Abstract
Purpose
Streptococcus gallolyticus subspecies gallolyticus (SGG) is an opportunistic pathogen causing invasive infections in the elderly often associated with colon neoplasia. The prevalence of SGG in the stools of patients with normal colonoscopy (control) was compared with patients with colorectal adenomas (CRA) or with carcinomas (CRC) from stages I to IV. The presence of the pks island encoding colibactin as well as other CRC-associated bacteria such as toxicogenic Bacteroides fragilis, Fusobacterium nucleatum, and Parvimonas micra was also investigated.
Patients and Methods
Fecal samples collected in France between 2011 and 2016 from patients with normal colonoscopy (n = 25), adenoma (n = 23), or colorectal cancer at different stages (n = 81) were tested by PCR for the presence of SGG, B. fragilis, F. nucleatum, P. micra, and the pks island. Relative quantification of SGG, F. nucleatum, and P. micra in stools was performed by qPCR.
Results
SGG prevalence was significantly increased in the CRC group. Our results also revealed i) a strong and significant increase of toxinogenic B. fragilis in patients with early-stage adenoma and of pks island at late-stage CRC and ii) increased levels of F. nucleatum and P. micra in the stools of CRC patients. Furthermore, the simultaneous detection of these five bacterial markers was only found in CRC patients.
Conclusions
Our results indicate that the prevalence or relative levels of CRC-associated bacteria vary during CRC development. Among them, B. fragilis (bft+) was singled out as the sole pathobiont detected at the early adenoma stage.
Keywords: Streptococcus gallolyticus, pks island, Bacteroides fragilis bft+, Parvimonas micra, Fusobacterium nucleatum, colon cancer, adenomas, adenocarcinomas
Introduction
The human colon is a complex symbiotic organ where bacteria achieve the highest density, referred to as the gut microbiome. Thousands of bacterial species colonizing the host mucosal surface are commensal symbionts, shaping the host immune system, participating in the host metabolism, and providing a barrier against colonization by potential pathogenic bacteria. Colorectal cancer (CRC) is the second most common cause of cancer mortality worldwide with more than 1.9 million new cases and more than 900,000 deaths per year (Bray et al., 2018; Xi and Xu, 2021). The etiology of CRC is heterogeneous and multifactorial. CRC arises from the accumulation of genetic and epigenetic alterations over a period of 10–15 years leading to a typical adenoma–carcinoma sequence (Fearon and Vogelstein, 1990). Recent progress in high-throughput sequencing revealed a link between the risk of colorectal cancer in humans and modifications of the composition of the gut microbiome referred to as dysbiosis (Sobhani et al., 2011; Nakatsu et al., 2015; Gagnière et al., 2016). In 1974, a seminal study demonstrated that germ-free rats develop much fewer tumors compared with the same rats with normal flora (Reddy et al., 1974). Equally important, mice transplanted with fecal microbiota from either patients with CRC or mice carrying the ApcMin mutation develop more intestinal polyps than mice receiving microbiota from healthy volunteers or from control animals (Wong et al., 2017). It is now widely accepted that colonic microbiota dysbiosis participates in the oncogenic process with a higher relative abundance of potentially pro-carcinogenic bacteria, including Fusobacterium nucleatum, Parvimonas micra, Escherichia coli pks+, toxinogenic Bacteriodes fragilis, and Streptococcus gallolyticus (Janney et al., 2020).
The first report linking S. gallolyticus subsp. gallolyticus (SGG) to CRC was published in 1951 (McCOY and Mason, 1951). In this early study, SGG was referred to as Streptococcus bovis biotype I. A reclassification of this large group of bacteria belonging to group D streptococci has been proposed based on molecular tools (Poyart et al., 2002; Schlegel et al., 2003). Streptococcus gallolyticus is now subdivided into three subspecies, subsp. gallolyticus, subsp. pasteurianus, and subsp. macedonicus. Only SGG is associated with CRC. Intestinal carriage of S. gallolyticus in humans has been estimated between 2.5% and 15% by culture techniques (Klein et al., 1977; Dubrow et al., 1991; Chirouze et al., 2013; Paritsky et al., 2015). However, a study enrolling 99 healthy volunteers in Germany using real-time PCR indicates a much higher detection rate (62.5%) (Dumke et al., 2017).
In this work, we aimed at determining the prevalence of SGG in the stools of French patients with adenomas or carcinomas at various stages as compared with a control group of patients with normal colonoscopies. In addition, we also tested the presence of other “suspects” such as B. fragilis bft+ (ETBF), E. coli pks+, F. nucleatum, and P. micra.
Patients and Methods
Recruitment of Participants and Collection of Samples
Patients referred to university hospitals for colonoscopy were enrolled in several cohorts. All individuals underwent colonoscopy due to symptoms or due to a positive fecal blood test (FOBT). All cohort studies were registered on ClinicalTrials.gov (NCT01270360). The study protocol was approved by the Ethics Committee of Comité de Protection des Personnes Paris Est-Henri Mondor (no. 10-006 in 2010). All participants signed an informed consent. The exclusion criteria for these cohorts were a history of colorectal surgery due to CRC, familial adenomatous polyposis, Lynch syndrome, infection, inflammatory bowel disease, and exposure to antibiotics during the 3 weeks preceding the colonoscopy. For more details, see (Sobhani et al., 2019) and Table S1 .
