Abstract
Past genotypic studies of Mycobacterium tuberculosis may have incorrectly estimated the importance of specific drug resistance mutations due to a number of sampling biases including an overrepresentation of multidrug-resistant (MDR) isolates. An accurate assessment of resistance mutations is crucial for understanding basic resistance mechanisms and designing genotypic drug resistance assays. We developed a rapid closed-tube PCR assay using fluorogenic reporter molecules called molecular beacons to detect reportedly common M. tuberculosis mutations associated with resistance to isoniazid and rifampin. The assay was used in a comparative genotypic investigation of two different study populations to determine whether these known mutations account for most cases of clinical drug resistance. We analyzed samples from a reference laboratory in Madrid, Spain, which receives an overrepresentation of MDR isolates similar to prior studies and from a community medical center in New York where almost all of the resistant isolates and an equal number of susceptible controls were available. The ability of the molecular beacon assay to predict resistance to isoniazid and rifampin was also assessed. The overall sensitivity and specificity of the assay for isoniazid resistance were 85 and 100%, respectively, and those for rifampin resistance were 98 and 100%, respectively. Rifampin resistance mutations were detected equally well in isolates from both study populations; however, isoniazid resistance mutations were detected in 94% of the isolates from Madrid but in only 76% of the isolates from New York (P = 0.02). In New York, isoniazid resistance mutations were significantly more common in the MDR isolates (94%) than in single-drug-resistant isolates (44%; P < 0.001). No association between previously described mutations in the kasA gene and isoniazid resistance was found. The first mutations that cause isoniazid resistance may often occur in sequences that have not been commonly associated with isoniazid resistance, possibly in other as yet uncharacterized genes. The molecular beacon assay was simple, rapid, and highly sensitive for the detection of rifampin-resistant M. tuberculosis isolates and for the detection of isoniazid resistance in MDR isolates.
The increasing prevalence of tuberculosis in many areas of the world, coupled with a rise in drug-resistant and multidrug-resistant (MDR) Mycobacterium tuberculosis strains, presents a major threat to global health (21). Nucleic acid amplification-based genotypic assays are potentially the most rapid and sensitive methods for the detection of drug resistance and are theoretically able to provide a same-day diagnosis from clinical samples. The utility of these assays is dependent on their ability to detect all common drug resistance mutations. The mutations that predominate in M. tuberculosis during in vitro selection of antibiotic-resistant strains may differ substantially from those that develop in human populations (18, 24). For this reason, it is essential to perform genotypic investigations with representative samples of clinical isolates. Previous studies have suggested that the DNA sequence of M. tuberculosis is extraordinarily well conserved and that mutations in the M. tuberculosis genome are almost always associated with drug resistance (18, 26). Between 12 and 75% of isoniazid-resistant strains have been found to contain mutations either in codon 315 of the katG gene or the inhA ribosomal binding site (8, 14, 15, 18, 19, 24, 28), and an additional 13 to 18% have contained mutations in the ahpC-oxyR intergenic region (11, 25, 28, 31), often in conjunction with katG mutations outside of codon 315 (25). Recently, 17% of resistant strains and 8.5% of resistant strains without other mutations have been reported to contain mutations in the kasA gene (16). Mutations in an 81-bp “core region” of the rpoB gene have been found in approximately 95% of all rifampin-resistant strains (10, 27). By contrast, drug-sensitive M. tuberculosis isolates have had almost uniformly wild-type DNA sequences in these regions.
Many of the prior genotypic drug resistance studies may have incorrectly estimated the relevance of specific mutations in clinical M. tuberculosis isolates due to biases introduced by their sampling methods. Most of these investigations included a disproportionate number of MDR strains (14, 15, 18, 28), collected nonrepresentative samples from around the world without uniform sampling strategies (11, 17, 18, 24, 25), or sequenced only a small fraction of the total resistant strains (8, 14). Most of the investigations also failed to include adequate numbers of susceptible control isolates. In order to perform large genetic studies, we developed a simple, rapid, and robust method for analyzing DNA sequences of moderate length using a new class of probes called molecular beacons (22, 29). Molecular beacons are ideally suited for genotypic assays, because they are able to detect amplicons as they are synthesized during real-time PCR and are able to discriminate between DNA sequences that differ from one another by as little as a single nucleotide substitution (22, 30). Amplicon detection is carried out in sealed reaction tubes, which simplifies analysis and eliminates an important source of assay contamination. Here, we designed a molecular beacon assay to compare the distribution of isoniazid and rifampin resistance mutations in two different study populations. Samples were obtained from a reference laboratory with a high incidence of multidrug resistance, similar to that in many prior studies, and from case and control subjects sampled for culture at a medical center where almost all of the resistant isolates from a community were available. This investigation enabled us to compare the distribution of specific resistance mutations in these two types of populations and to determine whether these mutations account for most cases of clinical drug resistance. The utility of the molecular beacon assay for the rapid detection of isoniazid- and rifampin-resistant M. tuberculosis was also assessed.
MATERIALS AND METHODS
Study setting, patients, and isolates.
