Abstract
Background.
Lung transplant patients are vulnerable to various forms of allograft injury, whether from acute rejection (AR) (encompassing acute cellular rejection [ACR] and antibody-mediated rejection [AMR]), chronic lung allograft dysfunction (CLAD), or infection (INFXN). Previous research indicates that donor-derived cell-free DNA (dd-cfDNA) is a promising noninvasive biomarker for the detection of AR and allograft injury. Our aim was to validate a clinical plasma dd-cfDNA assay for detection of AR and other allograft injury and to confirm and expand on dd-cfDNA and allograft injury associations observed in previous studies.
Methods.
We measured dd-cfDNA fraction using a novel single-nucleotide polymorphism-based assay in prospectively collected plasma samples paired with clinical-pathologic diagnoses. dd-cfDNA fraction was compared across clinical-pathologic cohorts: stable, ACR, AMR, isolated lymphocytic bronchiolitis, CLAD/neutrophilic-responsive allograft dysfunction (NRAD), and INFXN. Performance characteristics were calculated for AR and combined allograft injury (AR + CLAD/NRAD + INFXN) versus the stable cohort.
Results.
The study included 195 samples from 103 patients. Median dd-cfDNA fraction was significantly higher for ACR (1.43%, interquartile range [IQR]: 0.67%–2.32%, P = 5 × 10−6), AMR (2.50%, IQR: 2.06%–3.79%, P = 2 × 10−5), INFXN (0.74%, IQR: 0.46%–1.38%, P = 0.02), and CLAD/NRAD (1.60%, IQR: 0.57%–2.60%, P = 1.4 × 10−4) versus the stable cohort. Area under the receiver operator characteristic curve for AR versus stable was 0.91 (95% confidence interval [CI]: 0.83-0.98). Using a ≥1% dd-cfDNA fraction threshold, sensitivity for AR was 89.1% (95% CI: 76.2%-100.0%), specificity 82.9% (95% CI: 73.3%-92.4%), positive predictive value, 51.9% (95% CI: 37.5%-66.3%), and negative predictive value, 97.3% (95% CI: 94.3%-100%). For combined allograft injury area under the receiver operator characteristic curve was 0.76 (95% CI: 0.66-0.85), sensitivity 59.9% (95% CI: 46.0%-73.9%), specificity 83.9% (95% CI: 74.1%-93.7%), positive predictive value, 43.6% (95% CI: 27.6%-59.6%), and negative predictive value, 91.0% (95% CI: 87.9%-94.0%).
Conclusions.
These results indicate that our dd-cfDNA assay detects AR and other allograft injury. dd-cfDNA monitoring, accompanied by standard clinical assessments, represents a valuable precision tool to support lung transplant health and is appropriate for further assessment in a prospective randomized-controlled study.
Lung transplantation (LT) is the treatment of choice for patients with a broad spectrum of end-stage chronic pulmonary diseases.1,2 However, long-term lung transplant success rates are poor, largely due to the prevalent and pernicious complication of chronic lung allograft dysfunction (CLAD). The 5-y CLAD-free survival after lung transplantation remains a sobering 50% and about 75% of all lung transplant recipients develop CLAD within 10 y.3 Patients who experience acute cellular rejection (ACR), antibody-mediated rejection (AMR), or allograft dysfunction through other means (eg, infection) are at increased risk of allograft failure. Overall, complications from lung transplantation results in median survival of only 6.7 y. Disappointingly, 5-y lung allograft survival has improved only minimally over the past several decades as reported by the International Society for Heart and Lung Transplantation (ISHLT).4
Surveillance through bronchoscopic assessment with bronchoalveolar lavage (BAL) and transbronchial lung biopsies (TBBxs) is standard of care for lung transplant recipients. However, this procedure lacks sensitivity to detect rejection because of inadequate tissue sampling and inconsistency in histopathologic interpretation.5-7 Indeed, in the multicenter Lung Allograft Rejection Gene Expression Observational study, the interobserver pathologist agreement Kappa score was only 0.183 for ACR.6
Donor-derived cell-free DNA (dd-cfDNA) has emerged as a valuable plasma analyte for detection of allograft injury (acute rejection [AR]) after LT.8-10 De Vlaminck et al10 described the kinetics of dd-cfDNA after lung transplantation and elevation from baseline values during episodes of ACR, AMR, and CLAD. Intriguingly, Keller et al11 observed that higher early posttransplant dd-cfDNA fraction in the setting of primary graft dysfunction predicts an increased risk for CLAD development. Further, elevated dd-cfDNA fraction in association with HLA donor-specific antibodies (DSAs) was reported to precede the clinical-pathologic diagnosis of AMR by approximately 2.8 mo.8 Although such investigations provided insightful observations arguing for a role in implementing dd-cfDNA in care of patients, all required complete exome sequencing of both donor and recipient. This may pose a major impediment to real-world clinical utility, because of the inability to provide rapid turn-around of results to the clinician.
Other investigators have employed single-nucleotide polymorphism panel based dd-cfDNA tests to detect allograft injury, foregoing the need to genotype both donors and recipients. Khush et al12 used a case-control design in which dd-cfDNA tests were paired with histopathologic assessment and reported associations between plasma dd-cfDNA and ACR and CLAD. Significant associations with AMR or infection were not observed, possibly because of limited sample size.12 In a pivotal study offering the first real-world experiences implementing plasma dd-cfDNA for surveillance after LT, Keller et al13 reported high sensitivity and negative predictive value for the combined primary end point of acute lung allograft dysfunction (ie, either AR or infection events). However, the authors emphasized caution in interpretation of these results because of inherent limitations in study design.13
In advance of commencing enrollment into both a prospective, multicenter, noninferiority design, parallel cohort, randomized-controlled trial of dd-cfDNA surveillance versus standard of practice surveillance TBBx (Lung Allograft Monitoring using Blood Dd-cfDNA Assessments [LAMBDA] 001) and a longitudinal observational study for later development of CLAD (LAMBDA 002) (ClinicalTrials.gov modifier: NCT05170425), we endeavored to validate a clinical dd-cfDNA assay in LT recipients that had previously been validated in kidney transplant recipients.14 The test determines the fraction of dd-cfDNA in recipient plasma using a curated panel of >13 000 single-nucleotide polymorphisms and offers real-world utility with rapid turnaround time (approximately 72 h from phlebotomy) and no need for genome sequencing.
