Abstract
Motivation
Analysis of volatile organic compounds (VOCs) in exhaled breath by proton transfer reaction time-of-flight mass spectrometry (PTR-TOF-MS) is of increasing interest for real-time, non-invasive diagnosis, phenotyping and therapeutic drug monitoring in the clinics. However, there is currently a lack of methods and software tools for the processing of PTR-TOF-MS data from cohorts and suited for biomarker discovery studies.
Results
We developed a comprehensive suite of algorithms that process raw data from patient acquisitions and generate the table of feature intensities. Notably, we included an innovative two-dimensional peak deconvolution model based on penalized splines signal regression for accurate estimation of the temporal profile and feature quantification, as well as a method to specifically select the VOCs from exhaled breath. The workflow was implemented as the ptairMS software, which contains a graphical interface to facilitate cohort management and data analysis. The approach was validated on both simulated and experimental datasets, and we showed that the sensitivity and specificity of the VOC detection reached 99% and 98.4%, respectively, and that the error of quantification was below 8.1% for concentrations down to 19 ppb.
Availability and implementation
The ptairMS software is publicly available as an R package on Bioconductor (doi: 10.18129/B9.bioc.ptairMS), as well as its companion experiment package ptairData (doi: 10.18129/B9.bioc.ptairData).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Volatolomics is the study of volatile organic compounds (VOCs) emitted by a biological system (Amann et al., 2014), which can be found in several human matrices such as saliva, urine, skin, blood and exhaled breath. Recently, many studies have highlighted the potential of VOC analysis from exhaled breath for early diagnosis, disease phenotyping, therapeutic drug monitoring or toxicological analysis (Boots et al., 2015; Bruderer et al., 2019; Einoch Amor et al., 2019; Pereira et al., 2015; Rattray et al., 2014). One of the main advantages of breath analysis is its non-invasive nature (Devillier et al., 2017).
Mass spectrometry is a powerful method for the study of small volatile molecules (Rattray et al., 2014). Recently, ‘on-line’ technologies, where the patient blows directly into the mass spectrometer, have emerged as promising approaches for the real-time analysis at the point of care (Bruderer et al., 2019; Devillier et al., 2017). Such strategies are of major interest for the screening and monitoring of individual patients or cohorts (Trefz et al., 2013). The potential of proton transfer reaction coupled to time-of-flight mass spectrometry (PTR-TOF-MS; Blake et al., 2009; Herbig et al., 2009; Jordan et al., 2009) for biomedicine has been shown in applications such as emphysema, liver cirrhosis, chronic kidney disease and diabetes (Cristescu et al., 2011; Fernández del Río et al., 2015; Obermeier et al., 2017; Pleil et al., 2019). PTR-TOF-MS spectrometers provide limits of detection in the parts per billion by volume (ppbv) range and rely on VOCs ionization with a transfer of proton from a reagent ion (usually H3O), then subsequent detection of the resulting ions with time-of-flight (TOF)-MS. During data acquisition, which is very fast, the instrument continuously analyzes the air flowing through a buffer tube (i.e. ambient air by default) and the patient is asked to expire a few times into the tube. Each data file (in the HDF5 open format; Koziol, 2011) contains the ion intensities stored as a numerical matrix whose dimensions are the TOF bins (which can be converted to m/z values) and the acquisition time.
Two processing software are currently available for PTR-TOF-MS data, the commercial Ionicon Data Analyzer (IDA) released in 2020 based on the algorithms by Müller et al. (2013) and the open-source PTRwid (Holzinger, 2015). These software tools allow the analysis of high-resolution, TOF-MS data with the following characteristics: (i) single (or multiple for PTRwid) file analysis, (ii) internal m/z calibration, (iii) untargeted peak detection and deconvolution and (iv) quantification and suggestion of elemental composition. They are particularly suited for the analysis of very large files resulting from continuous environmental monitoring. However, there are specific needs for breath research in patient cohorts which have to be covered. For instance, the simultaneous analysis of multiple samples requires that peak lists from different samples may be aligned; in addition, the parallel processing of several files would be a time-sparing capability; furthermore, a correct distinction of the signals coming from the background and the expiratory phases is needed; finally, implementing a background correction of the ambient air composition as a function of time would be an asset for accurate peak detection and quantification (Beauchamp, 2011; Filipiak et al., 2012; Španěl et al., 2013).
We have therefore developed a suite of algorithms for the processing and analysis of PTR-TOF-MS data for untargeted breath analysis and biomarker discovery in patient cohorts. In particular, the penalized regression on a B-spline basis (P-splines) was used for adaptive temporal modeling (Eilers and Marx, 1996), and the coefficients in both m/z and time dimensions were jointly estimated with a two-dimensional (2D) tensor product. This approach enables to estimate all temporal trends without any parametric hypothesis, and to precisely separate peaks in the m/z dimension at each time. The temporal profiles are then used to correct the external contamination, using linear ambient air baseline removal and statistical testing of mean intensity in ambient air versus exhaled breath.
The whole workflow from the raw data files up to the table of peak intensities is implemented as the ptairMS package (doi: 10.18129/B9.bioc.ptairMS) available on Bioconductor (Gentleman et al., 2004). It includes specific features to facilitate routine clinical analysis (e.g. graphical user interface, quality control checks, sample metadata management, iterative inclusion of new acquisitions). In the following, we will first describe the methods used for each step of the workflow, and then present the results obtained with simulated, experimental, and clinical datasets.
2 Materials and methods
The suite of algorithms developed for the processing of PTR-TOF-MS data from exhaled breath, and implemented in the ptairMS R package, takes as input the name of the directory containing the raw files in HDF5 format, and ultimately generates the samples by variables table of peak intensities. The main steps of the workflow are summarized below and detailed in the following of Section 2. This workflow proposes innovative developments for the breathomics analysis of cohorts, including 2D processing and ambient air quantification and correction methods, which were implemented to previous literature on breath analysis.