One hundred forty-nine lysates of primary cultures of M. tuberculosis from unique patients from Madrid, Spain, and New York City were tested for drug resistance by the molecular beacon assay (Table 1). The Madrid isolates were from 56 of 68 (82.4%) consecutive patient samples from the Instituto Carlos III; 51 of the isolates were resistant to at least isoniazid and/or rifampin. This institute serves as a reference laboratory for Madrid and other Spanish medical centers and receives samples of predominantly drug-resistant M. tuberculosis isolates. The New York isolates consisted of isolates from 46 of the 53 (87%) consecutive patients from December 1989 through November 1997 with isoniazid- and/or rifampin-resistant M. tuberculosis infections treated at Montefiore Medical Center. An additional 47 fully susceptible control isolates from unique patients matched by year to resistant isolates from Montefiore Medical Center were also included in the study. Montefiore Medical Center is the largest provider of primary care to the 1.2 million residents of New York City's Bronx community. Except for a higher percentage of Hispanic patients, the demographic and clinical characteristics of patients with tuberculosis treated at Montefiore Medical Center are similar to those of all patients with tuberculosis in New York (1). Over the study period, the incidence of isoniazid resistance at Montefiore was 9.6%, and that of multidrug resistance was 8.1%. Twelve lysates from Madrid contained insufficient material to be included in the study, and seven cultures with drug-resistant isolates from New York could not be located. DNA fingerprints for 141 of the 149 lysates (94.6%) either were available from previous studies (1, 28) or were obtained during the current investigation.
TABLE 1.
Results of molecular beacon assay
| Drug, isolate, and susceptibility | No. of isolates
|
Sensitivity | 95% CIa for sensitivity | Specificity | 95% CI for specificity | ||
|---|---|---|---|---|---|---|---|
| Total | Assay positive | Assay negative | |||||
| Isoniazid | |||||||
| Total susceptible | 55 | 0 | 55 | 1.00 | 0.94–1.00b | ||
| Madrid | 7 | 0 | 7 | 1.00 | 0.59–1.00b | ||
| New York | 48 | 0 | 48 | 1.00 | 0.92–1.00b | ||
| Total resistant | 94 | 80 | 14 | 0.85 | 0.76–0.92 | ||
| Madrid | 49 | 46 | 3 | 0.94 | 0.83–0.99 | ||
| MDR | 43 | 41 | 2 | 0.95 | 0.84–0.99 | ||
| Single-drug resistantc | 6 | 5 | 1 | 0.83 | 0.36–0.99 | ||
| New York | 45 | 34 | 11 | 0.76 | 0.61–0.87 | ||
| MDR | 27 | 26 | 1 | 0.96 | 0.81–1.00b | ||
| Single-drug resistant | 18 | 8 | 10 | 0.44 | 0.22–0.69 | ||
| Rifampin | |||||||
| Total susceptible | 82 | 0 | 82 | 1.00 | 0.96–1b | ||
| Madrid | 15 | 0 | 15 | 1.00 | 0.78–1b | ||
| New York | 67 | 0 | 67 | 1.00 | 0.95–1b | ||
| Total resistant | 67 | 66 | 1 | 0.98 | 0.93–0.99 | ||
| Madrid | 41 | 40 | 1 | 0.98 | 0.87–0.99 | ||
| MDR | 41 | 40 | 1 | 0.98 | 0.87–0.99 | ||
| Single-drug resistantc | 0 | ||||||
| New York | 26 | 26 | 0 | 1.00 | 0.87–1.00b | ||
| MDR | 26 | 26 | 0 | 1.00 | 0.87–1.00b | ||
| Single-drug resistant | 0 | ||||||
CI, confidence interval.
One-tailed 97.5% confidence interval.
Ethambutol susceptibility not known.
Sample preparation, susceptibility testing, and DNA fingerprinting.
M. tuberculosis lysates were prepared by transferring colonies from a Lowenstein-Jensen slant into a 2-ml screw-cap tube containing 1 ml of H2O and boiling for 20 min. PCR assays were performed with 5 μl of each lysate. Conventional antibiotic susceptibility testing was performed at the respective institutions where the isolates were first detected by using the Centers for Disease Control and Prevention version of the proportion method (12). Resistance was defined as greater than 1% growth in the presence of 0.2 μg of isoniazid per ml, 2 μg of rifampin per ml, 5 μg of ethambutol per ml, or 2 μg of streptomycin per ml. From 1993 to 1997, the New York isolates were initially screened for drug resistance by the BACTEC method (7). By this method, drug resistance was defined as greater than 1% growth in the presence of 0.1 μg of isoniazid per ml, 2 μg of rifampin per ml, 2.5 μg of ethambutol per ml, or 2 μg of streptomycin per ml. Isolates found to be resistant by the BACTEC method were retested by the proportion method before being reported as resistant. Multidrug resistance was defined as resistance to at least two of the following drugs: isoniazid, rifampin, ethambutol, or streptomycin. Susceptibility to ethambutol was not known for the Madrid isolates. Both the Madrid and New York isolates were also analyzed by DNA fingerprinting as described previously (1, 28), and the Madrid isolates had previously been characterized by single-strand conformational polymorphism PCR (28). Mutant kasA amplicons were constructed by standard PCR-based site-directed mutagenesis (9). Specific mutations were confirmed by automated DNA sequencing.
