Abstract
Background:
Miscellaneous cardiovascular risk factors have been defined, but the contribution of environmental pollutants exposure on cardiovascular disease (CVD) remains underappreciated.
Objective:
We investigated the potential impact of typical environmental pollutant exposure on atherogenesis and its underlying mechanisms.
Methods:
We used human umbilical vein endothelial cells (HUVECs) and apolipoprotein E knockout () mice to investigate how 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone (PCB29-pQ, a toxic polychlorinated biphenyl metabolite) affects atherogenesis and identified early biomarkers of CVD associated with PCB29-pQ exposures. Then, we used long noncoding RNAs (lncRNAs) HDAC7-AS1–overexpressing mice and apolipoprotein E/caveolin 1 double-knockout () mice to address the role of these early biomarkers in PCB29-pQ–induced atherogenesis. Plasma samples from patients with coronary heart disease (CHD) were also used to confirm our findings.
Results:
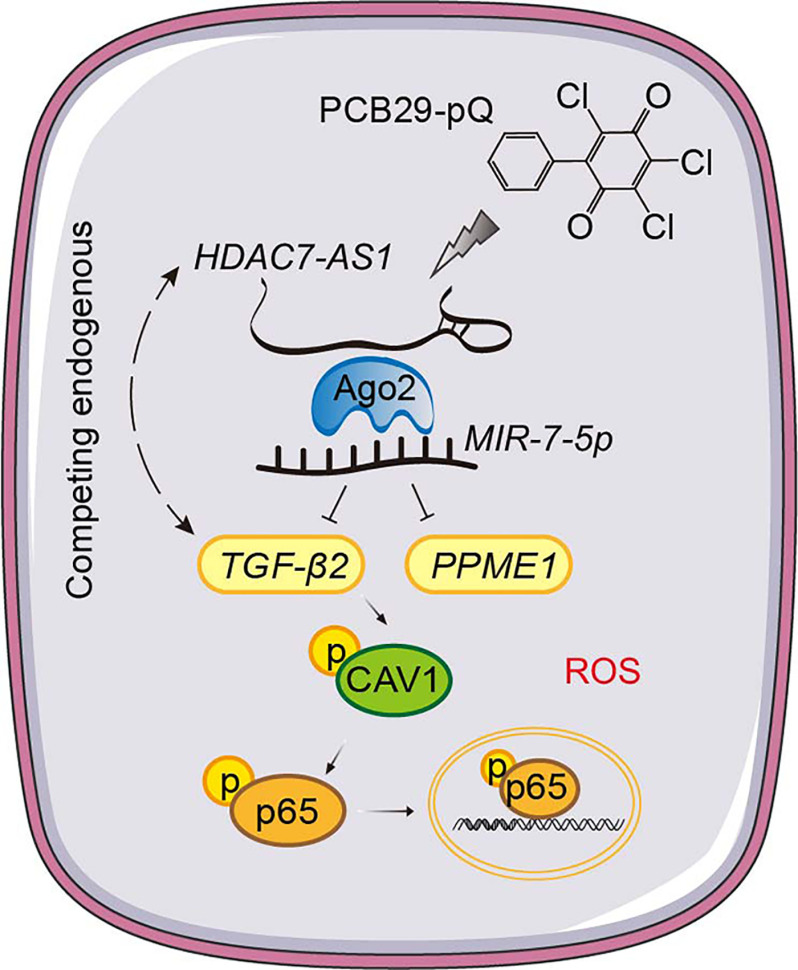
Our data indicate that lncRNA HDAC7-AS1 bound to MIR-7-5p via argonaute 2 in PCB29-pQ–challenged HUVECs. Our mRNA sequencing assay identified transforming growth () as a possible target gene of MIR-7-5p; HDAC7-AS1 sponged MIR-7-5p and inhibited the binding of to MIR-7-5p. The effect of PCB29-pQ–induced endothelial injury, vascular inflammation, development of plaques, and atherogenesis in mice was greater with MIR-7-5p–mediated inhibition, whereas HDAC7-AS1–overexpressing mice and mice showed the opposite effect. Consistently, plasma levels of HDAC7-AS1 and MIR-7-5p were found to be significantly associated individuals diagnosed with CHD.
Discussions:
These findings demonstrated that a mechanism-based, integrated-omics approach enabled the identification of potentially clinically relevant diagnostic indicators and therapeutic targets of CHD mediated by environmental contaminants using in vitro and in vivo models of HUVECs and and mice. https://doi.org/10.1289/EHP9833
Introduction
Polychlorinated biphenyls (PCBs) are typical environmental pollutants that have been widely used in industry in the last century. Approximately 1.5 to tons of PCBs remain in the environment globally (Liu et al. 2020), even after the strict prohibition of their production for industrial and consumer applications. PCBs are resistant to environmental degradation, which leads to their accumulation in the ecosystem. Moreover, lower chlorinated PCBs are still produced inadvertently and can be found in, for example, paint pigments (Anezaki and Nakano 2014; Hu and Hornbuckle 2010). PCBs can easily accumulate in the adipose tissue of the human body and metabolize into hydroxylated metabolites under the action of cytochrome P450 enzymes (Liu et al. 2020). In turn, with the aid of oxidative enzymes, hydroxylated PCBs further turn into their oxidized form, PCB quinones (Amaro et al. 1996). Our previous studies indicated that a PCB quinone metabolite (PCB29-pQ) has great potential to generate downstream reactive oxygen species (ROS), in particular, the hydroxyl radical, which is implicated its toxicity (Song et al. 2008, 2009).
Current epidemiological evidence shows that exposure to PCBs can increase the risk of hypertension (Peters et al. 2014; Yorita Christensen and White 2011), diabetes (Airaksinen et al. 2011; Codru et al. 2007), obesity (Ben Hassine et al. 2014; Donat-Vargas et al. 2014), and dyslipidemia (Ben Hassine et al. 2014; Donat-Vargas et al. 2014; Goncharov et al. 2008; Lee et al. 2011); all these factors are clinically relevant to the occurrence and development of cardiovascular diseases (CVDs). Furthermore, in a population-based cohort study, the dose of PCBs in serum was positively correlated with the risk of CVD (Raffetti et al. 2018).
CVDs are the leading cause of death worldwide and are responsible for nearly 30% of the global burden of deaths (Joseph et al. 2017). Risk factors, such as obesity (Van Gaal et al. 2006), diabetes (Zinman et al. 2015), hypertension (Fernández-Ruiz 2019), elevated blood cholesterol (Huynh 2020), smoking (Le Bras 2018), gender, and age (Emerging Risk Factors Collaboration et al. 2010), have been directly linked to the pathogenesis of CVD. However, the contribution of environmental pollutant exposure to CVD is still underappreciated (Cosselman et al. 2015; Olden 2004). Epidemiological investigations have established an association between environmental pollution and increased risk of CVD (Yusuf et al. 2020); however, the pollutant-driven etiology of CVD remains poorly characterized, in part because suitable approaches are missing to mechanistically link specific environmental exposures to clinically useful early biomarkers of CVD.
Few biomarkers are currently available to predict future coronary rupture; therefore, it is difficult to identify individuals with subclinical CVD (Blaha et al. 2016; Schulte et al. 2020). Omics-related techniques provide an unbiased approach and, therefore, have the potential to improve the early and specific diagnosis and prognosis of CVD. Noncoding RNAs (ncRNAs) are important in the physiology and pathology of CVD (Zhang et al. 2019). Many ncRNAs, for example, long noncoding RNAs (lncRNAs), are dysregulated in atherogenesis (Lorenzen and Thum 2016; Uchida and Dimmeler 2015). Similarly, growing evidence indicates that the functions of microRNAs (miRNAs) in vascular endothelial cells include modulating endothelial repair (Khalyfa et al. 2016), promoting cell proliferation (Chen and Gorski 2008) or apoptosis (Fish et al. 2008), and regulating inflammatory responses (Zhang et al. 2019). An important regulatory pattern of lncRNAs is to act as competing endogenous RNAs (ceRNAs), which can regulate gene expression by competitively binding miRNA (Salmena et al. 2011; Thomson and Dinger 2016). The interplay of lncRNAs and miRNA has attracted significant attention in CVD pathophysiology (Ballantyne et al. 2016). ncRNAs profile changes driven by PCBs have been reported (Chen et al. 2020; Shan et al. 2020; Wahlang et al. 2016); nevertheless, detailed molecular mechanisms of action of ncRNAs in PCB-mediated CVD remain to be investigated.
CVDs are multistage diseases and include initiation, progression/regression, and complications. Current knowledge indicates caveolin-1 (CAV1, a major structural protein of caveolae, which function in signaling and transport and regulating vascular reactivity) is involved in the action of CVDs. CAV1 binds with multiple proteins through its binding domain, and there is evidence for a role for CAV1 in atherosclerosis (Frank et al. 2009; Sowa 2012). Compared with ApoE knockout () mice, CAV1 deficiency has resulted in a sharp reduction in plaque size in ApoE/CAV1 double-knockout () mice (Frank et al. 2004), alleviated aggregation of low-density lipoprotein (LDL) into the arterial wall, promoted the production of nitric oxide, and alleviated the expression of leukocyte adhesion molecules, such as intercellular adhesion molecule 1 (ICAM-1), vascular cell adhesion molecule 1 (VCAM-1), and E-selectin (Sowa 2012).
In the present in vitro and in vivo study, human umbilical vein endothelial cells (HUVECs) and and mice were chosen to identify early biomarkers that are associate with PCB29-pQ exposure. In addition, these candidate ncRNAs and genes were confirmed in clinical samples from individuals diagnosed with coronary heart disease (CHD).
Methods
Reagents
Antibodies used in this study are listed in Table S1. The cell counting kit-8 (CCK-8) was purchased from Selleck Chemicals. 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI), protein () agarose beads, and 3,3′-diaminobenzidine tetrahydrochloride (DAB) were purchased from Beyotime Institute of Biotechnology. The Oil Red O solution and hematoxylin-eosin (HE) staining kit were purchased from Merck Life Science Co. Ltd. Vitamin C (VC), Vitamin E (VE), -acetyl-l-cysteine (NAC), and radio immunoprecipitation assay (RIPA) lysis buffer were purchased from Dingguo Biotechnology Co., Ltd. MIR-7-5p mimics, MIR-7-5p inhibitor, small interfering RNA (siRNA), protein phosphatase methylesterase 1 (PPME1) siRNA, and HDAC7-AS1 siRNA were synthesized by Shanghai Gene Pharma Co., Ltd. 2′,7′-Dichlorodihydrofluorescein diacetate (DCFH-DA) was purchased from Sigma-Aldrich Inc. Pyrrolidine dithiocarbamate ammonium (PDTC) was purchased from Beyotime Biotechnology Co., Ltd. PEGlated superoxide dismutase (PEG-SOD), PEGlated catalase (PEG-CAT), and glutathione monoethyl ester (GSH-MEE) were purchased from Sigma-Aldrich.
Cell Culture and Treatment
HUVECs and human monocytic cells (THP-1 cells) were purchased from KeyGEN Biotech Co., Ltd. HUVECs and THP-1 cells were grown in Roswell Park Memorial Institute 1640 medium supplemented with 10% fetal bovine serum (Zhejiang Tianhang Biotechnology Co., Ltd.), penicillin, and streptomycin at 37°C in a 5% incubator. In addition, the medium for THP-1 cells was supplemented with . HUVECs () were seeded in six-well culture plates overnight and exposed to PCB29-pQ for 24 h for subsequent experimental testing. HUVECs were pretreated with antioxidants ( VC, VE, NAC, PEG-SOD, PEG-CAT, or GSH-MEE) for 1 h, followed with PCB29-pQ exposure for 6 h for subsequent experimental testing. Parallel control cultures received equal volumes of dimethyl sulfoxide (DMSO). Each experiment was independently repeated at least three times.
miRNA, lncRNA, and mRNA Sequence Analysis
Six sets of HUVEC samples, comprising three biological replicates of PCB29-pQ and an equal volume of DMSO (control), were subjected to RNA sequence (RNA-seq) analysis. Total RNA was extracted by TRNzol universal reagent (Tiangen Biotech), ribosomal RNA was eliminated, and ncRNAs and mRNAs were retained via Illumina Ribo-Zero Gold (Human; Illumina). Enriched ncRNAs and mRNAs were fragmented into short fragments using fragmentation buffer (NEB E7490L; New England Biolabs) and reverse transcribed into complementary DNA (cDNA) with random primers, DNA polymerase I, ribonuclease H (RNase H) and deoxyribose nucleotide triphosphate (dNTP; New England Biolabs). In addition, cDNA fragments were purified using a QiaQuick polymerase chain reaction (PCR) extraction kit. cDNA library construction was performed by Genedenovo Biotechnology Co., Ltd. using DNA polymerase I, RNase H, dNTP, and buffer; cDNA fragments were connected to the Illumina sequencing adapter; and then polyadenylic acid was introduced. Uracil--glycosylase (UNG) was used to digest second-strand cDNA. Agarose gel (1.5%) electrophoresis was used for fragment size selection and PCR amplification. The PCR program (98°C 30 s; 98°C 10 s, 65°C 75 s, 12 cycles; 65°C 5 min; 4°C ) and reagents ( NEBNext Q5 Hot Start HiFi PCR Master Mix, of Universal PCR Primer and of Index (X) Primer) were performed by Genedenovo Biotechnology Co., Ltd. Finally, the library was sequenced using Illumina HiSeqTM 4000 by Gene Denovo Biotechnology Co., Ltd.
Identification of Differentially Expressed lncRNA, miRNA, and mRNA
The R package edgeR (version 3.3) (Gentleman et al. 2004; Oshlack et al. 2010; Robinson et al. 2010) (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) was used to identify differentially expressed transcripts across the samples or groups. We defined mRNA and miRNA with a fold difference of and and lncRNA with a fold difference of and a false discovery rate (FDR) for comparison as significant differentially expressed genes (DEGs).
miRNA Target Prediction
miRNA target gene prediction, RNAhybrid (version 2.1.2) (Krüger and Rehmsmeier 2006; Rehmsmeier et al. 2004) (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid), miRanda (version 3.3a) (Enright et al. 2003; John et al. 2004, 2006) (http://cbio.mskcc.org/microrna_data/miRanda-aug2010.tar.gz), and TargetScan (version 7.0) (Agarwal et al. 2015; Chiang et al. 2010; Friedman et al. 2009), (http://www.targetscan.org/vert_71/), and the intersection of results from the three software was selected as the array of predicted target genes of miRNA. Only miRNA target genes that were identified by all three methods were selected.
