Abstract
To understand the chemical basis of action for the PDR5-encoded multidrug resistance transporter of Saccharomyces cerevisiae, we compared the relative hypersensitivities of the wild-type (RW2802) and null mutant strains toward a series of tri-n-alkyltin compounds. These compounds differ from each other in a systematic fashion—either by hydrocarbon chain length or by anion composition. Using zone-of-inhibition and fixed-concentration assays, we found that the ethyl, propyl, and butyl compounds are strong PDR5 substrates, whereas the methyl and pentyl compounds are weak. We conclude that hydrophobicity and anion makeup are relatively unimportant factors in determining whether a tri-n-alkyltin compound is a good PDR5 substrate but that the dissociation of the compound and the molecular size are significant.
The yeast PDR5 gene encodes a 160-kDa protein that is a member of the ATP-binding cassette transport superfamily (1). Loss-of-function mutations in this gene create broad-spectrum hypersensitivity to a large array of chemically diverse inhibitors because of an inability to cause efflux of such compounds (11). Overexpression of the PDR5 gene product, in contrast, results in multidrug resistance (MDR) (1, 14). As is the case for most of the other MDR proteins encountered in eucaryotes, the chemical basis for the broad specificities of the PDR5 transporter remains unknown. Precise knowledge of the mechanism by which transporters recognize their substrates might have important clinical ramifications. Furthermore, it could help explain the basis for the interesting classes of MDR mutants with altered specificities that have been identified for yeast (3) and mammalian (6, 12) cells.
Most models of MDR action invoke the requirement of hydrophobicity (5). This idea is based upon the fact that several structurally related drugs that differ in their ability to be transported by the mammalian MDR transporter differ in their relative hydrophobicities, as measured by their water/octanol partitioning ratio (logP) (15, 18). This observation was used as evidence for proposing that the mammalian MDR protein is a flippase. Thus, a drug must be intercalated into the lipid bilayer before it interacts with a binding site on the efflux protein (5). To address various models of PDR5 substrate recognition, we analyzed the abilities of the wild-type and isogenic null mutant proteins to mediate resistance toward a family of structurally related tri-n-alkyltin compounds that interfere with mitochondrial ATPase activity (2) and differ systematically either by the length of the hydrocarbon chain or by the counterion. Zone-of-inhibition and fixed-concentration assays were used so that quantitative comparisons could be made with a high degree of accuracy. The results obtained were compared to measures of hydrophobicity, ion dissociation, and molecular size.
MATERIALS AND METHODS
Yeast strains.
The two isogenic strains of Saccharomyces cerevisiae used in this study were previously described (11). RW2802 contains a functional PDR5 gene, while JG436 has a large Tn5 insertion mutation in the promoter region and makes no detectable transcript (14). The SIN4 (DY150) and sin4::URA3 (DY1704) strains were generously provided by David Stillwell and are described elsewhere (7). Isogenic strains bearing snq2::URA3 disruptions were constructed by transforming RW2802 and JG436 to create PDR5 snq2::URA3 (JG545) and pdr5::Tn5 snq2::URA3 (JG546) mutants, respectively. To do this, 1 μg of pAE9 DNA, kindly provided by Scott Moye-Rowley, was linearized with SacI and SalI prior to transformation of yeast cells with a Gietz Lab Transformation Kit (Tetra Link, Amherst, N.Y.). The resulting chromosomal disruptions were verified by Southern hybridization. In addition, transformants were tested for hypersensitivity toward 4-nitroquinoline-n-oxide, which is characteristic of snq2 but not pdr5 mutants. The transformant strains selected exhibited this phenotype. Thus, both strains bearing snq2::URA3 disruptions were hypersensitive to 4-nitroquinoline-n-oxide when compared to the isogenic, wild-type strain (RW2802) and the pdr5 mutant strain (JG436). Once the constructions were verified, a single transformant of each strain (JG436, JG545, and JG546) was used in all of the experiments described below. For all of the assays, saturated cultures of yeasts grown in liquid medium were used.
Tri-n-alkyltin compounds and other inhibitors.
Triphenyltin chloride, tri-n-pentyltin chloride, and tri-n-butyltin chloride, acetate, and bromide were purchased from Aldrich (Milwaukee, Wis.). Tri-n-ethyltin chloride was obtained from Stem Chemicals (Newburyport, Mass.). Tri-n-methyltin chloride was purchased from Organometallics, Inc. (Hampstead, N.H.). Tri-n-propyltin chloride was obtained from Alfa (Ward Hill, Mass.). Chloramphenicol, clotrimazole, and cycloheximide were all purchased from Sigma Chemical Company (St. Louis, Mo.). No further purification of these compounds was done. The compounds were dissolved in dimethyl sulfoxide (DMSO) (Sigma). When DMSO was applied by itself in the concentrated form, no zone of inhibition was observed with any strain.