Molecular beacon assays.
Molecular beacons were synthesized from modified oligonucleotides. The quencher 4-(4′-dimethylaminophenylazo)benzoic acid (DABCYL; Molecular Probes, Eugene, Oreg.) was covalently linked to one arm, and either fluorescein, tetramethylrhodamine, or tetrachlorofluorescein (Molecular Probes) was linked to the other arm. The synthesis is described in a detailed protocol available on the Internet (http://www.molecular-beacons.org).
The sequences of the molecular beacons that were specific for M. tuberculosis sequences associated with isoniazid resistance were as follows (underlining identifies the arm sequences): katG, fluorescein-5′-CCGAGGCACCAGCGGCATCGACCTCGG-3′-DABCYL; inhA, fluorescein-5′-CGAGGCCGACAACCTATCGTCTCCCTCG-3′-DABCYL; ahpC1, fluorescein-5′-CGCTCGGGCAAAGGTGATATATCACGAGCG-3′-DABCYL; ahpC2, fluorescein-5′-CGATCGCGACATTCCATCGTGCCCGATCG-3′-DABCYL. The sequences of the molecular beacons that were used to investigate the association of mutations in kasA and isoniazid resistance were as follows: kasA66, tetrachlorofluorescein-5′-CCAGCCTCAAGGATCCGGTCGCTGG-3′-DABCYL; kasA269, tetrachlorofluorescein - 5′ - CCAGCTGCCGGTATCACCTCGCTGG - 3′ - DABCYL; kasA312, fluorescein-5′-CCAGGCAACGCGCACGGCACCCTGG-3′-DABCYL; kasA413, tetrachlorofluorescein-5′-CCAGGGCTTGCCTTCGGGCGTTACTCCCTGG-3′-DABCYL. The sequences of the molecular beacons specific for the M. tuberculosis rpoB core region and the 16S rRNA gene were described previously (22).
The sequences of the PCR primers for the synthesis of the katG amplicon were 5′-CGTCGGCGGTCACACTTTCGGTAAGA-3′ and 5′-TTGTCCCATTTCGTCGGGGTGTTCGT-3′; for the inhA amplicon they were 5′-GTGGACATACCGATTTCG-3′ and 5′-CTCCGGTAACCAGGAGTGAACGGG-3′; for the ahpC-oxyR amplicon they were 5′-AGCAGTGGCATGACTCTC-3′ and 5′-CGGCCGGCTAGCACCTCT-3′; for the kasA66 amplicon they were 5′-TCGCCGGACATCGAGAGCA-3′ and 5′-GGGGCCGCCCGCATTCATCA-3′; for the kasA269 amplicon they were 5′-ATCGCGGCGTTCTCCATGA-3′ and 5′-CGCGGGCGCCACCATAT-3′; for the kasA312 amplicon they were 5′-CGGTATCACCTCGGACGCCTTTCA-3′ and 5′-ATCGAGTGGCCCAGCGCAGACTTC-3′; and for the kasA413 amplicon they were 5′-CATCCGCGTCGCCGGTTGTGAT-3′ and 5′-CCCGCGATGTCGTGCTTCAGTA-3′. The sequences of the primers for the rpoB core region and 16S rRNA gene amplicons were described previously (22).
Assays were performed in sealed wells in a 96-position microtiter plate (Perkin-Elmer, Foster City, Calif.). Aliquots of clinical M. tuberculosis culture lysates were placed into nine wells, each containing PCR reagents, one of the nine fluorescein-labeled katG, ahpC, inhA, or rpoB molecular beacons, and the appropriate PCR primers. Each well also contained a tetramethylrhodamine-labeled molecular beacon specific for the 16S rRNA gene amplicon and the appropriate 16S primers. Except as described below, the PCR conditions were identical for all reactions and have been described previously (22). Amplifications were performed in an Applied Biosystems 7700 Prism spectrofluorometric thermal cycler (Perkin-Elmer). By labeling the molecular beacons with different fluorophores, it was possible to distinguish the individual emission spectra of the fluorescein and tetramethylrhodamine molecular beacons in the same reaction well. These fluorescent signals were measured independently during the 60-s annealing step of every PCR cycle and were plotted automatically for each sample. The molecular beacons were designed to hybridize only to targets that did not contain mutations associated with antibiotic resistance. With drug-susceptible M. tuberculosis, the molecular beacons were expected to hybridize to their targets in all nine wells and to fluoresce with increasing intensity as the amplicons were synthesized during the course of the PCR. Drug resistance was indicated by the absence of a characteristic increasing fluorescent signal during PCR amplification in a well in which the 16S rRNA control molecular beacon present in the same well fluoresced normally. The assays with the kasA molecular beacons were performed in a similar manner except that annealing temperatures were 63°C for kasA66 and kasA312 and 51°C for kasA269 and kasA413.
Normalized fluorescence intensities.
