Abstract
Background.
In 2018, the Centers for Disease Control and Prevention and the Vermont Department of Health investigated an outbreak of multidrug-resistant Shigella sonnei infections in a retirement community that offered a continuum of care from independent living through skilled nursing care. The investigation identified 24 culture-confirmed cases. Isolates were resistant to trimethoprim-sulfamethoxazole, ampicillin, and ceftriaxone, and had decreased susceptibility to azithromycin and ciprofloxacin.
Methods.
To evaluate clinical and microbiologic response, we reviewed inpatient and outpatient medical records for treatment outcomes among the 24 patients with culture-confirmed S. sonnei infection. We defined clinical failure as diarrhea (≥3 loose stools per day) for ≥1 day after treatment finished, and microbiologic failure as a stool culture that yielded S. sonnei after treatment finished. We used broth microdilution to perform antimicrobial susceptibility testing, and whole genome sequencing to identify resistance mechanisms.
Results.
Isolates contained macrolide resistance genes mph(A) and erm(B) and had azithromycin minimum inhibitory concentrations above the Clinical and Laboratory Standards Institute epidemiological cutoff value of ≤16 μg/mL. Among 24 patients with culture-confirmed Shigella infection, 4 were treated with azithromycin; all had clinical treatment failure and 2 also had microbiologic treatment failure. Isolates were susceptible to ciprofloxacin but contained a gyrA mutation; 2 patients failed treatment with ciprofloxacin.
Conclusions.
These azithromycin treatment failures demonstrate the importance of clinical breakpoints to aid clinicians in identifying alternative treatment options for resistant strains. Additionally, these treatment failures highlight a need for comprehensive susceptibility testing and systematic outcome studies, particularly given the emergence of multidrug-resistant Shigella among an expanding range of patient populations.
Keywords: antibiotic treatment failure, multidrug resistance, azithromycin, ciprofloxacin, Shigella sonnei
Multidrug-resistant (MDR) Shigella strains pose a threat to public health [1]. Most Shigella infections are self-limited, but antibiotics are indicated for severe illness and to limit transmission. Recommended antibiotics, such as azithromycin and ciprofloxacin, are often prescribed for treatment [2, 3]. Over the last decade, however, decreased susceptibility to both azithromycin and ciprofloxacin has steadily increased in the United States [4] and has spread globally, particularly among men who have sex with men [5].
In March 2021, the Clinical and Laboratory Standards Institute (CLSI) established clinical breakpoints for azithromycin resistance in Shigella isolates [6]. Previously, epidemiological cutoff values (ECVs) delineated wild-type and non-wild-type (NWT) isolates; although useful for surveillance, ECVs did not define resistance clinically, limiting the information available to healthcare providers to aid in management decisions. In recent years, the US Centers for Disease Control and Prevention (CDC) identified an increasing number of NWT Shigella isolates with azithromycin minimum inhibitory concentrations (MICs) that exceeded the CLSI ECV of ≤16 μg/mL [7]; in 2018, 34.0% of Shigella isolates reported to CDC’s National Antibiotic Resistance Monitoring System (NARMS) had decreased susceptibility to azithromycin [4]. Furthermore, CDC identified an increasing number of Shigella isolates that were susceptible to ciprofloxacin per CLSI criteria but harbored at least 1 quinolone resistance mechanism, raising concerns about the potential for fluoroquinolone treatment failure [7]. Among Shigella isolates reported to NARMS in 2018, 34.4% had decreased susceptibility to ciprofloxacin [4].
In October 2018, CDC assisted the Vermont Department of Health with investigating an outbreak of MDR Shigella sonnei infections in a retirement community that offered a continuum of care from independent living through skilled nursing care [8]. The investigation identified 75 cases, 24 of which were culture-confirmed, with onset from 9 October through 3 November 2018. Results of a case-control study indicated that illness was associated with dining at brunch and breakfast at the facility on 14 October. Based on these results and the identification of an early case among a staff member who prepared food while ill during 11–14 October, the investigation implicated food contaminated by a food handler as the most likely mode of transmission. Isolates were resistant to trimethoprim-sulfamethoxazole, ampicillin, and ceftriaxone, and had decreased susceptibility to azithromycin and ciprofloxacin. Among confirmed cases, antibiotic treatment and clinical course varied. Thus, this outbreak provided a unique opportunity to characterize treatment outcomes among a cohort of persons with MDR shigellosis.