Gene Ontology Analysis
Gene Ontology (GO) enrichment compares an input data set (in this case, all significantly enriched DEGs) to the genome background and categorizes the DEGs that correspond to biological function. All DEGs that mapped to GO terms in the GO database were analyzed using the online tool Gene Ontology (Ashburner et al. 2000; Gene Ontology Consortium 2021) (http://www.geneontology.org). The R package clusterProfiler (version 3.6.0) (Yu et al. 2012) (https://guangchuangyu.github.io/software/clusterProfiler/) was used for GO enrichment analysis. The number of DEGs associated with each unique GO term was calculated. We then applied a hypergeometric test included in R package clusterProfiler (version 3.6.0) to find GO entries for which DEGs were significantly enriched compared with the entire human reference genome genomes. The GO terms with a were considered significant.
Kyoto Encyclopedia of Genes and Genomes Analysis
Pathway-based analysis helps to further understand genes’ biological functions. The Kyoto Encyclopedia of Genes and Genomes (KEGG; release 94) (Kanehisa and Goto 2000; Kanehisa 2019; Kanehisa et al. 2021) (http://www.kegg.jp/kegg/kegg1.html) is the major public pathway-related database. Pathway enrichment analysis identified significantly enriched metabolic pathways or signal transduction pathways in DEGs, comparing with the whole genome background following exposure of HUVECs to PCB29-pQ.
Construction of ceRNA Network
The ceRNA network was constructed based on the ceRNA theory as follows. a) Expression correlations between mRNA and miRNA or lncRNA and miRNA were evaluated using the Spearman’s rank correlation coefficient (SCC). Pairs with were selected as the negatively coexpressed lncRNA–miRNA or mRNA–miRNA pairs. b) The correlations between lncRNA and mRNA expression were evaluated using the Pearson’s correlation coefficient (PCC). Pairs with were selected as coexpressed lncRNA–mRNA pairs. c) The hypergeometric cumulative distribution function test was used to identify common miRNA sponges of the target genes. Only gene pairs with a were selected for further study. For ceRNA prediction, mireap (version 2.0; http://sourceforge.net/projects/mireap/), miRanda (version 3.3a), and TargetScan (version 7.0) were used, and the intersection of the three methods was assumed as the ceRNA prediction result.
The relationship between ENST00000080059 (HDAC7) and ENST00000599515 (HDAC7-AS1) was predicted using RNAplex (version 2.5.0) (Lorenz et al. 2011, 2016; Tafer and Hofacker 2008) (http://www.tbi.univie.ac.at/RNA/RNAplex.1.html).
Animals and PCB29-pQ Exposure
Mice pairs deficient in ApoE or CAV1 (C57BL/6J genetic background) were obtained from Jackson Laboratory. mice were crossed with mice to generate mice. mice were mated to generate and mice on the C57BL/6J background, then or mice were mated to generate mice. All mice and mice used in the experiments were littermates. Genotyping was performed by PCR (KF960-C; Hangzhou Jingle Scientific Co., Ltd.), and the primer was produced by Sangon Biotech (Shanghai) Co., Ltd. The primers are shown Table S2, and the PCR program is shown in Table S3.
HDAC7-AS1–overexpressing vector (pEZ-M61-HDAC7-AS1) and control vector (pEZ-M61-NC) were constructed by GeneCopoeia Co. Ltd. For more stable HDAC7-AS1 overexpression in vivo, the adeno-associated virus (AAV) vectors were packaged into AAV-HDAC7-AS1 virus particles. AAV vector and virus particles were provided by GeneCopoeia Co. Ltd. The prepared AAV-HDAC7-AS1 virus particles were stored at . mice were intravenously (i.v.) injected with AAV-HDAC7-AS1 ( particles/mouse) via the tail vein 1 wk prior to PCB29-pQ exposure, following the published procedure (Chen et al. 2017; Kahles et al. 2018; Roche-Molina et al. 2015).
Male mice ( wk of age, in each group) were housed under standard environmental conditions (12 h light/dark cycle, 22°C), in accordance with the protocols approved by the Dalian Medical University Institutional Animal Care and Use Committee (AEE17059). The mice had a 1-wk acclimation, during which time they received standard chow containing 4% fat (Jiangsu Xietong Pharmaceutical Bio-Engineering Co., Ltd.) and tap water ad libitum. Then, the mice were fed a high-fat diet [68% basic forage, 1% cholesterol, 15% lard, 10% egg yolk powder, 6% milk powder (Jiangsu Xietong Pharmaceutical Bio-Engineering Co., Ltd.)]. The mice were injected with body weight of PCB29-pQ or equal volumes of corn oil intraperitoneally (i.p.) (Arsenescu et al. 2008, 2011) injection once a week for 12 continuous wk, with the first two injections during the first week (3 d apart). After the end of PCB29-pQ exposure, the body weight of each group was recorded and the mice were euthanized by cervical dislocation 7 d after the last exposure. Blood was collected via the enucleation of the eyeball and transferred into tubes containing sodium citrate (3.8% final) as the anticoagulant; subsequently, the plasma was separated by centrifugation, , 10 min at 4°C temperature, and stored at until further use. Then, the mice in each group were randomly divided into two subgroups with an equal number of animals. The fresh aortas of the first subgroup of the mice were snap frozen in liquid nitrogen and stored at for real-time quantitative PCR (RT-qPCR) or Western blotting assays. The fresh aortas of the second subgroup of the mice were fixed with 4% paraformaldehyde and embedded for gross Oil Red O or HE staining.
Protein Extraction and Western Blotting
Cells or snap-frozen aorta of or mice were collected, washed with phosphate-buffered saline (PBS), and lysed with RIPA lysis buffer. Cellular lysates were centrifuged at at 4°C for 10 min. The supernatant was collected, and protein concentrations were determined with a bicinchoninic acid assay kit (Dingguo Biotechnology Co., Ltd.). Proteins () were separated using 12.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) with 40% acrylamide (Genview), Tris-HCl (pH 6.8 or 8.8) (Genview), 10% SDS (Genview), 10% ammonium persulfate (Macklin), and -tetramethyl-ethylenediamine (Macklin). Subsequently, proteins were transferred onto nitrocellulose membranes (Cat. No. 40795444; Pall). The membranes were blocked with 5% bovine serum albumin (BSA) at 37°C for 2 h and then incubated with appropriate primary antibodies at 4°C overnight. After rinsing three times with Tris-buffered saline containing Tween-20 (TBST), the membranes were incubated with a secondary antibody at room temperature for 2 h. The membranes were visualized using a BeyoECL Star (Beyotime Biotechnology). Subsequently, Western blots were imaged using the Tanon 5200 system and quantified by ImageJ software (Schneider et al. 2012).
RT-qPCR
Cells or aorta of or mice were collected. Total RNA was isolated using TRNzol universal reagent, according to the protocol recommended by the manufacturer (Tiangen Biotech). For mRNA, the purified RNA was reverse transcribed into cDNA using a Hifair II first-strand cDNA synthesis SuperMix for RT-qPCR (gDNA digester plus) kit (Yeasen Biotech Co., Ltd.). Quantitative analysis of mRNA expression was performed using SYBR Green qPCR master mix (Bimake) in an RT-qPCR machine (LightCycler96; Roche). The relative mRNA expression was normalized to the levels. For miRNAs, total RNA was reverse transcribed into cDNA using a miRcute Plus miRNA First-Strand cDNA Synthesis Kit (Tiangen Biotech). Quantitative analysis of miRNA expression was performed via RT-qPCR using a miRcute Plus miRNA qPCR detection kit and primers specific for MIR-7-5p, MIR-9-5p, and MIR-24-3p. The relative miRNA expressions were normalized to the U6 levels. The housekeeping gene U6 primer (Cat. No. MPM00002) was purchased from Applied Biological Materials. Inc. The primer sequences for lncRNAs and mRNAs are shown in Tables S4 and S5. The RT-qPCR program is shown in Table S6. The mRNA levels were determined following the threshold cycle () protocol.
Cell Transfection
MIR-7-5p mimics (Cat. No. B01001), MIR-7-5p inhibitor (Cat. No. B03001), siRNA (Cat. No. A01001), PPME1 siRNA (Cat. No. A01001), and HDAC7-AS1 siRNA (Cat. No. A01001) were introduced into 30–50% confluent HUVECs using siRNA-mate transfection reagent (Shanghai Gene Pharma Co., Ltd.) according to the manufacturer’s protocol. siRNAs, mimic, and inhibitor target sequences are shown in Table S7. After transfection for 24 h, cells were exposed to PCB29-pQ for 24 h for subsequent experimental testing.
Ninety to 95% confluent HUVECs were transfected with pEZ-M61-HDAC7-AS1 or pEZ-M61-NC (GeneCopoeia Co. Ltd.) using EndoFectin Max transfection reagent (GeneCopoeia Co. Ltd.), according to the manufacturer’s protocol. After transfection for 24 h, cells were exposed to PCB29-pQ for 24 h for subsequent experimental testing. Transfection was performed independently for three times as biological replicates.
Dual-Luciferase Reporter Assay
HUVECs were plated onto 12-well plates 24 h before transfection. At 90% confluence, cells were co-transfected with of pEZX-MT05 reporter vector containing negative control–3′untranslated region (NC-3′ UTR), wild-type (WT), or mutated (MUT) 3′UTR of [or protein phosphatase methylesterase 1 (PPME1)] inserted downstream of Gaussia luciferase (Gluc)/secreted alkaline phosphatase (SEAP) genes produced by GeneCopoeia Co. Ltd. and of NC mimic or MIR-7-5p mimic using siRNA-mate transfection reagent (Shanghai Gene Pharma Co. Ltd.). After transfection for 48 h, Gluc and SEAP activities were measured by a Secrete-Pair dual luminescence kit, according to the manufacturer’s protocol (GeneCopoeia Co. Ltd.). The Gluc emits flash bioluminescence that was measured at using a microplate reader (SpectraMax iD3; Molecular Devices).
Biotin-Labeled miRNA Pull-Down Assay
Thirty to 50% confluent HUVECs ( cells) in six-well plates transfected with biotinylated negative control (Bio-NC) or biotinylated MIR-7-5p (Bio-MIR-7-5p) using siRNA-mate transfection reagent (Shanghai Gene Pharma Co., Ltd.), according to the manufacturer’s protocol. Bio-NC and Bio-MIR-7-5p were synthesized by Sangon Biotech (Shanghai) Co., Ltd. After 48 h of transfection, cells were lysed in RIPA lysis buffer (Beijing Dingguo Changsheng Biotechnology Co., LTD.). Cell lysates were incubated with M-280 streptavidin magnetic beads (Invitrogen) at 4°C overnight. To prevent nonspecific binding of RNA and protein complexes, the beads were coated with 5% RNase-free BSA and yeast transfer RNA (Sigma-Aldrich) at 4°C for 3 h, and washed with ice-cold lysis buffer () three times and a high-salt buffer [, containing 0.1% SDS, 1% Triton X-100, ethylenediaminetetraacetic acid, Tris-HCl (pH 8.0), and sodium chloride] once before use. The bound RNAs were purified using TRNzol universal reagent for further analysis.
RNA-Binding Protein Immunoprecipitation Assays
RNA-binding protein immunoprecipitation (RIP) assays were performed with a Magna RIP RNA-binding protein immunoprecipitation kit, according to the manufacturer’s instruction (Millipore). HUVECs ( cells) were seeded in plates and held overnight, then the cells were exposed to PCB29-pQ for 24 h. Cells were lysed using of complete RIP lysis buffer and harvested. Magnetic beads were conjugated to the argonaute 2 (Ago2) antibody using the Magna RIP RNA-binding protein immunoprecipitation kit, according to the manufacturer’s protocol (Cat. No. 17-700). The cell lysates were then incubated with RIP buffer containing magnetic beads conjugated to Ago2 antibody (Proteintech Group, Inc.) at 4°C overnight. The removal of unbounded antibody was performed by centrifugation and washing, according to the manufacturer’s protocol (Cat. No. 17-700). The co-precipitated RNAs were isolated using TRNzol universal reagent, and the following RNAs were detected by RT-qPCR (HDAC7-AS1, RP3-416H24.1, LINC01547, TUG1, MCM3AP-AS1, and FGD5-AS1).
Tube Formation Assay
Matrigel (BD Biosciences) was placed in 96-well plates () and allowed to gel at 37°C for 30 min. HUVECs were exposed to PCB29-pQ or an equal volume of DMSO for 24 h, then the cells were trypsinized using 0.25% porcine pancreas trypsin (Dimond Shanghai) and plated on Matrigel at a density of cells/well. After incubating at 37°C for 18 h, the formation of tubes was observed by an optical microscope (Olympus IX71). Tube formation was analyzed with ImageJ software.
Cell Viability Assay
Cell viability was assessed using a CCK-8 assay (Bimake). Briefly, HUVECs were seeded in 96-well plates at a density of cells/well. After attachment, the cells were transfected with corresponding MIR-7-5p mimics (), MIR-7-5p inhibitor (), siRNA (), PPME1 siRNA (), HDAC7-AS1 siRNA (), or pEZ-M61-HDAC7-AS1 () for 24 h and exposed to PCB29-pQ () or an equal volume of DMSO for an additional 24 h. Cells were incubated with 10% CCK-8 solution (diluted in serum-free medium) at 37°C for 2 h. The optical density (OD) was measured at using a microplate reader.
Apoptosis Analysis
HUVECs ( cells/well) were seeded in six-well culture plates overnight. HUVECs were transfected with corresponding MIR-7-5p mimics (), MIR-7-5p inhibitor (), siRNA (), PPME1 siRNA (), HDAC7-AS1 siRNA (), or pEZ-M61-HDAC7-AS1 () for 24 h and exposed to PCB29-pQ () or an equal volume of DMSO for an additional 24 h. Cell apoptosis was tested by using an Annexin V-fluorescein/propidium iodide (Annexin V-FITC/PI) apoptosis detection kit (Yeasen Biotech Co., Ltd.). The cells were washed twice with cold PBS, resuspended in binding buffer, and incubated with Annexin V-FITC for 10 min and PI for 5 min at room temperature. Fluorescence was determined using a flow cytometer (BD FACSMelody), and the percentage of apoptotic cells was calculated using BD Cell Quest software (BD Biosciences).
Immunofluorescence Staining
HUVECs ( cells/well) were seeded in confocal dishes (Cat. No. J40101; NEST) overnight. HUVECs were transfected with CAV1 siRNA (Cat. No. A01001) for 24 h and exposed to PCB29-pQ or an equal volume of DMSO for an additional 24 h. After PCB29-pQ exposure, HUVECs in the confocal dish were washed with PBS and fixed in 4% paraformaldehyde for 30 min at room temperature. Then, cells were permeabilized with Triton X-100 (1% in PBS) for 20 min. After blocking with 5% BSA at 4°C for 1 h, the cells were incubated with a rabbit phospho-nuclear factor-kappa B (; p65) (pSer536) primary antibody (Table S1) at 4°C overnight. Then, the cells were washed with PBS three times, and the cells were incubated with a secondary Alexa Fluor 488-conjugated Affineur goat anti-rabbit IgG () antibody (Table S1) for an additional 1 h at room temperature. After three washes with PBS, the cells were counterstained with DAPI for 10 min. Finally, the cells were observed under a confocal laser scanning microscopy (Nikon N-SIM) with wavelengths of 405 and .