We confirm and expand on results from previous studies by assessing the clinical validity of a dd-cfDNA test to assess lung allograft health, with a sufficiently large sample size to detect associations across the full spectrum of early and late rejection and infection. We include diagnostic cohorts based on histopathologic AR, including both ACR and AMR, infection events, and a cohort encompassing either CLAD or neutrophilic-responsive allograft dysfunction (NRAD) as characterized by BAL neutrophilia (≥15%) in the absence of histopathologic diagnoses of AR or infection. We present performance characteristics of the test for detecting AR and other allograft injury.
MATERIALS AND METHODS
Study Design, Setting, and Participants
We prospectively collected plasma samples at a single-center, The Ohio State University (OSU). Sequential outpatient bronchoscopy procedures with contemporaneous clinical data were required for a sample to be included in the study. Routine standard of practice procedures included: chest radiography, office-based spirometry, and routine laboratory and immunogenetic testing for HLA Class I and Class II DSA. Correlations of dd-cfDNA fraction with contemporaneous diagnostic test results (the latter used to assign samples to diagnostic cohorts) were determined, and the performance of dd-cfDNA as a surveillance tool for AR and allograft injury after lung transplantation was assessed. This case-control study was approved by the OSU Institutional Review Board (number 2020H0445). Informed patient consent was obtained for participation.
Bronchoscopy Procedures and dd-cfDNA Testing
Bronchoscopy procedures were performed as either routine surveillance or based on clinical indications in accordance with OSU standards of practice. Fiberoptic bronchoscopy procedures were performed using topical analgesia and general anesthesia in accordance with American College of Chest Physicians guidelines.15,16 BAL was performed per ISHLT guidelines17 with 2-aliquot instillations each of 60 mL sterile normal saline directed to the right middle lobe, lingula, or chest radiographic area of abnormality.
BAL was submitted for routine posttransplant microbiologic studies including cultures for bacteria, fungi and mycobacteria, and a multiplex respiratory viral polymerase chain reaction panel (QiaStat; Qiagen, Hilden, Germany). BAL fluid cytologic analysis for cell count differential was performed in accordance with the American Thoracic Society Clinical Practice Guideline.18
TBBxs were performed with histopathological grading. Adequacy of TBBx for determination of rejection was determined by pathologists with expertise in lung allograft histology, consonant with the ISHLT working formulation (2007).19 TBBx specimens were fixed in formalin and embedded into a paraffin block; 5-μm histologic sections were obtained by serial cutting at 2 levels and immunohistochemical staining performed according to standard criteria (hematoxylin and eosin as well as Gram staining) and, if indicated, for cytomegalovirus, herpes, Pneumocystis jirovecii, Verhoeff’s Elastica, Ziehl-Neelson, and Grocott staining.
Peripheral blood samples were obtained on the same day prior to each fiberoptic bronchoscopy procedure. Blood was collected in 10-mL cell-free DNA Streck tubes. Initial centrifugation was performed on Streck tubes at 2000 × g for 20 min at 22 °C followed by transfer of plasma to the corresponding 15-mL falcon tubes. A second centrifugation was next performed at 3220 × g for 30 min at 22 °C with transfer of plasma from the 15-mL falcon tubes to new 15-mL falcon tubes marked with deidentified sample numbers and storage at −80 °C. Deidentified plasma samples were batch analyzed using the Prospera test (Natera, Inc., Austin, TX) at Natera’s Clinical Laboratory Improvement Amendments-certified, College of American Pathologists-accredited laboratory in San Carlos, CA. Cell-free DNA (cfDNA) was amplified using a massively multiplexed polymerase chain reaction assay targeting a curated panel of >13 000 single-nucleotide polymorphisms designed to maximize variant frequency across ethnicities.14 For each sample, amplicons were sequenced by next-generation sequencing performed on the Illumina NextSeq500 on rapid run with an average of 14 to 15 million reads per sample and sequencing data was processed to estimate the fraction of dd-cfDNA (expressed as a percentage) in relation to total cfDNA.
Diagnostic Cohort Assignments
Treating clinicians, blinded to dd-cfDNA test results, assigned each sample to a clinical-diagnostic cohort based on results from contemporaneous bronchoscopy procedures and clinical information (pulmonary function tests, imaging, and physical exam). Samples from the same patient could be assigned to different diagnostic cohorts. Since the last ISHLT working formulation of AR (2007) had not yet definitively recognized AR as inclusive for AMR, here we defined AR as encompassing both ACR and AMR.19 ACR was graded based on the intensity of perivascular lymphocytic infiltrates in accordance with the revised (2007) ISHLT working formulation.19 Subacute Grade A1 rejection episodes (ie, absence of associated signs or symptoms and without physiologic allograft dysfunction) were not treated by the clinicians and not included in the AR cohort. AMR was classified as clinical (definite, probable, possible) or subclinical in accordance with the ISHLT (2016) consensus statement.18 Chronic rejection included CLAD, further phenotyped as either obstructive or restrictive physiology. NRAD was defined in accordance with ISHLT consensus statement criteria20 and included: forced expiratory volume-1 s <80% of maximal posttransplant value, absence of allograft infection and BAL neutrophil fraction ≥15%.20 The infection (INFXN) cohort was defined based on ISHLT working formulation for definitions of infections in cardiothoracic transplant or based on clinician assessment that antimicrobial treatment was warranted.21 Final groupings for clinical-pathologic cohorts were as follows: (1) AR: either ACR (ISHLT Grades A1, A2-A4) or AMR; (2) INFXN; (3) lymphocytic bronchiolitis (LB; Grades B1R-B2R); (4) CLAD/NRAD; and (5) stable (including samples with untreated, subacute ISHLT Grade A1 ACR on histopathology). dd-cfDNA samples associated with inadequate TBBx tissue procurement, lack of concurrently performed BAL, confounding or comorbid conditions (eg, aspiration pneumonitis, ischemic bronchitis/stricture, cancer), or obtained <28 d posttransplant were excluded from the study.