Patient Population
We first used a collection of 74 fecal samples from patients with normal colonoscopy (n = 25), colorectal adenoma (CRA, n = 23), and colorectal cancer (CRC, n = 26), collected at the Henri Mondor Hospital between 2011 and 2016 (VATNIMAD collection). CRA patient colonoscopies were indicative of early stages with benign tumors including small hyperproliferations and small and intermediate adenomas, but no severe dysplasia (precancerous polyps). Twenty-three CRC patients at stages I and II, according to the Union of International Cancer Control (UICC) classification (O’Sullivan et al., 2017), displayed various stages of the primary tumor but no signs of lymph node invasion or metastasis. We completed the initial collection by adding fecal samples from 37 patients with stage III CRC (DETECT, CCR, and ECKINOXE collections) and 18 patients with stage IV CRC (DETECT and CLIMAT collections). Finally, another 85 additional fecal samples of patients with normal colonoscopies (VATNIMAD and CCR collections) were included to measure the prevalence of S. gallolyticus subsp. gallolyticus in a larger control population.
Fecal Samples and Bacterial DNA Extraction
Whole fresh stools were collected in sterile boxes, and within 4 h, 10 g were frozen at –20°C, for analysis. All the fecal samples included here were chosen at baseline prior to therapy. Bacterial DNA was extracted from aliquots of feces. DNA isolation was performed using the Promega Wizard® Genomic DNA Purification Kit, following the modified protocol from Ahlroos and Tynkkynen in 2009 (Ahlroos and Tynkkynen, 2009). Total DNA was extracted from 150 mg of stools. Quantification of extracted DNA was measured with the “Qubit 2 Fluorometer”.
Detection of Bacteria by PCR
PCRs were conducted in a 2720 Thermal Cycler (Applied Biosystems, Foster City, USA) using primers specific for S. gallolyticus (sodA1/sodA2) (Sasaki et al., 2004), P. micra (16S rDNA) (Eick et al., 2011), B. fragilis (16S rDNA) (Tong et al., 2011), F. nucleatum (16S rDNA) (Boutaga et al., 2005), clbN implicated in colibactin polyketide synthesis (Johnson et al., 2008), and bft encoding a toxin of B. fragilis ( Table S2 ). Amplifications were carried out with the following thermal cycling profiles: 3 min at 95°C; 40 cycles of amplification consisting of 10 s at 94°C, 15 s at 55°C, and 25 s at 72°C; and 3 min at 72°C for the final extension. DNA fragments of correct sizes were identified on agarose gels.
Relative Quantification of Bacteria Species by qPCR
Quantitative real-time PCR was conducted in a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) using the same oligonucleotide primers except for S. gallolyticus (sodA5/sodA6) ( Table S2 ). DNA fragments of the expected size were identified by melt temperature peak analysis. Relative quantification was calculated using the following ΔCt formula: Rq = log10(2−(CtBacteria − CtAllBact)).
Identification of the Streptococcus gallolyticus Subspecies
The sensitivity of sodA primers was first evaluated by artificial contamination of a fecal sample with various amounts of SGG (from 1.25 * 109 to 1.25 * 102 CFU/100 mg feces). After DNA extraction of the stools, we found that sodA1/sodA2 primers allowed the detection of 1.25 * 102 CFU/100 mg feces (data not shown) and were used hereafter for SGG detection in our human stool collection.
Differences in the sodA DNA sequence were used to distinguish the three S. gallolyticus subspecies, which were fully sequenced in all S. gallolyticus-positive samples.
Statistical Analysis
The χ 2 test or the Fisher’s exact test was used to compare bacterial proportions between each group of patients. The non-parametric Mann–Whitney U test was used to compare differences in continuous variables between groups. Results were considered statistically significant when p-values were <0.05.
Results
Patient Characteristics
The characteristics of individuals included in this study are summarized in Table 1 and details can be found in Table S1 . The cohort was composed of 80 men (62%) and 49 women (38%). The average age was 63.5 ± 7.9 years and the mean body mass index (BMI) is 25.2 ± 5.8. The study included 25 individuals with normal colonoscopy (13 men, 12 women, ratio 1.1:1.0), 23 patients with benign adenomas (the CRA group consisting of 14 men and 9 women, ratio 1.6:1.0), 26 patients with colorectal cancer at early stages I/II (17 men, 9 women, ratio 1.9:1.0), 37 patients with CRC at stage III (24 men, 13 women, ratio 1.8:1.0), and 18 patients with CRC at stage IV (12 men, 6 women, ratio 2.0:1.0). We noted a higher incidence of CRC in men compared with the general population. The gender ratio (1.9:1.0) in the global CRC group (including stages I to IV) reflected the higher CRC incidence observed among men in Western Europe (Rawla et al., 2019).
Table 1.
Characteristics of patients included in this study.
| Total | Control | CRA | CRC | ||||
|---|---|---|---|---|---|---|---|
| Global | Stage I/II | Stage III | Stage IV | ||||
| Number of samples | 129 | 25 | 23 | 81 | 26 | 37 | 18 |
| Men (n) | 80 | 13 | 14 | 53 | 17 | 24 | 12 |
| Women (n) | 49 | 12 | 9 | 28 | 9 | 13 | 6 |
| Mean age (years) | 63.5 | 63.7 | 61.4 | 64.1 | 67.4 | 63.2 | 61.2 |
| Mean age: men (years) | 63.1 | 62.6 | 60.5 | 63.9 | 66.3 | 64.8 | 59.0 |
| Mean age: women (years) | 64.2 | 64.3 | 62.8 | 64.3 | 69.2 | 60.3 | 65.7 |
| Mean BMIa | 25.2 | 26.0 | 26.1 | 24.6 | 26.9 | 21.8 | 27.2 |
| Mean BMI: men | 24.8 | 24.9 | 26.1 | 24.4 | 25.7 | 22.3 | 26.5 |
| Mean BMI: women | 25.8 | 27.0 | 26.2 | 25.1 | 29.1 | 20.9 | 28.7 |
aBMI, body mass index.
