Abstract
Summary
The visualization and analysis of genomic repeats is typically accomplished using dot plots; however, the emergence of telomere-to-telomere assemblies with multi-megabase repeats requires new visualization strategies. Here, we introduce StainedGlass, which can generate publication-quality figures and interactive visualizations that depict the identity and orientation of multi-megabase tandem repeat structures at a genome-wide scale. The tool can rapidly reveal higher-order structures and improve the inference of evolutionary history for some of the most complex regions of genomes.
Availability and implementation
StainedGlass is implemented using Snakemake and available open source under the MIT license at https://mrvollger.github.io/StainedGlass/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Dot plot analyses are often used to reveal the underlying structure of complex repeats, including differences in sequence identity and orientation (Cabanettes and Klopp, 2018; Gibbs and McIntyre, 1970; Rice et al., 2000; Sonnhammer and Durbin, 1995). Advances in long-read sequencing technology, however, have recently made more complex repeat structures and genetic variation available for sequence analysis (Audano et al., 2019; Chaisson et al., 2015; Ebert et al., 2021). With increasingly contiguous assemblies of reference genomes (Rhie et al., 2021) and complete human chromosomes (Logsdon et al., 2021; Miga et al., 2020; Nurk et al., 2021), complete centromeres, tandem duplications and other heterochromatic arrays can now be systematically analyzed in their entirety. The size and complexity of these structures, often many megabase pairs in size, elude traditional dot plot analyses for three reasons: (i) current visualization methods are largely based on perfect or k-mer matches that do not lend themselves to the complex higher-order repeats found in centromeres (Willard and Waye, 1987) and the expected mismatches between these large repeats; (ii) dot plots offer limited resolution for tandem arrays consisting of megabases of sequence data, frequently reducing them to black squares that relay little information other than the size and presence of a repeat; and (iii) the number of possible pairwise matches increases rapidly when identifying exact matches in tandem arrays (e.g., in MUMmer) and this problem is exacerbated further when using a small minimum-match length for comparing more divergent arrays.
In this work, we present StainedGlass, which generates identity heatmaps based on sequence alignment rather than small k-mers using an easy, scalable and customizable workflow that allows for interactive use as well as publication-ready figures. The tool can be applied to study repeat structures at a genome-wide scale or focused at particular regions to characterize complex higher-order repeat structures. As part of our recent analysis of chromosome 8, we developed a prototype of this method that rendered the higher-order repeat structure of the 2 Mbp centromere as an identity heatmap. This prototype facilitated the discovery of higher-level symmetry and a layered organization in the centromere, which assisted in the development of a more refined model for centromere evolution, as well as the discovery of hotspots for copy number variation (Logsdon et al., 2021).
2 Materials and methods
To generate pairwise sequence identity heatmaps for StainedGlass, the input sequence is fragmented into windows of a configurable size (default 1 kbp). All possible pairwise alignments between the fragments are calculated using minimap2 (Li, 2018). The color gradient used in the heatmap is then determined by the sequence identities of the alignments, which are calculated as:
where M is the number of matches, X the number of mismatches, I the number of insertion events and D the number of deletion events. When there are multiple alignments between the same two sequence fragments, all alignments other than the one with the most matches are filtered out regardless of their sequence identity. This situation often arises when aligning tandem repeats where there can be multiple valid alignments between fragments, and this strategy assists in highlighting the most representative alignment.
StainedGlass generates two types of outputs: fixed resolution static figures as well as multi-resolution Cooler files (Abdennur and Mirny, 2020) suitable for interactive multiscale visualization using HiGlass (Kerpedjiev et al., 2018). Sequence identity in Cooler files is calculated using the method described above for the highest resolution and interpolated by averaging values for lower resolutions. The static figures are more appropriate for visualization of relatively small regions (30 Mbp or less) at publication quality (Fig. 1) while the HiGlass visualization is better suited for data exploration of whole-genome alignments, such as the sequence identity relationships among the short arms of human acrocentric chromosomes (interactive HiGlass browser) (Kerpedjiev et al., 2018; Nurk et al., 2021).