The ability of the assay to detect mutations in target amplicons without real-time measurements was also investigated. The fluorescence intensity resulting from each molecular beacon after 40 amplification cycles was normalized (Fnorm1) so that Fnorm1 = (F − Fmin)/(Fmax − Fmin), where Fmax and Fmin were the maximum and minimum fluorescence intensities of each molecular beacon, respectively, compared to those for the positive and negative controls. To account for initial differences in template concentration, all Fnorm1 values for a given isolate were normalized a second time (Fnorm2) to reflect their value relative to the largest Fnorm1 value for each isolate (Fnorm1(max)), so that Fnorm2 = Fnorm1/Fnorm1(max). All assays were performed and interpreted by investigators who were blinded to the antibiotic susceptibility profiles of each M. tuberculosis isolate.
Statistical methods.
Sensitivity and specificity were calculated for the molecular beacon assay by using conventional susceptibility testing as a “gold standard.” Exact 95% confidence intervals for proportions were obtained by using STATA, version 5.0. Two-tailed tests were used, except when sensitivity or specificity was 1.00, in which case a one-tailed test with 97.5% confidence was performed. For comparisons of the proportion of mutations between the Madrid and New York isolates, P values were calculated by the Fisher exact test.
RESULTS
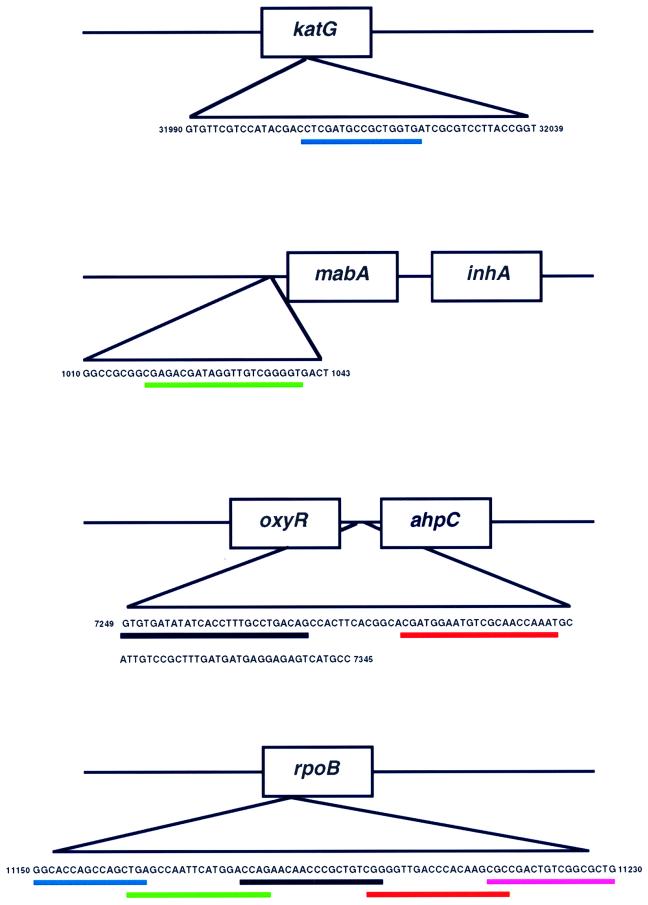
Mutations associated with isoniazid resistance are located on several widely separated regions of the M. tuberculosis genome. To test for isoniazid resistance, four molecular beacons were designed to hybridize to three different target amplicons (Fig. 1, top). One molecular beacon was complementary to codons 313 to 318 of the katG gene, one molecular beacon was complementary to the inhA ribosomal binding site, and two molecular beacons were complementary to two sequences that together spanned a selected area of the ahpC-oxyR intergenic region. In contrast to isoniazid, almost all mutations associated with resistance to rifampin occur in an 81-nucleotide “core” region of the M. tuberculosis rpoB gene. To test for rifampin resistance, five molecular beacons were used to hybridize to a single rpoB amplicon such that their probe sequences spanned the entire core region, with overlapping sequences of one to three nucleotides (Fig. 1, bottom). Real-time fluorescence analysis of molecular beacon hybridization during PCR amplification was carried out for all lysates. An isolate was determined to be drug resistant if one of the nine sample wells lacked a characteristic increasing fluorescent signal and the 16S rRNA control molecular beacon present in the same well fluoresced normally.
FIG. 1.
Localization of molecular beacons within their target genes. The accession number of each sequence in the GenBank database is as follows: katG, accession no. Z97193; inhA, accession no. Z79701; oxyR-ahpC, accession no. Z81451; rpoB, accession no. Z95972. Underlines identify the molecular beacon target sequences.
The overall sensitivity of the molecular beacon assay for the detection of isoniazid resistance was 85%, and the overall sensitivity of the molecular beacon assay for the detection of rifampin resistance was 98%. The specificity of the assay was 100% for both antibiotics (Table 1). These findings agreed with those of previous genetic investigations. However, when the results for isoniazid resistance were stratified on the basis of sample population, there was a marked heterogeneity in the assay's ability to detect isoniazid resistance. Mutations associated with isoniazid resistance were detected in 94% of the isolates from Madrid, but these mutations were detected in only 76% of the isolates from New York (P = 0.02). Stratification on the basis of drug resistance pattern revealed that this disparity was not due to differences among the MDR isolates. Isoniazid resistance mutations were detected in 94% of the MDR isolates from Madrid and 96% of the MDR isolates from New York (P = 1.0). A difference was noted among isolates that were resistant only to isoniazid (single-drug-resistant isolates). Isoniazid resistance mutations were detected in 5 of 6 (83%) of the single-drug-resistant isolates from Madrid but were detected in only 8 of 18 (44%) of the single-drug-resistant isolates from New York (P = 0.17). Analysis restricted to New York isolates revealed that detectable isoniazid resistance mutations were significantly more common in the MDR isolates (94%) than in the single-drug-resistant isolates (44%) (P < 0.001). These findings were not a consequence of a limited number of strains that contained unusual mutations. DNA fingerprinting studies determined that the isoniazid-resistant isolates comprised at least 53 different strains, including at least 9 different strains that were resistant only to isoniazid and that had no identifiable mutations.