Statistical Analysis
The distribution of dd-cfDNA was expressed as a median with 25% to 75% interquartile range (IQR) for each diagnostic cohort, with normality assessed by the Kolmogorov-Smirnov test. As lack of normality of dd-cfDNA fraction was detected in the cohorts, comparisons across cohorts were performed using the nonparametric, Mann-Whitney U test. Area under the receiver operator characteristic curve (AUROC) was calculated for both AR and combined allograft injury (defined here as encompassing AR, INFXN, or CLAD/NRAD and excluding LB—see discussion) versus the stable cohort for several dd-cfDNA fraction thresholds using a bootstrapping approach, with 100 000 iterations.22 The bootstrapping approach was used to account for potential dependence between samples that came from the same patient. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated using a dd-cfDNA threshold based on our analysis and comparison to the literature.9 Calculations for PPV and NPV used the study cohort prevalence for AR. In supplementary analyses, we calculated AUROC and accuracy measures for AMR, ACR, and combined rejection (ACR +AMR + CLAD/NRAD) versus the stable cohort. dd-cfDNA fraction was also compared for single lung transplant (SLT) versus bilateral lung transplant (BLT) in stable and AR cohorts (Mann-Whitney U test). We used a multinomial logistic regression model to assess the influence of time since LT (categorized at 1 <6 mo, 6 <12 mo, and ≥12 mo) on the association between dd-cfDNA fraction (entered into the model as a continuous variable) and diagnostic cohort—wherein the multinomial dependent variable included AR, CLAD/NRAD, INFXN, and stable). Differences in cellular profile distribution across diagnostic cohorts was compared using the Kruskal-Wallis test (again due to the lack of normality of these distributions). If the result was significant, Dunn test was used to test for statistical differences between specific cohorts. All statistical analyses were performed with Python V3.9 Statistical Software.
RESULTS
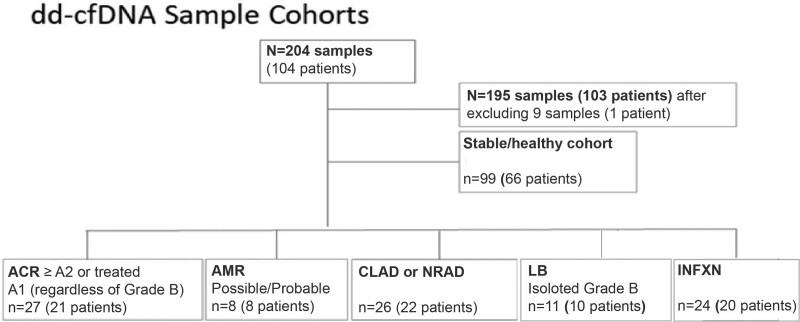
A total of 204 dd-cfDNA samples from 104 LT recipients with paired clinical-pathologic data were collected at OSU from September 2020 through June 2021. Nine samples were excluded from further analysis leaving 195 samples from 103 patients (Figure 1). Reasons for exclusion were lack of adequate TBBx tissue sample (n = 4), lack of BAL microbiology (n = 1), bronchial anastomosis complications (n = 2), aspiration pneumonitis (n = 1), or ambiguous dd-cfDNA fraction related to prior bone marrow transplant (n = 1). Samples were assigned to the following cohorts: ACR (n = 27), AMR (n = 8), CLAD/NRAD (n = 26), INFXN (n = 24), isolated LB (n = 11), and stable (n = 99). For all cohorts combined, the median age was 62.0 (IQR: 55.5–68.0) y and 60% of the patients were male. Interstitial lung disease (category D in Table 1) was the most common indication for transplantation (48.2%). A total of 21.4% of recipients received SLT. Overall, 14.9% of TBBxs were performed for cause and 85.1% performed as routine surveillance. For the AR cohort, these proportions were 22.9% and 77.1%, respectively (Table 1).
FIGURE 1.
Flow chart showing details of diagnostic cohorts. ACR, acute cellular rejection; AMR, antibody-mediated rejection; CLAD, chronic lung allograft dysfunction; dd-cfDNA, donor-derived cell-free DNA; INFXN, infection; LB, lymphocytic bronchiolitis; NRAD, neutrophilic-responsive allograft dysfunction.
TABLE 1.
Patient demographics for clinical-pathologic diagnostic cohorts
| AR | CLAD/NRAD | INFXN | LB | Stable | Total | |
|---|---|---|---|---|---|---|
| Samples (patients) | 35 (29) | 26 (22) | 24 (20) | 11 (10) | 99 (66) | 195 (103) |
| Median age, y (IQR)a | 59 (55.0–67.0) | 60 (55.5–65.8) | 61.5 (56.2–65.2) | 60 (52.5–67.2) | 62 (55.2–68.8) | 62 (55.5–68.0) |
| Gender, n (%)a,b | ||||||
| Male | 20 (69) | 16 (73) | 14 (70) | 3 (30) | 35 (53) | 62 (60) |
| Female | 9 (31) | 6 (27) | 6 (30) | 7 (70) | 31 (47) | 41 (40) |
| Race/ethnicity (%)a | ||||||
| White/Caucasian | 27 (93.1) | 18 (81.8) | 19 (95.0) | 10 (100) | 62 (93.9) | 96 (93.2) |
| Black/AA | 2 (6.9) | 4 (18.2) | 1 (5.0) | 0 (0) | 3 (4.5) | 6 (5.8) |
| Hispanic/Latino | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1.5) | 1 (1.0) |
| Asian | 0 (0) | 0 (0) | 0(0) | 0 (0) | 0 (0) | 0(0) |
| Median time post-LT, d (IQR)a | 194 (39.0–293.0) | 385.5 (198.3–1133.0) | 110.5 (79.0–316.0) | 205 (155.0–369.0) | 184 (95.5–304.0) | 198 (91.5–357.5) |
| Reason for TBBxc | ||||||
| For cause | 8 (22.9) | 14 (53.8) | 3 (12.5) | 0 (0.0) | 4 (4.0) | 29 (14.9) |
| Surveillance | 27(77.1) | 12 (46.2) | 21 (87.5) | 11 (100) | 95 (96.0) | 166 (85.1) |
| Native diseasec (LAS categorical,d %) | ||||||
| A (obstructive) | 12 (34.3) | 10 (38.5) | 12 (50.0) | 9 (81.8) | 41 (41.4) | 84 (43.1) |
| B (vascular) | 2 (5.7) | 2 (7.7) | 1 (4.2) | 0 (0) | 7 (7.1) | 12 (6.2) |
| C (suppurative) | 1 (2.9) | 2 (7.7) | 0 (0) | 0 (0) | 2 (2.0) | 5 (2.6) |
| D (restrictive) | 20 (57.1) | 12 (46.2) | 11 (45.8) | 2 (18.2) | 49 (49.5) | 94 (48.2) |
| Transplant type, n (%)a,b | ||||||
| Single | 2 (6.9) | 4 (18.2) | 7 (35.0) | 2 (20) | 14 (21.2) | 22 (21.4) |
| Bilateral | 27 (93.1) | 18 (81.8) | 13 (65.0) | 8 (80.0) | 52 (78.8) | 81 (78.6) |
aValues reflect patient counts.
