Abstract
Motivation
The R programming language is one of the most widely used programming languages for transforming raw genomic datasets into meaningful biological conclusions through analysis and visualization, which has been largely facilitated by infrastructure and tools developed by the Bioconductor project. However, existing plotting packages rely on relative positioning and sizing of plots, which is often sufficient for exploratory analysis but is poorly suited for the creation of publication-quality multi-panel images inherent to scientific manuscript preparation.
Results
We present plotgardener, a coordinate-based genomic data visualization package that offers a new paradigm for multi-plot figure generation in R. Plotgardener allows precise, programmatic control over the placement, esthetics and arrangements of plots while maximizing user experience through fast and memory-efficient data access, support for a wide variety of data and file types, and tight integration with the Bioconductor environment. Plotgardener also allows precise placement and sizing of ggplot2 plots, making it an invaluable tool for R users and data scientists from virtually any discipline.
Availability and implementation
Package: https://bioconductor.org/packages/plotgardener, Code: https://github.com/PhanstielLab/plotgardener, Documentation: https://phanstiellab.github.io/plotgardener/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The increasing size, complexity and sheer volume of multi-omic datasets has created a dire need for tools to efficiently visualize, interpret and communicate the underlying biological signals present in these data. Towards this end, genome browsers, including the UCSC Genome Browser and IGV, have revolutionized our ability to investigate genomic data in a rapid and intuitive fashion, using a stacked linear representation of a wide variety of data types and annotations (Abeel et al., 2012; Carver et al., 2009; Chelaru et al., 2014; Flicek et al., 2011; Freese et al., 2016; Kent et al., 2002; Thorvaldsdóttir et al., 2013; Zhou et al., 2011). Recently, more specialized browsers like Juicebox (Durand et al., 2016) and HiGlass (Kerpedjiev et al., 2018) have increased the ability to visualize non-linear data types, such as 3D chromatin contact frequency (Djekidel et al., 2017; Wang et al., 2018). Furthermore, an ever-increasing array of programmatic libraries and browser APIs now allow code-based, integrated data analysis and construction of browser tracks, which has improved reproducibility and automation (Durinck et al., 2009; Hahne and Ivanek, 2016; Lawrence et al., 2009; Wickham, 2016; Yin et al., 2012).
While these tools have been transformative for data exploration, they are largely based on single-panel figures and vertical stacking of genomic tracks and are often ill-suited for the generation of complex multi-panel figures that include both genomic and non-genomic plot types. Such complex figures are often critical for evaluating the underlying biology and are almost always used to present multi-omic data in publications. Thus, a tool specifically designed to programmatically create and arrange publication-quality multi-panel figures is critical to extend the rigor, reproducibility and clarity of scientific data visualizations.
Currently existing R packages like patchwork (Pedersen, 2020), cowplot (Wilke, 2020), gridExtra (Auguie, 2017), egg (Auguie, 2019), multipanelfigure (Graumann and Cotton, 2018) and Sushi (Phanstiel et al., 2014) can be used to arrange multi-panel plots. However, these layout packages use relative positioning to place plots and are limited to standard grid-style layouts, giving users little control over precise sizing and arrangement. Furthermore, these packages arrange and align entire plot panels as opposed to internal plot elements like axes. Figures generated with these tools often need finishing in graphic design software such as Adobe Illustrator (Adobe Inc., 2019), Inkscape (Inkscape Project, 2020), PowerPoint (Microsoft Corporation, 2018) and Keynote (Apple Inc., n.d.). In addition to the cost of purchasing proprietary graphic design software and the steep learning curve often associated with their use, generating multi-panel figures with these software requires non-programmatic, manual user interactions, a labor intensive process that decreases reproducibility.
Here, we introduce plotgardener, an R package for absolute coordinate-based plot placement and sizing of complex multi-panel plots. This paradigm gives users precise control over size, placement, typefaces, font sizes and virtually all plot esthetics without the need for graphic design software. Plotgardener (i) supports a vast array of genomic data types, (ii) allows precise placement and sizing of genomic and non-genomic figures, (iii) is tightly integrated with the Bioconductor environment (Gentleman et al., 2004) and (iv) is optimized for speed and user-experience. The code is open source, extensively documented and freely available via GitHub and Bioconductor.