Isolates varied by population as to the number and type of mutations that were present (Table 2). The Madrid isolates were more likely to contain inhA mutations (P < 0.001) or multiple mutations (P = 0.094) and were less likely to contain katG mutations (P < 0.012). Overall, the MDR isolates were more likely than the single-drug-resistant isolates to contain multiple mutations associated with isoniazid resistance (20 versus 4%; P = 0.104). Multiple mutations were defined as mutations detected in at least two of the three katG, inhA, and ahpC-oxyR amplicons or single mutations detected in the ahpC-oxyR region (which do not in themselves confer isoniazid resistance) that are associated with additional mutations in katG outside of codon 315 (25).
TABLE 2.
Mutations detected in isoniazid-susceptible and isoniazid-resistant isolates by molecular beacon assay
| Mutation | No. (%) of isolates
|
P | |||
|---|---|---|---|---|---|
| Madrid isolates
|
New York isolates
|
||||
| Susceptible (n = 7) | Resistant (n = 49) | Susceptible (n = 48) | Resistant (n = 45) | ||
| katG codon 315 alone | 15 (30.6) | 26 (57.8) | <0.012 | ||
| inhA ribosomal binding site alone | 21 (42.9) | 4 (8.8) | <0.001 | ||
| ahpC-oxyR intragenic region alone | 4 (8.2) | 2 (4.4) | NSa | ||
| katG plus inhA | 1 (2) | 1 (2.2) | NS | ||
| katG plus ahpC-oxyR | 1 (2) | 1 (2.2) | NS | ||
| inhA plus ahpC-oxyR | 5 (10.2) | NS | |||
| kasA codon 66 | NS | ||||
| kasA codon 269 | 5 (10.4) | 5 (11.1) | NS | ||
| kasA codon 312 | NS | ||||
| kasA codon 413 | NS | ||||
| No mutation detected | NAb | 3 (6.1) | NA | 11 (24.4) | 0.02 |
NS, not significant.
NA, not applicable.
In order to investigate whether mutations in the kasA gene account for additional resistance to isoniazid, we constructed molecular beacons to rapidly screen all of the Madrid and New York M. tuberculosis isolates for the presence of mutations in kasA codons 66, 269, 312, and 413, previously reported to be associated with isoniazid resistance (16). No mutations were found in codon 66, 312, or 413. Ten isolates were found to contain the G269S mutation; however, five of these isolates were fully isoniazid susceptible, suggesting that this mutation is not responsible for isoniazid resistance. The five susceptible isolates with G269S mutations originated in New York and comprised four different strains, as defined by DNA fingerprinting. Two of the susceptible kasA strains with the G269S mutation were available for repeat susceptibility testing. Both strains were susceptible to ≤0.1 μg of isoniazid per ml.
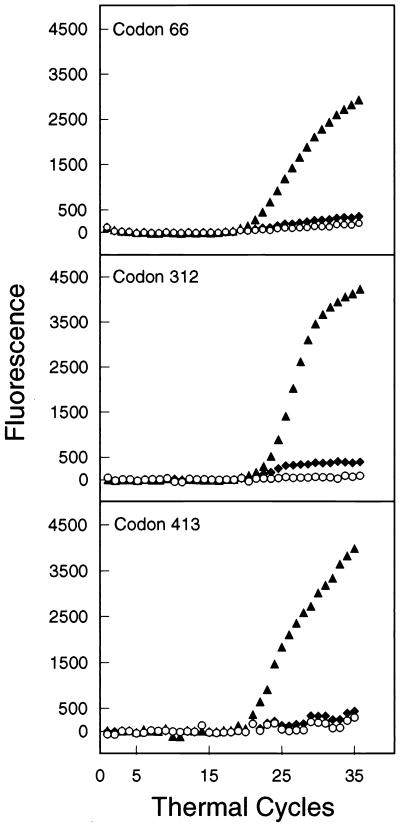
The molecular beacon assay identified mutations in kasA codon 269 and all katG, inhA, ahpC-oxyR, and rpoB target sequences. The assay was also able to distinguish between wild-type and mutant kasA codon 66, 312, and 413 sequences when they were tested in control reactions (Fig. 2). These results indicate that all of the molecular beacons were able to detect the appropriate mutations. DNA sequencing of a subset of the isolates that were identified as resistant by the molecular beacon assay confirmed the presence of the expected mutations and deletions. DNA sequencing of the complete katG gene, inhA operon, and ahpC-oxyR genes of two of the isoniazid-resistant isolates from Madrid that were misidentified as isoniazid susceptible by the molecular beacon assay failed to reveal any mutations in these genes. The sequences of the five susceptible kasA G269S mutants were also confirmed by automated DNA sequence analysis. The single rifampin-resistant isolate that was misidentified as rifampin susceptible originated from an isolate known to consist of a mixture of drug-resistant and drug-susceptible organisms on culture.