METHODS
The Vermont Department of Health Laboratory cultured stool specimens, isolated S. sonnei, performed whole genome sequencing (WGS), and submitted isolates to NARMS at CDC for antimicrobial susceptibility testing [9]. We defined resistance according to CLSI breakpoints when available. The term “decreased susceptibility to azithromycin” is used to describe isolates with an MIC above the ECV for S. sonnei of ≤16 μg/mL established by CLSI in 2015 (also referred to as NWT isolates), as clinical breakpoints had not yet been established in 2018. The term “decreased susceptibility to ciprofloxacin” is used to describe isolates with an MIC of 0.12–0.25 μg/mL and the presence of a resistance mechanism detected by WGS (the 2020 CLSI interpretive standards define ciprofloxacin susceptible as ≤0.25 μg/mL, intermediate as 0.5–1 μg/mL, and resistant as ≥1 μg/mL [10]).
Isolate assemblies generated using shovill 1.0.9 with the SPAdes assembler (https://github.com/tseemann/shovill) were screened for resistance genes in the ResFinder database [11] using staramr 0.4.0 (https://github.com/phac-nml/staramr) with 90% identity and 50% cutoff thresholds. Mutations involved in resistance (as outlined in the PointFinder database for Escherichia [12]) were screened using ARIBA 2.12.0 directly from reads [13]. Isolate relatedness was assessed by core genome multilocus sequence typing (cgMLST) and a phylogenetic tree was constructed using BioNumerics version 7.6 (Applied Maths, http://www.applied-maths.com).
To evaluate clinical and microbiologic response, we reviewed inpatient and outpatient medical records for treatment outcomes among the 24 patients with culture-confirmed S. sonnei infection. We defined clinical failure as diarrhea (≥3 loose stools per day) for ≥1 day after treatment finished, and microbiologic failure as a stool culture that yielded S. sonnei after treatment finished.
RESULTS
Among the 24 patients with culture-confirmed S. sonnei infection, the median age was 84.5 years (range, 42–99 years); 18 were retirement community residents, 3 were staff members, and 3 were visitors. Four patients were hospitalized and 2 died; both deceased patients had severe illness associated with comorbid conditions and shigellosis was not considered the primary cause of death.
Isolates were highly related to each other (within 0–4 alleles) by cgMLST (Figure 1A). All isolates harbored resistance genes and showed phenotypic resistance to ampicillin and ceftriaxone (blaCTX-M-27), trimethoprim-sulfamethoxazole (sul2, dfrA1), streptomycin (aph(3”)-Ib, aph(6)-Id, aadA1), and tetracycline (tet(A)). Isolates also showed decreased susceptibility to azithromycin (MIC >32 μg/mL) conferred by mph(A) and erm(B) genes. Isolates carried an S83L mutation in gyrA that confers resistance to nalidixic acid and decreased susceptibility to ciprofloxacin; however, they were susceptible to ciprofloxacin (MIC = 0.12 μg/mL) by CLSI criteria [10] (Figure 1B).
Figure 1.