Detection of Streptococcus gallolyticus subsp. gallolyticus in Human Stools
Detection of S. gallolyticus was performed by PCR using oligonucleotide primers specific for the sodA gene followed by DNA sequencing for subspecies identification, i.e., gallolyticus, pasteurianus, or macedonicus as previously described (Sasaki et al., 2004).
SGG was detected at about 30% in normal (n = 25) and CRA (n = 23) stools and at about 50% in CRC stools from stages I/II to IV (n = 81) ( Table 2 ). Since our initial control cohort sample was low (n = 25), we tested the prevalence of SGG in two other cohorts composed of 50 (30 women, 20 men) and 35 (24 women, 11 men) individuals with normal colonoscopy. We found 28 positive samples over a total of 85 samples (32.9%). This result is in perfect agreement with our initial result (8/25, 32%) ( Table 2 ). Taken together, the prevalence of SGG in our control population (36 positive over 110) is 32.5%.
Table 2.
Detection of Streptococcus gallolyticus subsp. gallolyticus (SGG), Fusobacterium nucleatum (FN), Bacteroides fragilis, enterotoxigenic B. fragilis (ETBF), Parvimonas micra (PM), and pks island in the stools from CRC, CRA, and control patients using the PCR assay.
| Positive detection of: | Total (N = 129) | Control (N = 25) | CRA (N = 23) | CRC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Global | CRC I+II | CRC III | CRC IV | |||||||||||
| (N = 81) | (N = 26) | (N = 37) | (N = 18) | |||||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | |
| SGG | 56 | 43.4 | 8 | 32.0 | 7 | 30.4 | 41 | 50.6 | 13 | 50.0 | 19 | 51.4 | 9 | 50.0 |
| FN | 82 | 63.6 | 12 | 48.0 | 13 | 56.5 | 57 | 70.4 | 16 | 61.5 | 28 | 75.7 | 13 | 72.2 |
| BF | 81 | 62.8 | 12 | 48.0 | 19 | 82.6 | 50 | 61.7 | 16 | 61.5 | 25 | 67.6 | 9 | 50.0 |
| ETBF | 44 | 34.1 | 6 | 24.0 | 13 | 56.5 | 25 | 30.9 | 9 | 34.6 | 12 | 32.4 | 4 | 22.2 |
| PM | 103 | 79.8 | 18 | 72.0 | 15 | 65.2 | 70 | 86.4 | 22 | 84.6 | 32 | 86.5 | 16 | 88.9 |
| pks island | 53 | 41.1 | 6 | 24.0 | 7 | 30.4 | 40 | 49.4 | 11 | 42.3 | 16 | 43.2 | 13 | 72.2 |
The results in Table 2 together with those obtained above show that the detection rate of SGG is not different in CRA stools (7/23; 30.4%) but is increased significantly (p = 0.02) in CRC samples at all stages (41/81, 50.6%) as compared with the global control group (36/110, 32.5%).
Prevalence of Bacteroides fragilis, Fusobacterium nucleatum, and Parvimonas micra
Detection of B. fragilis, F. nucleatum, and P. micra was carried out on the same samples by PCR and shown in Table 2 .
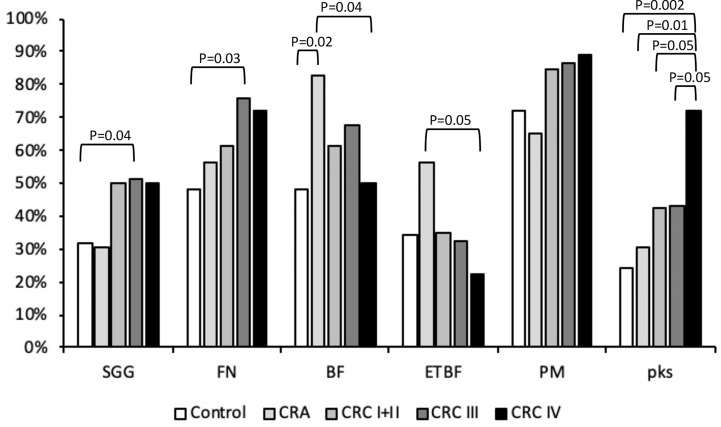
Bacteroides fragilis is a member of Bacteroidetes, a major phylum of the human gut microbiota. Bacteroides fragilis is considered as a commensal bacterium, whereas enterotoxigenic B. fragilis (ETBF) synthesizing fragilysin (bft +) has been strongly associated with CRC. Bacteroides fragilis was detected in 62.8% of our stool collection ( Table 2 ). It was detected in 19/23 CRA samples (83%) but only in approximately 62% of the CRC samples and 48% of the control stools ( Table 2 ), indicating a solid association between B. fragilis and CRA (CRA vs. control, p = 0.02). To determine the type (non-toxinogenic or toxinogenic) of B. fragilis present in our samples, detection of the bft gene encoding B. fragilis toxin was carried out by PCR using specific primers ( Table S2 ). As shown in Table 2 , the presence of ETBF was significantly increased in the stools from CRA patients (56.5%) as compared with the control (24%, p < 0.05) and CRC groups (31%, p = 0.06) ( Table 2 , Figure 1 ). Interestingly, ETBF detection decreased during the evolution of CRC, from 34.6% in CRC stage I/II to 22.2% in CRC stage IV ( Table 2 , Figure 1 ), with a significant statistical difference (p < 0.05) between CRA and CRC stage IV.
Figure 1.