2 Philosophy
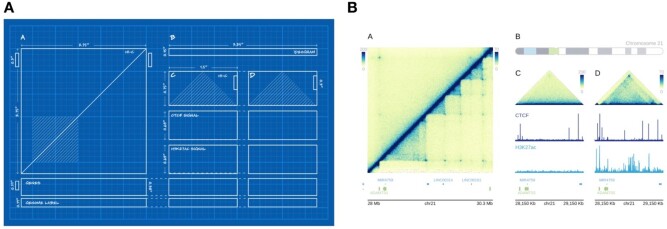
The defining feature of plotgardener that separates it from virtually all other genomic visualization tools is that it allows exact sizing and placement of plots using an absolute, coordinate-based plotting system (Fig. 1). Each plot, axis and annotation is placed independently according to user-specified positions and dimensions. Each plot or feature extends from edge to edge of the defined coordinates, allowing for precise control and perfect alignment of plots. Rulers and guidelines can be temporarily added for ease of plotting and then removed prior to file generation. Adding additional plots does not shift or resize existing ones, so figures can be built incrementally and adjusted without affecting other figure panels, allowing rapid and easy construction of publication-quality multi-panel figures.
Fig. 1.
Plotgardener uses a coordinate-based plotting system to size and arrange plots. (a) Blueprint outline of a multi-omic figure to be created with specified dimensions and placements on a defined page. (b) Multi-panel, multi-omic figure programmatically created with plotgardener using the sizing and placement coordinates from (a). The plotgardener functions used to create this figure include pageCreate, plotHicSquare, annoHeatmapLegend, plotGenes, annoGenomeLabel, plotIdeogram, plotHicTriangle, plotSignal and plotText. Code to reproduce this plot is included in the plotgardener package
3 Data types
Plotgardener can display a vast array of genomic data types which can be provided as either external files or R data classes. Plotgardener has 45 functions for plotting and annotating diverse genomic data types, including genome sequences, gene/transcript annotations, chromosome ideograms, signal tracks, GWAS Manhattan plots, genomic ranges (e.g. peaks, reads, contact domains, etc.), paired ranges (e.g. paired-end reads, chromatin loops, structural rearrangements, QTLs, etc.) and 3D chromatin contact frequencies. Plotgardener automatically recognizes and reads compressed, indexed file types including ‘.bam’, ‘.bigwig’ and ‘.hic’, allowing for rapid and memory-efficient reading and plotting of large genomic data. Supplementary Figure S1 displays the runtime required to read and plot various types of genomic data. Even with file sizes exceeding 50 GBs, plotgardener can read and plot data in under a second. Multiple classes of R objects are supported, including ‘data.frame’, ‘data.table’, ‘tibble’, ‘Granges’ and ‘GInteractions’. Plotgardener automatically detects whether the input is a file path or an R object and handles them accordingly, providing a seamless and flexible experience for the user.
4 Bioconductor integration
Plotgardener is tightly integrated with the Bioconductor ecosystem (Gentleman et al., 2004), making it compatible with many existing workflows. It has 29 built-in genomes and associated annotations but can easily accommodate custom genomes and annotations using Bioconductor TxDb (Lawrence et al., 2013), OrgDb (Pagès et al., 2021) and Bsgenome (Pagès, 2021) packages and/or objects. Plotgardener leverages these annotation resources on behalf of the user to obtain and plot chromosome sizes, gene and transcript structures and nucleotide sequences. By preconfiguring the genome builds and associated feature data, plotgardener allows users to focus their attention on layout and to quickly visualize their data rather than spending time and effort on curation and organization of sequences and genome annotations.
5 User experience
Plotgardener includes a variety of user-friendly features to maximize ease of use for both novices and experienced R programmers. We describe just some of these features here. Parameters can be set within each function call or passed in a pgParams object for more efficient code. Genomic coordinates can be set either by supplying the chromosome, start and end position or by providing a gene name (e.g. IL1B), reference genome name (e.g. ‘hg19’) and optional base pair window around the gene (e.g. 50 000 bp). Resolution of Hi-C contact matrices, signal tracks and gene tracks are automatically determined based on the genomic range being plotted, but can be overwritten if desired. When genomic regions are too large to label all genes, plotGenes and plotTranscripts will choose which genes/transcripts to label based on frequency of appearance in publications. Users can provide their own priorities or select individual genes to highlight with text and colors. A ‘colorby’ function allows users to flexibly color genomic features by quantitative and qualitative attributes. Plotgardener is open source, version controlled and extensively documented via articles and vignettes (https://phanstiellab.github.io/plotgardener/).
6 ggplot and beyond
In addition to its included functions for plotting and annotating genomic data, plotgardener allows for the absolute sizing and placement of non-genomic plots within a plotgardener page. Users can make multi-panel figures seamlessly by integrating and aligning plotgardener and non-plotgardener plots or create coordinate-based layouts entirely composed of external plot types and objects. For example, plotgardener was used to arrange and add text annotations to the ggplot2 plot objects featured in Supplementary Figure S2. Plotgardener intuitively sizes, arranges and overlays plots, text and geometric objects to make complex figure arrangements beyond basic grid-style or relative layouts. We are actively developing the package and potential future additions include more plotting functions, templates for common arrangements, convenient functions for multiplotting, enhanced ggplot2 integration and more.