-
Processing of each file
Internal calibration of the m/z axis
Determination of expiration limits
-
Untargeted peak detection and quantification in exhaled breath
Detecting peaks on the average total ion spectrum
Estimating the temporal evolution for each peak
Quantifying
Ambient inhaled air correction
Statistical testing of intensity differences between ambient air and expiration phases
-
Alignment between samples followed by quality control
Aligning features between samples
Filtering features based on reproducibility within the whole cohort or sample classes
Filtering features based on the P-value from the test in (1.c)
Imputation of missing values
Putative annotation (including isotopes)
Export of the peak table and metadata
Peak table update when new files are included in the input directory
2.1 Processing of each file
2.1.1 Calibration
Calibration converts the TOF values recorded by the mass spectrometer into m/z values: (Brown and Gilfrich, 1991). To estimate the parameters (a, b), the Levenberg–Marquardt algorithm is used, with couples (tof, m/z) of reference peaks without overlap (Cappellin et al., 2010; Müller et al., 2013). For exhaled breath, we suggest using the following peaks: the primary ion isotope (m/z 21.022), dinitrogen (m/z 29.013) and the acetone isotope (m/z 60.053). External calibration ions such as iodobenzene (m/z 203.943), and diiodobenzene (m/z 330.850) can also be used for calibration in instruments with internal permeation devices. As a drift over time is observed due to low changes of temperature, calibration is performed periodically (e.g. every minute) to update the (a, b) values. The shift is subsequently estimated for each m/z as a function of time by linear interpolation.
2.1.2 Expiration detection
Determination of expiration limits and background (ambient air) is a very important step for the analysis, as boundaries will be used for quantification and for the statistical test for features selection in Section 2.1.3. Classically, a raw data ion trace is used to automatically detect expiration. Herbig et al. (2009) propose to use acetone (m/z 59.049), CO2 (m/z 44.997) or humidity with the water cluster isotope (m/z 39.033) as ion traces. We used the same method as described by Schwoebel et al. (2011) and Trefz et al. (2013), to automatically detect expiration and inhalation phases on an ion trace. In addition, we designed a specific panel from our graphical interface to the visualization (and possible manual modification) of the expiration limits (as described in Section 3.3 below).
2.1.3 Untargeted peak detection and quantification in exhaled breath
Raw data consist in a numerical matrix of TOF counts, whose dimensions are bins (m/z between 0 and 500 Da), and s (depending on the acquisition time). After m/z calibration, data are processed sequentially within bands centered at each nominal mass within an interval of ±0.6 Da (since VOCs are of low molecular weight, <500 Da, peak m/z are clustered around nominal masses; Cappellin et al., 2011; Müller et al., 2011), and covering the full time range. The following steps are then applied: (i) peaks are detected in the mass axis on the sum spectrum, (ii) their temporal evolution is estimated by a tensor product with P-splines, (iii) statistical tests are performed to identify if VOCs come from exhaled breath or ambient air and (iv) their average intensity in expiration phases are quantified in ppb (Fig. 1).
Fig. 1.
Main steps of the pre-processing algorithms for a single PTR-TOF-MS raw file containing six expirations. (A) Peak detection in the m/z dimension with a parametric peak shape after baseline correction. (B) Two-dimensional penalized regression, with a tensor product between the mixture of peak functions from Step (A) and a P-spline basis. The penalty parameter for the time axis is estimated by the generalized cross-validation criterion. Crosses indicate knot locations (i.e. where the coefficients are estimated). The fitted splines for Peak 1 at m/z 39.02 (respectively, Peak 2 at m/z 39.03) are shown in red (respectively blue). (C) Estimation of the temporal evolutions by summing each modeled peak from Step (B) along the time dimension. Two unilateral t-tests are applied to compare expiration and ambient air intensities. If expiration values are significantly greater (respectively, lower) than ambient air, as for Peak 2 (respectively, Peak 1), the feature is considered as originating from ‘expiration’ (respectively, ‘ambient air’)
Peak detection on the average spectrum in 1D: The peak picking algorithm in the m/z dimension is mainly based on Müller et al. (2013). Due to the medium resolution of the instrument (5000 to 10 000), a parametric peak function is required for peak separation. The described estimation of the peak shape starts from the 10% envelop quantile of the normalized and filtered raw spectrum between a given intensity range, and performs an iterative peak detection on the residuals to deconvolve the peaks (Holzinger, 2015; Müller et al., 2013). We also included three alternative parametric functions which may be useful for TOF peak shapes, namely the asymmetric sech2, gaussian and lorentzian functions (Lange et al., 2007). The best peak function is selected automatically according to the R2 criterion on the calibration peaks. To sum up, the different steps of the peak detection on the average ion spectrum around each nominal mass are (Fig. 1A):
Baseline removal (Ryan et al., 1988)
Estimation of the noise threshold and autocorrelation within the ‘off-peak’ interval (Müller et al., 2011)
Savitzky–Golay signal filtering by using optimal windows, followed by detection of local maxima by using the first and second derivatives (Savitzky and Golay, 1964; Vivo Truyols and Schoenmakers, 2006)
Peak deconvolution, by using a peak function of the mass m and depending on the parameters μ (peak center), σ (peak width) and h (peak height):
Iterative residual analysis, which stops as soon as one of the following criteria is met: R2 > R2min (default: 0.995), noise auto-correlation < autocorMax (default: 0.3), the maximum number of iterations is reached (default: 4), the maximum number of detected peaks is reached (default: 7) (Müller et al., 2013)
Estimation of the temporal evolution with penalized signal regression using P-splines in 2D: To estimate the temporal evolution of each peak, we used a 2D regression approach (Marx and Eilers, 2005), which consists of a tensor product between P-splines and the previously estimated m/z peak functions (Fig. 1B). B-splines (basis splines) are polynomial basis functions spread all over a set of knots (de Boor, 1978; Dierckx, 1995). P-splines (penalized B-splines) are B-splines with a difference penalty applied to the coefficients to control the smoothness, and thus overfitting (Eilers and Marx, 1996). The P-spline approach is very powerful to model any profile without a priori knowledge of the data and to provide interpretable coefficients (Eilers and Marx, 2021; Wood, 2006). It has been used in many applications and theoretical works (Eilers et al., 2015), such as data smoothing (Currie and Durban, 2002), Bayesian statistics (Gressani and Lambert, 2021) and machine learning with generalized additive models (Brezger and Lang, 2006; Wood, 2006). To model interactions in multiple dimensions, the tensor product provides a straightforward generalization of this basis (Sidiropoulos et al., 2017). Here, we therefore used tensor product modeling to achieve a fast deconvolution of peaks in both m/z and time dimensions simultaneously, as described below. Raw data are processed sequentially within bands around detected peaks (the 1% quantile of the estimated mixture of peak functions is used to define the m/z bounds), and covering the full acquisition time. In a preliminary step, the baseline in the m/z dimension is estimated at each time point by linear regression between the two m/z boundaries and is subsequently removed, and the calibration shift estimated in Section 2.1.1 is corrected by linear interpolation. Let us then denote , and , the functions representing the acquisition time and the m/z profiles, respectively, with being the function of peak i estimated in the previous section, and with being cubic B-spline functions for the set of knots . The 2D model is obtained by writing each peak coefficient hi in the B-spline basis: , with .