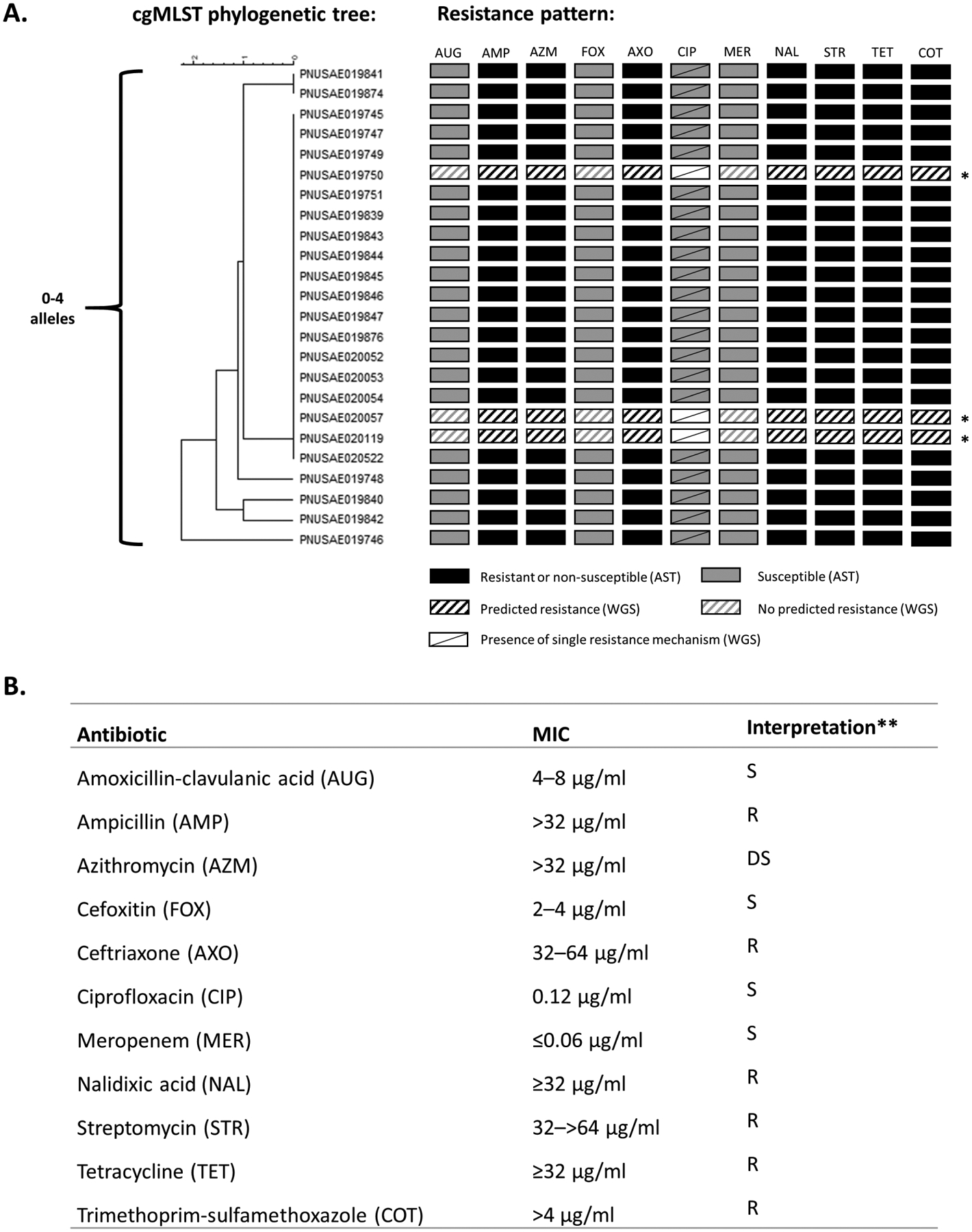
Core genome multilocus sequence typing (cgMLST) phylogenetic tree and antibiotic susceptibility of Shigella sonnei outbreak isolates—Vermont, 2018. A, cgMLST phylogenetic tree containing isolates from 24 patients with culture-confirmed S. sonnei infection. Resistance patterns by antibiotic susceptibility testing (AST) and/or identification of resistance mechanisms by whole genome sequencing (WGS) are shown. B, AST results and interpretation for outbreak isolates. *AST was not performed for these isolates; only predicted resistance based on WGS is shown. **Interpretation of minimum inhibitory concentrations (MICs) is according to Clinical and Laboratory Standards Institute (CLSI) breakpoints when available. Decreased susceptibility to azithromycin is defined by an MIC above the epidemiological cutoff value (ECV) for Shigella sonnei of ≤16 μg/mL established by CLSI in 2015. The ECV should not be used to predict clinical effectiveness. Abbreviations: AMP, ampicillin; AST, antibiotic susceptibility testing; AUG, amoxicillin-clavulanic acid; AXO, ceftriaxone; AZM, azithromycin; cgMLST, core genome multilocus sequence typing; CIP, ciprofloxacin; COT, trimethoprim-sulfamethoxazole; DS, decreased susceptibility; FOX, cefoxitin; MER, meropenem; MIC, minimum inhibitory concentration; NAL, nalidixic acid; R, resistant; S, susceptible; STR, streptomycin; TET, tetracycline; WGS, whole genome sequencing.