For immunofluorescence analysis of paraffin sections, animal tissue sections at thickness were permeabilized with 0.5% Triton X-100 at room temperature for 45 min and blocked with 5% BSA for 1 h at room temperature. Then, paraffin sections of the aortic root were incubated with endothelial cell marker CD31, von Willebrand factor (vWF), , PPME1, ICAM-1, or VCAM-1 antibodies (Table S1) at 4°C overnight. Then, the sections were incubated with the corresponding secondary antibody for 2 h at room temperature. Finally, the sections were stained with DAPI for 10 min and analyzed using a confocal laser scanning microscopy (Nikon N-SIM) at wavelengths of 405, 488, and . Images (pixels: ) were analyzed with ImageJ Coloc2 plug-in software to determine the PCC (automated thresholding, Point Spread Function 3, Costes’ randomizations 10), which uses the Costes’ significance test (Costes et al. 2004). Here, the colocalization rate was expressed using the PCC as a statistical measure, revealing a linear correlation between the two fluorescence intensities.
ROS Production Assay
ROS production in the cells were determined using DCFH-DA. HUVECs were seeded in six-well plates at cells/well for 24 h, then the cells were exposed to PCB29-pQ for 6 h with or without pretreatment with antioxidants VC (), VE (), and NAC () for 1 h. Next, the cells were trypsinized and loaded with DCFH-DA diluted in serum-free Roswell Park Memorial Institute 1640 medium at 37°C for 30 min. After incubation, the cells were washed three times with PBS. Fluorescence intensities were determined using a flow cytometer (BD FACSMelody).
Monocyte Adhesion
HUVECs were seeded in six-well plates at cells/well for 24 h, treated with PCB29-pQ for 6 h, and washed with PBS. 80–90% confluent THP-1 cells grown in six-well plates were trypsinized and loaded with the fluorescent probe calcein-AM (Cat. No. 148504-34-1; Beijing Fanbo Biochemicals Co., Ltd.) at 37°C for 30 min and then washed with PBS centrifugation at for 10 min. A total of Calcein-AM–loaded THP-1 cells were added to the HUVECs in each well and incubated for 3 h. Unbound monocytes were washed twice with PBS, and attached fluorescent monocytes were counted under a fluorescence microscope (Olympus IX71) at a wavelength of .
Aortic Oil Red O Staining
Atherosclerotic lesions were measured by staining the entire aorta with Oil Red O. The aortas were incubated in 4% paraformaldehyde overnight, dissecting scissors were used to cut the blood vessel longitudinally along the vessel wall, and the aortas were incubated with Oil Red O at 37°C for 1 h. Finally, the aortas were decolorized with 75% ethanol until the normal tissue became milky white and cleaned with distilled water. The plaque formation was observed using a digital camera (Canon, Japan), and the atherosclerotic lesion area was measured using ImageJ.
HE Staining and Immunohistochemistry
Paraffin sections of the aortic root were prepared at approximately thickness. The aorta sections were washed with PBS, incubated with hematoxylin for 5 min, and rinsed with water. The sections were differentiated with 1% hydrochloric acid for 30 s. After bluing up with water for 10 min, the sections were stained with eosin for 10 s. Then, the sections were soaked once in 85% ethanol, twice in 95% ethanol, twice in anhydrous ethanol, and twice in xylene (5 min each time) to dehydrate the tissue. The sections were dried and fixed with neutral balsam. The images were acquired using a microscope (OLYMPUS IX71).
For the immunohistochemistry analysis of the paraffin sections, the animal tissue sections were permeabilized with 0.5% Triton X-100 at room temperature for 45 min and blocked with 5% BSA for 1 h at room temperature. Then, paraffin sections of the aortic root were incubated with monocyte/macrophage marker CD68 antibodies (Table S1) at 4°C overnight. Then, the sections were incubated with the corresponding secondary antibody for 2 h at room temperature. Finally, the sections were analyzed using a microscope (Olympus IX71).
Terminal Deoxynucleotidyl Transferase Deoxyuridine Triphosphate Nick End Labeling Staining
Paraffin sections of the aortic root were immersed in PBS at room temperature for 5 min and rinsed twice. Then, the sections were treated with citric acid buffer for 5 min and rinsed twice with PBS. Next, sections were incubated with 3% hydrogen peroxide in the dark at room temperature for 10 min and rinsed twice with PBS. Finally, each section was incubated with terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling (TUNEL) reaction buffer (Servicebio) in the dark at 37°C for 1 h and rinsed 3 times with PBS. The nuclei were stained with DAPI. The sections were analyzed using a confocal laser scanning microscopy (Nikon N-SIM) at wavelengths of 405, 488, and .
Fluorescence in Situ Hybridization
MIR-7-5p detection probe 5′-UGG AAG ACU AGU GAU UUU GUU GUA ACA AAA UCA CUA GUC UUC CAU U-3′ was designed and synthesized by Servicebio. HDAC7-AS1 detection probe 5′-TGA GCT CAG ACT GAA GGA TCT TAG TAA GTG TGG AGA TGG GAC ACA AAC AAA GGT AGG CGA AGC GGA A-3′ was designed and synthesized by Shanghai Gene Pharma Co., Ltd. Paraffin sections of aortic roots were dehydrated in xylene. The sections were covered with hybridization buffer (Cat. No. G3016-3; Servicebio) and placed on an electric heating plate for 5 min. Hybridization with the probes was performed at 55°C for 16 h. Then, the sections were washed with SDS (Servicebio) twice at room temperature and SDS (Servicebio) twice at 55°C. After 5% BSA blocking for 1 h, the sections were incubated with a DIG-488 antibody (Servicebio) at room temperature for 1 h. The sections were washed twice with PBS, and DAPI staining solution (Servicebio) was added dropwise to the sections followed by incubation in the dark for 10 min. Samples were washed with PBS, and antifluorescence quenching mounting tablets (Beyotime) were added dropwise to mount the slides. The samples were visualized by a confocal laser scanning microscopy (Nikon N-SIM) at wavelengths of 405 and .
Human Blood Samples
This prospective single-institution study was approved and validated by the ethics committee of Beijing Friendship Hospital, Capital Medical University (2020-P2-165-01). The investigation conformed to the principles outlined in the Declaration of Helsinki. All participants gave their signed informed consent to the study. We explored patients who underwent initial diagnostic coronary angiography between September 2019 and January 2020 from Beijing Friendship Hospital, Capital Medical University. CHD was demonstrated by at least 70% stenosis in a major epicardial artery or 50% stenosis in the left main coronary artery, and control participants claimed chest discomfort but had normal coronary arteries on angiography. Individuals who had abnormal hematopoietic function, autoimmune diseases, malignancies, chronic or acute infections, severe heart failure, or advanced liver or renal diseases were excluded. All participants underwent physical examination and laboratory assessment [including fasting blood glucose (FBG), HbA1c; lipid profile, including total cholesterol (TC), triglycerides, high- and LDL-cholesterol (HDL- and LDL-C); renal function; and liver function, uric acid and high-sensitivity C-reactive protein (hs-CRP)]. Their current medical history was also collected. The clinical characteristics of study participants are shown in Table S8.
Fasting blood samples () were collected from 77 CHD patients and 50 control participants on the morning of their first visit. The level of hemolysis in the plasma samples was assessed before RNA extraction, as previously suggested by Kirschner et al. (2013). Plasma was separated from the blood by centrifugation at for 15 min to remove residual blood cells. After centrifugation, plasma samples were transferred to RNase/DNase-free tubes and stored at before RNA extraction. Total RNA for RT-qPCR analysis was extracted by TRNzol universal reagent (Tiangen Biotech). Hemolysis was assessed as described. Free hemoglobin absorbance was measured at using a multifunctional microplate reader. The procedure was repeated three to five times, and the average OD was calculated. No hemolysis occurred in these plasma samples and plasma samples with .
Statistical Analysis
Data from at least three independent experiments are presented as the deviation (SD). Statistical analyses were performed using GraphPad Prism (version 8.0; GraphPad Software) and SPSS 21.0 statistical software (SPSS Inc.). The data were compared using unpaired Student’s -test or one-way analysis of variance (ANOVA) followed by Tukey’s post hoc analysis or two-way ANOVA followed by Tukey’s or Sidak’s post hoc test. In addition, categorical variables were shown as a number with percentage and analyzed using the chi-square test. The SCC was used to assess the correlation between two indicated factors. Independent factors for predicting CHD were calculated by logistic regression; variables—including gender, diabetes, current smoker, LDL-cholesterol (LDL-C), FBG, albumin, hs-CRP, urea, creatinine, interleukin 1 beta (), interleukin 6 (IL-6), tumor necrosis factor-alpha (), , PPME1, HDAC7-AS1, and MIR-7-5p showing in univariate analysis—were included in a multivariate model. A was considered statistically significant.
Results
Differentially Expressed lncRNA and miRNA Identification
In an in vitro HUVECs model, cells treated with PCB29-pQ exhibited 23 differentially regulated lncRNAs and 38 differentially expressed miRNAs compared with the control group (Figure 1A,B; the exact data are presented in Excel Tables S1 and S2). The differentially expressed miRNAs were selected for GO and KEGG pathways analysis. Thirty-four GO terms and 20 KEGG pathways were identified (Figures S1 and S2; the exact data are presented in Excel Tables S3 and S4). The lncRNA and miRNA sequencing data were next validated using RT-qPCR. HUVECs exposed to PCB29-pQ displayed lower expression levels of lncRNAs ENST00000417835 (FGD5-AS1), ENST00000444998 (MCM3AP-AS1), ENST00000519077 (TUG1), ENST00000546686 (RP3-416H24.1), and ENST00000599515 (RP5-1057I20.4) compared with the control group (Figure S3a). Among the detectable differential expressions of miRNAs, MIR-7-5p showed the highest expression level after exposure to PCB29-pQ (Figure S3b). ENST00000397841 (LINC01547) showed no differences in expression between PCB29-pQ and control groups. The expression levels of ENST00000411824 (SLCO4A1-AS1), ENST00000414676 (NRSN2-AS1), ENST00000565916 (AC002550.5), and ENST00000626479 (SLFNL1-AS1) were higher in PCB29-pQ–treated cells, contrary to the sequencing results. ENST00000599515 (RP5-1057I20.4) was the most robust lowest expression lncRNA.
Figure 1.
![Figure 1A is a heatmap, plotting T C O N S underscore 00101945; E N S T 00000565916; E N S T 00000599515; E N S T 00000546686; E N S T 00000411824; T C O N S underscore 00014579; E N S T 00000519077; T C O N S underscore 00085732; E N S T 00000414676; E N S T 00000397841; E NS T 00000626479; E N S T 00000417835; E N S T 00000444998; T C O N S underscore 00081338; E N S T 00000369605; E N S T 00000589924; T C O N S underscore 00024105; E N S T 00000418546; E N S T 00000610057; E N S T 00000607166; E N S T 00000574306; T C O N S underscore 00047136; and E N S T 00000570416 (y-axis) across control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone (x-axis). A scale depicting fold change is ranging from negative 1.5 to 1.5 in increments of 0.5. Figure 1B is a heatmap, plotting M I R-3412-lowercase y; H S A-M I R-4709-5 lowercase p; M I R-9-lowercase y; M I R-5854-lowercase y; M I R-9861-lowercase x; M I R-7462-lowercase x; M I R-4106-lowercase y; M I R-2985-lowercase y; M I R-4913-lowercase y; H S A-M I R-499 A-5 lowercase p; M I R-590-lowercase y; M I R-8276-lowercase y; M I R-5872-lowercase y; H S A-M I R-1343-3 lowercase p; M I R-450-lowercase y; M I R-8358-lowercase y; M I R-4561-lowercase y; M I R-363-lowercase y; M I R-556-lowercase y; M I R-342-lowercase x; M I R-1551-lowercase x; M I R-7457-lowercase y; M I R-3200-lowercase y; M I R-1254-lowercase x; H S A-M I R-589-3 lowercase p; M I R-574-lowercase x; H S A-M I R-1248; H S A-M I R-3150 A-5 lowercase p; H S A-M I R-1249-5 lowercase p; H S A-M I R-219 A-5 lowercase p; H S A-M I R-6514-5 lowercase p; H S A-M I R-3661; M I R-589-lowercase y; M I R-24-lowercase x; M I R-7-lowercase x; H A S-M I R-7-5 lowercase p; M I R-625-lowercase y; and M I R-7974-lowercase y (y-axis) across control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone (x-axis). A scale depicting fold change is ranging from negative 1.5 to 1.5 in increments of 0.5. Figure 1C is a bar graph, plotting relative long non-coding ribonucleic acid enrichment, ranging from 0 to 5 in unit increments (y-axis) across H D A C 7-A S 1, R P 3-416 H 24.1, L I N C 01547, T U G 1, M C M 3 A P-A S 1, and F G D -A S 1 (x-axis) for anti-Immunoglobulin G and Anti-Argonaute 2. Figure 1D is a bar graph, plotting relative H D A C 7-A S 1 level, ranging from 0 to 20 in increments of 5 (y-axis) across bio-N C and bio-M I R-7-5 lowercase p (x-axis).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/de69/8963524/ac651cefa1ea/ehp9833_f1.jpg)
High-throughput lncRNA and miRNA sequence analysis and relevant RT-qPCR results from HUVECs treated with PCB29-pQ. HUVECs were exposed to PCB29-pQ for 24 h. Heat maps of the differentially expressed (A) lncRNAs and (B) miRNAs with a fold change and in the three parallel PCB29-pQ–treated groups () compared with the relative control group (), as determined by high-throughput lncRNA and miRNA sequence analysis. “ to 1.5” represent fold change of lncRNAs and miRNAs expression value. The exact data are presented in Excel Table, S1 and S2. (C) RT-qPCR analysis of the RNA-binding protein immunoprecipitation assays conducted using anti-Ago2 or anti-IgG antibody. Data are presented as (). Expression of each lncRNA in the anti-Ago2 samples was normalized to its expression in the anti-IgG control sample. The red box represents the most significantly up-regulated lncRNA in all differentially expressed lncRNAs. (D) The expression of HDAC7-AS1 in HUVECs transfected with Bio-MIR-7-5p in comparison with the control group. HUVECs were transfected with Bio-NC or Bio-MIR-7-5p for 48 h without the treatment of PCB29-pQ. Cells were harvested for a biotin-based pull-down assay. HDAC7-AS1 expression was analyzed by RT-qPCR. Data are presented as (). -Values were determined by two-way ANOVA followed by Tukey’s or Sidak’s post hoc and unpaired Student’s -test. The exact mean and SD-values are presented in Table S13. Note: ANOVA, analysis of variance; Ago2, argonaute 2; Bio, biotinylated; HUVECs, human umbilical vein endothelial cells; IgG, immunoglobulin G; lncRNA, long non-coding RNA; miRNA, microRNA; NC, negative control; PCB29-pQ, 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone; RT-qPCR, real-time quantitative polymerase chain reaction; SD, standard deviation.