bCounts from individual categories do not add to total as samples from the same patient can be classified to different diagnostic categories.
cValues reflect sample counts.
dLAS ISHLT Native Lung Disease Category.
AR, acute rejection; CLAD/NRAD, chronic lung allograft dysfunction or neutrophilic-responsive allograft dysfunction; INFXN, infection; IQR, interquartile range; ISHLT, International Society of Heart and Lung Transplantation; LAS, lung allocation score; LB, lymphocytic bronchiolitis; LT, lung transplantation; TBBx, transbronchial lung biopsiy.
Diagnostic Cohort Comparisons
The ACR cohort contained both treated Grade A1 (n = 17) and Grade A2 (n = 10), with assignment to this cohort made regardless of concurrent histopathologic Grade B status. The AMR cohort samples classified as possible or probable were associated with HLA Class I (n = 2), Class II (n = 5), and concurrent HLA Class I and Class II (n = 1) de novo DSA.
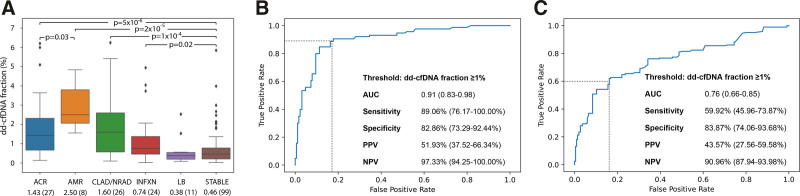
The median time post-LT for the AR and stable cohorts was 194.0 (IQR: 39.0–293.0) and 184.0 (IQR: 95.5–304.0) d, respectively (Table 1) (P = 0.39). The median dd-cfDNA fractions of 1.43% (IQR: 0.67%–2.32%) and 2.50% (IQR: 2.06%–3.79%) for ACR and AMR, respectively, were both significantly higher than that of the stable cohort of 0.46% (IQR: 0.21%–0.78%) (ACR versus stable P = 5 × 10−6, AMR versus stable P = 2 × 10−5, Figure 2A; Figure S1, SDC, http://links.lww.com/TXD/A416). The dd-cfDNA fraction for AMR was approximately 2-fold higher than ACR (P = 0.03) (Figure 2A; Figure S1, SDC, http://links.lww.com/TXD/A416).
FIGURE 2.
Performance characteristics of donor-derived cell-free DNA (dd-cfDNA) to discriminate clinical-pathologic cohorts. A, Relationship between dd-cfDNA fraction (%) (Y-axis) and clinical-pathologic diagnostic cohorts (X-axis): acute cellular rejection (ACR), antibody-mediated rejection (AMR), chronic lung allograft dysfunction or neutrophilic-responsive allograft dysfunction (CLAD/NRAD), allograft infection (INFXN), isolated lymphocytic bronchiolitis (LB), and stable cohort (STABLE). The box and whisker plots show median (horizontal line), interquartile range (IQR, box), minimum and maximum whiskers (first quartile − 1.5 × IQR, third quartile + 1.5 × IQR) and outliers (dots). The X-axis also shows median and (n). B, Receiver operator characteristic (ROC) curve for acute rejection (AR) cohort (ACR + AMR) vs the stable cohort. C, ROC curve for combined allograft injury (ACR, AMR, CLAD/NRAD, and INFXN) vs the stable cohort. Positive predictive value (PPV) and negative predictive value (NPV) calculated using cohort prevalence of 17.2%.
All isolated LBs were Grade B1R. The median time since transplant was 205 (IQR: 155.0–369.0) d (Table 1). LB, in the absence of either concurrent AR or INFXN, demonstrated median dd-cfDNA fraction of 0.38% (IQR: 0.20%–0.55%), which was not statistically different from the stable cohort (P = 0.56, Figure S1, SDC, http://links.lww.com/TXD/A416).
Median time posttransplant for the INFXN cohort was 110.5 (IQR: 79.0–316.0) d (Table 1). This cohort had a mild but statistically significant elevation in median dd-cfDNA fraction of 0.74% (IQR: 0.46%–1.38%) compared with the stable cohort (P = 0.02, Figure 2A; Figure S1, SDC, http://links.lww.com/TXD/A416). Isolated BAL pathogens in the INFXN cohort included: Pseudomonas aeruginosa (N = 9), Staphylococcus aureus (N = 3), Streptococcus viridans (N = 2), Enterobacter cloacae complex (N = 1), Enterococcus (N = 3), Cytomegalovirus (N = 1), Adenovirus (N = 1), Enterovirus/Rhinovirus (N = 6), and Mycobacterium mucogenicum (N = 1) (27 infection events in total as >1 infection could be counted per sample).