Fifteen patients received a single course of ciprofloxacin (500–750mg orally [PO] twice daily for 3 days), 5 received multiple courses of antibiotics, and 4 did not receive an antibiotic (Figure 2). Specifically, 4 patients (patients 2, 5, 6, and 11) received azithromycin; patient 2 received 500mg azithromycin intravenously (IV) daily for 2 days, and patients 5 and 11 received 500mg azithromycin PO daily for 4 days. Patient 6 received 250–500mg azithromycin PO daily for 5 days; this patient started the azithromycin course 1 day before diarrhea began as treatment for suspected pneumonia and concomitant infections. All 4 patients had subsequent clinical failure; diarrhea continued for a median of 3.5 days (range, 1–11 days) after finishing azithromycin. Two of these 4 patients also had microbiologic failure; 1 (patient 2) had 4 stool cultures that yielded S. sonnei ranging from 7 to 32 days after treatment and the other (patient 5) had 5 stool cultures that yielded S. sonnei ranging from 2 to 23 days after treatment.
Figure 2.
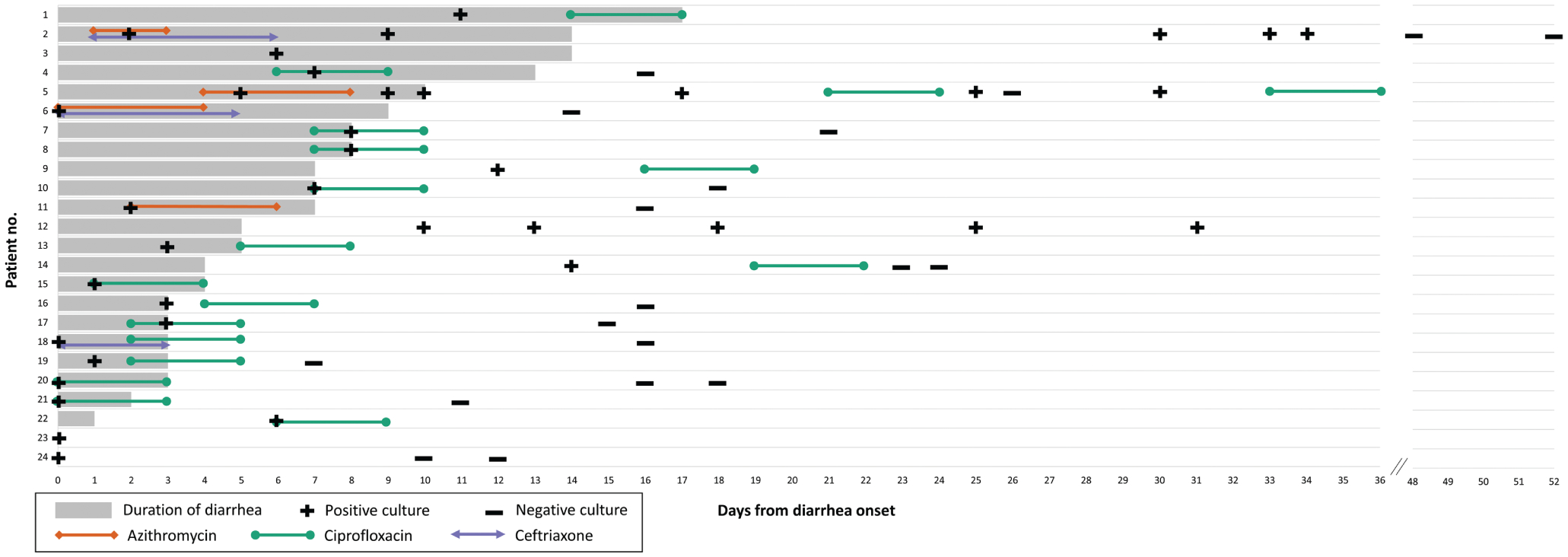
Diarrhea duration, treatment, and testing of 24 patients with Shigella sonnei infection—Vermont, 2018. This figure shows the duration of diarrhea (horizontal bars) for each patient, overlaid with timing of culture results for S. sonnei and antibiotic treatment with azithromycin, ciprofloxacin, and/or ceftriaxone.