The RIP assay was conducted to determine whether these down-regulated lncRNAs were associated with RNA-induced silencing complex (RISC). Ago2 protein was confirmed to be significantly enriched in the input group in control HUVECs (Figure S3c). HDAC7-AS1 (located on the antisense strand of HDAC7; Figure S4a) and LINC01547 lncRNAs were significantly enriched in the anti-Ago2 immunoprecipitation samples relative to the IgG control (Figure 1C). siRNA and an overexpression vector were then used to knock-down and overexpress HDAC7-AS1 in HUVECs, respectively (Figure S4b,c). After treatment of the cells with PCB29-pQ for 24 h, however, mRNA expression of HDAC7, which located on the antisense strand of lncRNA HDAC7-AS1, was comparable in HDAC7-AS1–silenced, HDAC7-AS1–overexpressed, and control groups (Figure S4d,e), suggesting that HDAC7-AS1 does not regulate the genes on its antisense strand. The fluorescence in situ hybridization (FISH) assay showed that after treatment with PCB29-pQ for 24 h, HDAC7-AS1 was located in the nucleus and cytoplasm of HUVECs (Figure S5), suggesting that HDAC7-AS1 may act as a ceRNA. HUVECs were transfected with Bio-NC or Bio-MIR-7-5p without PCB29-pQ exposure to confirm the association of HDAC7-AS1 with MIR-7-5p. The biotin-labeled miRNA pull-down assay showed higher HDAC7-AS1 expression level in HUVECs transfected with Bio-MIR-7-5p, compared with the control group (Figure 1D). Furthermore, a total of five HDAC7-AS1 binding sites to MIR-7-5p were identified by mireap, miRanda, and TargetScan (Table S9).
The Function of HDAC7-AS1 and MIR-7-5p in Vitro and in Vivo
mRNA sequencing (mRNA-Seq) analysis was performed to identify differentially expressed genes after PCB29-pQ exposure in HUVECs (Figure S6), and 306 differentially expressed genes were detected (the exact data are presented in Excel Table S5). RNAhybrid, miRanda, and TargetScan were used to predict possible target genes of MIR-7-5p. Correlation analysis between DEGs and predicted target genes was performed. Negatively correlated miRNA–gene pairs are shown in Table S10. and PPME1 were selected from this list to investigate their regulatory association with MIR-7-5p. Lower mRNA and protein levels of and PPME1 were observed in HUVECs exposed to PCB29-pQ (Figure 2A,B). Consistently, transfection with a MIR-7-5p mimic group showed lower levels of mRNA and protein expressions of and PPME1, compared with the control group (Figure 2C,D). Transfection with a MIR-7-5p inhibitor caused higher levels of and PPME1 protein expression in PCB29-pQ–exposed cells, and or PPME1 siRNA-treated groups displayed lower levels of and PPME1, respectively (Figure S7).
Figure 2.
The expression of HDAC7-AS1, MIR-7-5p, and in HUVECs exposed to PCB29-pQ. HUVECs were treated with PCB29-pQ for 24 h. (A) mRNA expression of and PPME1 were detected by RT-qPCR against housekeeping gene . Data are presented as (). (B) (Left panel) Protein levels of and PPME1 were detected by Western blotting and quantified by ImageJ software (Schneider et al. 2012) (Right panel). was used as an internal loading control. Data are presented as (). (C,D) HUVECs were transfected with NC mimic or MIR-7-5p mimic for 48 h without PCB29-pQ exposure. (C) mRNA expression levels of and PPME1 were detected by RT-qPCR against housekeeping gene U6. Data are presented as (). (D) (Left panel) Protein levels of and PPME1 were detected by Western blotting. was used as an internal loading control. (Right panel) The relative protein and PPME1 expression levels were quantified by ImageJ software. Data are presented as (). (E) Predicted MIR-7-5p binding sites in and PPME1 3′UTR. RNAhybrid, miRanda, and TargetScan were used, and the intersection of results from the three software was selected as the predicted target genes of miRNA. (F,G) Luciferase reporter plasmids containing (WT)-3′UTR or PPME1 (WT)-3′UTR co-transfected with MIR-7-5p mimic and pEZ-M61-NC or pEZ-M61-HDAC7-AS1 () into HUVECs without the treatment of PCB29-pQ. The luciferase activity represents mean value of Gaussia luciferase (Gluc) activity measure at by multifunctional microplate reader. Data are presented as (). -Values were determined by two-way ANOVA followed by Tukey’s post hoc test and one-way ANOVA followed by Tukey’s post hoc test. The exact mean and SD-values are presented in Table S14. Note: a.u., arbitrary units; ANOVA, analysis of variance; HUVECs, human umbilical vein endothelial cells; NC, negative control; PCB29-pQ, 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone; PPME1, protein phosphatase methylesterase 1 (gene); RT-qPCR, real-time quantitative polymerase chain reaction; TGF, transforming growth factor; UTR, untranslated region; SD, standard deviation; WT, wild type.
According to the predictions with RNAhybrid, miRanda, and TargetScan, both and PPME1 contained a target site for MIR-7-5p (Figure 2E). A MIR-7-5p mimic and the reporter plasmids containing NC-3′UTR, WT, or MUT 3′UTR of and PPME1 were co-transfected with HUVECs for 48 h. A dual-luciferase reporter assay showed lower luciferase activity in response to the MIR-7-5p mimic compared with the NC mimic that was observed in cells transfected with the WT or PPME1 UTR but not in cells transfected with the respective MUT 3′UTR (Figure S8a,b). Moreover, luciferase reporter plasmids containing NC and WT or MUT binding sites of HDAC7-AS1 were transfected into HUVECs. The luciferase activity in WT, but not in the NC or MUT HDAC7-AS1 groups, was significantly lowered by co-transfection with the MIR-7-5p mimic (Figure S8c). Then, we tested whether HDAC7-AS1 interfered with the binding of MIR-7-5p to its target mRNAs. Luciferase reporter plasmids containing the 3′UTR of or PPME1 were co-transfected with the MIR-7-5p mimic and the HDAC7-AS1–overexpressing plasmid into HUVECs. The luciferase activity was lower with MIR-7-5p mimic transfection and was restored with HDAC7-AS1 overexpression (Figure 2F,G).
Next, we determined the effect of PCB29-pQ on the development of atherosclerotic lesions in a model of mice by staining the aortas with HE. Male mice fed a high-fat diet were exposed to body weight PCB29-pQ via i.p. injection once a week for 12 continuous wk. The aortic plaque area in mice exposed to PCB29-pQ was significantly larger than that in the control group (Figure 3A). Aortic root sections were stained with CD31 to visualize their endothelial cells, and apoptotic rates were quantified by TUNEL. The proportion of TUNEL-positive cells in the aortic root of the PCB29-pQ group was substantially higher compared with the control group (Figure 3B).
Figure 3.
![Figure 3A is a set of one stained tissue and one error bar graph. The stained tissue has one column, namely, H E and two rows, namely, control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone. The bar graph, plotting plaque area (relative to aortic section), ranging from 0 to 60 in increments of 20 across control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone (x-axis). Figure 3B is a set of one stained tissue and one error bar graph. The stained tissue has three columns, namely, T U N E L, C D 31 plus D A P I, and merge and two rows, namely, control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone. The bar graph, plotting Pearson’s correlation coefficient, ranging from 0.0 to 1.5 in increments of 0.5 (y-axis) across control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone. Figure 3C is a set of eight flow cytometry plots. The first four flow cytometry plots, plotting Propidium Iodide, including control, ranging as 10 begin superscript 0 end superscript, 10 begin superscript 1 end superscript, 10 begin superscript 2 end superscript, 10 begin superscript 3 end superscript, 10 begin superscript 4 end superscript, 10 begin superscript 5 end superscript, and 10 begin superscript 6 end superscript (y-axis) across Annexin-V-F I T C, ranging as 10 begin superscript 0 end superscript, 10 begin superscript 1 end superscript, 10 begin superscript 2 end superscript, 10 begin superscript 3 end superscript, 10 begin superscript 4 end superscript, 10 begin superscript 5 end superscript, and 10 begin superscript 6 end superscript (x-axis) for N C inhibitor, including positive, negative, negative, negative; N C small interfering ribonucleic acid, including positive, positive, negative, negative; M I R-7-5 lowercase p inhibitor, including negative, positive, positive, positive; transforming growth factor lowercase beta-2 small interfering ribonucleic acid, including negative, negative, positive, negative; and Protein Phosphatase Methylesterase 1 small interfering ribonucleic acid, including negative, negative, negative, positive. The second flow cytometry plots, plotting Propidium Iodide, including 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, ranging as 10 begin superscript 0 end superscript, 10 begin superscript 1 end superscript, 10 begin superscript 2 end superscript, 10 begin superscript 3 end superscript, 10 begin superscript 4 end superscript, 10 begin superscript 5 end superscript, and 10 begin superscript 6 end superscript (y-axis) across Annexin-V-F I T C, ranging as 10 begin superscript 0 end superscript, 10 begin superscript 1 end superscript, 10 begin superscript 2 end superscript, 10 begin superscript 3 end superscript, 10 begin superscript 4 end superscript, 10 begin superscript 5 end superscript, and 10 begin superscript 6 end superscript (x-axis) for N C inhibitor, including positive, negative, negative, negative; N C small interfering ribonucleic acid, including positive, positive, negative, negative; M I R-7-5 lowercase p inhibitor, including negative, positive, positive, positive; transforming growth factor lowercase beta-2 small interfering ribonucleic acid, including negative, negative, positive, negative; and Protein Phosphatase Methylesterase 1 small interfering ribonucleic acid, including negative, negative, negative, positive. Figure 3D is a bar graph, plotting cell viability (percentage), ranging from 0 to 150 in increments 50 (y-axis) across N C inhibitor, including positive, negative, negative, negative; N C small interfering ribonucleic acid, including positive, positive, negative, negative; M I R-7-5 lowercase p inhibitor, including negative, positive, positive, positive; transforming growth factor lowercase beta-2 small interfering ribonucleic acid, including negative, negative, positive, negative; and Protein Phosphatase Methylesterase 1 small interfering ribonucleic acid, including negative, negative, negative, positive (x-axis) for control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone. Figure 3E is a set of five flow cytometry plots, plotting Bromodeoxyuridine, ranging as 10 begin superscript 0 end superscript, 10 begin superscript 1 end superscript, 10 begin superscript 2 end superscript, 10 begin superscript 3 end superscript, 10 begin superscript 4 end superscript, 10 begin superscript 5 end superscript, and 10 begin superscript 6 end superscript (y-axis) across Propidium Iodide, ranging as 10 begin superscript 4 end superscript, 10 begin superscript 5 end superscript, 10 begin superscript 6 end superscript, and 10 begin superscript 7 end superscript (x-axis) for N C inhibitor, including positive, positive, negative, negative, negative; N C small interfering ribonucleic acid, including positive, positive, negative, negative, negative; M I R-7-5 lowercase p inhibitor, including negative, negative, positive, positive, positive; transforming growth factor lowercase beta-2 small interfering ribonucleic acid, including negative, negative, negative, positive, negative; Protein Phosphatase Methylesterase 1 small interfering ribonucleic acid, including negative, negative, negative, negative, positive; and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, including negative, positive, positive, positive, positive.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/de69/8963524/1d6ef62e17bf/ehp9833_f3.jpg)
Endothelial injury and atherogenesis in mice exposed to PCB29-pQ and apoptotic rate in HUVECs exposed to PCB29-pQ. Male mice were fed a Western high-fat diet for 12 wk and tap water ad libitum. The mice were injected with body weight of PCB29-pQ or equal volumes of corn oil by i.p. injection once a week for 12 continuous wk, with the first two injections during the first week (3 d apart). (A) (Left panel) representative HE staining of aortic root cross sections. Scale bar: . (). Black arrows represent locations of plaque. (Right panel) Plaque area in the aortic root was quantified by ImageJ software (Schneider et al. 2012). Data are presented as (). -Values were determined by unpaired Student’s -test. (B) (Left panel) Apoptosis in aortic root cross sections was determined by TUNEL. Immunofluorescence staining was performed to observe TUNEL-positive foci (green), endothelial cell marker CD31 (red), and nucleus marker DAPI (blue). Scale bar: . (). White arrows represent the colocalization of TUNEL (green) and CD31 (red). (Right panel) Co-localization of TUNEL and CD31 was analyzed by Pearson’s correlation coefficient. -Values were determined by unpaired Student’s -test. (C–E) After co-transfection with NC inhibitor or MIR-7-5p inhibitor and or PPME1 siRNA () for 24 h, HUVECs were treated with PCB29-pQ for 24 h. (C) HUVECs apoptosis was evaluated by Annexin V-FITC/PI with a flow cytometer (). (D) Cell viability was measured using a CCK-8 kit. Data are presented as (). (E) Cell proliferation was measured by BrdU/PI double staining. -Values were determined by two-way ANOVA followed by Tukey’s post hoc test. The exact mean and SD-values are presented in Table S15. Note: ANOVA, analysis of variance; , apolipoprotein E knockout; BrdU, 5-bromo-2′-deoxyuridine; CCK-8, cell counting kit-8; CD31, endothelial cell marker; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride; FITC, fluorescein; HE, hematoxylin-eosin (stain); HUVECs, human umbilical vein endothelial cells; i.p., intraperitoneal; i.v., intravenous; IL, interleukin; NC, negative control; PCB29-pQ, 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone; PI, propidium iodide; PPME1, protein phosphatase methylesterase 1 (gene); SD, standard deviation; siRNA, small interfering RNA; TGF, transforming growth factor; TUNEL, terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling.
The ability for HUVECs to form tubes was then evaluated. Cells exposed to PCB29-pQ exhibited significantly less tube formation than control cells, and PCB29-pQ–exposed cells treated with a MIR-7-5p inhibitor exhibited significantly higher tube formation ability than their PCB29-pQ plus NC inhibitor control counterparts (Figure S9). We subsequently investigated the role of and PPME1 in MIR-7-5p–mediated endothelial injury during PCB29-pQ exposure. A lower percentage of apoptosis was observed in HUVECs coexposed to the MIR-7-5p inhibitor and PCB29-pQ. This effect was abolished by and PPME1 siRNA transfection (Figure 3C). and PPME1 siRNAs transfection groups showed lower cell viability (Figure 3D) compared with the MIR-7-5p inhibitor transfection and PCB29-pQ–treated cells; we therefore examined cell proliferation by 5-bromo-2′-deoxyuridine/propidium iodide (BrdU/PI) assay. The PCB29-pQ group showed remarkably lower number of BrdU-positive cells (S phase) compared with control group (Figure 3E). However, treatment with the MIR-7-5p inhibitor recovered cell proliferation by showing a higher number of BrdU-positive cells, and and PPME1 silencing abrogated its effect. In parallel, co-transfection with the MIR-7-5p inhibitor and or PPME1 siRNA reversed the effects of the MIR-7-5p inhibitor and showed lower cell viability compared with MIR-7-5p inhibitor transfection and PCB29-pQ–treated cells.