The median time posttransplant for the CLAD/NRAD cohort was 385.5 (IQR: 198.3–1133.0) d (Table 1). Median dd-cfDNA fraction for this cohort (1.60%; IQR: 0.57%–2.60%) was significantly higher than the stable cohort (P = 0.0001, Figure 2A; Figure S1, SDC, http://links.lww.com/TXD/A416). Cell count fractional median BAL neutrophilia showed significant variation across cohorts (P = 1 × 10−5). Direct comparison of specific cohorts found significant differences for AR (median 4.0%; IQR: 2.0%–18.0%), CLAD/NRAD (median 9.0%; IQR: 3.0%–19.0%), and INFXN (median 8.5%; IQR: 1.5%–36.2%) relative to the stable cohort (median 2.0%; IQR: 1.0%–4.0%; P < 0.05 for all comparisons) (Table 2). The eosinophil fraction also showed significant variation across cohorts (P = 0.03), with a small but significant elevation in the CLAD cohort compared with the stable cohort (P = 0.04) (Table 2). To our knowledge, these are the first reported results of analyses comparing BAL cellular profiles with dd-cfDNA fractions after lung transplant.
TABLE 2.
Bronchoalveolar lavage cellular profiles for clinical-pathologic diagnostic cohorts (medians and IQR)
| Acute rejection | CLAD/NRAD | INFXN | LB | Stable | Total | |
|---|---|---|---|---|---|---|
| Neutrophils (%) | 4.0 | 9.0 | 8.5 | 2.0 | 2.0 | 3.0 |
| (IQR) | (2.0-18.0) | (3.0-19.0) | (1.5-36.2) | (1.0-3.5) | (1.0-4.0) | (1.0-8.0) |
| Lymphocytes (%) | 3.0 | 4.0 | 2.0 | 3.0 | 3.0 | 3.0 |
| (IQR) | (1.0-6.0) | (1.0-13.0) | (1.0-3.8) | (2.0-7.0) | (1.0-5.0) | (1.0-6.0) |
| Eosinophils (%) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| (IQR) | (0.0-0.0) | (0.0-1.0) | (0.0-0.0) | (0.0-0.0) | (0.0-0.0) | (0.0-0.0) |
CLAD/NRAD, chronic lung allograft dysfunction or neutrophilic-responsive allograft dysfunction; INFXN, infection; IQR, interquartile range; LB, lymphocytic bronchiolitis.
The AMR cohort had higher dd-cfDNA fraction than individual allograft injury-related cohorts with significant differences found for AMR versus ACR (P = 0.03), AMR versus INFXN (P = 0.001), and AMR versus LB (P = 0.001) (Figure 2A; Figure S1, SDC, http://links.lww.com/TXD/A416). In addition to AMR, LB had significantly lower dd-cfDNA fraction than other injury-related cohorts (ACR versus LB: P = 0.01, CLAD versus LB: P = 0.01) (Figure S1, SDC, http://links.lww.com/TXD/A416). No other significant differences were found in comparisons of cohorts with graft injury, but modest sample sizes for some cohorts limited the power for detecting small differences in dd-cfDNA fraction.
Multinomial Logistic Model Including dd-cfDNA Fraction and Time Since Transplant as Covariates
We found that time after LT significantly influenced the risk of CLAD/NRAD (P = 0.0005) but not any of the other dependent variables (AR, INFXN, stable) representing diagnostic category that were included in the multinomial model (P = 0.55–0.95). As with our unadjusted analyses presented above, dd-cfDNA fraction was associated with diagnostic cohort in the time-adjusted model (AR: P = 2.2 × 10−6; CLAD/NRAD: P = 1.7 × 10−5; INFXN: P = 0.01, using stable as the reference category for the dependent multinomial variable). Overall accuracy of the model was 0.59 and the likelihood-ratio test for comparison to the intercept only model was significant (P = 5.1 × 10−10).
dd-cfDNA Test Performance
A dd-cfDNA threshold of ≥1.0% was chosen to discriminate both AR and combined allograft injury from the stable cohort as this cutoff was consonant with prior publications from the GRAfT consortium9 and test performance characteristics in the range of 1.0% to 1.25% were similar (Table S1, SDC, http://links.lww.com/TXD/A416). AUROC and performance characteristics for our primary comparison groups (AR versus stable and combined allograft injury versus stable) are shown in Figure 2B and C. For the AR versus stable cohort the AUROC was 0.91 (95% confidence interval [CI]: 0.83%-0.98%). The sensitivity and specificity to detect AR was 89.1% (95% CI: 76.2%-100.0%) and 82.9% (95% CI: 73.3%-92.4%), respectively. PPV was calculated as 51.9% (95% CI: 37.5%-66.3%), and NPV 97.3% (95% CI: 94.3%-100.0%) using the study cohort prevalence for AR of 17.2%.23 For combined allograft injury (ACR + AMR + CLAD/NRAD + INFXN) AUROC was 0.76 (95% CI: 0.66-0.85), sensitivity 59.9% (95% CI: 46.0%-73.9%), and specificity 83.9% (95% CI: 74.1%-93.7%). Results for additional receiver operating characteristic analyses—ACR versus stable, AMR versus stable, and combined rejection (ACR + AMR + CLAD/NRAD) versus stable—are shown in Table S2 (SDC, http://links.lww.com/TXD/A416).
dd-cfDNA Fraction Lower in SLT
We explored differences in dd-cfDNA fractions in SLT and BLT patients in stable and AR cohorts. The median dd-cfDNA fraction in the stable cohort was 2.7-fold higher for BLT (0.56%, IQR: 0.31%–0.87%, n = 76) than SLT (0.21%, IQR: 0.11%–0.37%, n = 23) (P < 0.0001). The median dd-cfDNA fraction for the 4 episodes of AR in the SLT group (0.74%, IQR: 0.19%–1.31%, n = 4) was lower than in BLT (1.98%, IQR: 1.10%–2.87%, n = 31). Performance of the cohort was calculated using a 2× factor for dd-cfDNA in SLT patients, as in prior studies24 and performance numbers (Table S3, SDC, http://links.lww.com/TXD/A416) were similar to that from our main analysis (Table S1, SDC, http://links.lww.com/TXD/A416; Figure 2B and C).