Presence of Streptococcus gallolyticus (SGG), Fusobacterium nucleatum (FN), enterotoxigenic Bacteroides fragilis (ETBF), and Parvimonas micra (PM) pks gene in feces. The presence of bacterial DNA was tested by PCR using specific primers ( Table 1 ). Statistical significance was calculated using the χ 2 test or Fisher’s exact test (when n < 5). The real p-value is indicated for groups displaying significant differences.
Fusobacterium is a Gram-negative aerobic bacillus commensal of the human mouth and gastrointestinal and urogenital tracts. Fusobacterium nucleatum was detected by PCR and its presence steadily increased: 48% in the control group, 56.5% in the CRA samples, and 70.4% in the CRC group ( Table 2 and Figure 1 ) (CRC III vs. controls, p < 0.05). Fusobacterium nucleatum detection increased, although this was not statistically significant, at later stages of CRC (III/IV) as compared with earlier stages I/II (74.5% vs. 61.5%, p = 0.2, NS).
Parvimonas micra is a Gram-positive anaerobic coccus of the oral flora associated with CRC (Wu et al., 2013; Yu et al., 2017). As shown in Table 2 and Figure 1 , we could not detect any significant difference in the carriage of P. micra in the stools of our control, CRA, and CRC patients. However, a slight increase of the bacterial load in the CRC group (86.4%) was observed as compared with the control (72%) or CRA (65.2%) groups (p = 0.09, NS).
Prevalence of the pks Island
Colibactin is a genotoxin causing DNA double-strand breaks in eukaryotic cells. It is synthesized by a non-ribosomal polyketide synthase (pks) assembly line consisting of 19 genes (clbA to clbS) located on a 54-kb genomic island. Detection of the pks island was performed by PCR of the clbN gene often used as a marker for the 3′ region of this island (Johnson et al., 2008). Overall, clbN was detected with a higher prevalence in CRC stools (49.4%) compared with the non-symptomatic (control 24%, p < 0.005) or CRA groups (30.4%, p < 0.01) ( Table 2 and Figure 1 ). Analysis of pks prevalence in our global cohort revealed a progressive increase during CRC evolution peaking at the latest stage (72.2% CRC stage IV) as compared with earlier stages, i.e., CRC stage I/II (42.3%, p < 0.05) and CRC stage III (43.2%, p < 0.05) ( Table 2 and Figure 1 ).
To test the association between the presence of a specific bacterial species and CRC, we used the χ 2 test for trends with the presence/absence of bacteria as the binary variable and controls, adenoma, CRC I/II, CRC III, and CRC IV as the ordered categorical variables. Results indicate a statistical significance for F. nucleatum (p < 0.05) and the pks island (p < 0.005).
Relative Quantification of Streptococcus gallolyticus, Fusobacterium nucleatum, and Parvimonas micra in Stools
We next decided to determine the relative quantity of three bacteria in our CRC cohort. Real-time qPCR assays were performed by using primers specific for S. gallolyticus, F. nucleatum, or P. micra ( Table S2 ) and universal 16S rRNA primers (Allbact, Table S2 ) to determine the total number of bacteria present in the stool sample. Relative quantification of SGG revealed no differences in abundance in control, CRA, and CRC cohort, nor during CRC development ( Figure 2A ). In contrast, the respective abundance of F. nucleatum ( Figure 2B ) and P. micra ( Figure 2C ) was statistically increased in CRC patients as compared with CRA or controls. Detailed results indicated that the relative abundance of F. nucleatum and P. micra increased during the development of cancer, notably at later stages ( Figures 2B, C ).
Figure 2.
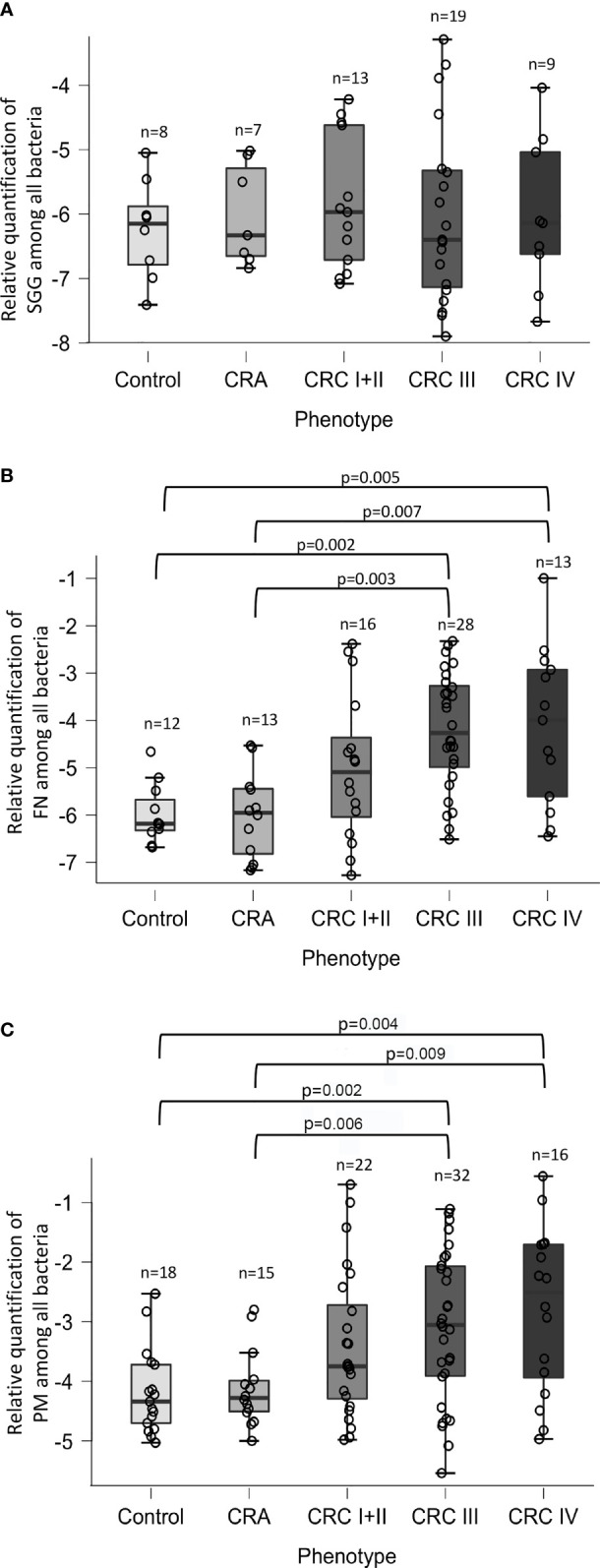
Relative quantification of Streptococcus gallolyticus (SGG), Fusobacterium nucleatum (FN), and Parvimonas micra (PM). The levels of (A) SGG, (B) FN, and (C) PM in control, adenomatous (CRA), or patients at each stage of colorectal cancer (stage I or II, stage III or IV) were calculated as a relative quantification of bacteria among total bacteria [log10(2−(CqBacteria − CqAllBacteria))] using the 16S rRNA gene as a reference. Statistical significance was calculated using the non-parametric Mann–Whitney U test. The real p-value is indicated for groups displaying significant differences.