In summary, plotgardener provides a new paradigm for generating complex publication-quality figures of both genomic and non-genomic data types, making it an invaluable tool for R users and data scientists from virtually any discipline.
7 Materials and methods
7.1 Visualization methods
Plotgardener is an open-source extension for R, building its visualization functions from primitive graphical functions in the grid package (R Core Team, 2021). Each plot and annotation is drawn within its own defined graphical region, or viewport, and then placed on a larger plotgardener page. These viewports give the power to specify the size and placement of plot containers and clip data to precise genomic and data axis measurements. To obtain large, reference genomic annotation data, plotgardener integrates and utilizes packages and data objects through Bioconductor.
7.2 Gene and transcript label publication frequency mining
Annotations for genes in PubMed articles were obtained from the PubTator text mining tool (Wei et al., 2013) and counted for each unique gene ID. Publication frequencies were matched via gene ID to Bioconductor transcript database (TxDb) gene IDs for the 29 built-in plotgardener genomes.
7.3 Evaluating runtimes of plotgardener plotting functions
To calculate plotgardener plotting runtimes, we used the R package microbenchmark (Mersmann, 2019). plotHicSquare, plotSignal, plotGenes and plotRanges functions were timed for various genomic region sizes and resolutions. Each condition was timed on 20 random genomic regions generated by BedtoolsR (Patwardhan et al., 2019).
Supplementary Material
Acknowledgements
The authors thank Hyejung Won and Jason Stein for helpful discussions and feedback. We thank Muhammad Saad Shamim and Neva Durand for assistance with the strawr package.
Funding
This work was supported by National Institutes of Health grants [R35-GM128645 to D.H.P.]. N.E.K. and E.S.D. were supported by the National Institutes of Health training grant T32-GM067553. S.M.P. is supported by the National Science Foundation GRFP DGE-1650116. M.I.L. was supported by National Institutes of Health grants R01-MH118349 and R01-HG009937.
Conflict of Interest: none declared.
Data availability
Various publicly available datasets are included with a Supplementary plotgardenerData package and were used to demonstrate the functionalities of plotgardener. Hi-C datasets from the GM12878 and IMR90 cell lines were downloaded from GEO (Barrett et al., 2013) under the accession code GSE63525. CTCF ChIP-seq signal files for the GM12878 and IMR90 cell lines were downloaded from the ENCODE portal (ENCODE Project Consortium, 2012) with accession codes ENCFF312KXX and ENCFF603PYX. H3K27ac ChIP-seq signal files for the GM12878 and IMR90 cell lines were downloaded from the NIH Roadmap Epigenomics Project (Bernstein et al., 2010) with reference epigenome identifiers E116 and E017. COVID-19 case data was downloaded from The COVID Tracking Project (https://covidtracking.com/). State population data and state COVID-19 vaccination data were downloaded from the Johns Hopkins Centers for Civic Impact COVID-19 GitHub repository (https://github.com/govex/COVID-19/).
Contributor Information
Nicole E Kramer, Curriculum in Bioinformatics and Computational Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Eric S Davis, Curriculum in Bioinformatics and Computational Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Craig D Wenger, Independent Scholar, Issaquah, WA 98207, USA.
Erika M Deoudes, Thurston Arthritis Research Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Sarah M Parker, Curriculum in Bioinformatics and Computational Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Michael I Love, Department of Biostatistics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Douglas H Phanstiel, Curriculum in Bioinformatics and Computational Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Thurston Arthritis Research Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Department of Cell Biology and Physiology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Curriculum in Genetics and Molecular Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
References
- Abeel T. et al. (2012) GenomeView: a next-generation genome browser. Nucleic Acids Res., 40, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adobe Inc. (2019) Adobe Illustrator (CC 2019 (23.0.3)) [Computer software]. https://adobe.com/products/illustrator (3 December 2021, date last accessed).
- Apple Inc. (n.d.) Keynote (Version 11.0.1) [MacOS]. https://www.apple.com/keynote/.
- Auguie B. (2017) gridExtra: miscellaneous Functions for “Grid” Graphics. https://CRAN.R-project.org/package=gridExtra (3 December 2021, date last accessed).
- Auguie B. (2019) egg: extensions for “ggplot2”: custom Geom, Custom Themes, Plot Alignment, Labelled Panels, Symmetric Scales, and Fixed Panel Size. https://CRAN.R-project.org/package=egg (3 December 2021, date last accessed).