DISCUSSION
In our study, we analyzed the plasma dd-cfDNA fraction across a spectrum of acute to chronic lung allograft rejection and infection events. Our principal finding is that elevated dd-cfDNA is strongly associated with AR. Further, our dd-cfDNA test detected both forms of AR, with median dd-cfDNA fraction approximately 3-fold elevated in the ACR cohort and 5-fold elevated for AMR, relative to the stable healthy LT cohort. This is in line with results from a previous study that used a donor-recipient based genome sequencing test.25 The performance of this dd-cfDNA assay to differentiate AR from stable cases was excellent, with high AUROC (0.91), sensitivity (89%), specificity (83%), PPV (52%), and NPV (97%) using a 1% dd-cfDNA cutoff, whereas prevalence of AR in our study was similar to that reported in Scientific Registry of Transplant Recipients data.23
Second, the dd-cfDNA fraction of the cohort characterized by airway neutrophilia, representing the likely continuum of NRAD and CLAD,26 was approximately 3-fold elevated compared with the stable cohort. We believe that an assessment of allograft injury by dd-cfDNA longitudinal surveillance may ultimately represent a valuable biomarker by indicating the presence of NRAD, CLAD, or risk factors such as INFXN.27 An extensive review by Kennedy et al28 provides a compelling argument for an association of BAL neutrophilia and interleukin (IL)-8 with development of CLAD.28 A novel finding in our study was a highly significant association between cohort type and BAL cellular profile, with cohorts with elevated dd-cfDNA levels having higher BAL neutrophilia (ie, AR, INFXN, CLAD/NRAD). We speculate that increased airway neutrophilia and associated IL-8 and IL-17 elaboration may contribute to resultant graft injury, reflected by elevated dd-cfDNA.12 Alternatively, circulating dd-cfDNA per se, whereas acting by damage-associated molecular pattern signaling and pattern recognition receptors, such as Toll-like receptor-9, may further contribute to innate immune system activation, cytokine elaboration, and cell-death pathways.29
Third, the diagnostic cohort representing allograft infection (in the absence of rejection) demonstrated a mild but statistically significant elevation in dd-cfDNA fraction. A previous study using a panel-based single-nucleotide polymorphism test did not observe this association.12 However, in another study from the GRAfT consortium and National Heart, Lung, and Blood Institute, Jang et al25 reported that median dd-cfDNA level during putative infection was elevated when associated with abnormal histopathologic findings (1.55%) or physiologic impairment (1.61%) but not when both were absent (0.53%). Our findings can also be considered consonant with a recent study by Bazemore et al30 where in variability in plasma dd-cfDNA fraction related more to the identity of the specific isolated BAL pathogens and their associated risk for CLAD rather than their mere presence.
Fourth, we observed that median dd-cfDNA fraction in the isolated LB (ISHLT Grade: B1R) cohort was not statistically distinguishable from the stable cohort. LB has been demonstrated to be an independent risk factor for bronchiolitis obliterans syndrome and would be expected to correlate with cellular injury.31 We were surprised not to see elevated dd-cfDNA fraction in patients with LB, and suspect that limited sample size and variability in extent of cellular injury and heterogeneity in histopathology may explain our finding.32
Fifth, dd-cfDNA was also found to predict a combined measure for allograft injury, which included AR, CLAD/NRAD, and INFXN (we did not include LB since this cohort’s dd-cfDNA levels were not statistically different from stable samples and LB is considered a nonspecific pattern as an isolated histologic finding). Performance characteristics for our combined injury cohort indicated that dd-cfDNA fraction distinguished allograft dysfunction from the stable cohort with an AUROC of 0.76. Using a dd-cfDNA threshold of ≥1.0%, the sensitivity and specificity were 60% and 84%, respectively. Performance of dd-cfDNA using other thresholds is presented in Table S1 (SDC, http://links.lww.com/TXD/A416).
We investigated whether time since transplant might confound the association between dd-cfDNA fraction and allograft injury, by including time as a covariate in a logistic regression model with a multinomial dependent variable that included AR, INFXN, CLAD/NRAD, and stable cohort membership. There was a statistically significant association between time since transplant and CLAD/NRAD, which should be expected since CLAD is generally a late complication following transplant. However, associations between dd-cfDNA and allograft injury were shown to be robust as significant associations for AR, CLAD/NRAD, and INFXN observed in our primary analysis remained, despite inclusion of the time covariate in the model.
Seeking to understand the effect of SLT versus BLT on dd-cfDNA levels, we explored the merits of implementing a correction or multiplier for SLT samples. Prior reports recommending a 2x-multiplier for dd-cfDNA were theoretically based on anticipated differences in total lung mass for SLT versus BLT allografts.9,10,12,33 Intuitively, correction for SLT may be considered as appropriate, however, in nontransplant studies after surgical pneumonectomy the potential for lung hypertrophy and hyperplasia has been well established.34,35 We found that the dd-cfDNA fraction was significantly higher (2.7-fold) in BLT than SLT in stable patients. Although insufficient sample size for AR in SLT samples limited our analysis, we conclude that implementing a 2x-multiplier for SLT samples would be appropriate in order to optimize sensitivity for these types of patients. Future analyses derived from robust multicenter studies would be appropriate. Here, the performance of dd-cfDNA when using the 2x-multiplier for SLT patients was similar to the performance for our main (uncorrected) analyses.