FIG. 2.
Mutational analysis during real-time PCR with molecular beacons complementary to wild-type kasA sequences. Characteristic sequence-dependent hybridization curves are shown for wild-type amplicons (▴), for mutant amplicons constructed by site-directed PCR mutagenesis (⧫), and for control reaction mixtures to which no template DNA was added (○).
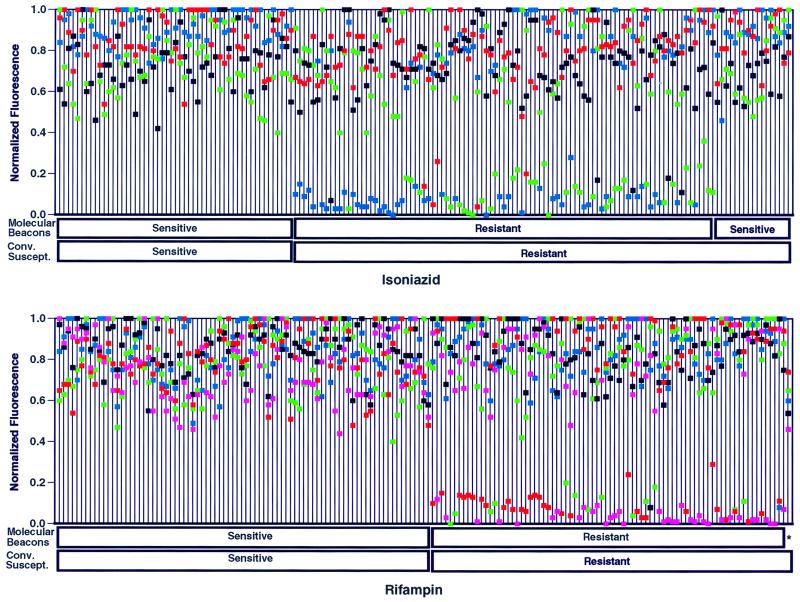
We investigated the ability of the results of the molecular beacon assay to be interpreted without real-time analysis. The fluorescence of each molecular beacon at the end of the PCR was measured, and normalized fluorescence intensity values were plotted for each M. tuberculosis isolate (Fig. 3). The results show that normalized fluorescence intensity was bimodal, with the majority of values either above 60% or less than 20%. When plotted in this manner, all of the isolates that had been determined to be resistant on the basis of real-time fluorescent measurements had at least one molecular beacon that fluoresced below 38% of the normalized maximum.
FIG. 3.
Detection of isoniazid-resistant (upper panel) and rifampin-resistant (lower panel) M. tuberculosis isolates from culture lysates by conventional susceptibility testing and molecular beacon assays. The results for each M. tuberculosis isolate are indicated on a single line, on which the normalized fluorescence intensity values for each molecular beacon for that isolate are plotted. The results for the 149 study lysates plus an additional 16 duplicate lysates of isolates from study patients are shown. An isolate is scored as drug susceptible if all of the normalized fluorescence intensity values for that isolate are greater than 0.38. Isoniazid susceptibility results are plotted for molecular beacons that are specific for katG (blue), inhA (green), and oxyR-ahpC (black and red) target amplicons. Rifampin susceptibility results are plotted for molecular beacons complementary to different target sequences within the core region of the rpoB gene (blue, green, black, red, and pink). The colors used to identify the results obtained with each molecular beacon are the same as those used to identify the molecular beacons in Fig. 1. An asterisk identifies the sole rifampin-resistant isolate identified as sensitive by the molecular beacon assay.
DISCUSSION
Understanding the genetic events that lead to drug resistance in clinical M. tuberculosis isolates is important for the development of genetic assays, elucidation of the mechanisms of action of antimicrobial agents, and the design of novel antibiotics that are active against drug-resistant strains. The results of this investigation suggest that the frequency of mutations responsible for many cases of isoniazid-resistant tuberculosis may have been incorrectly estimated by prior studies. It suggests that the overall drug resistance profile and, therefore, the clinical history of an M. tuberculosis isolate are important factors in determining which isoniazid resistance mutations are present. The differences that were observed between isolates from a reference laboratory and those from a community medical center emphasize the importance of appropriate sample selection for future genetic studies. Although 95.7% of the total MDR M. tuberculosis isolates tested were associated with the specific katG, inhA, and ahpC mutations targeted in the current assay, only 44% of the New York strains that were resistant only to isoniazid had identifiable mutations. Molecular beacon analysis for the kasA mutations that were thought to be associated with isoniazid resistance (16) did not improve the sensitivity of the assay. The kasA codon 269 mutations were equally present in isoniazid-susceptible and isoniazid-resistant isolates, and no other kasA mutations were present in any of the isolates tested.