The βij coefficients are estimated according to the P-splines theory, by minimizing the following penalized regression, where the penalty is applied only to the time dimension:
| (1) |
where is the second order difference, i (resp. j) represents the knots location of mass (respectively, time) axis, m (respectively, t) represents the index of mass (respectively, time) axis, and Y is the raw data matrix of dimensions M × T after baseline removal and calibration shift correction.
The choice of the knot locations and the penalty coefficient λ are very important, since too many knots may lead to over fitting, and too few knots may result in under fitting. Classically, knots are uniformly distributed over the data range in order to facilitate the interpretation of the penalty applied to the successive knot differences (Eilers and Marx, 1996). In our case, however, (i) exhaled breath phases are the main focus of our quantification and (ii) inhaled air phases are generally constant. We therefore propose to target the knot locations mainly around the expiration phases (Supplementary Fig. S1). This allows to reduce the dimension of the model, and thus the computational time, while maintaining a good fit (Supplementary Table S1). Alternatively, a uniform distribution of the knots along the time axis may be selected, in case the user has no a priori knowledge about the temporal profile of the compound. The optimal λ value is selected with grid search using the generalized cross-validation criterion (Eilers and Marx, 2010).
Quantification: For each peak i, quantification (in counts per extraction) is first performed at each time point t by summing the 2D model along the m/z dimension: . This results in a temporal series , with T being the acquisition duration (Fig. 1C).
These amounts of VOC i at each time point are then normalized and converted to absolute quantities as follows. First, since the intensities provided by the instrument at each time point are in fact the sum of a fixed number of internal acquisitions, the are normalized (as counts of ions per second; cps) by dividing by the integrated internal time period and by multiplying by the single ion pulse voltage (Müller et al., 2014). To obtain the concentration, the latter values are then normalized by the reagent ion (H3O) intensities, the reaction rate coefficient between the VOC and H3O, and the residence time of the primary ions in the drift tube (normalized cps, ncps; Cappellin et al., 2012). The final normalization by the density of the air in the reaction chamber gives the absolute concentration of the VOC, expressed in part per billion (ppb).
The absolute concentration of VOC i in exhaled breath is obtained by averaging all corresponding to the time points t within the expiration phases.
Ambient inhaled air correction: To correct the ambient inhaled air level in exhaled breath, we propose to subtract the ambient air baseline of the temporal profile of each detected VOC, using a polynomial fit (default degree 3) computed on the ambient air time points. This method is based on the concept of ‘alveolar gradient’, introduced by Phillips (1997). Note that the subtraction step may be omitted in particular cases, as detailed in the discussion (a specific parameter is included in the software tool).
Statistical testing of intensity differences between expiration and ambient air phases: Two unilateral statistical tests (t-tests) are used to compare intensities within and between expirations (i.e. exhaled breath and ambient air). Compounds with intensities that are significantly higher (respectively, lower) within expiration phases are considered to be from exhaled breath (respectively, from ambient air). If none of the tests is significant, the compound is labeled as ‘constant’ (e.g. in the case of internal ions generated by the instrument).
2.2 Alignment
Once the peak lists have been extracted from each file, alignment of the features between the samples is performed by using a kernel Gaussian density (Delabrière et al., 2017; Smith et al., 2006). Two quality control steps may then be applied to select features (i) with a high reproducibility between samples (alternatively between classes of samples), and/or (ii) labeled as ‘exhaled breath’ in the majority of samples (by thresholding the P-value of the statistical tests described above).
2.3 Imputation
Imputation of missing values is performed by re-rerunning the peak detection algorithm on the raw data with updated constraints in the m/z dimension, namely without any minimum intensity threshold and with a restricted m/z width for the peak center.
2.4 Annotation and isotope detection
Putative annotations are computed by matching the measured ion masses to an internal table extracted from the Human Breathomics Database (Kuo et al., 2020). Isotope annotations are suggested on the basis of three criteria: m/z difference value, correlation of the temporal profiles within the sample, and correlation of the intensities between the samples.
2.5 Software implementation
All algorithms were written in R (R Core Team, 2021), and implemented as the ptairMS package (https://doi.org/10.18129/B9.bioc.ptairMS), freely available on the Bioconductor platform (Gentleman et al., 2004). The companion ptairData experiment package (https://doi.org/10.18129/B9.bioc.ptairData), also available on Bioconductor, contains the raw files from two datasets from exhaled breath and bacteria culture head space, respectively, as well as the simulated raw data file described in the following Section 3.
The main ptairMS methods are described in Supplementary Figure S2. Briefly, a ptrSet object is built by providing the name of the directory containing the HDF5 raw files. This object is then completed at each step of the processing. In addition, the ptrSet may be updated by adding new raw files to the directory, or by providing new sample metadata. The ptairMS output contains the table of peak intensities as well as the sample and variable metadata, which can be exported as three tabular files, or as a single ExpressionSet object, for subsequent statistical analysis.
3 Results
We developed a suite of algorithms for the preprocessing of PTR-TOF-MS data files and the untargeted analysis of exhaled breath from cohorts. Our workflow consists of the following main modules: peak detection, expiratory phases detection, temporal estimation, VOCs quantification and alignment between samples (Supplementary Fig. S2). It has been implemented in R as the ptairMS package, which is freely available on the Bioconductor repository. The package includes a Shiny graphical interface to facilitate data management and analysis by the end-user.
