A New Conversion Method from (−)-Limonene to Nepetalactones. Chem. Pharm. Bull 36, 172–177 (1988). [Google Scholar]
- 36.von Bühler C; Le-Huu P; Urlacher VB Cluster Screening: An Effective Approach for Probing the Substrate Space of Uncharacterized Cytochrome P450s. ChemBioChem 14, 2189–2198 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Bakkes PJ; Riehm JL; Sagadin T; Rühlmann A; Schubert P; Biemann S; Girhard M; Hutter MC; Bernhardt R; Urlacher VB Engineering of Versatile Redox Partner Fusions That Support Monooxygenase Activity of Functionally Diverse Cytochrome P450s. Sci. Rep 7, 9570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez de Felipe F; Hugenholtz J Purification and Characterisation of the Water Forming NADH-Oxidase from Lactococcus Lactis. Int. Dairy J 11, 37–44 (2001). [Google Scholar]
- 39.Spaans SK; Weusthuis RA; Van Der Oost J; Kengen SWM NADPH-Generating Systems in Bacteria and Archaea. Front. Microbiol 6, 742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mordhorst S; Andexer JN Round, Round We Go – Strategies for Enzymatic Cofactor Regeneration. Nat. Prod. Rep 37, 1316–1333 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Bologna FP; Andreo CS; Drincovich MF Escherichia Coli Malic Enzymes: Two Isoforms with Substantial Differences in Kinetic Properties, Metabolic Regulation, and Structure. J. Bacteriol 189, 5937–5946 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morlock LK; Böttcher D; Bornscheuer UT Simultaneous Detection of NADPH Consumption and H2O2 Production Using the Ampliflu™ Red Assay for Screening of P450 Activities and Uncoupling. Appl. Microbiol. Biotechnol 102, 985–994 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Br L; Mo K; Gr T; G S; Cem S; Dm L; Se O Uncoupled Activation and Cyclization in Catmint Reductive Terpenoid Biosynthesis. Nat. Chem. Biol 15, 71–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olson EJ; Hartsough LA; Landry BP; Shroff R; Tabor JJ Characterizing Bacterial Gene Circuit Dynamics with Optically Programmed Gene Expression Signals. Nat. Methods 11, 449–455 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Chavez A; Scheiman J; Vora S; Pruitt BW; Tuttle M; Iyer EPR; Lin S; Kiani S; Guzman CD; Wiegand DJ; Ter-Ovanesyan D; Braff JL; Davidsohn N; Housden BE; Perrimon N; Weiss R; Aach J; Collins JJ; Church GM Highly Efficient Cas9-Mediated Transcriptional Programming. Nat. Methods 12, 326–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zalatan JG; Lee ME; Almeida R; Gilbert LA; Whitehead EH; La Russa M; Tsai JC; Weissman JS; Dueber JE; Qi LS; Lim WA Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell 160, 339–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee YJ; Hoynes-O’Connor A; Leong MC; Moon TS Programmable Control of Bacterial Gene Expression with the Combined CRISPR and Antisense RNA System. Nucleic Acids Res. 44, 2462–2473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwander T; Schada von Borzyskowski L; Burgener S; Cortina NS; Erb TJ A Synthetic Pathway for the Fixation of Carbon Dioxide in Vitro. Science 354, 900–904 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.