HDAC7-AS1–Overexpressed HUVECs and Mice
Because our findings suggest a direct interaction between HDAC7-AS1 and MIR-7-5p, we hypothesize that HDAC7-AS1 may play a protective role in the effect of PCB29-pQ on cardiovascular outcomes. This hypothesis was tested in HDAC7-AS1–overexpressing HUVECs and mice. Among cells treated with PCB29-pQ, the HDAC7-AS1–overexpressing group had a remarkably lower MIR-7-5p level compared with cells transfected with pEZ-M61-NC (Figure 4A). In contrast, among HDAC7-AS1–overexpressing PCB29-pQ–treated HUVECs, those transfected with the MIR-7-5p mimic showed higher MIR-7-5p (Figure 4A) and HDAC7-AS1 (Figure 4B) levels than those transfected with the NC mimic. In PCB29-pQ–exposed cells, the HDAC7-AS1–overexpressing group had higher mRNA (Figure 4C,D) and protein (Figure 4E) levels of and PPME1, compared with the relative pEZ-M61-NC group. This effect was abrogated by transfection with the MIR-7-5p mimic. The relative protein and PPME expression levels, quantified by ImageJ software, are shown in Figure 4F,G. Indeed, HDAC7-AS1 overexpression partially rescued the PCB29-pQ–induced loss of cell viability (Figure 4H), higher level of apoptosis (Figure S10a), and lower level of cell proliferation (Figure S10b); the MIR-7-5p mimic was unable to rescue the loss of cell viability but partially abrogated apoptosis. In addition, higher MIR-7-5p expression level (Figure S11a) and lower HDAC7-AS1 (Figure S11b), (Figure S11c), and PPME1 (Figure S11d) expression levels were achieved after PCB29-pQ–treated HUVECs were transfected with HDAC7-AS1 siRNA compared with the PBC29-pQ–treated NC siRNA cells. Consistent results were obtained at the protein level (Figure S11e–g). Thus, lower cell viability (Figure S11h) and higher apoptotic rate in PCB29-pQ–challenged HUVECs transfected with HDAC7-AS1 siRNA were observed (Figure S11i), compared with the PBC29-pQ–treated NC siRNA cells. However, co-transfection with the MIR-7-5p inhibitor partially reversed the effect of HDAC7-AS1 siRNA on these outcomes in PCB29-pQ–treated cells (Figure S11j).
Figure 4.
![Figures 4A, 4B, 4C, 4D, 4F, 4G, and 4H are bar graphs, Relative M I R-7-5 lowercase p level, ranging from 0 to 3 in unit increments and 90 to 120 in increments of 10; Relative H D A C 7-A S 1 level, ranging from 0 to 2 in unit increments and 10 to 14 in unit increments; relative transforming growth factor lowercase beta-2 level, ranging from 0.0 to 1.5 in increments of 0.5; relative Phosphatase Methylesterase 1 level, ranging from 0.0 to 1.5 in increments of 0.5; transforming growth factor lowercase beta-2 per lowercase beta actin, ranging from 0.0 to 1.5 in increments of 0.5; Protein Phosphatase Methylesterase 1 per lowercase beta actin (fold of control), ranging from 0.0 to 1.5 in increments of 0.5; and Cell viability (percentage), ranging from 0 to 150 in increments of 50 (y-axis) across N C mimic, including positive, positive, positive, negative; p E Z-M 61-N C, including positive, positive, negative, negative; M I R-7-5 lowercase p mimic, including negative, negative, negative, positive; and p E Z-M 61 H D A C 7-A S 1, including negative, negative, positive, positive; and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, including negative, negative, positive, positive (x-axis), respectively. Figure 4E is a Western blot for transforming growth factor lowercase beta-2, Phosphatase Methylesterase 1, and lowercase beta-actin across N C mimic, including positive, positive, positive, negative; p E Z-M 61-N C, including positive, positive, negative, negative; M I R-7-5 lowercase p mimic, including negative, negative, negative, positive; and p E Z-M 61 H D A C 7-A S 1, including negative, negative, positive, positive; and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, including negative, negative, positive, positive.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/de69/8963524/398411c31825/ehp9833_f4.jpg)
Endothelial injury and protein and mRNA expressions of and PPME1 in PCB29-pQ–exposed HUVECs transfected with an HDAC7-AS1 overexpression vector or a MIR-7-5p mimic. After co-transfection with NC mimic or MIR-7-5p mimic for 24 h and pEZ-M61-NC or pEZ-M61-HDAC7-AS1 () for 6 h, then, HUVECs were treated with PCB29-pQ for 24 h. RNA expression of (A) MIR-7-5p, (B) HDAC7-AS1, (C) and (D) PPME1 were detected by RT-qPCR. was used as a housekeeping gene for all RNAs except MIR-7-5p, for which U6 was used. The primer information is shown in Tables S4 and S5. Data are presented as (). (E) Protein levels of PPME1 and were detected by Western blotting. was used as an internal loading control. The relative protein (F) and (G) PPME1 expression levels were quantified by ImageJ software (Schneider et al. 2012). -Values were determined by one-way ANOVA, followed by Tukey’s post hoc test. Data are graphed relative to the cells exposed to NC mimic, pEZ-M61-NC, and vehicle control. (H) Cell viability was measured using a CCK-8 kit. Data are presented as (). -Values were determined by one-way ANOVA, followed by Tukey’s post hoc test. The exact mean and SD-values are presented in Table S16. Note: ANOVA, analysis of variance; CCK-8, cell counting kit-8; HUVECs, human umbilical vein endothelial cells; NC, negative control; PCB29-pQ, 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone; PPME1, protein phosphatase methylesterase 1 (gene); RT-qPCR, real-time quantitative polymerase chain reaction; SD, standard deviation; TGF, transforming growth factor.
To confirm the role of HDAC7-AS1 in atherogenesis, we injected AAV-HDAC7-AS1 into the tail vein of mice to create an HDAC7-AS1 overexpression mouse model. mice that received AAV vector treatment were used as the negative control (AAV-NC). Mice were then exposed to body weight PCB29-pQ once a week for 12 continuous wk via i.p. injection. Oil Red O and HE staining in the mouse aorta showed that the HDAC7-AS1 AAV group had a smaller area of detected aortic plaques compared with those injected with AAV-NC (Figure 5A; Figure S12a). TC and LDL-C levels were higher in mice exposed to PCB29-pQ and injected with AAV-NC compared with the AAV-HDAC7-AS1/PCB29-pQ coexposure group (Figure S12b,c). FISH was performed to evaluate the co-localization of HDAC7-AS1 and CD31. The proportion of HDAC7-AS1–expressing aortic root sections that co-localized with CD31 was lower than mice exposed to the vehicle. By contrast, mice treated with AAV-HDAC7-AS1 and exposed to PCB29-pQ exhibited a similar proportion of HDAC7-AS1 FISH staining on aortic root endothelial cells as those of control mice (Figure 5B,C). RT-qPCR results supported the fact that mice exposed to PCB29-pQ had lower HDAC7-AS1 levels, and this effect was reversed by HDAC7-AS1 AAV injection in vivo (Figure 5D). Meanwhile, MIR-7-5p expression in the PCB29-pQ group was attenuated by HDAC7-AS1 AAV treatment (Figure 5E). Correspondingly, the lower expression of and PPME1 in PCB29-pQ–treated cells compared with control were rescued by treatment with HDAC7-AS1 AAV (Figure 5F,G).
Figure 5.
Atherosclerosis and inflammation in mice exposed to PCB29-pQ accompanied by AAV-HDAC7-AS1 treatment. Male mice were i.v. injected with AAV-HDAC7-AS1 via the tail vein ( particles/mouse) to create an HDAC7-AS1–overexpressed mice model. Male mice that received AAV vector treatment were considered as AAV-NC mice. Control and HDAC7-AS1 overexpressed mice were fed a Western high-fat diet for 12 wk and tap water ad libitum. Mice were injected with body weight of PCB29-pQ or equal volumes of corn oil by i.p. injection once a week for 12 continuous wk, with the first two injections during the first week (3 d apart). (A) (Left panel) Representative images of Oil Red O staining of aortas. (). (Right panel) The relative plaque area in the aorta via Oil Red O staining was quantified by ImageJ software (Schneider et al. 2012). Data are presented as (). (B) HDAC7-AS1 in aortic root cross sections was determined by FISH. The slices were stained with HDAC7-AS1 probe (red), endothelial cell marker CD31 (green), and nucleus marker DAPI (blue), respectively. Scale bar: . (). Mice aorta RNA was extracted by TRNzol universal reagent. (C) Co-localization of HDAC7-AS1 and CD31 was analyzed by the PCC. RNAs expression of (D) HDAC7-AS1, (E) MIR-7-5p, (F) , (G) PPME1, (H) , (I) IL-6, and (J) in mice aorta was determined by RT-qPCR. was used as the housekeeping gene for all RNAs except MIR-7-5p, for which U6 was used. The primer information is shown in Tables S3 and S4. Data are presented as (). -Values were determined by one-way ANOVA, followed by Tukey’s post hoc test. (K) (Left panel) The expressions of p-p65 in aortic root cross sections were determined by double immunostaining with antibodies against p-p65 (red), CD31 (green), and DAPI (blue), respectively. Scale bar: . (Right panel) Co-localization of p-p65 and CD31 was analyzed by the PCC. Data are presented as (). The exact mean and SD-values are presented in Table S17. Note: AAV, adeno-associated virus (vector); ANOVA, analysis of variance; , apolipoprotein E knockout; CD31, endothelial cell marker; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride; FISH, fluorescence in situ hybridization (assay); HE, hematoxylin-eosin (stain); i.p., intraperitoneal; i.v., intravenous; IL, interleukin; NC, negative control; p-p65, phosphorylated phospho-nuclear factor-kappa B (; PCB29-pQ, 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone; PCC, Pearson’s correlation coefficient; PPME1, protein phosphatase methylesterase 1 (gene); RT-qPCR, real-time quantitative polymerase chain reaction; SD, standard deviation; TGF, transforming growth factor; TNF, tumor necrosis factor.
Not surprisingly, PCB29-pQ–treated, HDAC7-AS1 AAV-injected mice showed significantly lower , IL-6, and pro-inflammatory cytokines than the PCB29-pQ–treated AAV-NC control group (Figure 5H,J). Furthermore, as indicated by immunofluorescence assay, HDAC7-AS1–overexpressing mice showed lower proportions of cells that co-stained with phosphorylated p-65 (p-p65) and CD31 in both PCB29-pQ and control cells (Figure 5K).
Evaluation of a Role for CAV1 in CVD-Associated Outcomes in HUVECs and and Mice
Because of suggested negative associations between and the major caveolae structural protein CAV1 (Gvaramia et al. 2013; Moreno-Càceres et al. 2016; Razani et al. 2001), we next evaluated the role of CAV1 in the mechanism(s) of PCB29-pQ–mediated CVD outcomes and measures using both in vitro and in vivo models. First, HUVECs were transfected with negative control siRNA (NC siRNA) or CAV1 siRNA for 24 h, then treated with PCB29-pQ for up to 24 h. PCB29-pQ–treated HUVECs displayed a higher CAV1 phosphorylation level at tyrosine 14 (Tyr14) but not total CAV1 level (Figure S13a–c). Consistent with our previous results, protein levels of were lower in PCB29-pQ–challenged cells compared with control, and this was antagonized by CAV1 siRNA (Figure S13b,d). , IL-6, and pro-inflammatory cytokines levels in cells exposed to PCB29-pQ were slightly higher than cells treated with CAV1 siRNA and exposed to PCB29-pQ (Figure S13b,e–h).
We further evaluated the phosphorylation level of CAV1 in the HDAC7-AS1–overexpressing mouse model. Compared with the mice injected with AAV-NC, mice with HDAC7-AS1 overexpression had a significantly lower proportion of CD31-positive cells that are also p-CAV1 (Tyr14) positive in both PCB29-pQ and control groups (Figure S14).
To further explore the role of CAV1 in PCB29-pQ–induced inflammation, mice were generated by crossing mice with mice (both on the C57BL/6J background; genotype was confirmed by genomic PCR; Figure S15). mice exposed to PCB29-pQ showed a larger formation of atherosclerotic plaques compared with the vehicle-treated animals, and the area of atherosclerotic plaques was significantly lower in mice (both PCB29-pQ–treated and control mice) (Figure 6A). However, the CAV1 knockout did not completely neutralize the pro-atherosclerosis effect of PCB29-pQ given that the PCB29-pQ–treated mice exhibited a slightly larger, although not significantly different, area of plaque formation compared with the control-treated mice. mice demonstrated a significantly larger area of plaque at the aortic wall in mice treated with PCB29-pQ than in vehicle-treated animals of the same genotype. Conversely, the knockout of CAV1 affected the expansion of plaque area in mice, which had significantly less plaque area than their counterparts after exposure to PCB29-pQ (Figure 6B). Similarly, PCB29-pQ–exposed mice showed a higher accumulation of macrophages at the aortic wall than control mice (Figure S16a), as well as higher TC and LDL-C levels (Figure S16b,c), but this effect was partially abrogated in mice. The pro-inflammatory effects of PCB29-pQ in the arterial wall were then compared in and mice. The expressions of p-p65 and p-CAV1 were significantly higher in the arterial wall of PCB29-pQ–treated mice than control animals, whereas the expression levels of p-p65 and p-CAV1 were both lower in mice than in mice (Figure 6C).
Figure 6.