Zone-of-inhibition assays.
The degree of resistance toward the tri-n-alkyltin compounds was determined quantitatively by use of a previously described zone-of-inhibition assay (14). Each determination (see Table 1) is the average for at least three samples. For each sample, 0.2 ml of culture grown in yeast-peptone-dextrose medium (about 107 cells) was mixed with 4 ml of 1% agar (melted in doubly deionized water and autoclaved prior to use), and the mixture was plated on yeast-peptone-glycerol (YPG) medium (containing, per liter, 20 g of peptone, 10 g of yeast extract, 20 ml of glycerol, and 10 g of agar, except for testing resistance to clotrimazole and cycloheximide, for which 20 g of dextrose was used instead of glycerol). YPG plates were incubated for 72 h at 30°C before measurement. Dextrose plates were scored at 48 h following incubation at 30°C. YPG plates were used for the tri-n-alkyltin compounds because these inhibitors act on the mitochondrial ATPase and glycerol is nonfermentable. Dextrose plates were used for nonmitochondrial inhibitors because they permit faster scoring. Very similar results are obtained, however, when glycerol plates are used (J. Golin, unpublished observations).
TABLE 1.
Zones of inhibition with tri-n-alkyltin compounds for RW2802 (PDR5) and JG436 (pdr5)a
| Chloride compound | Concn (mmol) | Zone of inhibition (cm) for:
|
Ratio of zone for JG436 to zone for RW2802 | |
|---|---|---|---|---|
| RW2802 | JG436 | |||
| Trimethyltin | 0.5 | 2.0 ± 0.1 | 2.0 ± 0.1 | 1.0 ± 0.1 |
| 1.0 | 2.5 ± 0.1 | 2.5 ± 0.1 | 1.0 ± 0.1 | |
| 3.0b | 3.5 ± 0.1 | 3.7 ± 0.2 | 1.0 ± 0.0 | |
| Triethyltin | 0.2 | 2.2 ± 0.3 | 2.8 ± 0.2 | 1.3 ± 0.2 |
| 1.6 | 2.5 ± 0.2 | 3.3 ± 0.1 | 1.3 ± 0.1 | |
| 20 | 3.5 ± 0.2 | 4.1 ± 0.2 | 1.2 ± 0.1 | |
| Tripropyltin | 0.005 | 1.8 ± 0.1 | 3.3 ± 0.1 | 1.8 ± 0.1 |
| 0.02b | 2.4 ± 0.4 | 3.8 ± 0.8 | 1.6 ± 0.4 | |
| 0.05 | 3.3 ± 0.0 | 5.0 ± 0.1 | 1.5 ± 0.1 | |
| Tributyltin | 0.36c | 2.0 ± 0.3 | 2.7 ± 0.4 | 1.4 ± 0.3 |
| 1.08 | 2.5 ± 0.1 | 3.2 ± 0.1 | 1.3 ± 0.1 | |
| 3.6b | 3.3 ± 0.0 | 4.5 ± 0.2 | 1.4 ± 0.1 | |
| Tripentyltin | 0.24b | 2.2 ± 0.1 | 2.5 ± 0.1 | 1.1 ± 0.1 |
| 0.48d | 2.3 ± 0.3 | 2.6 ± 0.4 | 1.1 ± 0.2 | |
Zones and ratios are given as means ± standard errors of the means.
Average of two assays.
Average of three assays.
Average of four assays.
Fixed-concentration assays.
To make medium with a known concentration of inhibitor, YPG medium was prepared as described above and autoclaved. Following sterilization, a particular tri-n-alkyltin inhibitor was dissolved in DMSO to a desired concentration, and the mixture was added to medium that had been cooled to 60°C in a water bath. The medium was then poured into petri dishes. To test cells for the ability to grow on medium with a particular concentration of inhibitor, overnight cultures of cells were diluted to 5 × 106 cells. Two samples (10 μl) of the dilution were applied to the petri dishes. The dishes were incubated for 48 h at 30°C and scored for the presence or absence of growth.
Calculation of logP.
The logP was calculated by use of the ClogP program obtained from BioByte Corporation (Claremont, Calif.). The method used to arrive at ClogP is described elsewhere (10). It should be noted that for the tri-n-alkyltin compounds tested there is good agreement between ClogP and experimentally determined values (see Table 5) (17).