We observed that M. tuberculosis isolates resistant only to isoniazid were less likely than MDR isolates to contain mutations in the gene targets investigated. This finding suggests that many clinical isolates develop isoniazid resistance by a stepwise accumulation of mutations, with the first mutation occurring in sequences or genes that have not been commonly associated with resistance. Multidrug resistance is strongly associated with patients who have had prior treatment for tuberculosis and thereby repeated exposure to isoniazid (5). We suggest that prolonged treatment with isoniazid under circumstances that permit resistance to develop results in a progressive accumulation of mutations, ultimately leading to katG and/or inhA mutations in virtually all MDR strains. The accumulated mutations may be important for achieving higher levels of isoniazid resistance or for maintaining full virulence in the human host. This hypothesis does not exclude the possibility that full resistance can also develop in a single step, as is observed in laboratory mutants. An association between high-level isoniazid resistance (MIC, >1.0 μg/ml) and mutations in the gene targets investigated would have supported the hypothesis that mutations accumulate over time. However, information on high-level resistance was not available for many of the isolates. This association will be explored in future studies.
This study demonstrates that the use of molecular beacons provides a simple and rapid method for the identification of rifampin-resistant M. tuberculosis isolates and for the identification of isoniazid resistance in the setting of multidrug resistance. Given the increasing worldwide incidence of drug-resistant tuberculosis, the search for assays for the rapid detection of drug resistance has taken on a new sense of urgency. Unacceptable delays in the diagnosis of drug-resistant tuberculosis can occur when conventional culture-based susceptibility tests are used because of the slow growth of the M. tuberculosis bacillus. More rapid approaches that use genetic analysis for the detection of mutations associated with drug resistance have been proposed (3, 4, 6, 20, 23, 28). However, these methods have restricted applicability to clinical laboratories and are limited principally to the detection of rifampin resistance. Molecular beacon assays have significant advantages over other techniques. Amplification, molecular beacon hybridization, and analysis are all performed simultaneously in sealed wells. The entire assay, including analysis, can be completed in under 3 h. Unlike many other nucleic amplification tests, the molecular beacon assay was 100% specific. This was likely due to the closed-well protocol, which eliminates amplicon carryover. With the identification of additional targets for isoniazid resistance, the assay should perform well for the detection of isoniazid resistance in single-drug-resistant isolates.
The molecular beacon assay requires expensive equipment that is not yet widely available. However, it is likely that this type of equipment will become more commonplace in diagnostic laboratories as the broad applicability of closed-tube PCR assays is demonstrated (2, 13). In this study, drug resistance was predicted equally well by measuring the normalized fluorescence intensities of completed PCRs and by real-time PCR analysis. These findings suggest that less complex fluorescence viewers that will be able to analyze molecular beacon assays after PCR is completed can be developed. In summary, the molecular mechanisms of isoniazid resistance require further study with representative populations. With the identification of the full complement of resistance mutations, molecular beacon assays will be able to rapidly and simply identify resistance to isoniazid and rifampin, the two principal antituberculosis drugs.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI37015, AI45244, 5T32, AI07501, and HL43521 and was aided by a grant from the New York Lung Association.
We thank S. Tyagi, S. A. E. Marras, and M. H. Levi for advice and assistance and S. T. Cole for the collection and initial characterization of the Madrid strains.
REFERENCES
- 1.Alland D, Kalkut G E, Moss A R, McAdam R A, Hahn J A, Bosworth W, Drucker E, Bloom B R. Transmission of tuberculosis in New York City, an analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med. 1994;330:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 2.Bassler H A, Flood S J, Livak K J, Marmaro J, Knorr R, Batt C A. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. App Environ Microbiol. 1995;61:3724–3728. doi: 10.1128/aem.61.10.3724-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Beenhouwer H, Lhiang Z, Jannes G, Mijs W, Machtelinckx L, Rossau R, Traore H, Portaels F. Rapid detection of rifampin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tuberc Lung Dis. 1995;76:425–430. doi: 10.1016/0962-8479(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 4.Felmlee T A, Liu Q, Whelen A C, Williams D, Sommer S S, Persing D H. Genotypic detection of Mycobacterium tuberculosis rifampin resistance: comparison of single-strand conformation polymorphism and dideoxy fingerprinting. J Clin Microbiol. 1995;33:1617–1623. doi: 10.1128/jcm.33.6.1617-1623.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frieden T R, Sterling T, Pablos-Mendez A, Kilburn J O, Cauthen G M, Dooley S W. The emergence of drug-resistant tuberculosis in New York City. N Engl J Med. 1993;328:521–526. doi: 10.1056/NEJM199302253280801. . [Erratum, 329:148.] [DOI] [PubMed] [Google Scholar]