3.1 Quantification and untargeted VOCs detection in a standardized gas mixture
The quality of VOC detection and absolute quantification by ptairMS was first assessed with the analysis of a reference gas containing a mixture of VOCs in known amounts: 14 compounds with 8 distinct masses and 18 isotopes (TO-14 standard gas mixture, Restek; see the detailed list of expected molecules in the Supplementary Table S3). Ten dilutions of the gas mixture were measured in six replicates, with or without applying an activated charcoal filter (Supelpure HC hydrocarbon trap, Sigma-Aldrich, Saint-Quentin-Fallavier, France) on the ambient air input (three replicates each). During each acquisition, the aspiration of the reference gas was switched on and off three times to mimic ‘expiration’ profiles. Sample analysis was performed with a PTR-Qi-TOF (Ionicon, Innsbrück, Austria).
The 60 raw files were pre-processed by ptairMS in less than 15 min (on a quad-core laptop). A total of 314 (respectively, 180) compounds were detected in the absence (respectively, presence) of the charcoal filter. In particular, 45 compounds were selected after sample alignment in at least 90% of one dilution factor, and in the simulated ‘expiration’ phases of at least 90% of all samples (Fig. 2A), according to the statistical test implemented in ptairMS to compare intensities between simulated expiration and ambient air phases (see Section 2).
Fig. 2.

ptairMS analysis of a reference VOC mixture. (A) Heatmap of the log2 concentrations in ppb of the 45 selected VOCs before the imputation step. (B) The sum of the 45 compounds concentrations for each replicate (dashed line) as a function of the concentration factor. The expected total concentration is shown as a red line
Importantly, all the expected compounds were detected, as well as their isotopes, with an m/z error inferior to 20 ppm, and an average coefficient of linearity R2 with the concentration factor of 0.999. The 19 additional detected features most likely correspond to fragments from these VOCs, since some are below the expected concentration (Supplementary Fig. S3). To evaluate the quantification, we computed the difference between the sum of the 45 compound concentrations and the expected concentration, which was less than 8.1% for the concentrations above 19 ppb (Table 1 and Fig. 2B). The coefficient of variation (CV) between replicates was <5% (Table 1), even in the absence of charcoal filter, which demonstrates that the ambient air intensity is well subtracted from the exhaled breath signal in ptairMS.
Table 1.
Mean absolute percentage error (MAPE) and coefficient of variation (CV) between replicates of the ptairMS processed data from the reference gas mixture acquisitions
| Expected ppb per compound | MAPE (%) | CV (%) |
|---|---|---|
| 47.9 | 4.3 | |
| 8.1 | 3.4 | |
| 2.5 | 2.8 |
3.2 VOCs temporal profile classification and comparison to the state of the art on simulated data
The performance of the present and previously described software (Holzinger, 2015; Müller et al., 2013) were compared using simulated data from PTR-TOF-MS exhaled breath analysis. First, temporal evolutions were extracted from a large in-house database of patient acquisitions (>10 000 expiration and ambient air profiles), after normalization and Savitzky–Golay smoothing. Second, peak clusters were generated around nominal masses 21 to 400, with an asymmetric sech2 peak shape distribution. Peaks parameters were randomly selected for each nominal mass: i.e. the asymmetry coefficient the peak width, the number of overlapping peaks (1 to 3), the peak proximity, the intensity of the highest peak, the ratio of neighboring peaks, and the class of temporal profile (‘expiration’, ‘ambient air’ or ‘constant’). The exact m/z value of the first peak was selected from the formula library CxHyOzNt used by PTRwid (Holzinger, 2015). Finally, background noise was added by using a Poisson stochastic process (Gundlach-Graham et al., 2018), with a Gaussian distribution to model the single ion Pulse-Height. The random drawing of each parameters is detailed in the Supplementary Table S2, and the code used for the simulation, as well as a representative simulated data file in the HDF5 format, are included in the ptairData R/Bioconductor companion package.
Ten simulated files, containing a total of 7028 peaks, were processed with ptairMS (version 0.1), PTRwid (version 002 IDL) and IDA (version beta 0.9.4.8). ptairMS, which is the only software allowing simultaneous multiple file processing, enabled to process the 10 files in <10 min. Mass calibration was performed using the peaks at m/z 21.022, 203.943 and 330.84 for the three software, intentionally simulated without overlap at the exact masses. The calibration stability period was set to the acquisition duration, since no calibration shift was added. To ensure a good estimation of the peak shape for the three software, we simulated more single peaks in the intensity range set for the calculation of the peak shape. Finally, the ‘sensitivity’ parameter for IDA peak detection was decreased to 25%, in order to limit the number of false positives. The other parameters from each software tool were kept to default values.
Results of the comparison are shown in Table 2. The best precision of peak detection and mass accuracy were obtained with ptairMS, and the peak detection recall was slightly lower than IDA (98.40% versus 98.49%). The mass accuracy depends only on peak detection, since no mass deviation was included in the simulation. Of note, the reported mass accuracy for PTRwid was computed before calibration: indeed, the masses from the simulated multiple peaks may not match with the internal chemical formula library used by PTRwid for calibration, especially for masses >300 Da (the mass accuracy for PTRwid after calibration was 20 ppm).
Table 2.
Comparison of peak detection and quantification by ptairMS, PTRwid and IDA on 10 simulated files (7028 peaks)
| Software | ptairMS | PTRwid | IDA |
|---|---|---|---|
| Mass accuracy (ppm) | 3 | 12a | 5 |
| Peak detection precision (%) | 99.99 | 98.87 | 97.30 |
| Peak detection recall (%) | 98.40 | 87.19 | 98.49 |
| MAPE (%) | 4.96 | 14.65 | 5.38 |
| Expiration sensitivity (%) | 98.53 | 91.45 | 94.52 |
| Expiration specificity (%) | 99.01 | 86.31 | 97.03 |
| Global accuracy (%) | 99.12 | 86.73 | 95.31 |
Note: The precision (respectively, recall) of peak detection is the proportion of detected peaks which correspond to actual simulated peaks (respectively, the proportion of actual simulated peaks which were detected by the software tools). The mean absolute percentage error (MAPE) is used to assess the quality of the temporal profile estimation. Expiration sensitivity, specificity and accuracy refer to the classification of VOC origin as exhaled breath (vs. ambient air). For each metric, the best performance is shown in bold.
The reported mass accuracy for PTRwid was computed before calibration as explained in the text.