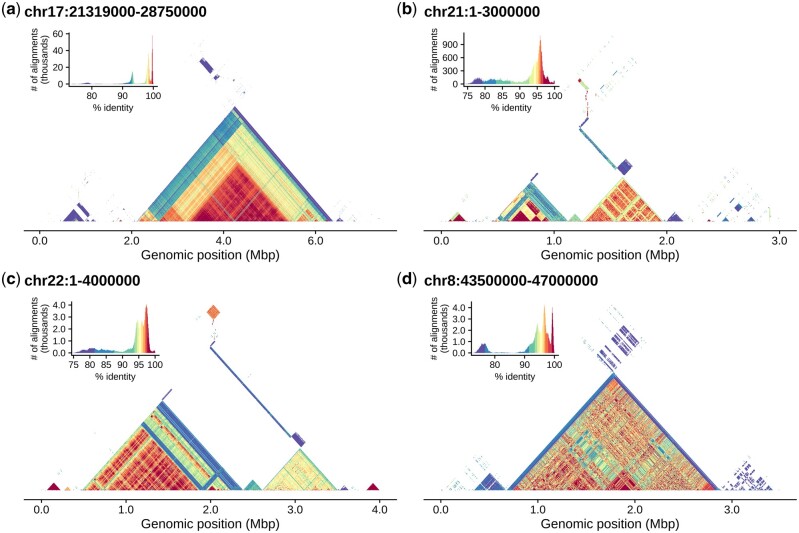
Fig. 1.
Tandem arrays in T2T-CHM13 visualized with StainedGlass. This figure was generated entirely by StainedGlass and has not been edited or refined in any way. (a) The chromosome 17 centromere dot plot reveals several different higher-order repeat arrays, which are made apparent by the colored percent identity. (b, c) The beginning of the acrocentric short arms of chromosomes 21 and 22. (d) Highlights the centromeric array on chromosome 8 first presented by Logsdon et al. (2021). An interactive browser of this data is available at https://resgen.io/paper-data/T2T/views/MtjcVgrlQmymnHIvdck5-g
The tool is made available using Snakemake (Köster and Rahmann, 2012, 2018; Mölder et al., 2021), which allows for reproducible and scalable data analyses. The stability of new changes is automatically tested with each new addition using continuous integration via GitHub actions. Finally, StainedGlass is a Snakemake standard compliant workflow and therefore has automated usage documentation on the Snakemake website.
3 Usage and examples
StainedGlass is run using the Snakemake workflow language. To run StainedGlass, clone the repository from https://github.com/mrvollger/StainedGlass and follow the installation instructions. From the cloned directory, the alignments can then be generated with StainedGlass using the following command:
snakemake --use-conda --cores 24 --config fasta={path.to.fasta} sample={sample.prefix.for.output}
All configuration options are described in config/README.md and parameters can be specified on the command line as above or by modifying the configuration file (config/config.yaml). The jobs that will be run can also be previewed without executing the pipeline by adding -n to the command line. To generate static identity heatmaps from the StainedGlass alignments, add make_figures to the Snakemake command. For an example of the output of this command (see Fig. 1).
snakemake --use-conda --cores 24 --config fasta={path.to.fasta} sample={sample.prefix.for.output} make_figures
StainedGlass can also be used to make Cooler files (Abdennur and Mirny, 2020) that can be loaded into HiGlass for whole-genome visualizations. An example of this interactive browser for the telomere-to-telomere assembly of CHM13 can be found at resgen.io (Nurk et al., 2021).
snakemake --use-conda --cores 24 --config fasta={path.to.fasta} sample={sample.prefix.for.output} cooler
The resulting Cooler files can then be easily visualized on a local browser using HiGlass.
pip install higlass-manage
higlass-manage view results/{sample}.{\d+}.{\d+}.mcool
To further demonstrate the utility of StainedGlass, we applied the program to the recently published Arabidopsis assembly from Naish et al. and include this analysis in Supplementary Materials (Naish et al., 2021).