Of the 15 patients who received ciprofloxacin, 2 had treatment failure; both received 500mg ciprofloxacin PO twice daily for 3 days. One (patient 4) had clinical failure; diarrhea continued for 4 days after finishing ciprofloxacin. A stool culture obtained 4 days after diarrhea resolved did not yield S. sonnei. The other (patient 5) was a staff member whose symptoms resolved before ciprofloxacin treatment was given in an attempt to eradicate stool carriage for return-to-work requirements; this patient had microbiologic failure with a stool culture yielding S. sonnei 2 days after treatment and again 7 days after treatment. The patient received a second course of ciprofloxacin following the last culture and had a negative culture-independent diagnostic test result on day 36; no additional stool specimens were obtained. Among the 13 patients who did not have treatment failures to ciprofloxacin, 12 received 500mg PO twice daily for 3 days and 1 received 750mg PO twice daily for 3 days.
Three patients received ceftriaxone 1000–2000mg IV daily for 35 days. Two (patients 2 and 6) had clinical failure; diarrhea continued for 4 and 8 days after finishing ceftriaxone, respectively. Both patients were concurrently treated with azithromycin, as previously described. One (patient 2) also had microbiologic failure with stool culture yielding S. sonnei 4 days after finishing ceftriaxone. A third patient, who did not have treatment failure, received ceftriaxone in combination with ciprofloxacin.
DISCUSSION
Few studies have described treatment outcomes in patients with MDR S. sonnei. Given rising rates of decreased susceptibility to azithromycin among US Shigella isolates [4], data on azithromycin failures in patients whose isolates yielded azithromycin MICs above the ECV (NWT isolates) have important implications for public health. Generally, azithromycin is an effective oral antibiotic for susceptible Shigella infections; randomized controlled trials of patients with wild-type strains (ie, strains with azithromycin MICs at or below the ECV) have demonstrated high efficacy in treatment of shigellosis in Bangladesh (82% success in 34 children with Shigella strains with MICs <1.5 μg/mL) [14] and in Paraguay (93% success in 36 adults with Shigella strains with a clear zone of ≥17mm around a 15 μg disk) [15]. By comparison, none of our 4 patients infected with an NWT strain had a successful response to treatment. Azithromycin treatment failures for NWT Shigella isolates have also been described in France, [16] the Netherlands [17], and Bangladesh [18]. The azithromycin treatment failures from this outbreak highlight the importance of clinical breakpoints since ECVs cannot be used to predict clinical outcome. In March 2021, CLSI established azithromycin clinical breakpoints for Shigella [6], allowing clinical laboratories to report resistance and aiding clinicians in identifying alternative treatment options for resistant strains.
We also observed ciprofloxacin treatment failures in the outbreak isolate, a ciprofloxacin-susceptible strain harboring a gyrA resistance mechanism. CLSI ciprofloxacin breakpoints for Shigella are extrapolated from those for the Enterobacterales family (excluding Salmonella); Shigella isolates with a ciprofloxacin MIC of ≤0.25 μg/mL are characterized as susceptible to ciprofloxacin, isolates with an MIC of 0.5 μg/mL are characterized as intermediate, and isolates with an MIC ≥1 μg/mL are characterized as resistant [10]. Clinical laboratories have limited ability to differentiate the ciprofloxacin MIC values within the susceptible range of ≤0.25 μg/mL. Shigella isolates without a quinolone resistance mechanism typically have a ciprofloxacin MIC of ≤0.015 μg/mL [7], whereas isolates in our study had a ciprofloxacin MIC of 0.12 μg/mL. It is possible that fluoroquinolone treatment of a Shigella infection with a ciprofloxacin MIC in this reduced susceptibility range (0.12–0.25 μg/mL) may be associated with a worse clinical outcome or could increase the risk of transmission [7]. Randomized controlled trials in Bangladesh have shown very high efficacy for ciprofloxacin treatment of susceptible Shigella strains; among 78 adults with Shigella strains with an MIC ≤0.004 μg/mL, no treatment failures were reported [19], and among 60 adults with Shigella strains with an MIC ≤0.031 μg/mL, 3 clinical failures and no microbiologic failures were reported [20]. Thus, the 2 clinical and microbiologic failures among our cohort of 15 patients treated with a commonly recommended regimen of ciprofloxacin are notable and warrant further investigation of outcomes among patients with isolates in the reduced susceptibility range. These 2 failures occurred among patients who received 500mg PO twice daily for 3 days; the sole patient who received a 750mg regimen of ciprofloxacin did not have treatment failure. While no conclusions can be drawn from this small sample, they do highlight a need for investigation of high-dose fluoroquinolone treatment of Shigella strains with decreased susceptibility to ciprofloxacin, though any potential benefits of these treatment courses should be weighed against the risks of adverse effects [21].