Quantification was further evaluated on the peaks which were well detected by all software. The mean absolute percentage error between the estimated temporal evolution and the input of the simulation was 4.96% for ptairMS and 14.65% (respectively, 5.38%) for PTRwid (respectively, IDA). Finally, we compared the ability to discriminate the compounds from exhaled breath and ambient air, based on two unilateral t-tests comparing the intensities in the two acquisition phases (see Section 2.1.3). ptairMS was shown to detect the expiration profiles with the highest sensitivity and specificity, with a global accuracy of 99% (compared to 87% and 95% for PTRwid and IDA; Table 2). As illustrated in Figure 3 on two simulated peaks with close m/z values, an exogenous VOC (i.e. with a constant profile) at m/z 82.034 was erroneously classified as ‘expiration’ by PTRwid and IDA but not by ptairMS, as a result of a less precise temporal estimation of the two first software tools. Altogether, these results demonstrate that ptairMS is well suited for biomarker research by breath analysis.
Fig. 3.
Estimation of the temporal profile by ptairMS, compared to the PTRwid and IDA software on simulated data. Right: raw simulated data of two overlapping peaks (as shown in 2D), and the corresponding total mass spectrum. In this particular example, the VOC at m/z 82.02 (respectively, m/z 82.03) was simulated by using an ‘expiration’ (respectively, a ‘constant’) temporal profile. Left: temporal profiles estimated by the three software (solid colored lines), compared to the simulated profile (ground truth shown as black dots), for the two peaks (top: m/z 82.02 and bottom: m/z 82.03). As observed with the peak at m/z 82.03, the temporal estimations from PTRwid and IDA lead to an erroneous classification of the VOC as expiration or ambient air
3.3 Application to real datasets
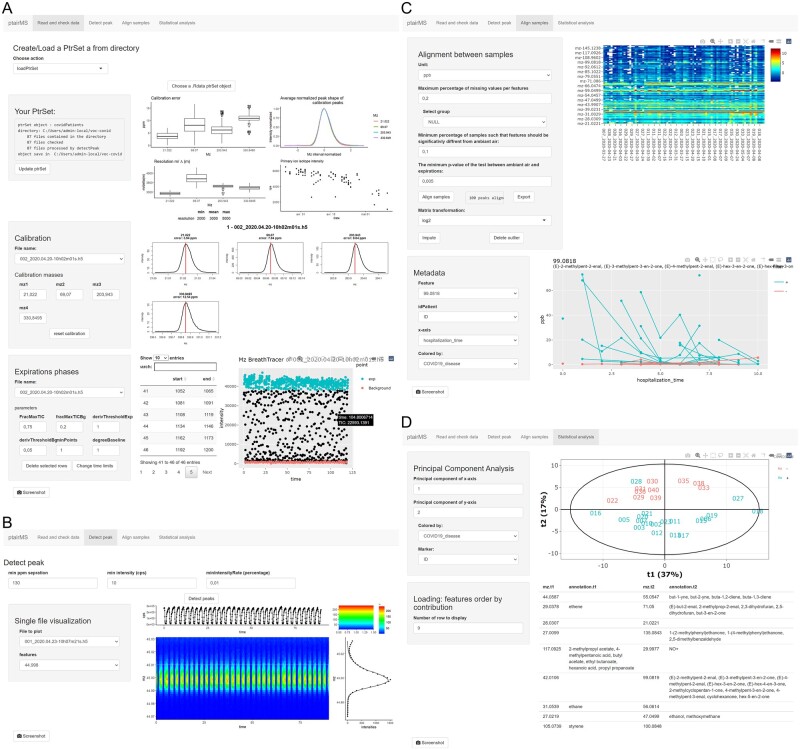
The ptairMS software has been designed for biomarker discovery in large clinical cohorts. First, it is fast (<1 min for a 3–5 min acquisition), and files can be processed with parallel computing and in a batch mode. Second, studies can be readily incremented with new files (e.g. if new patients are included): only the processing of these new files and the final alignment between samples are performed to update the peak table of the whole cohort. Third, the whole workflow can be run interactively through a graphical user interface, which provides visualizations (expiration phases, peaks in the raw data, peak table, individual VOCs), quality controls (calibration, resolution, peak shape and evolution of the reagent ions with time), and exploratory data analysis (Fig. 4). A detailed documentation including several use cases is included in the package.
Fig. 4.
The ptairMS graphical user interface to monitor the processing and exploratory analysis of cohorts, as illustrated with the COVID-19 dataset (Grassin-Delyle et al., 2021). (A) The ‘Read and check data’ tab enables to open the data (either from a new study or to update an existing one), and to perform the calibration and the detection of expirations, and provides optimal parameter values for the peak shape and the resolution. (B) The ‘Detect peak’ tab provides single file visualizations of the raw data, of the detected peaks, and of the temporal profiles. (C) The ‘Align samples’ tab displays the final peak table as well as the individual features colored according to the sample metadata. (D) The ‘Statistical Analysis’ tab displays the score plot from the Principal Component Analysis of the peak table [only the first time point of each patient is shown here, as in Grassin-Delyle et al. (2021)], and the list of features with their putative annotations, in decreasing order of loading values
ptairMS is already used in routine in the clinic to process the acquisitions from freely breathing patients in some breath research centers using PTR-Qi-TOF MS. Files from a distinct PTR-TOF 8000 instrument (Ionicon) (Trefz et al., 2013; Vita et al., 2015) were also successfully processed with ptairMS (Supplementary Figs S4 and S5). These results highlight the ability of the algorithms to adapt to various resolutions, time bin periods, peak shapes and temporal profiles.
4 Discussion
We have developed an innovative workflow for the fast processing of PTR-TOF-MS data from exhaled breath. The suite of algorithms includes untargeted peak detection and deconvolution in the mass dimension, expiration phases detection, estimation of the temporal evolution of the peak intensity during the acquisition and quantification. Compared to the two existing software, it enables for the first time to conduct the analysis of clinical cohorts, with parallel file processing, incremental addition of new patient files, quality control of the acquisitions along clinical trials, alignment between the samples, and final statistical tests to discard exogenous VOCs. The full workflow was implemented in the R package ptairMS which is publicly available on the Bioconductor repository and includes a detailed tutorial. Raw files from two experimental datasets, as well as one simulated file, are provided in the companion ptairData package. The public availability of all data and source code will therefore be of high value for the reproducibility of the analyzes, and the benchmark of software tools (Wilkinson et al., 2016).