![Figure 6A is a set of one representative image and one error bar graph. The representative image depicts the Oil Red O staining of aortas in control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone each include Apolipoprotein E negative negative and Apolipoprotein E negative negative per Caveolin-1 negative negative. The error bar graph, plotting plaque area (percentage of whole aorta), ranging from 0 to 30 in increments of 10 (y-axis) across control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone (x-axis) for Apolipoprotein E negative negative and Apolipoprotein E negative negative per Caveolin-1 negative negative. Figure 6B is a set of one stained tissue and one error bar graph. The stained tissues has two columns, namely, control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone and two rows, namely, Apolipoprotein E negative negative and Apolipoprotein E negative negative per Caveolin-1 negative negative. The error bar graph, plotting plaque area (percentage of aortic section), ranging from 0 to 30 in increments of 10 (y-axis) across control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone (x-axis) for Apolipoprotein E negative negative and Apolipoprotein E negative negative, Apolipoprotein E negative negative per Caveolin-1 negative negative. Figure 6C is a set of one Western blot and two bar graphs. The Western blot has two columns, namely, control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone and five rows, namely, p-p 65, p 65, p-Caveolin-1, Caveolin-1, and lowercase beta-actin. The two bar graphs, plotting p-p 65 to p 65 ratio, ranging from 0 to 4 in unit increments and p-Caveolin-1 to Caveolin-1 ratio, ranging from 0 to 5 in unit increments (y-axis) across control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone (x-axis) for Apolipoprotein E negative negative and Apolipoprotein E negative negative, Apolipoprotein E negative negative per Caveolin-1 negative negative. Figures 6D and 6E are set of one stained tissue and one error bar graph. The stained tissues have four columns, namely, Apolipoprotein E negative negative and Apolipoprotein E negative negative per Caveolin-1 negative negative each in control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone and have three rows, namely, von Willebrand factor plus D A P I, Intercellular Adhesion Molecule 1 plus D A P I, and merge. The error bar graphs, plotting Pearson’s correlation coefficient, ranging from 0.0 to 1.5 in increments of 0.5 (y-axis) across control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone (x-axis) for Apolipoprotein E negative negative and Apolipoprotein E negative negative per Caveolin-1 negative negative. Figures 6F and 6G are error bar graphs, plotting level of interleukin-6 (picograms per milliliter), ranging from 0 to 1,000 in increments of 200 and level of tumor necrosis factor lowercase alpha (picograms per milliliter), ranging from 0 to 2,500 in increments of 500 (y-axis) across control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone (x-axis) for Apolipoprotein E negative negative and Apolipoprotein E negative negative per Caveolin-1 negative negative.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/de69/8963524/3ffd2e4258bd/ehp9833_f6.jpg)
Inflammation and atherogenesis in and mice exposed to PCB29-pQ. mice were crossed with mice to generate mice. and mice were fed a Western high-fat diet for 12 wk and tap water ad libitum. Mice were injected with body weight of PCB29-pQ or equal volumes of corn oil by i.p. injection once a week for 12 continuous wk, with the first two injections during the first week (3 d apart). (A) (Left panel) Representative images of Oil Red O staining of aortas. (Right panel) The relative plaque area in the aorta via Oil Red O staining was quantified by ImageJ software (Schneider et al. 2012). Data are presented as (). (B) Representative images of HE staining. Scale bar: . The relative plaque area in the aorta was quantified by calculating the ratio of atherosclerotic plaque to the surface of the entire aorta. Data are presented as (). (C) Protein levels of p-p65 and p-CAV1 in the aorta were analyzed by Western blotting. was used as an internal loading control. Proteins were quantified by ImageJ software. (). (D) (Left panel) ICAM-1 at the aortic root section in each group of mice was detected by double immunostaining with antibodies against ICAM-1 (red), endothelial cell marker vWF (green), and nucleus marker DAPI (blue), respectively. Scale bar: . (Right panel) Co-localization of vWF and ICAM-1 was analyzed using the PCC. Data are presented as (). (E) (Left panel) VCAM-1 at the aortic root section in each group of mice was detected by double immunostaining with antibodies against VCAM-1 (red), endothelial cell marker vWF (green), and nucleus marker DAPI (blue), respectively. Scale bar: . (Right panel) Co-localization of vWF and VCAM-1 was analyzed by the PCC. Data are presented as (). ELISA examination of (F) IL-6 and (G) levels in and mice serum. Data are presented as (). -Values were determined by two-way ANOVA, followed by Tukey’s post hoc test. The exact mean and SD-values are presented in Table S18. Note: ANOVA, analysis of variance; , apolipoprotein E knockout; , caveolin 1 knockout; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride; ELISA, enzyme-linked immunosorbent assay; HE, hematoxylin-eosin (stain); i.p., intraperitoneal; ICAM-1, intercellular adhesion molecule 1; IL, interleukin; p-p65, phosphorylated phospho-nuclear factor-kappa B (; PCB29-pQ, 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone; PCC, Pearson’s correlation coefficient; SD, standard deviation; TNF, tumor necrosis factor; VCAM-1, vascular cell adhesion molecule 1; vWF, von Willebrand factor.
In the early stage of atherosclerosis, dysfunctional endothelial cells released adhesion proteins, such as ICAM-1 and VCAM-1, promoted the migration of monocytes to the arterial wall (Bourdillon et al. 2000; Kasper et al. 1996). PCB29-pQ–treated mice had a distinctly lower proportion of vWF-positive cells that were also ICAM-1 and VCAM-1 positive in the aortic root plaque compared with their similarly treated counterparts (Figure 6D,E). PCB29-pQ exposure resulted in higher serum levels of IL-6 and in mice, compared with mice (Figure 6F,G).
Consistent with the in vivo study, cellular protein levels of , IL-6, , ICAM-1, and VCAM-1 were higher in a time-dependent manner in HUVECs after stimulation with PCB29-pQ (Figure S17a–f). In addition, the number of human THP-1 monocytes adhering to HUVECs was significantly higher after PCB29-pQ exposure (Figure S17g). Consistently, lower levels of nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-alpha () and higher levels of p-p65 were found upon PCB29-pQ challenge (Figure S17h–j). However, HUVECs pretreated with a p65 inhibitor, PDTC, for 1 h (Figure S17k–n) and CAV1 siRNA (Figure S18a–f) both abrogated the effect of PCB29-pQ. CAV1 siRNA treatment also showed lower adhesion molecules and pro-inflammatory cytokines release, attenuated degradation, as suggested by higher levels of , p65 phosphorylation (Figure S18g–i) and p65 nucleus translocation, as suggested by the lower proportion of DAPI-stained nuclei co-staining with p-p65 (Figure S18j).
Moreover, HUVECs with 1 h of antioxidants (VC, VE, or NAC) pretreatment had lower protein expression of pro-inflammatory genes (Figure S19a–g) and ROS levels (Figure S19h) compared with PCB29-pQ–treated cells alone. Similarly, pretreatment with antioxidants (VC, VE, or NAC; Figure S20a) or antioxidant enzymes (PEG-SOD, PEG-CAT, or GSH-MEE; Figure S20b) resulted in lower p-CAV1 levels compared with PCB29-pQ–treated cells alone.
HDAC7-AS1 and MIR-7-5p Levels in Patients with CHD
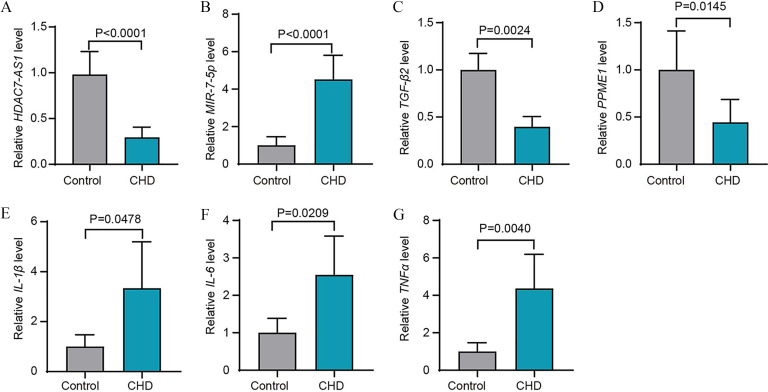
ncRNAs can serve as biomarkers and therapeutics in various CVDs (Gangwar et al. 2018). Thus, it is of interest to identify novel and potentially functional ncRNAs in patients with clinical CVDs. To further explore the significance of HDAC7-AS1, MIR-7-5p, , and PPME1 in atherosclerosis, the expression levels of these molecules were measured in the plasma of patients with CHD. The characteristics and clinical results of this study population are summarized in Table S8. RNA was extracted from the plasma samples of patients with CHD () and controls () for RT-qPCR analysis. The plasma of patients with clinical CHD was characterized by low expression of HDAC7-AS1 (Figure 7A), high expression of MIR-7-5p (Figure 7B), and low expressions of (Figure 7C) and PPME1 (Figure 7D), respectively.
Figure 7.
Plasma HDAC7-AS1, MIR-7-5p, , PPME1, , IL-6, and levels in patients with CHD. Plasma RNA was extracted by TRNzol universal reagent. CHD group () contains patients with coronary artery stenosis, and the control group () contains individuals with coronary artery stenosis. RT-qPCR analysis of (A) HDAC7-AS1, (B) MIR-7-5p, (C) , (D) PPME1, (E) , (F) IL-6, and (G) expressions. 18s was used as a housekeeping gene for all analyses with the exception of MIR-7-5p, for which U6 was used. The primer information is shown in Tables S4 and S5. Data are presented as . -Value determined by unpaired Student’s -test. The exact mean and SD-values are presented in Table S19. Note: CHD, coronary heart disease; IL, interleukin; PPME1, protein phosphatase methylesterase 1 (gene); RT-qPCR, real-time quantitative polymerase chain reaction; SD, standard deviation; TGF, transforming growth factor; TNF, tumor necrosis factor.
The mRNAs expressions of , IL-6, and were measured in the plasma samples (Figure 7E–G). Interestingly, negative correlations were found between MIR-7-5p and HDAC7-AS1 (, ), MIR-7-5p and (, ), MIR-7-5p and PPME1 (, ) in patients with CHD, whereas a positive correlation was found between MIR-7-5p and hs-CRP (, ) (Figure S21 and Table S11). Moreover, after adjusting for gender, diabetes, current smoker, LDL-C, FBG, albumin, hs-CRP, urea, creatinine, , IL-6, , , and PPME1, multivariable logistic regression models showed that MIR-7-5p had a statistically significant association with CHD [, ; 95% confidence interval: 1.223, 1.966 (Table S12)].
Discussion
Despite great improvements in CVD interventions and therapies, CVD remains the leading cause of death worldwide (Danaei et al. 2013; Roth et al. 2020). Smoking, physical activity, diet, weight, cholesterol, blood pressure, and glucose control were well-documented risk factors for CVD (Virani et al. 2020). In our opinion, the current biomarkers do not link the CVD risks caused by long-term exposure of environmental pollutions. There remains a critical need to characterize environmental risk factors for CVD and to develop novel diagnostic and therapeutic approaches for exposed populations. Many individuals with CVD are asymptomatic until plaque rupture (Halliday et al. 2010); therefore, it is difficult to identify patients with subclinical CVD, especially considering the substantial heterogeneity in the definitions of patients and controls. Omics-related techniques provide an unbiased approach to improve the early, specific diagnosis and prognosis of CVD (Turner et al. 2019). CHD is the major complication and acute phase of atherosclerosis, typically resulting in chest pain or heart damage (Hansson 2005). In the present study, we found that the expression MIR-7-5p was significantly altered in patients with CHD, which suggests the use of MIR-7-5p as a biomarker for detecting the different entities of CVD. This potential biomarker of CVD was identified based on in vitro and in vivo mechanistic studies investigating a metabolite of PCBs.
Different cohort studies have confirmed an association between PCB exposure and CVD risk (Åkesson et al. 2019; Bergkvist et al. 2016; Raffetti et al. 2018). PCBs are readily oxidized to hydroxylated, dihydroxylated, and quinone metabolites (Perkins et al. 2016). In our previous study using HUVECs, PCB29-pQ stimulated endothelial hyperpermeability, leading to endothelial barrier dysregulation (Zhang et al. 2015). As the major components of the vascular endothelium, endothelial cells played an important role in vascular homeostasis and functions (Haybar et al. 2019; Sun et al. 2016). Dysfunction or activation of endothelium is critical for atherogenesis, thus providing novel biomarkers for early diagnosis (Gimbrone and García-Cardeña 2013). In this study, our data suggest that PCB29-pQ–induced endothelial injury in vitro and in vivo manifest as the induction of HUVECs apoptosis and inhibition of vascular tube formation by endothelial cells. Simultaneously, abnormal morphology of the aortic intima and increased apoptosis were found in mice challenged with PCB29-pQ.
The lncRNA/miRNA/mRNA networks are important regulators in CVD etiology (Feinberg and Moore 2016). Here, we have established a PCB29-pQ–challenged endothelial cell model and analyzed the expression profile of lncRNAs and miRNAs via high-throughput sequencing technology. HDAC7-AS1 expression was significantly lower with PCB29-pQ exposure. RNA-Seq results suggested the binding of and PPME1 with MIR-7-5p. Our results suggest that MIR-7-5p induced endothelial cell apoptosis through the target genes and PPME1. Overexpression of HDAC7-AS1 blocked PCB29-pQ–induced apoptosis and inhibition of proliferation in HUVECs, and these effects were abrogated by the overexpression of MIR-7-5p. Consistently, mice with HDAC7-AS1 overexpression had less PCB29-pQ–induced atherosclerosis. These results suggest that HDAC7-AS1 sequestered MIR-7-5p as a ceRNA and participated in PCB29-pQ–induced endothelial injury and atherosclerosis. Please note that PCB concentration used in the animals was higher than PCB serum levels in the general population (Murphy et al. 2010; Wang et al. 2018; Zheng et al. 2016). Therefore, further investigations on the linkage of occupational PCB exposure, candidate biomarkers, and CVD risks need to be conducted.
CAV1 is a major structural protein of caveolae on the cellular membrane. CAV1 offers a scaffold for inflammatory signaling, which functions as a vital mediator of atherosclerosis (Ramírez et al. 2019). A previous review has demonstrated that receptors were internalized via caveolae (Chen 2009). Interestingly, several studies have suggested negative associations between and CAV1 (Gvaramia et al. 2013; Moreno-Càceres et al. 2016; Razani et al. 2001). The distinct role of CAV1 in endothelial cell inflammation has been established (Ramírez et al. 2019), and our data indicate that the negative CAV1 regulator is involved in PCB29-pQ–induced endothelial cell inflammation. Therefore, an mouse model was generated to characterize the systemic inflammatory status upon PCB29-pQ challenge. Previous studies in mice have suggested that silencing of CAV1 suppresses vascular inflammation and atherosclerosis (Ramírez et al. 2019; Zhang et al. 2020). Moreover, CAV1 was reported to be involved in functional changes in endothelial cells following exposure to environmental toxicants. For instance, PCB77-induced up-regulation of monocyte chemoattractant protein-1 required CAV1 in an atherosclerotic mouse model (Majkova et al. 2009), and porcine and mouse endothelial cells were protected from PCB126-induced inflammatory response (Petriello et al. 2014). These observations suggest the involvement of CAV1 in PCB-induced inflammation. Phosphorylation of CAV1 is an early signal transduction event induced by oxidative stress (Nah et al. 2017). A similar study has demonstrated that PCBs increased the phosphorylation of CAV1 in human-derived endothelial cells (Lim et al. 2007), consistent with our current finding that PCB29-pQ activates CAV1 phosphorylation and influences CAV1-related signaling. Notably, the biological significance of CAV1 phosphorylation was controversial because both pro-survival and pro-death effects of CAV1 have both been reported in vitro and in vivo (Quest et al. 2013).