Moreover, 2 of the 3 patients treated with ceftriaxone, to which the outbreak strain was resistant, had poor response to treatment; both patients also received azithromycin. Although ceftriaxone is often considered the treatment of choice for invasive shigellosis, increasing resistance to third-generation cephalosporins [22], as seen in this outbreak strain and described in Asia [23], may limit options for treatment. The existence of such strains underscores a need for awareness of MDR Shigella among medical providers.
This study is subject to several limitations. First, the clinical outcome data were observational and obtained from medical records. Antibiotics were prescribed at the discretion of each treating physician and treatment was not standardized; furthermore, most physicians did not have access to susceptibility results to inform treatment decisions. Cultures were not collected at systematic time points or frequently enough to assess the overall duration of shedding among all patients. We were unable to evaluate the effect of patient characteristics, such as age and comorbidities, on treatment outcomes. Given the advanced age of these patients and that all received inpatient or outpatient healthcare that resulted in stool submission and culture confirmation of Shigella infection, infections may have been more severe than Shigella infections on average in the greater population. Finally, our definition of clinical failure cannot distinguish between ongoing shigellosis and antibiotic-associated diarrhea (eg, from dysbiosis). Testing for other pathogens, such as Clostridium difficile, was not systematically conducted; however, no instances of C. difficile colitis or other infections suspected to cause diarrhea were reported.
The observed outcomes highlight a need for comprehensive susceptibility testing and systematic outcome studies, particularly given the emergence of MDR Shigella among an expanding range of patient populations. Further investigation is needed to explore patient outcomes following fluoroquinolone treatment for Shigella isolates that have a single resistance mechanism to ciprofloxacin and an MIC in the susceptible range. Antibiotic susceptibility testing, dependent on clinical breakpoints, is important for clinical decision making, especially in medically vulnerable populations where prompt, appropriate treatment can help prevent adverse health outcomes. While the extensive resistance demonstrated by outbreak isolates (resistant to trimethoprim-sulfamethoxazole, ampicillin, and ceftriaxone; decreased susceptibility to azithromycin and ciprofloxacin) was relatively unusual in the United States in 2018, representing only 2.4% of Shigella isolates reported to NARMS [4], resistance or decreased susceptibility to azithromycin or ciprofloxacin is not uncommon, and rates of MDR infections have consistently increased in recent years [4]. When treating a patient whose Shigella infection warrants antibiotic treatment, clinicians should always test for antibiotic susceptibility. Because resistance to all 3 agents routinely reported by clinical laboratories (ampicillin, trimethoprim-sulfamethoxazole, and fluoroquinolones) is increasingly common [4], it may be necessary to consult an infectious disease specialist to discuss additional susceptibility testing and appropriate treatment options for patients infected with MDR strains.
Acknowledgments.
The authors thank Patsy Kelso, Keeley Weening, Cheryl Achilles, Valarie Devlin (Vermont Department of Health) Kaitlin Tagg, Jean Whichard, Jason Folster, Hayat Caidi, Eshaw Vidyaprakash, Nancy Strockbine, Haley Martin, Michelle Gleason, Gabriella Veytsel, Sarah Collier, and Beth Karp (Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging Zoonotic and Infectious Diseases, Centers for Disease Control and Prevention [CDC]) for assistance with data collection, laboratory testing, and interpretation of findings.