The quality of the untargeted peak detection and absolute quantification was assessed by using a standardized gas mixture: all compounds were detected by ptairMS with an m/z precision lower than 20 ppm, an intensity error below 8.1% (for compounds with concentrations >19 ppb), an average R2 coefficient with the concentration factor of 0.999, and a CV <5%, thus demonstrating the performance of the detection and quantification. However, it is important to note that the standardized gas used does not reflect breath matrices. In practice, humidity saturation of exhaled breath biases the VOC quantification in PTR-MS instruments, with divergent behavior for different substance classes (Trefz et al., 2018). This effect also impacts the proposed correction of the ambient air level (which consists in subtracting the ambient air baseline from the temporal profile estimated for each VOC). Since the exhaled breath and ambient air have different concentrations of humidity, O2, and CO2, the direct subtraction should not therefore be considered as an absolute quantification, but rather as a relative concentration, which can be used to compare patients. To further compute accurate concentration differences between inspiratory and expiratory phases, adequate humidity-adapted calibrations are required (Trefz et al., 2018).
Since the estimation of the temporal profiles is a key aspect of breath analysis, we have developed a 2D model based on P-spline regression. Compared to the existing software which are well suited for single-file, large data from environmental monitoring, we demonstrate that ptairMS is very convenient for breath analysis, achieving highest sensitivity and accurate quantification. It should be noted that the temporal estimation of the peak intensities relies on the m/z values previously computed on the total ion spectrum (i.e. these m/z values are not re-evaluated at each time point) which allows a fast computation. While alternative approaches may be considered for the combined estimation of location and intensity of the peaks in 2D (such as Bayesian methods or non-linear optimization; Barat et al., 2007; Binette et al., 2020; He et al., 2014), the ptairMS algorithms already provides precise m/z and intensity estimations, in a computation time (<1 min) which is compatible with the real-time patient analysis.
The classification of the VOC origin between exhaled breath and ambient air was shown to be improved with ptairMS (due to the 2D modeling), with an accuracy up to 99%. The control of external factors such as the ambient air (Trefz et al., 2013), but also the dioxygen concentration (Trefz et al., 2019), the patient medication, or specific diets, is of critical importance in breath analysis (Hanna et al., 2019). ptairMS therefore checks the sample reproducibility after alignment to avoid some of these unwanted variations. In all cases, attention should be paid during the design of the study to the matching of patients and sampling conditions between the groups of interest.
Importantly, ptairMS automatically suggests optimal values for the parameters, such as the resolution and the peak shape (as evaluated on the calibration peaks), but also the location of spline knots (at higher densities within the expiration phases) and the penalization for the 2D regression (based on generalized cross-validation). This enables to adapt the processing to specific instruments (e.g. with distinct resolutions) but also to various biological matrices (e.g. with different time profiles). As an example, ptairMS was used to process files from both PTR-TOF 8000 and PTR-Qi-TOF instruments (Ionicon Analytik). Files from other vendors (e.g. Tofwerk) should be processed accordingly, since they are in the same open source HDF5 format, which is a data storage format of choice within the MS community (Askenazi et al., 2017). Beyond exhaled breath, ptairMS was successfully applied to atmospheric air data (hospital room and corridor air), headspace analysis from mycobacteria (see the package tutorial) and truffles (Vita et al., 2015; Supplementary Fig. S5).
A graphical interface was developed to facilitate data analysis and result interpretation by experimenters (e.g. clinicians). It covers the processing of raw data up to the exploratory data analysis of the cohort, with interactive tables and graphics. Since clinical studies may last several months, or even years, the interface includes a dedicated panel for the real-time control of instrument parameters to avoid unwanted effects resulting from drift in temperature, pressure, or variations in the amount of reagent ion. Incremental addition of new patient files is also possible without the need to reprocess all of the previous acquisitions. New features in future implementations will include visualizations (such as the superposition of multiple temporal profiles for several patients), and statistical testing of clinical metadata for each detected VOC. Finally, a putative annotation of the compounds and their isotopes based on the m/z values is provided to facilitate interpretation. To achieve higher confidence levels of 2 or 1 for the most interesting VOCs, complementary experiments with hyphenated techniques such as GC-MS are required (Ibrahim et al., 2019; Nardi-Agmon et al., 2016; Wilde et al., 2019).
Recently, ptairMS was successfully applied to intubated, mechanically ventilated patients, and enabled to discover a biomarker signature of four VOCs for the diagnosis of coronavirus disease-19 infection (Grassin-Delyle et al., 2021). In addition, it is routinely used for clinical trials in centers performing exhaled breath research, not only for online patient analysis, but also for the off-line analysis of breath collected in sampling bags, allowing the analysis of samples from multisite patients.
Altogether, these results demonstrate the value of the ptairMS software as a key resource in breathomics for real-time analysis at the point of care and in biomarker discovery studies, with a high clinical potential for the phenotyping of health and disease, therapeutic drug monitoring, toxicological studies and precision medicine (Fernández del Río et al., 2015; Ibrahim et al., 2019; Jung et al., 2021; Löser et al., 2020; Zhou et al., 2017).
Supplementary Material
Acknowledgements
We thank all the staff from the Exhalomics platform (Foch Hospital, Suresnes) for providing PTR-MS data. We thank Pierrick Roger and Paul Zheng for their help with the volatolomics database, the raw data format and the package design. We thank Dr Phillip Trefz for kindly providing acquisition files for the PTR-TOF-MS 8000 instrument. We also thanks Dr Vincent Le Moine and Pr Jean-Louis Herrmann for providing bacteria cultures.
Funding
The study was funded by the Agence Nationale de la Recherche (SoftwAiR project, ANR-18-CE45-0017).
Conflict of Interest: none declared.
Contributor Information
Camille Roquencourt, Département Métrologie Instrumentation & Information (DM2I), CEA, LIST, Laboratoire Sciences des Données et de la Décision, F-91191 Gif-Sur-Yvette, France.