- 6.Gingeras T R, Ghandour G, Wang E, Berno A, Small P M, Drobniewski F, Alland D, Desmond E, Holodniy M, Drenkow J. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- 7.Heifets L B. Rapid automated methods (BACTEC system) in clinical mycobacteriology. Semin Respir Infect. 1986;1:242–249. [PubMed] [Google Scholar]
- 8.Heym B, Honore N, Truffot-Pernot C, Banerjee A, Schurra C, Jacobs W R, Jr, van Embden J D, Grosset J H, Cole S T. Implications of multidrug resistance for the future of short-course chemotherapy of tuberculosis: a molecular study. Lancet. 1994;344:293–298. doi: 10.1016/s0140-6736(94)91338-2. [DOI] [PubMed] [Google Scholar]
- 9.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 10.Kapur V, Li L L, Iordanescu S, Hamrick M R, Wanger A, Kreiswirth B N, Musser J M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994;32:1095–1098. doi: 10.1128/jcm.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley C L, Rouse D A, Morris S L. Analysis of ahpC gene mutations in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:2057–2058. doi: 10.1128/aac.41.9.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent P R, Kubica G P. Public health mycobacteriology: a guide for the level III laboratory. U.S. Atlanta, Ga: Department of Health and Human Services; 1985. [Google Scholar]
- 13.Kostrikis L G, Tyagi S, Mhlanga M M, Ho D D, Kramer F R. Spectral genotyping of human alleles. Science. 1998;279:1228–1229. doi: 10.1126/science.279.5354.1228. [DOI] [PubMed] [Google Scholar]
- 14.Marttila H J, Soini H, Huovinen H P, Viljanen M K. katG mutations in isoniazid-resistant Mycobacterium tuberculosis isolates recovered from Finnish patients. Antimicrob Agents Chemother. 1996;40:2187–2189. doi: 10.1128/aac.40.9.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marttila H J, Soini H, Eerola E, Vyshnevskaya E, Vyshnevskiy B I, Otten T F, Vasilyef A V, Viljanen M K. A Ser315Thr substitution in KatG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob Agents Chemother. 1998;42:2443–2445. doi: 10.1128/aac.42.9.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mdluli K, Slayden R A, Zhu Y, Ramaswamy S, Pan X, Mead D, Crane D D, Musser J M, Barry C E., III Inhibition of a Mycobacterium tuberculosis beta-ketoacyl ACP synthase by isoniazid. Science. 1998;280:1607–1610. doi: 10.1126/science.280.5369.1607. [DOI] [PubMed] [Google Scholar]
- 17.Morris S, Bai G H, Suffys P, Portillo-Gomez L, Fairchok M, Rouse D. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J Infect Dis. 1995;171:954–960. doi: 10.1093/infdis/171.4.954. [DOI] [PubMed] [Google Scholar]
- 18.Musser J M, Kapur V, Williams D L, Kreiswirth B N, van Soolingen D, van Embden J D. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J Infect Dis. 1996;173:196–202. doi: 10.1093/infdis/173.1.196. [DOI] [PubMed] [Google Scholar]
- 19.Nachmakin E, Kang C, Weinstein M P. Detection of resistance to isoniazid, rifampin and streptomycin in clinical isolates of Mycobacterium tuberculosis by molecular methods. Clin Infect Dis. 1997;24:894–900. doi: 10.1093/clinids/24.5.894. [DOI] [PubMed] [Google Scholar]
- 20.Nash K A, Gaytan A, Inderlied C B. Detection of rifampin resistance in Mycobacterium tuberculosis by use of a rapid, simple, and specific RNA/RNA mismatch assay. J Infect Dis. 1997;176:533–536. doi: 10.1086/517283. [DOI] [PubMed] [Google Scholar]
- 21.Pablos-Mendez A, Raviglione M C, Laszlo A, Binkin N, Rieder H L, Bustreo F, Cohn D L, Lambregts-van Weezenbeek C S, Kim S J, Chaulet P, Nunn P. Global surveillance for antituberculosis-drug resistance, 1994–1997. N Engl J Med. 1998;338:1641–1649. doi: 10.1056/NEJM199806043382301. [DOI] [PubMed] [Google Scholar]
- 22.Piatek A S, Tyagi S, Pol A C, Telenti A, Miller L P, Kramer F R, Alland D. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat Biotech. 1998;16:359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 23.Rossau R, Traore H, De Beenhouwer H, Mijs W, Jannes G, De Rijk P, Portaels F. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob Agents Chemother. 1997;41:2093–2098. doi: 10.1128/aac.41.10.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouse D A, Li Z, Bai G H, Morris S L. Characterization of the katG and inhA genes of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1995;39:2472–2477. doi: 10.1128/aac.39.11.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sreevatsan S, Pan X, Zang Y, Deretic V, Musser J M. Analysis of the oxyR-ahpC region in isoniazid-resistant and susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob Agents Chemother. 1997;41:600–606. doi: 10.1128/aac.41.3.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1998;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 28.Telenti A, Honore N, Bernasconi C, March J, Ortega A, Heym B, Takiff H E, Cole S T. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at a reference laboratory level. J Clin Microbiol. 1997;35:719–723. doi: 10.1128/jcm.35.3.719-723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tyagi S, Kramer F R. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotech. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 30.Tyagi S, Bratu D P, Kramer F R. Multicolor molecular beacons for allele discrimination. Nat Biotech. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 31.Wilson T M, Collins D M. ahpC, a gene involved in isoniazid resistance of the Mycobacterium tuberculosis complex. Mol Microbiol. 1996;19:1025–1034. doi: 10.1046/j.1365-2958.1996.449980.x. [DOI] [PubMed] [Google Scholar]