- Barrett T. et al. (2013) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res., 41, D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E. et al. (2010) The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol., 28, 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T. et al. (2009) DNAPlotter: circular and linear interactive genome visualization. Bioinformatics, 25, 119–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelaru F. et al. (2014) Epiviz: interactive visual analytics for functional genomics data. Nat. Methods, 11, 938–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djekidel M.N. et al. (2017) HiC-3DViewer: a new tool to visualize Hi-C data in 3D space. Quant. Biol., 5, 183–190. [Google Scholar]
- Durand N.C. et al. (2016) Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst., 3, 99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S. et al. (2009) GenomeGraphs: integrated genomic data visualization with R. BMC Bioinf., 10, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P. et al. (2011) Ensembl 2011. Nucleic Acids Res., 39, D800–D806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese N.H. et al. (2016) Integrated genome browser: visual analytics platform for genomics. Bioinformatics, 32, 2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C. et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol., 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann J., Cotton R. (2018) multipanelfigure: simple assembly of multiple plots and images into a compound figure. J.Stat. Softw. Code Snippets, 84, 1–10. [Google Scholar]
- Hahne F., Ivanek R. (2016) Visualizing genomic data using GVIZ and bioconductor. In: Mathé E., Davis S. (eds.) Statistical Genomics: Methods and Protocols. Springer, New York, pp. 335–351. [DOI] [PubMed] [Google Scholar]
- Project I. (2020) Inkscape (Version 0.92.5) [Computer software]. https://inkscape.org (3 December 2021, date last accessed).
- Kent W.J. et al. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P. et al. (2018) HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol., 19, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M. et al. (2009) rtracklayer: an R package for interfacing with genome browsers. Bioinformatics, 25, 1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M. et al. (2013) Software for computing and annotating genomic ranges. PLoS Comput. Biol., 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersmann O. (2019) microbenchmark: accurate Timing Functions. https://CRAN.R-project.org/package=microbenchmark (3 December 2021, date last accessed).
- Microsoft Corporation (2018) Microsoft PowerPoint (2019 (16.0)) [Computer software]. https://office.microsoft.com/PowerPoint.
- Pagès H. (2021) BSgenome: software infrastructure for efficient representation of full genomes and their SNPs. https://bioconductor.org/packages/BSgenome (3 December 2021, date last accessed).
- Pagès H. et al. (2021) AnnotationDbi: manipulation of SQLite-based annotations in Bioconductor. https://bioconductor.org/packages/AnnotationDbi (3 December 2021, date last accessed).
- Patwardhan M.N. et al. (2019) Bedtoolsr: an R package for genomic data analysis and manipulation. J. Open Source Softw., 4, 1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen T.L. (2020) patchwork: the Composer of Plots. https://CRAN.R-project.org/package=patchwork (3 December 2021, date last accessed).
- Phanstiel D.H. et al. (2014) Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics, 30, 2808–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2021) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ (3 December 2021, date last accessed). [Google Scholar]
- Thorvaldsdóttir H. et al. (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinf., 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2018) The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol., 19, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.-H. et al. (2013) PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res., 41, W518–W522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York. https://ggplot2.tidyverse.org (3 December 2021, date last accessed). [Google Scholar]
- Wilke C.O. (2020) cowplot: streamlined Plot Theme and Plot Annotations for “ggplot2.” https://CRAN.R-project.org/package=cowplot (3 December 2021, date last accessed).
- Yin T. et al. (2012) ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol., 13, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. et al. (2011) The Human Epigenome Browser at Washington University. Nat. Methods, 8, 989–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Various publicly available datasets are included with a Supplementary plotgardenerData package and were used to demonstrate the functionalities of plotgardener. Hi-C datasets from the GM12878 and IMR90 cell lines were downloaded from GEO (Barrett et al., 2013) under the accession code GSE63525. CTCF ChIP-seq signal files for the GM12878 and IMR90 cell lines were downloaded from the ENCODE portal (ENCODE Project Consortium, 2012) with accession codes ENCFF312KXX and ENCFF603PYX. H3K27ac ChIP-seq signal files for the GM12878 and IMR90 cell lines were downloaded from the NIH Roadmap Epigenomics Project (Bernstein et al., 2010) with reference epigenome identifiers E116 and E017. COVID-19 case data was downloaded from The COVID Tracking Project (https://covidtracking.com/). State population data and state COVID-19 vaccination data were downloaded from the Johns Hopkins Centers for Civic Impact COVID-19 GitHub repository (https://github.com/govex/COVID-19/).