Overall, the present study suggests the involvement of specific ncRNAs in the ROS-driven pro-inflammatory cascade. We hypothesize that upon PCB29-pQ exposure, the signal axis induces endothelial cell apoptosis and inflammatory cytokine release (Figure 8). We further hypothesize that HDAC7-AS1 alleviates the degradation of the target genes and PPME1 via up-regulation of MIR-7-5p induced by PCB29-pQ, which in turn ameliorates endothelial cell apoptosis, inflammatory cytokine release, and, ultimately, atherogenesis. Overall, our approach identified a network of HDAC7-AS1/MIR-7-5p pairs in the inflammatory setting and demonstrated the negative associations between HDAC7-AS1 and MIR-7-5p in clinical samples from patients with CHD. These findings provided insight into the potential use of plasma levels of HDAC7-AS1, MIR-7-5p, and as diagnostic biomarkers and therapeutic targets in PCB-induced atherosclerosis. Importantly, the translational framework established in this study is amenable to high-throughput screening approaches and can be used to identify biomarkers of CVD that are associated with other environmental toxicants or environmental factors.
Figure 8.
The proposed mechanism of PCB29-pQ–disturbed axis on atherogenesis. Note: Ago2, argonaute 2; CAV1, caveolin 1; p, phosphorylated; p65, phospho-nuclear factor-kappa B (; PCB29-pQ, 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone; PPME1, protein phosphatase methylesterase 1 (gene); ROS, reactive oxygen species; TGF, transforming growth factor.
Supplementary Material
Acknowledgments
The authors thank S. Wu for assistance with statistical analysis. This work was supported by the National Natural Science Foundation of China (21976145 and 22176206). All study data are included in the article and Supplemental Materials. The RNA-sequence analysis data have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive and are accessible through BioProject accession number PRJNA679564.
References
- Agarwal V, Bell GW, Nam JW, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:e05005, PMID: , 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airaksinen R, Rantakokko P, Eriksson JG, Blomstedt P, Kajantie E, Kiviranta H. 2011. Association between type 2 diabetes and exposure to persistent organic pollutants. Diabetes Care 34(9):1972–1979, PMID: , 10.2337/dc10-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åkesson A, Donat-Vargas C, Berglund M, Glynn A, Wolk A, Kippler M. 2019. Dietary exposure to polychlorinated biphenyls and risk of heart failure—a population-based prospective cohort study. Environ Int 126:1–6, PMID: , 10.1016/j.envint.2019.01.069. [DOI] [PubMed] [Google Scholar]
- Amaro AR, Oakley GG, Bauer U, Spielmann HP, Robertson LW. 1996. Metabolic activation of PCBs to quinones: reactivity toward nitrogen and sulfur nucleophiles and influence of superoxide dismutase. Chem Res Toxicol 9(3):623–629, PMID: , 10.1021/tx950117e. [DOI] [PubMed] [Google Scholar]
- Anezaki K, Nakano T. 2014. Concentration levels and congener profiles of polychlorinated biphenyls, pentachlorobenzene, and hexachlorobenzene in commercial pigments. Environ Sci Pollut Res Int 21(2):998–1009, PMID: , 10.1007/s11356-013-1977-2. [DOI] [PubMed] [Google Scholar]
- Arsenescu V, Arsenescu RI, King V, Swanson H, Cassis LA. 2008. Polychlorinated biphenyl-77 induces adipocyte differentiation and proinflammatory adipokines and promotes obesity and atherosclerosis. Environ Health Perspect 116(6):761–768, PMID: , 10.1289/ehp.10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenescu V, Arsenescu R, Parulkar M, Karounos M, Zhang X, Baker N, et al. 2011. Polychlorinated biphenyl 77 augments angiotensin II-induced atherosclerosis and abdominal aortic aneurysms in male apolipoprotein E deficient mice. Toxicol Appl Pharmacol 257(1):148–154, PMID: , 10.1016/j.taap.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25(1):25–29, PMID: , 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantyne MD, McDonald RA, Baker AH. 2016. lncRNA/microRNA interactions in the vasculature. Clin Pharmacol Ther 99(5):494–501, PMID: , 10.1002/cpt.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Hassine S, Hammami B, Ben Ameur W, El Megdiche Y, Barhoumi B, El Abidi R, et al. 2014. Concentrations of organochlorine pesticides and polychlorinated biphenyls in human serum and their relation with age, gender, and BMI for the general population of Bizerte, Tunisia. Environ Sci Pollut Res Int 21(10):6303–6313, PMID: , 10.1007/s11356-013-1480-9. [DOI] [PubMed] [Google Scholar]
- Bergkvist C, Berglund M, Glynn A, Julin B, Wolk A, Åkesson A. 2016. Dietary exposure to polychlorinated biphenyls and risk of myocardial infarction in men—a population-based prospective cohort study. Environ Int 88:9–14, PMID: , 10.1016/j.envint.2015.11.020. [DOI] [PubMed] [Google Scholar]
- Blaha MJ, Yeboah J, Al Rifai M, Liu K, Kronmal R, Greenland P. 2016. Providing evidence for subclinical CVD in risk assessment. Glob Heart 11(3):275–285, PMID: , 10.1016/j.gheart.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdillon MC, Poston RN, Covacho C, Chignier E, Bricca G, McGregor JL. 2000. ICAM-1 deficiency reduces atherosclerotic lesions in double-knockout mice (ApoE–/–/ICAM-1–/–) fed a fat or a chow diet. Arterioscler Thromb Vasc Biol 20(12):2630–2635, PMID: , 10.1161/01.atv.20.12.2630. [DOI] [PubMed] [Google Scholar]
- Chen N, Shan Q, Qi Y, Liu W, Tan X, Gu J. 2020. Transcriptome analysis in normal human liver cells exposed to 2, 3, 3′, 4, 4′, 5-hexachlorobiphenyl (PCB 156). Chemosphere 239:124747, PMID: , 10.1016/j.chemosphere.2019.124747. [DOI] [PubMed] [Google Scholar]
- Chen Q, Zhai H, Li X, Ma Y, Chen B, Liu F, et al. 2017. Recombinant adeno-associated virus serotype 9 in a mouse model of atherosclerosis: determination of the optimal expression time in vivo. Mol Med Rep 15(4):2090–2096, PMID: , 10.3892/mmr.2017.6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG. 2009. Endocytic regulation of TGF-β signaling. Cell Res 19(1):58–70, PMID: , 10.1038/cr.2008.315. [DOI] [PubMed] [Google Scholar]
- Chen Y, Gorski DH. 2008. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood 111(3):1217–1226, PMID: , 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 24(10):992–1009, PMID: , 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codru N, Schymura MJ, Negoita S, Akwesasne Task Force on the Environment, Rej R, Carpenter DO. 2007. Diabetes in relation to serum levels of polychlorinated biphenyls and chlorinated pesticides in adult Native Americans. Environ Health Perspect 115(10):1442–1447, PMID: , 10.1289/ehp.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosselman KE, Navas-Acien A, Kaufman JD. 2015. Environmental factors in cardiovascular disease. Nat Rev Cardiol 12(11):627–642, PMID: , 10.1038/nrcardio.2015.152. [DOI] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86(6):3993–4003, PMID: , 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaei G, Singh GM, Paciorek CJ, Lin JK, Cowan MJ, Finucane MM, et al. 2013. The global cardiovascular risk transition: associations of four metabolic risk factors with national income, urbanization, and Western diet in 1980 and 2008. Circulation 127(14):1493–1502, PMID: , 10.1161/CIRCULATIONAHA.113.001470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donat-Vargas C, Gea A, Sayon-Orea C, Carlos S, Martinez-Gonzalez MA, Bes-Rastrollo M. 2014. Association between dietary intakes of PCBs and the risk of obesity: the SUN project. J Epidemiol Community Health 68(9):834–841, PMID: , 10.1136/jech-2013-203752. [DOI] [PubMed] [Google Scholar]
- Emerging Risk Factors Collaboration; Sarwar N, Gao P, Seshasai SRK, Gobin R, Kaptoge S, et al. 2010. Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. Lancet 375(9733):2215–2222, PMID: , 10.1016/S0140-6736(10)60484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. 2003. MicroRNA targets in Drosophila. Genome Biol 5(1):R1, PMID: , 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg MW, Moore KJ. 2016. MicroRNA regulation of atherosclerosis. Circ Res 118(4):703–720, PMID: , 10.1161/CIRCRESAHA.115.306300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Ruiz I. 2019. Systolic and diastolic hypertension independently predict CVD risk. Nat Rev Cardiol 16(10):578–579, PMID: , 10.1038/s41569-019-0248-4. [DOI] [PubMed] [Google Scholar]
- Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, et al. 2008. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 15(2):272–284, PMID: , 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank PG, Lee H, Park DS, Tandon NN, Scherer PE, Lisanti MP. 2004. Genetic ablation of caveolin-1 confers protection against atherosclerosis. Arterioscler Thromb Vasc Biol 24(1):98–105, PMID: , 10.1161/01.ATV.0000101182.89118.E5. [DOI] [PubMed] [Google Scholar]
- Frank PG, Pavlides S, Lisanti MP. 2009. Caveolae and transcytosis in endothelial cells: role in atherosclerosis. Cell Tissue Res 335(1):41–47, PMID: , 10.1007/s00441-008-0659-8. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KKH, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1):92–105, PMID: , 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwar RS, Rajagopalan S, Natarajan R, Deiuliis JA. 2018. Noncoding RNAs in cardiovascular disease: pathological relevance and emerging role as biomarkers and therapeutics. Am J Hypertens 31(2):150–165, PMID: , 10.1093/ajh/hpx197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. 2021. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 49(D1):D325–D334, PMID: , 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5(10):R80, PMID: , 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimbrone MA Jr, García-Cardeña G. 2013. Vascular endothelium, hemodynamics, and the pathobiology of atherosclerosis. Cardiovasc Pathol 22(1):9–15, PMID: , 10.1016/j.carpath.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncharov A, Haase RF, Santiago-Rivera A, Morse G, Akwesasne Task Force on the Environment; McCaffrey RJ, et al. 2008. High serum PCBs are associated with elevation of serum lipids and cardiovascular disease in a Native American population. Environ Res 106(2):226–239, PMID: , 10.1016/j.envres.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gvaramia D, Blaauboer ME, Hanemaaijer R, Everts V. 2013. Role of caveolin-1 in fibrotic diseases. Matrix Biol 32(6):307–315, PMID: , 10.1016/j.matbio.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Halliday A, Harrison M, Hayter E, Kong X, Mansfield A, Marro J, et al. 2010. 10-year stroke prevention after successful carotid endarterectomy for asymptomatic stenosis (ACST-1): a multicentre randomised trial. Lancet 376(9746):1074–1084, PMID: , 10.1016/S0140-6736(10)61197-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson GK. 2005. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med 352(16):1685–1695, PMID: , 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- Haybar H, Shahrabi S, Rezaeeyan H, Shirzad R, Saki N. 2019. Endothelial cells: from dysfunction mechanism to pharmacological effect in cardiovascular disease. Cardiovasc Toxicol 19(1):13–22, PMID: , 10.1007/s12012-018-9493-8. [DOI] [PubMed] [Google Scholar]
- Hu D, Hornbuckle KC. 2010. Inadvertent polychlorinated biphenyls in commercial paint pigments. Environ Sci Technol 44(8):2822–2827, PMID: , 10.1021/es902413k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh K. 2020. Non-HDL cholesterol levels linked with long-term risk of CVD. Nat Rev Cardiol 17(3):132–133, PMID: , 10.1038/s41569-019-0329-4. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. 2004. Human microRNA targets. PLoS Biol 2(11):e363, PMID: , 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Sander C, Marks DS. 2006. Prediction of human microRNA targets. Methods Mol Biol 342:101–113, PMID: , 10.1385/1-59745-123-1:101. [DOI] [PubMed] [Google Scholar]
- Joseph P, Leong D, McKee M, Anand SS, Schwalm JD, Teo K, et al. 2017. Reducing the global burden of cardiovascular disease, part 1: the epidemiology and risk factors. Circ Res 121(6):677–694, PMID: , 10.1161/CIRCRESAHA.117.308903. [DOI] [PubMed] [Google Scholar]
- Kahles F, Liberman A, Halim C, Rau M, Möllmann J, Mertens RW, et al. 2018. The incretin hormone GIP is upregulated in patients with atherosclerosis and stabilizes plaques in ApoE–/– mice by blocking monocyte/macrophage activation. Mol Metab 14:150–157, PMID: , 10.1016/j.molmet.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. 2019. Toward understanding the origin and evolution of cellular organisms. Protein Sci 28(11):1947–1951, PMID: , 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. 2021. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 49(D1):D545–D551, PMID: , 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28(1):27–30, PMID: , 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper HU, Schmidt A, Roessner A. 1996. Expression of the adhesion molecules ICAM, VCAM, and ELAM in the arteriosclerotic plaque. Gen Diagn Pathol 141(5–6):289–294, PMID: . [PubMed] [Google Scholar]
- Khalyfa A, Kheirandish-Gozal L, Bhattacharjee R, Khalyfa AA, Gozal D. 2016. Circulating microRNAs as potential biomarkers of endothelial dysfunction in obese children. Chest 149(3):786–800, PMID: , 10.1378/chest.15-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MB, Edelman JJB, Kao SCH, Vallely MP, van Zandwijk N, Reid G. 2013. The impact of hemolysis on cell-free microRNA biomarkers. Front Genet 4:94, PMID: , 10.3389/fgene.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J, Rehmsmeier M. 2006. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34(Web Server issue):W451–W454, PMID: , 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras A. 2018. Public health: light smoking and CVD risk. Nat Rev Cardiol 15(3):136, PMID: , 10.1038/nrcardio.2018.10. [DOI] [PubMed] [Google Scholar]
- Lee DH, Steffes MW, Sjödin A, Jones RS, Needham LL, Jacobs DR Jr.. 2011. Low dose organochlorine pesticides and polychlorinated biphenyls predict obesity, dyslipidemia, and insulin resistance among people free of diabetes. PLoS One 6(1):e15977, PMID: , 10.1371/journal.pone.0015977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim EJ, Smart EJ, Toborek M, Hennig B. 2007. The role of caveolin-1 in PCB77-induced eNOS phosphorylation in human-derived endothelial cells. Am J Physiol Heart Circ Physiol 293(6):H3340–H3347, PMID: , 10.1152/ajpheart.00921.2007. [DOI] [PubMed] [Google Scholar]
- Liu J, Tan Y, Song E, Song Y. 2020. A critical review of polychlorinated biphenyls metabolism, metabolites, and their correlation with oxidative stress. Chem Res Toxicol 33(8):2022–2042, PMID: , 10.1021/acs.chemrestox.0c00078. [DOI] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. 2011. ViennaRNA package 2.0. Algorithms Mol Biol 6:26, PMID: , 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Hofacker IL, Stadler PF. 2016. RNA folding with hard and soft constraints. Algorithms Mol Biol 11:8, PMID: , 10.1186/s13015-016-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen JM, Thum T. 2016. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol 12(6):360–373, PMID: , 10.1038/nrneph.2016.51. [DOI] [PubMed] [Google Scholar]