Presence of Several Bacterial Markers in CRC Stools
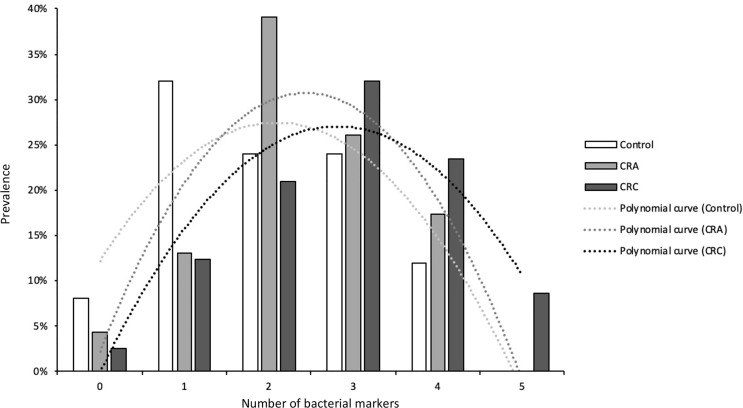
As shown in Figure 3 , a clear shift is observed in the number of suspicious bacterial markers detected in patients, increasing progressively with the evolution of the disease. This result clearly indicates that multiple opportunistic bacteria contribute to the progression of CRC. Hence, the development of potential fecal diagnostic tools for CRC detection should be based on at least five bacterial markers [SGG, F. nucleatum (FN), Parvimonas micra (PM), ETBF, and pks).
Figure 3.
The number of suspects bacterial markers found in the three groups: control, CRA, and CRC patients.
Discussion
Genetic and epigenetic modifications are of major concern in CRC development. It is now well established that dysbiosis of the gut microbiota contributes to the progression of colonic carcinogenesis. The development of new detection methods (16S rRNA gene sequencing, metagenomics, transcriptomics, proteomics, metabolomics) has led to an incredible improvement in defining the composition and function of the intestinal microbiome. However, microbiome variations along the colonic axis and between intraluminal and mucosal microbe communities have made the creation of a complete species catalog difficult (Sobhani et al., 2011). Nevertheless, several bacteria such as colibactin-producing E. coli, ETBF, SGG, and two oral bacteria, F. nucleatum and P. micra, appear to be strongly associated with colorectal cancer (Reddy et al., 1974; Fearon and Vogelstein, 1990). The molecular mechanisms underlying the promotion of colonic cell proliferation have not been completely elucidated until now. A bacterial “driver–passenger model” has been proposed in which CRC would be initiated by “driver” bacteria that cause changes in the tumor microenvironment allowing the colonization by “passenger” bacteria that could promote CRC progression (Tjalsma et al., 2012).
We found a prevalence of 32.5% for SGG in a French cohort of 110 patients with normal colonoscopies, which contrasts with the prevalence of 62.5% in the stools of 99 healthy volunteers in Germany (Dumke et al., 2017). Age and geographical distribution affecting microbiota composition could be the main factors explaining the difference between these two studies. Indeed, the mean age was 48.4 ± 14.9 in Dumke’s study, whereas it was 63.5 ± 7.9 years in our study. Our results demonstrating a higher prevalence but not a higher charge of SGG in CRC independent of the stage of CRC are more in favor of a “passenger” role for S. gallolyticus subsp. gallolyticus in CRC. This result is in line with our previous study indicating that SGG is a passenger bacterium taking advantage of tumoral conditions to outcompete commensals of the gut microbiota such as Enterococcus faecalis (Aymeric et al., 2018).
In agreement with metagenomics data showing a strong enrichment of F. nucleatum (FN) in the microbiota of CRC patients, we also observed a significant increase of FN in the stools of patients with CRC, especially at later stages. The bacterial adhesin Fap2 has been shown to bind specifically to colon tumoral tissues expressing high levels of Gal-Gal-NAc (Abed et al., 2016), while a recent study points to a role of FN in the adenoma–carcinoma transition (Bundgaard-Nielsen et al., 2019). Of note, a study on the Iranian population reports high numbers of F. nucleatum, S. bovis/gallolyticus, ETBF, and F. nucleatum in tubular adenomas and polyps, especially in villous/tubulovillous polyps, as compared with the samples from normal, hyperplastic, and sessile serrated polyps by quantitative real-time PCR (Rezasoltani et al., 2018).