Footnotes
Publisher's Disclaimer: Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. Atlanta, GA: CDC, 2019. [Google Scholar]
- 2.Shane AL, Mody RK, Crump JA, et al. 2017 Infectious Diseases Society of America clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis 2017; 65:e4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Academy of Pediatrics. Shigella infections. In: Pickering LK, Baker CJ, Kimberlin DW, Long SS, eds. Red Book: 2012 Report of the Committee on Infectious Diseases Vol. 2012. Elk Grove Village, IL: American Academy of Pediatrics, 2012:6457. [Google Scholar]
- 4.Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System (NARMS) Now: human data. Available at: https://www.cdc.gov/narmsnow. Accessed 20 April 2021. [Google Scholar]
- 5.Baker KS, Dallman TJ, Ashton PM, et al. Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study. Lancet Infect Dis 2015; 15:91321. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 31st ed. CLSI supplement M100. Wayne, PA: CLSI, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. HAN00411: update—CDC recommendations for managing and reporting Shigella infections with possible reduced susceptibility to ciprofloxacin. Atlanta, GA: CDC, 2018. [Google Scholar]
- 8.Strysko J, Fialkowski V, Marsh Z, et al. Notes from the field: outbreak of multidrug-resistant Shigella sonnei infections in a retirement community—Vermont, OctoberNovember 2018. MMWR Morb Mortal Wkly Rep 2019; 68:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. Antibiotics tested by NARMS. Available at: https://www.cdc.gov/narms/antibiotics-tested.html. Accessed 13 December 2019.
- 10.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 30th ed. CLSI supplement M100 Wayne, PA: CLSI, 2020. [Google Scholar]
- 11.Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67:26404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 2017; 72:27648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt M, Mather AE, Sánchez-Busó L, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom 2017; 3:e000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan WA, Seas C, Dhar U, Salam MA, Bennish ML. Treatment of shigellosis: V. Comparison of azithromycin and ciprofloxacin. A double-blind, randomized, controlled trial. Ann Intern Med 1997; 126:697703. [DOI] [PubMed] [Google Scholar]
- 15.Basualdo W, Arbo A. Randomized comparison of azithromycin versus cefixime for treatment of shigellosis in children. Pediatr Infect Dis J 2003; 22:3747. [PubMed] [Google Scholar]
- 16.Boumghar-Bourtchai L, Mariani-Kurkdjian P, Bingen E, et al. Macrolide-resistant Shigella sonnei. Emerg Infect Dis 2008; 14:12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassing RJ, Melles DC, Goessens WH, Rijnders BJ. Case of Shigella flexneri infection with treatment failure due to azithromycin resistance in an HIV-positive patient. Infection 2014; 42:78990. [DOI] [PubMed] [Google Scholar]
- 18.Houpt ER, Ferdous T, Ara R, et al. Clinical outcomes of drug-resistant shigellosis treated with azithromycin in Bangladesh. Clin Infect Dis 2021; 72:17938. [DOI] [PubMed] [Google Scholar]
- 19.Bennish ML, Salam MA, Khan WA, Khan AM. Treatment of shigellosis: III. Comparison of one- or two-dose ciprofloxacin with standard 5-day therapy. A randomized, blinded trial. Ann Intern Med 1992; 117:72734. [DOI] [PubMed] [Google Scholar]
- 20.Bennish ML, Salam MA, Haider R, Barza M. Therapy for shigellosis. II. Randomized, double-blind comparison of ciprofloxacin and ampicillin. J Infect Dis 1990; 162:7116. [DOI] [PubMed] [Google Scholar]
- 21.Owens RC Jr, Ambrose PG. Antimicrobial safety: focus on fluoroquinolones. Clin Infect Dis 2005; 41(Suppl 2):S14457. [DOI] [PubMed] [Google Scholar]
- 22.Collins JP, Friedman CR, Birhane MG, et al. Evidence of failure of oral third-generation cephalosporin treatment for Shigella sonnei infection. Open Forum Infect Dis 2020; 7:ofaa113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang Z, Zhang J, Ran L, et al. The changing epidemiology of bacillary dysentery and characteristics of antimicrobial resistance of Shigella isolated in China from 20042014. BMC Infect Dis 2016; 16:685. [DOI] [PMC free article] [PubMed] [Google Scholar]