4 Conclusion
We developed StainedGlass, a visualization tool for large genomic repeats that displays the higher-order structure and sequence relationship of these complex tandem repeat regions. The underlying Snakemake workflow makes StainedGlass both reproducible and scalable at the whole-genome level. StainedGlass produces static output visualizations, which are publication ready, and has additional options for interactive data exploration with HiGlass.
Supplementary Material
Acknowledgements
The authors thank April Lo, Michelle Noyes and Tonia Brown for help in editing this manuscript and G. A. Logsdon for esthetic suggestions.
Funding
This work was supported, in part, by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (A.M.P.) and grants from the U.S. National Institutes of Health [NIH grants 5R01HG002385, 5U01HG010971 and 1U01HG010973 to E.E.E.]. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Conflict of Interest: E.E.E. is an SAB member of Variant Bio, Inc; P.K. owns and receives income from Reservoir Genomics LLC.
Data availability
The code underlying this article is available on GitHub [https://github.com/mrvollger/StainedGlass] and archived on Zenodo [https://doi.org/10.5281/zenodo.5576601]. The assembly used by StainedGlass to generate Figure 1 was published by Nurk et al. (2021), and is available through NCBI under the accession GCA_009914755.3.
Contributor Information
Mitchell R Vollger, Department of Genome Sciences, University of Washington School of Medicine, Seattle, WA 98195, USA.
Peter Kerpedjiev, Reservoir Genomics LLC, Oakland, CA 94619, USA.
Adam M Phillippy, Genome Informatics Section, Computational and Statistical Genomics Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Evan E Eichler, Department of Genome Sciences, University of Washington School of Medicine, Seattle, WA 98195, USA; Howard Hughes Medical Institute, University of Washington, Seattle, WA 98195, USA.
References
- Abdennur N., Mirny L.A. (2020) Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics, 36, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audano P.A. et al. (2019) Characterizing the major structural variant alleles of the human genome. Cell, 176, 663–675.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanettes F., Klopp C. (2018) D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ, 6, e4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M.J.P. et al. (2015) Resolving the complexity of the human genome using single-molecule sequencing. Nature, 517, 608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert P. et al. (2021) Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science, 372, eabf7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A.J., McIntyre G.A. (1970) The diagram, a method for comparing sequences. Its use with amino acid and nucleotide sequences. Eur. J. Biochem., 16, 1–11. [DOI] [PubMed] [Google Scholar]
- Kerpedjiev P. et al. (2018) HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol., 19, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J., Rahmann S. (2012) Snakemake—a scalable bioinformatics workflow engine. Bioinformatics, 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Köster J., Rahmann S. (2018) Snakemake-a scalable bioinformatics workflow engine. Bioinformatics, 34, 3600. [DOI] [PubMed] [Google Scholar]
- Li H. (2018) Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics, 34, 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon G.A. et al. (2021) The structure, function and evolution of a complete human chromosome 8. Nature, 593, 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga K.H. et al. (2020) Telomere-to-telomere assembly of a complete human X chromosome. Nature, 585, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mölder F. et al. (2021) Sustainable data analysis with Snakemake. F1000Res, 10, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naish M. et al. (2021) The genetic and epigenetic landscape of the Arabidopsis centromeres. Science, 374, eabi7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S. et al. (2021) The complete sequence of a human genome. bioRxiv, 2021.05.26.445798. https://doi.org/10.1101/2021.05.26.445798.
- Rhie A. et al. (2021) Towards complete and error-free genome assemblies of all vertebrate species. Nature, 592, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P. et al. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L.L., Durbin R. (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene, 167, GC1–GC10. [DOI] [PubMed] [Google Scholar]
- Willard H.F., Waye J.S. (1987) Hierarchical order in chromosome-specific human alpha satellite DNA. Trends Genet., 3, 192–198. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code underlying this article is available on GitHub [https://github.com/mrvollger/StainedGlass] and archived on Zenodo [https://doi.org/10.5281/zenodo.5576601]. The assembly used by StainedGlass to generate Figure 1 was published by Nurk et al. (2021), and is available through NCBI under the accession GCA_009914755.3.