Limitations to our study included a single-center experience with limited sample size, particularly for some of the diagnostic cohorts. Additionally, our study lacked longitudinal dd-cfDNA data, precluding an analysis of intrasubject coefficient of variation or dd-cfDNA kinetics during courses of treatment for clinical events such as AR or infection. Finally, a vexing challenge to appropriate dd-cfDNA validation relates to the inherent lack of a definitive gold standard for clinical-pathological adjudication. Reliance on TBBx histopathology in isolation for assignment to diagnostic cohorts is limited by issues of sensitivity, specificity, and interobserver pathologist interpretation.36 It is possible that misclassification between diagnostic cohorts could have influenced results of our analyses. Future studies incorporating outcome measures may shed additional light on the relationship of dd-cfDNA, AR and graft injury, and histopathology, not to mention more speculative markers such as intragraft gene expression profile signatures37 and epigenetic signatures such as DNA methylation.38 Nevertheless, our study design and sample size permits insights regarding dd-cfDNA fractions and performance of a clinical assay across diverse types of LT allograft injury and health. These data provide a path forward in advance of an anticipated launch of a prospective, multicenter, noninferiority design, parallel cohort, randomized-controlled trial of dd-cfDNA surveillance versus standard of practice surveillance TBBx (LAMBDA 001) in LT and a companion longitudinal observational study to assess development of CLAD (LAMBDA 002) (ClinicalTrials.gov modifier: NCT05170425). Based on our current data and that of GRAfT,13,25 an algorithm implementing a 2x correction multiplier of dd-cfDNA in SLT samples, should be included in these future trials and further assessed.
We conclude that plasma dd-cfDNA is a promising precision biomarker in lung transplantation that can complement routine clinical assessments for evaluating lung allograft health.
Supplementary Material
Footnotes
This work was funded by an investigator-initiated grant from Natera Inc. and, in part, by CTSA Grant number UL1TR002733.
A.B., B.G.Z., D.J.R., E.A., G.F., J.S., N.L., M.O., R.S., Z.P.D., Y.-A.C., and P.R.B. are full-time employees at Natera Inc. with stocks or options to own stocks in the company. B.C.K. serves as a consultant to and on the speaker bureau for CareDx, Inc. and has received research funding from CareDx, Natera, and Zambon. J.P.R. has received research funding from Natera. The other authors declare no conflicts of interest.
A.B., B.C.K., B.G.Z., D.J.R., E.A., J.P.R., M.O., N.L., P.R.B., R.S., and Z.P.D. contributed to study design. B.C.K., J.P.R., and J.S. contributed to data acquisition. A.B., B.C.K., B.G.Z., D.J.R., A.W., G.F., E.A., J.S., J.P.R., M.B., M.O., N.L., R.S., and Y.C. contributed to data analysis and interpretation. B.C.K., D.J.R., G.F., J.P.R., and Z.P.D. contributed to drafting of the manuscript. A.W., B.C.K., B.G.Z., D.J.R., G.F., J.P.R., J.S., M.B., M.O., P.R.B., Y.C., and Z.P.D. were involved in revising the manuscript. B.C.K., D.J.R., J.P.R., and P.R.B. provided final approval.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantationdirect.com).
REFERENCES
- 1.Blumenthal NP, Petty MG, McCorkle R. Missing domains of lung transplant patient selection. Prog Transplant. 2017;27:90–97. [DOI] [PubMed] [Google Scholar]
- 2.Weill D, Benden C, Corris PA, et al. A consensus document for the selection of lung transplant candidates: 2014–an update from the Pulmonary Transplantation Council of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2015;34:1–15. [DOI] [PubMed] [Google Scholar]
- 3.Chambers DC, Cherikh WS, Goldfarb SB, et al. ; International Society for Heart and Lung Transplantation. The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: thirty-fifth adult lung and heart-lung transplant report-2018; Focus theme: multiorgan transplantation. J Heart Lung Transplant. 2018;37:1169–1183. [DOI] [PubMed] [Google Scholar]
- 4.Chambers DC, Cherikh WS, Harhay MO, et al. ; International Society for Heart and Lung Transplantation. The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: thirty-sixth adult lung and heart-lung transplantation Report-2019; Focus theme: donor and recipient size match. J Heart Lung Transplant. 2019;38:1042–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy L, Huszti E, Pal P, et al. The impact of inadequate (“AX”) transbronchial biopsies on post-lung transplant CLAD or death. Transplantation. 2021;105:390–395. [DOI] [PubMed] [Google Scholar]
- 6.Arcasoy SM, Berry G, Marboe CC, et al. Pathologic interpretation of transbronchial biopsy for acute rejection of lung allograft is highly variable. Am J Transplant. 2011;11:320–328. [DOI] [PubMed] [Google Scholar]
- 7.Loor K, Culebras M, Sansano I, et al. Optimization of transbronchial cryobiopsy in lung transplant recipients. Ann Thorac Surg. 2019;108:1052–1058. [DOI] [PubMed] [Google Scholar]
- 8.Agbor-Enoh S, Jackson AM, Tunc I, et al. Late manifestation of alloantibody-associated injury and clinical pulmonary antibody-mediated rejection: evidence from cell-free DNA analysis. J Heart Lung Transplant. 2018;37:925–932. [DOI] [PubMed] [Google Scholar]
- 9.Agbor-Enoh S, Wang Y, Tunc I, et al. Donor-derived cell-free DNA predicts allograft failure and mortality after lung transplantation. Ebiomedicine. 2019;40:541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Vlaminck I, Martin L, Kertesz M, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A. 2015;112:13336–13341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller M, Bush E, Diamond JM, et al. Use of donor-derived-cell-free DNA as a marker of early allograft injury in primary graft dysfunction (PGD) to predict the risk of chronic lung allograft dysfunction (CLAD). J Heart Lung Transplant. 2021;40:488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khush KK, De Vlaminck I, Luikart H, et al. Donor-derived, cell-free DNA levels by next-generation targeted sequencing are elevated in allograft rejection after lung transplantation. ERJ Open Res. 2021;7:00462–02020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller M, Sun J, Mutebi C, et al. Donor-derived cell-free DNA as a composite marker of acute lung allograft dysfunction in clinical care. J Heart Lung Transplant. 2022In press. [DOI] [PubMed] [Google Scholar]