The role of P. micra in CRC remains unknown, but it has been shown that it can promote intestinal carcinogenesis in APCmin/+ mice (Yu et al., 2019). Here, we demonstrated that the relative abundance of P. micra and F. nucleatum is statistically higher in CRC stools, as previously shown (Liang et al., 2017; Dai et al., 2018; Xu et al., 2020; Löwenmark et al., 2020). These results indicate that bacterial quantification is an important element for stool-based CRC diagnostics.
Enterotoxigenic B. fragilis is a gut pathobiont producing a potent toxin (BFT) that cleaves the adherens junction protein E-cadherin, altering the intestinal barrier and causing diarrhea. Alteration of E-cadherin/beta-catenin interactions activates the Wnt pathway leading to cell proliferation. Indeed, ETBF has been proposed to be a driver bacterium (Tjalsma et al., 2012). Our results strongly support this hypothesis since the prevalence of BFT was strongly increased in CRA stool samples as compared with the control or CRC groups ( Figure 1 ). Interestingly, bft presence decreased progressively during later stages of CRC indicating a transient role for this bacterium during transition from adenoma to carcinoma. However, these results need to be confirmed in a larger cohort especially with more samples from patients with colorectal adenomas.
Colibactin is another bacterial toxin strongly associated with CRC. The structure of colibactin and its DNA cross-links has been solved recently (Xue et al., 2019), and importantly, colibactin DNA-damage signatures have been found in CRC patients (Dziubańska-Kusibab et al., 2020). Colibactin is produced by the pks island found essentially in group B2 E. coli, but also in other Enterobacteriaceae such as Klebsiella pneumoniae, Enterobacter aerogenes, Citrobacter koseri, and group B1 E. coli isolates. We found that the prevalence of the clbN gene in DNA extracted from patient stools was mainly high in CRC samples, especially at stage IV. These results suggest a role for colibactin at terminal stages in the development of CRC. It is worth mentioning that bacterial biofilms composed mainly of E. coli pks+ and ETBF are found in the mucosa of patients with familial adenomatous polyposis (FAP) and that a synergistic effect of the two toxins (bft and pks) driving the development of colon tumor was demonstrated in ApcMinD716/+ (Dejea et al., 2018). Additional results from this study also indicate that Bacteroides precedes and promotes the adherence of pks+ E. coli, which agrees with our data.
Altogether our results support the notion that colorectal cancer development is not caused by a single pathogen but is probably driven by several pathobionts acting together or sequentially during the evolution of CRC. In line with this idea, detection and quantification of five suspect markers in fecal samples may be necessary to develop a robust non-invasive diagnostic tool (Yu et al., 2017; Liang et al., 2017).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The study protocol was approved by the Ethics Committee of Comité de Protection des Personnes Paris Est-Henri Mondor (no. 10-006 in 2010). All participants signed an informed consent. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
BP, JL-H, and VD performed the experimental work. EB and IS provided the patients’ stools. IS and PS supervised the writing of the manuscript. PT-C helped in funding acquisition. JL-H and SD designed the study. BP and SD analyzed the data and organized and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Institut National contre le Cancer (INCA, grant PLBIO16-025) and the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (grant no. ANR-10-LABX-62- IBEID). We also thank SNFGE, the French Society of Gastroenterology, for their financial support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are very grateful to Violeta Moya Alvarez for her expertise in statistical analysis. We thank Tarek Msadek for his expert editing skills. We wish to thank the patients and their families for contributing to these studies by giving consent and stool samples and for allowing us to use their clinical information; we thank URC, the research units of Henri Mondor and Saint Antoine hospitals from APHP, and all persons who managed clinical and sampling monitoring; we thank CRB (platform of biological resources) of Henri Mondor Hospital for holding the samples; we also thank all persons who contributed to the achievement of various studies mentioned here by their acronym: VATNIMAD, ECKINOXE, CLIMAT, DETECT, and CCR1; we also thank PHRC, a government program, for clinical research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.794391/full#supplementary-material
References
- Abed J., Emgård J. E. M., Zamir G., Faroja M., Almogy G., Grenov A., et al. (2016). Fap2 Mediates Fusobacterium Nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 20, 215–225. doi: 10.1016/j.chom.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlroos T., Tynkkynen S. (2009). Quantitative Strain-Specific Detection of Lactobacillus Rhamnosus GG in Human Faecal Samples by Real-Time PCR. J. Appl. Microbiol. 106, 506–514. doi: 10.1111/j.1365-2672.2008.04018.x [DOI] [PubMed] [Google Scholar]