Stanislas Grassin-Delyle, Département des maladies des voies respiratoires, Hôpital Foch, Exhalomics, Suresnes 92150, France; Département de Biotechnologie de la Santé, Université Paris-Saclay, UVSQ, INSERM, Infection et inflammation, Montigny le Bretonneux 78180, France; FHU SEPSIS (Saclay and Paris Seine Nord Endeavour to PerSonalize Interventions for Sepsis), Garches 92380, France.
Etienne A Thévenot, Département Médicaments et Technologies pour la Santé (MTS), Université Paris-Saclay, CEA, INRAE, MetaboHUB, F-91191 Gif sur Yvette, France.
References
- Amann A. et al. (2014) The human volatilome: volatile organic compounds (VOCs) in exhaled breath, skin emanations, urine, feces and saliva. J. Breath Res., 8, 034001. [DOI] [PubMed] [Google Scholar]
- Askenazi M. et al. (2017) The arc of Mass Spectrometry Exchange Formats is long, but it bends toward HDF5: plain HDF5 as a mass spectrometry exchange format. Mass Spectrom. Rev., 36, 668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barat E. et al. (2007). A nonparametric bayesian approach for PET reconstruction. In: 2007 IEEE Nuclear Science Symposium Conference Record. IEEE, Honolulu, HI, USA, pp. 4155–4162.
- Beauchamp J. (2011) Inhaled today, not gone tomorrow: pharmacokinetics and environmental exposure of volatiles in exhaled breath. J. Breath Res., 5, 037103. [DOI] [PubMed] [Google Scholar]
- Binette O. et al. (2020) Bayesian Closed Surface Fitting Through Tensor Products. J. Mach. Learn. Res., 26.
- Blake R.S. et al. (2009) Proton-transfer reaction mass spectrometry. Chem. Rev., 109, 861–896. [DOI] [PubMed] [Google Scholar]
- Boots A.W. et al. (2015) Exhaled molecular fingerprinting in diagnosis and monitoring: validating volatile promises. Trends Mol. Med., 21, 633–644. [DOI] [PubMed] [Google Scholar]
- Brezger A., Lang S. (2006) Generalized structured additive regression based on Bayesian P-splines. Comput. Stat. Data Anal., 50, 967–991. [Google Scholar]
- Brown R., Gilfrich N. (1991) Design and performance of a matrix-assisted laser desorption time-of-flight mass spectrometer utilizing a pulsed nitrogen laser. Anal. Chim. Acta, 248, 541–552. [Google Scholar]
- Bruderer T. et al. (2019) On-line analysis of exhaled breath: focus review. Chem. Rev., 119, 10803–10828. [DOI] [PubMed] [Google Scholar]
- Cappellin L. et al. (2010) Improved mass accuracy in PTR-TOF-MS: another step towards better compound identification in PTR-MS. Int. J. Mass Spectrom., 290, 60–63. [Google Scholar]
- Cappellin L. et al. (2011) On data analysis in PTR-TOF-MS: from raw spectra to data mining. Sens. Actuators B Chem., 155, 183–190. [Google Scholar]
- Cappellin L. et al. (2012) On quantitative determination of volatile organic compound concentrations using proton transfer reaction time-of-flight mass spectrometry. Environ. Sci. Technol., 46, 2283–2290. [DOI] [PubMed] [Google Scholar]
- Cristescu S.M. et al. (2011) Screening for emphysema via exhaled volatile organic compounds. J. Breath Res., 5, 046009. [DOI] [PubMed] [Google Scholar]
- Currie I.D., Durban M. (2002) Flexible smoothing with P-splines: a unified approach. Stat. Modelling, 2, 333–349. [Google Scholar]
- de Boor C. (1978) A Practical Guide to Splines. Applied Mathematical Sciences, Springer, New York.
- Delabrière A. et al. (2017) proFIA: a data preprocessing workflow for flow injection analysis coupled to high-resolution mass spectrometry. Bioinformatics, 33, 3767–3775. [DOI] [PubMed] [Google Scholar]
- Devillier P. et al. (2017) Metabolomics in the diagnosis and pharmacotherapy of lung diseases. Curr. Pharm. Des., 23, [DOI] [PubMed] [Google Scholar]
- Dierckx P. (1995) Curve and Surface Fitting with Splines. Monographs on numerical analysis p. 5.
- Eilers P., Marx B. (2021). Practical Smoothing: The Joys of P-Splines. Cambridge University Press, Cambridge. [Google Scholar]
- Eilers P.H.C., Marx B.D. (1996) Flexible smoothing with B-splines and penalties. Stat. Sci., 11, 89–121. [Google Scholar]
- Eilers P.H.C., Marx B.D. (2010) Splines, knots, and penalties. Wiley Interdiscip. Rev. Comput. Stat., 2, 637–653. [Google Scholar]
- Eilers P.H.C. et al. (2015) Twenty Years of P-Splines. SORT (Statistics and Operations Research Transactions). 39. 149–186.