- 14.Sigdel TK, Archila FA, Constantin T, et al. Optimizing detection of kidney transplant injury by assessment of donor-derived cell-free DNA via massively multiplex PCR. J Clin Med. 2018;8:E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sampsonas F, Kakoullis L, Karampitsakos T, et al. Bronchoscopy during the COVID-19 pandemic: effect on current practices and strategies to reduce procedure-associated transmission. Expert Rev Respir Med. 2021;15:773–779. [DOI] [PubMed] [Google Scholar]
- 16.Wahidi MM, Jain P, Jantz M, et al. American College of Chest Physicians consensus statement on the use of topical anesthesia, analgesia, and sedation during flexible bronchoscopy in adult patients. Chest. 2011;140:1342–1350. [DOI] [PubMed] [Google Scholar]
- 17.Martinu T, Koutsokera A, Benden C, et al. ; Bronchoalveolar Lavage Standardization Workgroup. International Society for Heart and Lung Transplantation consensus statement for the standardization of bronchoalveolar lavage in lung transplantation. J Heart Lung Transplant. 2020;39:1171–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine DJ, Glanville AR, Aboyoun C, et al. Antibody-mediated rejection of the lung: a consensus report of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2016;35:397–406. [DOI] [PubMed] [Google Scholar]
- 19.Stewart S, Fishbein MC, Snell GI, et al. Revision of the 1996 working formulation for the standardization of nomenclature in the diagnosis of lung rejection. J Heart Lung Transplant. 2007;26:1229–1242. [DOI] [PubMed] [Google Scholar]
- 20.Verleden GM, Glanville AR, Lease ED, et al. Chronic lung allograft dysfunction: definition, diagnostic criteria, and approaches to treatment-A consensus report from the Pulmonary Council of the ISHLT. J Heart Lung Transplant. 2019;38:493–503. [DOI] [PubMed] [Google Scholar]
- 21.Husain S, Mooney ML, Danziger-Isakov L, et al. ; ISHLT Infectious Diseases Council Working Group on Definitions. A 2010 working formulation for the standardization of definitions of infections in cardiothoracic transplant recipients. J Heart Lung Transplant. 2011;30:361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Efron B, Tibshirani R. Bootsrap methods for standard errors confidence intervals and other measures of statistical accuracy. Stat Sci. 1986;1:54–77. [Google Scholar]
- 23.Velapour M, Lehr CJ, Skeans MA, et al. OPTN/SRTR 2019 annual data report: lung. Am J Transplant. 2021;21(suppl 2):441–520. [DOI] [PubMed] [Google Scholar]
- 24.Agbor-Enoh S, Tunc I, De Vlaminck I, et al. Applying rigor and reproducibility standards to assay donor-derived cell-free DNA as a non-invasive method for detection of acute rejection and graft injury after heart transplantation. J Heart Lung Transplant. 2017;36:1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jang MK, Tunc I, Berry GJ, et al. Donor-derived cell-free DNA accurately detects acute rejection in lung transplant patients, a multicenter cohort study. J Heart Lung Transplant. 2021;40:822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neurohr C, Huppmann P, Samweber B, et al. ; Munich Lung Transplant Group. Prognostic value of bronchoalveolar lavage neutrophilia in stable lung transplant recipients. J Heart Lung Transplant. 2009;28:468–474. [DOI] [PubMed] [Google Scholar]
- 27.Verleden SE, Vandermeulen E, Ruttens D, et al. Neutrophilic reversible allograft dysfunction (NRAD) and restrictive allograft syndrome (RAS). Semin Respir Crit Care Med. 2013;34:352–360. [DOI] [PubMed] [Google Scholar]
- 28.Kennedy VE, Todd JL, Palmer SM. Bronchoalveolar lavage as a tool to predict, diagnose and understand bronchiolitis obliterans syndrome. Am J Transplant. 2013;13:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuji N, Agbor-Enoh S. Cell-free DNA beyond a biomarker for rejection: biological trigger of tissue injury and potential therapeutics. J Heart Lung Transplant. 2021;40:405–413. [DOI] [PubMed] [Google Scholar]
- 30.Bazemore K, Rohly M, Permpalung N, et al. Donor derived cell free DNA% is elevated with pathogens that are risk factors for acute and chronic lung allograft injury. J Heart Lung Transplant. 2021;40:1454–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glanville AR, Aboyoun CL, Havryk A, et al. Severity of lymphocytic bronchiolitis predicts long-term outcome after lung transplantation. Am J Respir Crit Care Med. 2008;177:1033–1040. [DOI] [PubMed] [Google Scholar]
- 32.Ross DJ, Marchevsky A, Kramer M, et al. “Refractoriness” of airflow obstruction associated with isolated lymphocytic bronchiolitis/bronchitis in pulmonary allografts. J Heart Lung Transplant. 1997;16:832–838. [PubMed] [Google Scholar]
- 33.Sayah D, Weigt SS, Ramsey A, et al. Plasma donor-derived cell-free DNA levels are increased during acute cellular rejection after lung transplant: pilot data. Transplant Direct. 2020;6:e608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laros CD, Westermann CJ. Dilatation, compensatory growth, or both after pneumonectomy during childhood and adolescence. A thirty-year follow-up study. J Thorac Cardiovasc Surg. 1987;93:570–576. [PubMed] [Google Scholar]
- 35.Hsia CC, Herazo LF, Ramanathan M, et al. Cardiopulmonary adaptations to pneumonectomy in dogs. IV. Membrane diffusing capacity and capillary blood volume. J Appl Physiol (1985). 1994;77:998–1005. [DOI] [PubMed] [Google Scholar]
- 36.Bhorade SM, Husain AN, Liao C, et al. Interobserver variability in grading transbronchial lung biopsy specimens after lung transplantation. Chest. 2013;143:1717–1724. [DOI] [PubMed] [Google Scholar]
- 37.Halloran K, Parkes MD, Timofte I, et al. Molecular T-cell–mediated rejection in transbronchial and mucosal lung transplant biopsies is associated with future risk of graft loss. J Heart Lung Transplant. 2020;39:1327–1337. [DOI] [PubMed] [Google Scholar]
- 38.Andargie TE, Tsuji N, Seifuddin F, et al. Cell-free DNA maps COVID-19 tissue injury and risk of death and can cause tissue injury. JCI Insight. 2021;6:147610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.