- Majkova Z, Smart E, Toborek M, Hennig B. 2009. Up-regulation of endothelial monocyte chemoattractant protein-1 by coplanar PCB77 is caveolin-1-dependent. Toxicol Appl Pharmacol 237(1):1–7, PMID: , 10.1016/j.taap.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Càceres J, Mainez J, Mayoral R, Martín-Sanz P, Egea G, Fabregat I. 2016. Caveolin-1-dependent activation of the metalloprotease TACE/ADAM17 by TGF-β in hepatocytes requires activation of Src and the NADPH oxidase NOX1. FEBS J 283(7):1300–1310, PMID: , 10.1111/febs.13669. [DOI] [PubMed] [Google Scholar]
- Murphy LE, Gollenberg AL, Buck Louis GM, Kostyniak PJ, Sundaram R. 2010. Maternal serum preconception polychlorinated biphenyl concentrations and infant birth weight. Environ Health Perspect 118(2):297–302, PMID: , 10.1289/ehp.0901150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nah J, Yoo SM, Jung S, Jeong EI, Park M, Kaang BK, et al. 2017. Phosphorylated CAV1 activates autophagy through an interaction with BECN1 under oxidative stress. Cell Death Dis 8(5):e2822, PMID: , 10.1038/cddis.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olden K. 2004. Genomics in environmental health research—opportunities and challenges. Toxicology 198(1–3):19–24, PMID: , 10.1016/j.tox.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Oshlack A, Robinson MD, Young MD. 2010. From RNA-seq reads to differential expression results. Genome Biol 11(12):220, PMID: , 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins JT, Petriello MC, Newsome BJ, Hennig B. 2016. Polychlorinated biphenyls and links to cardiovascular disease. Environ Sci Pollut Res Int 23(3):2160–2172, PMID: , 10.1007/s11356-015-4479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JL, Fabian MP, Levy JI. 2014. Combined impact of lead, cadmium, polychlorinated biphenyls and non-chemical risk factors on blood pressure in NHANES. Environ Res 132:93–99, PMID: , 10.1016/j.envres.2014.03.038. [DOI] [PubMed] [Google Scholar]
- Petriello MC, Han SG, Newsome BJ, Hennig B. 2014. PCB 126 toxicity is modulated by cross-talk between caveolae and Nrf2 signaling. Toxicol Appl Pharmacol 277(2):192–199, PMID: , 10.1016/j.taap.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quest AFG, Lobos-González L, Nuñez S, Sanhueza C, Fernández JG, Aguirre A, et al. 2013. The caveolin-1 connection to cell death and survival. Curr Mol Med 13(2):266–281, PMID: , 10.2174/156652413804810745. [DOI] [PubMed] [Google Scholar]
- Raffetti E, Donato F, Speziani F, Scarcella C, Gaia A, Magoni M. 2018. Polychlorinated biphenyls (PCBs) exposure and cardiovascular, endocrine and metabolic diseases: a population-based cohort study in a North Italian highly polluted area. Environ Int 120:215–222, PMID: , 10.1016/j.envint.2018.08.022. [DOI] [PubMed] [Google Scholar]
- Ramírez CM, Zhang X, Bandyopadhyay C, Rotllan N, Sugiyama MG, Aryal B, et al. 2019. Caveolin-1 regulates atherogenesis by attenuating low-density lipoprotein transcytosis and vascular inflammation independently of endothelial nitric oxide synthase activation. Circulation 140(3):225–239, PMID: , 10.1161/CIRCULATIONAHA.118.038571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B, Zhang XL, Bitzer M, von Gersdorff G, Böttinger EP, Lisanti MP. 2001. Caveolin-1 regulates transforming growth factor (TGF)-β/SMAD signaling through an interaction with the TGF-β type I receptor. J Biol Chem 276(9):6727–6738, PMID: , 10.1074/jbc.M008340200. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10(10):1507–1517, PMID: , 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140, PMID: , 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche-Molina M, Sanz-Rosa D, Cruz FM, García-Prieto J, López S, Abia R, et al. 2015. Induction of sustained hypercholesterolemia by single adeno-associated virus-mediated gene transfer of mutant hPCSK9. Arterioscler Thromb Vasc Biol 35(1):50–59, PMID: , 10.1161/ATVBAHA.114.303617. [DOI] [PubMed] [Google Scholar]
- Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, et al. 2020. Global burden of cardiovascular diseases and risk factors, 1990–2019: update from the GBD 2019 study. J Am Coll Cardiol 76(25):2982–3021, PMID: , 10.1016/j.jacc.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146(3):353–358, PMID: , 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH image to ImageJ: 25 years of image analysis. Nat Methods 9(7):671–675, PMID: , 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte C, Barwari T, Joshi A, Zeller T, Mayr M. 2020. Noncoding RNAs versus protein biomarkers in cardiovascular disease. Trends Mol Med 26(6):583–596, PMID: , 10.1016/j.molmed.2020.02.001. [DOI] [PubMed] [Google Scholar]
- Shan Q, Qu F, Chen N. 2020. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and polychlorinated biphenyl coexposure alters the expression profile of microRNAs in the liver associated with atherosclerosis. Biomed Res Int 2020:2652756, PMID: , 10.1155/2020/2652756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Buettner GR, Parkin S, Wagner BA, Robertson LW, Lehmler HJ. 2008. Chlorination increases the persistence of semiquinone free radicals derived from polychlorinated biphenyl hydroquinones and quinones. J Org Chem 73(21):8296–8304, PMID: , 10.1021/jo801397g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Wagner BA, Witmer JR, Lehmler HJ, Buettner GR. 2009. Nonenzymatic displacement of chlorine and formation of free radicals upon the reaction of glutathione with PCB quinones. Proc Natl Acad Sci U S A 106(24):9725–9730, PMID: , 10.1073/pnas.0810352106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa G. 2012. Caveolae, caveolins, cavins, and endothelial cell function: new insights. Front Physiol 2:120, PMID: , 10.3389/fphys.2011.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HJ, Hou B, Wang X, Zhu XX, Li KX, Qiu LY. 2016. Endothelial dysfunction and cardiometabolic diseases: role of long non-coding RNAs. Life Sci 167:6–11, PMID: , 10.1016/j.lfs.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Tafer H, Hofacker IL. 2008. RNAplex: a fast tool for RNA–RNA interaction search. Bioinformatics 24(22):2657–2663, PMID: , 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- Thomson DW, Dinger ME. 2016. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17(5):272–283, PMID: , 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- Turner AW, Wong D, Khan MD, Dreisbach CN, Palmore M, Miller CL. 2019. Multi-omics approaches to study long non-coding RNA function in atherosclerosis. Front Cardiovasc Med 6:9, PMID: , 10.3389/fcvm.2019.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Dimmeler S. 2015. Long noncoding RNAs in cardiovascular diseases. Circ Res 116(4):737–750, PMID: , 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- Van Gaal LF, Mertens IL, De Block CE. 2006. Mechanisms linking obesity with cardiovascular disease. Nature 444(7121):875–880, PMID: , 10.1038/nature05487. [DOI] [PubMed] [Google Scholar]
- Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, et al. 2020. Heart disease and stroke statistics—2020 update: a report from the American Heart Association. Circulation 141(9):e139–e596, PMID: , 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- Wahlang B, Petriello MC, Perkins JT, Shen S, Hennig B. 2016. Polychlorinated biphenyl exposure alters the expression profile of microRNAs associated with vascular diseases. Toxicol In Vitro 35:180–187, PMID: , 10.1016/j.tiv.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Yuan H, Jin J, Li P, Ma Y, Wang Y. 2018. Polychlorinated biphenyl concentrations in pooled serum from people in different age groups from five Chinese cities. Chemosphere 198:320–326, PMID: , 10.1016/j.chemosphere.2018.01.103. [DOI] [PubMed] [Google Scholar]
- Yorita Christensen KL, White P. 2011. A methodological approach to assessing the health impact of environmental chemical mixtures: PCBs and hypertension in the National Health and Nutrition Examination Survey. Int J Environ Res Public Health 8(11):4220–4237, PMID: , 10.3390/ijerph8114220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287, PMID: , 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf S, Joseph P, Rangarajan S, Islam S, Mente A, Hystad P, et al. 2020. Modifiable risk factors, cardiovascular disease, and mortality in 155 722 individuals from 21 high-income, middle-income, and low-income countries (PURE): a prospective cohort study. Lancet 395(10226):795–808, PMID: , 10.1016/S0140-6736(19)32008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Feng S, Bai H, Zeng P, Chen F, Wu C, et al. 2015. Polychlorinated biphenyl quinone induces endothelial barrier dysregulation by setting the cross talk between VE-cadherin, focal adhesion, and MAPK signaling. Am J Physiol Heart Circ Physiol 308(10):H1205–H1214, PMID: , 10.1152/ajpheart.00005.2015. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ramírez CM, Aryal B, Madrigal-Matute J, Liu X, Diaz A, et al. 2020. Cav-1 (caveolin-1) deficiency increases autophagy in the endothelium and attenuates vascular inflammation and atherosclerosis. Arterioscler Thromb Vasc Biol 40(6):1510–1522, PMID: , 10.1161/ATVBAHA.120.314291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang L, Wang Y, Ding H, Xue S, Qi H, et al. 2019. MicroRNAs or long noncoding RNAs in diagnosis and prognosis of coronary artery disease. Aging Dis 10(2):353–366, PMID: , 10.14336/AD.2018.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Yu LH, Chen SJ, Hu GC, Chen KH, Yan X, et al. 2016. Polychlorinated biphenyls (PCBs) in human hair and serum from e-waste recycling workers in Southern China: concentrations, chiral signatures, correlations, and source identification. Environ Sci Technol 50(3):1579–1586, PMID: , 10.1021/acs.est.5b04955. [DOI] [PubMed] [Google Scholar]
- Zinman B, Wanner C, Lachin JM, Fitchett D, Bluhmki E, Hantel S, et al. 2015. Empagliflozin, cardiovascular outcomes, and mortality in type 2 diabetes. N Engl J Med 373(22):2117–2128, PMID: , 10.1056/NEJMoa1504720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



![Figures 2A and 2C are bar graphs, plotting relative messenger ribonucleic acid level, ranging from 0.0 to 1.5 in increments of 0.5 (y-axis) across Transforming growth factor lowercase beta-2 and Protein Phosphatase Methylesterase 1 (x-axis) for control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone and for N C mimic and M I R-7-5 lowercase p mimic, respectively. Figures 2B and 2D are each set of one Western blot and one bar graph. The Western blot has two columns, namely, control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, and N C mimic and M I R-7-5 lowercase p mimic, respectively, and three rows, namely, Transforming growth factor lowercase beta-2, Protein Phosphatase Methylesterase 1, and lowercase beta-actin. The bar graphs are plotting protein per lowercase beta actin (fold of control), ranging from 0.0 to 1.5 in increments of 0.5 (y-axis) across Transforming growth factor lowercase beta-2 and Protein Phosphatase Methylesterase 1 (x-axis) for control and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, and N C mimic and M I R-7-5 lowercase p mimic, respectively. Figure 2E depicts the M I R-7-5 lowercase p binding sites in transforming growth factor lowercase beta-2 and Protein Phosphatase Methylesterase 1 three prime untranslated region. They used results of ribonucleic acid hybrid, miRanda, and TargetScan from three different software predicting target genes of micro ribonucleic acid. Figures 2F and 2G are bar graphs, plotting luciferase activity (arbitrary units), ranging from 0 to 40 in increments of 10 and 0 to 10 in increments of 2 (y-axis) across N C mimic, including positive, negative, negative; 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, including positive, positive, negative; M I R-7-5 lowercase p mimic, including negative, positive, positive; and p E Z-M 61 H D A C 7-A S 1, including negative, negative, positive (x-axis) for transforming growth factor lowercase beta-2 and Protein Phosphatase Methylesterase 1.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/de69/8963524/c0faec16e1c1/ehp9833_f2.jpg)
![Figure 5A is a set of one representative image and one bar graph. The representative image depicts the Oil Red O staining of aortas in Apolipoprotein E negative negative, including A A V-N C, A A V-N C plus 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, A A V-H D A C 7-A S 1, and A A V-H D A C 7-A S 1 plus 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone. The bar graph, plotting plaque area (percentage of whole aorta), 0 to 40 in increments of 10 (y-axis) across Apolipoprotein E negative negative, including A A V-N C, including positive, positive, negative, negative; A A V- H D A C 7-A S 1, including negative, negative, positive, positive; and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, including negative, positive, negative, positive (x-axis). Figure 5B is a stained tissue having four columns, namely, A A V-N C, A A V-N C plus 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, A A V-H D A C 7-A S 1, and A A V-H D A C 7-A S 1 plus 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone for Apolipoprotein E negative negative and four rows, namely, H D A C 7-A S 1, C D 31, D A P I, and Merge. Figures 5C to 5J are error bar graphs, plotting Pearson’s correlation coefficient, ranging from 0.0 to 1.0 in increments of 0.2; Relative H D A C 7-A S 1 level, ranging from 0 to 25 in increments of 5; Relative M I R-7-5 lowercase p level, ranging 0 to 8 in increments of 2; Relative transforming growth factor lowercase beta-2 level, ranging from 0 to 5 in unit increments; Relative Phosphatase Methylesterase 1 level, ranging from 0 to 5 in unit increments; Relative interleukin-1 lowercase beta level, ranging from 0 to 8 in increments of 2; Relative interleukin-6 level, ranging from 0 to 6 in increments of 2; and Relative tumor necrosis factor lowercase alpha level, ranging from 0 to 8 in increments of 2 (y-axis) across Apolipoprotein E negative negative, including A A V-N C, including positive, positive, negative, negative; A A V- H D A C 7-A S 1, including negative, negative, positive, positive; and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, including negative, positive, negative, positive (x-axis), respectively. Figure 5K is a set of one stained tissue and one bar graph. The stained tissue has four columns, namely, A A V-N C, A A V-N C plus 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, A A V-H D A C 7-A S 1, and A A V-H D A C 7-A S 1 plus 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone for Apolipoprotein E negative negative and four rows, namely, p-p 65, C D 31, D A P I, and Merge. The bar graph, plotting Pearson’s correlation coefficient, ranging from 0.0 to 1.5 in increments of 0.5 (y-axis) across Apolipoprotein E negative negative, including A A V-N C, including positive, positive, negative, negative; A A V- H D A C 7-A S 1, including negative, negative, positive, positive; and 2,3,5-trichloro-6-phenyl-[1,4]-benzoquinone, including negative, positive, negative, positive (x-axis).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/de69/8963524/79f4f51009de/ehp9833_f5.jpg)