- Einoch Amor R. et al. (2019) Breath analysis of cancer in the present and the future. Eur. Respir. Rev., 28, 190002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández del Río R. et al. (2015) Volatile biomarkers in breath associated with liver cirrhosis—comparisons of pre- and post-liver transplant breath samples. EBioMedicine, 2, 1243–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipiak W. et al. (2012) Dependence of exhaled breath composition on exogenous factors, smoking habits and exposure to air pollutants. J Breath Res, 6, 036008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C. et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol., 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassin-Delyle S. et al. (2021) Metabolomics of exhaled breath in critically ill COVID-19 patients: a pilot study. EBioMedicine, 63, 103154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressani O., Lambert P. (2021) Laplace approximations for fast Bayesian inference in generalized additive models based on P-splines. Comput. Stat. Data Anal., 154, 107088. [Google Scholar]
- Gundlach-Graham A. et al. (2018) Monte Carlo simulation of low-count signals in time-of-flight mass spectrometry and its application to single-particle detection. Anal. Chem., 90, 11847–11855. [DOI] [PubMed] [Google Scholar]
- Hanna G.B. et al. (2019) Accuracy and methodologic challenges of volatile organic compound-based exhaled breath tests for cancer diagnosis: a systematic review and meta-analysis. JAMA Oncol., 5, e182815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. et al. (2014) A spline filter for multidimensional nonlinear state estimation. Signal Process., 102, 282–295. [Google Scholar]
- Herbig J. et al. (2009) On-line breath analysis with PTR-TOF. J. Breath Res., 3, 027004. [DOI] [PubMed] [Google Scholar]
- Holzinger R. (2015) PTRwid: a new widget tool for processing PTR-TOF-MS data. Atmos. Meas. Tech., 8, 3903–3922. [Google Scholar]
- Ibrahim W. et al. (2019) Assessment of breath volatile organic compounds in acute cardiorespiratory breathlessness: a protocol describing a prospective real-world observational study. BMJ Open, 9, e025486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A. et al. (2009) A high resolution and high sensitivity proton-transfer-reaction time-of-flight mass spectrometer (PTR-TOF-MS). Int. J. Mass Spectrom., 286, 122–128. [Google Scholar]
- Jung Y.J. et al. (2021) Advanced diagnostic technology of volatile organic compounds real time analysis from exhaled breath of gastric cancer patients using proton-transfer-reaction time-of-flight mass spectrometry. Front. Oncol., 11, 560591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol Q. (2011). HDF5. In: Padua D. (ed.) Encyclopedia of Parallel Computing. Springer US, Boston, MA, pp. 827–833. [Google Scholar]
- Kuo T.-C. et al. (2020) Human breathomics database. Database (Oxford), 2020, baz139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange E. et al. (2007) A geometric approach for the alignment of liquid chromatography—mass spectrometry data. Bioinformatics, 23, i273–i281. [DOI] [PubMed] [Google Scholar]
- Löser B. et al. (2020) Changes of exhaled volatile organic compounds in postoperative patients undergoing analgesic treatment: a prospective observational study. Metabolites, 10, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx B.D., Eilers P.H. (2005) Multidimensional penalized signal regression. Technometrics, 47, 13–22. [Google Scholar]
- Müller M. et al. (2011) Enhanced spectral analysis of C-TOF aerosol mass spectrometer data: iterative residual analysis and cumulative peak fitting. Int. J. Mass Spectrom., 306, 1–8. [Google Scholar]
- Müller M. et al. (2013) A new software tool for the analysis of high resolution PTR-TOF mass spectra. Chemom. Intell. Lab. Syst., 127, 158–165. [Google Scholar]
- Müller M. et al. (2014) Detector aging induced mass discrimination and non-linearity effects in PTR-TOF-MS. Int. J. Mass Spectrom., 365–366, 93–97. [Google Scholar]
- Nardi-Agmon I. et al. (2016) Exhaled breath analysis for monitoring response to treatment in advanced lung cancer. J. Thorac. Oncol., 11, 827–837. [DOI] [PubMed] [Google Scholar]
- Obermeier J. et al. (2017) Exhaled volatile substances mirror clinical conditions in pediatric chronic kidney disease. PLoS One, 12, e0178745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J. et al. (2015) Breath analysis as a potential and non-invasive frontier in disease diagnosis: an overview. Metabolites, 5, 3–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. (1997) Method for the collection and assay of volatile organic compounds in breath. Anal. Biochem., 247, 272–278. [DOI] [PubMed] [Google Scholar]
- Pleil J.D. et al. (2019) Advances in proton transfer reaction mass spectrometry (PTR-MS): applications in exhaled breath analysis, food science, and atmospheric chemistry. J. Breath Res., 13, 039002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2021) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/. [Google Scholar]
- Rattray N.J. et al. (2014) Taking your breath away: metabolomics breathes life in to personalized medicine. Trends Biotechnol., 32, 538–548. [DOI] [PubMed] [Google Scholar]
- Ryan C.G. et al. (1988) SNIP, A Statistics Sensitive Background Treatment for the Quantitative Analysis of the Pixe Spectra in Geoscience Application. Nucl. Instrum. Methods. Phys. Res. B., 34, 396–402. [Google Scholar]
- Savitzky A., Golay M.J.E. (1964) Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem., 36, 1627–1639. [Google Scholar]
- Schwoebel H. et al. (2011) Phase-resolved real-time breath analysis during exercise by means of smart processing of PTR-MS data. Anal. Bioanal. Chem., 401, 2079–2091. [DOI] [PubMed] [Google Scholar]
- Sidiropoulos N.D. et al. (2017) Tensor decomposition for signal processing and machine learning. IEEE Trans. Signal Process., 65, 3551–3582. [Google Scholar]
- Smith C.A. et al. (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem., 78, 779–787. [DOI] [PubMed] [Google Scholar]
- Španěl P. et al. (2013) A quantitative study of the influence of inhaled compounds on their concentrations in exhaled breath. J. Breath Res., 7, 017106. [DOI] [PubMed] [Google Scholar]
- Trefz P. et al. (2013) Continuous real time breath gas monitoring in the clinical environment by proton-transfer-reaction-time-of-flight-mass spectrometry. Anal. Chem., 85, 10321–10329. [DOI] [PubMed] [Google Scholar]
- Trefz P. et al. (2018) Effects of humidity, CO2 and O2 on real-time quantitation of breath biomarkers by means of PTR-ToF-MS. J. Breath Res., 12, 026016. [DOI] [PubMed] [Google Scholar]
- Trefz P. et al. (2019) Effects of elevated oxygen levels on VOC analysis by means of PTR-ToF-MS. J. Breath Res., 13, 046004. [DOI] [PubMed] [Google Scholar]
- Vita F. et al. (2015) Volatile organic compounds in truffle (Tuber magnatum Pico): comparison of samples from different regions of Italy and from different seasons. Sci. Rep., 5, 12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivo Truyols G., Schoenmakers P.J. (2006) Automatic selection of optimal savitzky golay smoothing. Anal. Chem., 78, 4598–4608. [DOI] [PubMed] [Google Scholar]
- Wilde M.J. et al. (2019) Breath analysis by two-dimensional gas chromatography with dual flame ionisation and mass spectrometric detection—method optimisation and integration within a large-scale clinical study. J. Chromatogr. A, 1594, 160–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.D. et al. (2016) The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data, 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood S.N. (2006) Generalized Additive Models: an introduction with R. Chapman & Hall/CRC, Boca Raton, FL. p. 397.
- Zhou W. et al. (2017) Exhaled breath online measurement for cervical cancer patients and healthy subjects by proton transfer reaction mass spectrometry. Anal. Bioanal. Chem., 409, 5603–5612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.