- Aymeric L., Donnadieu F., Mulet C., du Merle L., Nigro G., Saffarian A., et al. (2018). Colorectal Cancer Specific Conditions Promote Streptococcus Gallolyticus Gut Colonization. Proc. Natl. Acad. Sci. U.S.A. 115, E283–E291. doi: 10.1073/pnas.1715112115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutaga K., van Winkelhoff A. J., Vandenbroucke-Grauls C. M. J. E., Savelkoul P. H. M. (2005). Periodontal Pathogens: A Quantitative Comparison of Anaerobic Culture and Real-Time PCR. FEMS Immunol. Med. Microbiol. 45, 191–199. doi: 10.1016/j.femsim.2005.03.011 [DOI] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Bundgaard-Nielsen C., Baandrup U. T., Nielsen L. P., Sørensen S. (2019). The Presence of Bacteria Varies Between Colorectal Adenocarcinomas, Precursor Lesions and Non-Malignant Tissue. BMC Cancer 19, 399. doi: 10.1186/s12885-019-5571-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirouze C., Patry I., Duval X., Baty V., Tattevin P., Aparicio T., et al. (2013). Streptococcus Bovis/Streptococcus Equinus Complex Fecal Carriage, Colorectal Carcinoma, and Infective Endocarditis: A New Appraisal of a Complex Connection. Eur. J. Clin. Microbiol. Infect. Dis. 32, 1171–1176. doi: 10.1007/s10096-013-1863-3 [DOI] [PubMed] [Google Scholar]
- Dai Z., Coker O. O., Nakatsu G., Wu W. K. K., Zhao L., Chen Z., et al. (2018). Multi-Cohort Analysis of Colorectal Cancer Metagenome Identified Altered Bacteria Across Populations and Universal Bacterial Markers. Microbiome 6, 70. doi: 10.1186/s40168-018-0451-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejea C. M., Fathi P., Craig J. M., Boleij A., Taddese R., Geis A. L., et al. (2018). Patients With Familial Adenomatous Polyposis Harbor Colonic Biofilms Containing Tumorigenic Bacteria. Science. 101, 721–725 doi: 10.1126/science.aah3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrow R., Edberg S., Wikfors E., Callan D., Troncale F., Vender R., et al. (1991). Fecal Carriage of Streptococcus Bovis and Colorectal Adenomas. Gastroenterology 101, 721–725. doi: 10.1016/0016-5085(91)90531-O [DOI] [PubMed] [Google Scholar]
- Dumke J., Vollmer T., Akkermann O., Knabbe C., Dreier J. (2017). Case-Control Study: Determination of Potential Risk Factors for the Colonization of Healthy Volunteers With Streptococcus Gallolyticus Subsp. Gallolyticus. PloS One 12. doi: 10.1371/journal.pone.0176515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziubańska-Kusibab P. J., Berger H., Battistini F., Bouwman B. A. M., Iftekhar A., Katainen R., et al. (2020). Colibactin DNA-Damage Signature Indicates Mutational Impact in Colorectal Cancer. Nat. Med. 26, 1063–1069. doi: 10.1038/s41591-020-0908-2 [DOI] [PubMed] [Google Scholar]
- Eick S., Straube A., Guentsch A., Pfister W., Jentsch H. (2011). Comparison of Real-Time Polymerase Chain Reaction and DNA-Strip Technology in Microbiological Evaluation of Periodontitis Treatment. Diagn. Microbiol. Infect. Dis. 69, 12–20. doi: 10.1016/j.diagmicrobio.2010.08.017 [DOI] [PubMed] [Google Scholar]
- Fearon E. R., Vogelstein B. (1990). A Genetic Model for Colorectal Tumorigenesis. Cell 61, 759–767. doi: 10.1016/0092-8674(90)90186-i [DOI] [PubMed] [Google Scholar]
- Gagnière J., Raisch J., Veziant J., Barnich N., Bonnet R., Buc E., et al. (2016). Gut Microbiota Imbalance and Colorectal Cancer. World J. Gastroenterol. 22, 501–518. doi: 10.3748/wjg.v22.i2.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janney A., Powrie F., Mann E. H. (2020). Host-Microbiota Maladaptation in Colorectal Cancer. Nature 585, 509–517. doi: 10.1038/s41586-020-2729-3 [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Johnston B., Kuskowski M. A., Nougayrede J.-P., Oswald E. (2008). Molecular Epidemiology and Phylogenetic Distribution of the Escherichia Coli Pks Genomic Island. J. Clin. Microbiol. 46, 3906–3911. doi: 10.1128/JCM.00949-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. S., Recco R. A., Catalano M. T., Edberg S. C., Casey J. I., Steigbigel N. H. (1977). Association of Streptococcus Bovis With Carcinoma of the Colon. New Engl. J. Med. 297, 800–802. doi: 10.1056/NEJM197710132971503 [DOI] [PubMed] [Google Scholar]
- Liang Q., Chiu J., Chen Y., Huang Y., Higashimori A., Fang J., et al. (2017). Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin. Cancer Res. 23, 2061–2070. doi: 10.1158/1078-0432.CCR-16-1599 [DOI] [PubMed] [Google Scholar]
- Löwenmark T., Löfgren-Burström A., Zingmark C., Eklöf V., Dahlberg M., Wai S. N., et al. (2020). Parvimonas Micra as a Putative Non-Invasive Faecal Biomarker for Colorectal Cancer. Sci. Rep. 10, 15250. doi: 10.1038/s41598-020-72132-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCOY W. C., Mason J. M. (1951). Enterococcal Endocarditis Associated With Carcinoma of the Sigmoid; Report of a Case. J. Med. Assoc. State Ala 21, 162–166. [PubMed] [Google Scholar]
- Nakatsu G., Li X., Zhou H., Sheng J., Wong S. H., Wu W. K. K., et al. (2015). Gut Mucosal Microbiome Across Stages of Colorectal Carcinogenesis. Nat. Commun. 6, 8727. doi: 10.1038/ncomms9727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan B., Brierley J., Byrd D., Bosman F., Kehoe S., Kossary C., et al. (2017). The TNM Classification of Malignant Tumours—Towards Common Understanding and Reasonable Expectations. Lancet Oncol. 18, 849–851. doi: 10.1016/S1470-2045(17)30438-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paritsky M., Pastukh N., Brodsky D., Isakovich N., Peretz A. (2015). Association of Streptococcus Bovis Presence in Colonic Content With Advanced Colonic Lesion. World J. Gastroenterol. 21, 5663–5667. doi: 10.3748/wjg.v21.i18.5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart C., Quesne G., Trieu-Cuot P. (2002). Taxonomic Dissection of the Streptococcus Bovis Group by Analysis of Manganese-Dependent Superoxide Dismutase Gene (Soda) Sequences: Reclassification of “Streptococcus Infantarius Subsp. Coli” as Streptococcus Lutetiensis Sp. Nov. And of Streptococcus Bovis Biotype 11.2 as Streptococcus Pasteurianus Sp. Nov. Int. J. Syst. Evol. Microbiol. 52, 1247–1255. doi: 10.1099/00207713-52-4-1247 [DOI] [PubMed] [Google Scholar]
- Rawla P., Sunkara T., Barsouk A. (2019). Epidemiology of Colorectal Cancer: Incidence, Mortality, Survival, and Risk Factors. Prz Gastroenterol. 14, 89–103. doi: 10.5114/pg.2018.81072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy B. S., Weisburger J. H., Narisawa T., Wynder E. L. (1974). Colon Carcinogenesis in Germ-Free Rats With 1,2-Dimethylhydrazine and N-Methyl-N’-Nitro-N-Nitrosoguanidine. Cancer Res. 34, 2368–2372. [PubMed] [Google Scholar]
- Rezasoltani S., Asadzadeh Aghdaei H., Dabiri H., Akhavan Sepahi A., Modarressi M. H., Nazemalhosseini Mojarad E. (2018). The Association Between Fecal Microbiota and Different Types of Colorectal Polyp as Precursors of Colorectal Cancer. Microb. Pathog. 124, 244–249. doi: 10.1016/j.micpath.2018.08.035 [DOI] [PubMed] [Google Scholar]
- Sasaki E., Osawa R., Nishitani Y., Whiley R. A. (2004). Development of a Diagnostic PCR Assay Targeting the Mn-Dependent Superoxide Dismutase Gene (Soda) for Identification of Streptococcus Gallolyticus. J. Clin. Microbiol. 42, 1360–1362. doi: 10.1128/jcm.42.3.1360-1362.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel L., Grimont F., Ageron E., Grimont P. A. D., Bouvet A. (2003). Reappraisal of the Taxonomy of the Streptococcus Bovis/Streptococcus Equinus Complex and Related Species: Description of Streptococcus Gallolyticus Subsp. Gallolyticus Subsp. Nov., S. Gallolyticus Subsp. Macedonicus Subsp. Nov. And S. Gallolyticus Subsp. Pasteurianus Subsp. Nov. Int. J. Syst. Evol. Microbiol. 53, 631–645. doi: 10.1099/ijs.0.02361-0 [DOI] [PubMed] [Google Scholar]
- Sobhani I., Bergsten E., Couffin S., Amiot A., Nebbad B., Barau C., et al. (2019). Colorectal Cancer-Associated Microbiota Contributes to Oncogenic Epigenetic Signatures. Proc. Natl. Acad. Sci. U.S.A. 116, 24285–24295. doi: 10.1073/pnas.1912129116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhani I., Tap J., Roudot-Thoraval F., Roperch J. P., Letulle S., Langella P., et al. (2011). Microbial Dysbiosis in Colorectal Cancer (CRC) Patients. PloS One 6, 215–219. doi: 10.1371/journal.pone.0016393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H., Boleij A., Marchesi J. R., Dutilh B. E. (2012). A Bacterial Driver-Passenger Model for Colorectal Cancer: Beyond the Usual Suspects. Nat. Rev. Microbiol. 10, 575–582. doi: 10.1038/nrmicro2819 [DOI] [PubMed] [Google Scholar]
- Tong J., Liu C., Summanen P., Xu H., Finegold S. M. (2011). Application of Quantitative Real-Time PCR for Rapid Identification of Bacteroides Fragilis Group and Related Organisms in Human Wound Samples. Anaerobe 17, 64–68. doi: 10.1016/j.anaerobe.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Wong S. H., Zhao L., Zhang X., Nakatsu G., Han J., Xu W., et al. (2017). Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 153, 1621–1633.e6. doi: 10.1053/j.gastro.2017.08.022 [DOI] [PubMed] [Google Scholar]
- Wu N., Yang X., Zhang R., Li J., Xiao X., Hu Y., et al. (2013). Dysbiosis Signature of Fecal Microbiota in Colorectal Cancer Patients. Microb. Ecol. 66, 462–470. doi: 10.1007/s00248-013-0245-9 [DOI] [PubMed] [Google Scholar]
- Xi Y., Xu P. (2021). Global Colorectal Cancer Burden in 2020 and Projections to 2040. Transl. Oncol. 14, 101174. doi: 10.1016/j.tranon.2021.101174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M., Kim C. S., Healy A. R., Wernke K. M., Wang Z., Frischling M. C., et al. (2019). Structure Elucidation of Colibactin and its DNA Cross-Links. Science 365, eaax2685. doi: 10.1126/science.aax2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yang M., Wang D., Zhang S., Yan S., Zhu Y., et al. (2020). Alteration of the Abundance of Parvimonas Micra in the Gut Along the Adenoma−Carcinoma Sequence. Oncol. Lett. 20, 1–1. doi: 10.3892/ol.2020.11967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Feng Q., Wong S. H., Zhang D., Liang Q. Y., Qin Y., et al. (2017). Metagenomic Analysis of Faecal Microbiome as a Tool Towards Targeted Non-Invasive Biomarkers for Colorectal Cancer. Gut 66, 70–78. doi: 10.1136/gutjnl-2015-309800 [DOI] [PubMed] [Google Scholar]
- Yu J., Zhao L., Zhao R., Long X., Coker O. O., Sung J. J. Y. (2019). The Role of Parvimonas Micra in Intestinal Tumorigenesis in Germ-Free and Conventional APCmin/+ Mice. JCO 37, 531–531. doi: 10.1200/JCO.2019.37.4_suppl.531 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.