TABLE 5.
logP values and ratios of zones of inhibition with pertinent inhibitors for RW2802 (PDR5) and JG436 (pdr5)
| Compound | ClogP | MlogPa | Zone of inhibition (cm)b for:
|
Ratio of zone for JG436 to zone for RW2802 | |
|---|---|---|---|---|---|
| RW2802 | JG436 | ||||
| Tri-n-alkyltin | |||||
| Trimethyltin chloride | −0.512 | −0.292 | 1.0 ± 0.1 | ||
| Triethyltin chloride | 1.075 | 0.568 | 1.3 ± 0.1 | ||
| Tripropyltin chloride | 2.662 | 2.716 | 1.6 ± 0.1 | ||
| Tributyltin chloride | 4.249 | 3.11 | 1.4 ± 0.1 | ||
| Tripentyltin chloride | 5.836 | 1.2 ± 0.1 | |||
| Other inhibitors | |||||
| Cycloheximide | −0.49 | 2.1 ± 0.1 | 3.0 ± 0.1 | 1.4 ± 0.1 | |
| Lincomycin | −0.12 | 1.7 ± 0.1 | 2.9 ± 0.1 | 1.7 ± 0.1 | |
| Chloramphenicol | 0.69 | 2.1 ± 0.1 | 4.3 ± 0.2 | 2.0 ± 0.1 | |
| Triphenyltin chloride | 3.56 | 2.2 ± 0.1 | 2.6 ± 0.1 | 1.2 ± 0.1 | |
| Clotrimazole | 5.05 | 2.2 ± 0.1 | 3.4 ± 0.2 | 1.6 ± 0.1 | |
Measured logP. See reference 16.
Concentrations of inhibitors in assays were as follows: triphenyltin chloride, 0.08 mmol; cycloheximide, 0.035 mmol; chloramphenicol, 0.123 mmol; lincomycin, 0.148 mmol; and clotrimazole, 0.015 mmol. Values for lincomycin are from an experiment described in reference 13.
Calculation of dissociation constants.
The dissociation constants (Kd) of the tri-n-alkyltin chlorides were calculated from the variation of the conductances of the compounds as a function of the concentrations (9). The conductances were measured with a YSI model 33 conductance meter. The conductance cell and solutions were kept at a constant temperature (25°C). The inverse of the equivalent conductance (1/Λ) was plotted against the specific conductance (L). The intercept is the inverse of the equivalent conductance (1/Λo) at infinite dilution. The Kd is found from the slopes of the linear portions of the lines: slope = (1/Λ0)2 (1,000/Kd). The conductance of each compound is the average of at least five independent measurements.
Multivariate analysis.
Multivariate analysis of the determinants of the ratios was performed by use of the ordinary least-squares method. The regression package was the TSP package (TSP International, Palo Alto, Calif.). Explanatory variables included the number of alkyl carbons in the tri-n-alkyltin chlorides (CARBON), ClogP, the molecular volumes (MV) and total surface areas (TSA) of the molecules (13), and the measured Kd of the compounds. The specific form of the regression equation is as follows: ratio = C + B1 CARBON + B2 ClogP + B3 MV + B4 TSA + B5 Kd. The number of observations was four.
RESULTS
Effect of hydrocarbon length on PDR5-modified drug resistance.
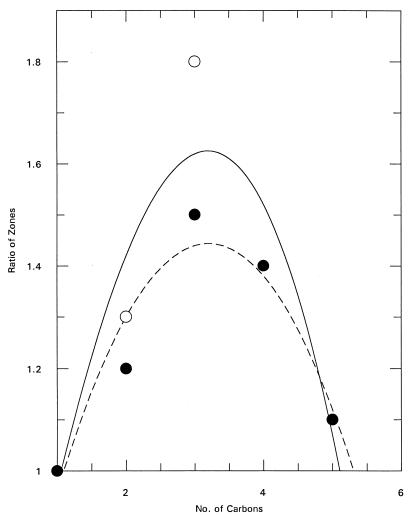
Zone-of-inhibition assays were carried out with various concentrations of tri-n-alkyltin compounds containing groups from methyl to pentyl. The goal was to find concentrations of different inhibitors that would give similar zone diameters for the wild-type strain. These concentrations were then used to determine the zone diameters for the mutant strain. These results are shown in Table 1. The data were collected for a range of inhibitor concentrations (usually at least 10-fold). An easy comparison of the two strains can be made by dividing the zone diameter found for the mutant by that found for the wild type. A ratio of 1.0 indicates no difference in the zones between the wild-type and mutant strains. In general, the reproducibilities of the zones are fairly good. For the most part, as the molar concentration increases, the diameter of the zone also increases in the expected nonlinear fashion, although a few exceptions are observed. These data can be used to generate curves for zones of lowest and highest concentrations and are shown in Fig. 1. In both cases, the peak ratio is highest with the propyl compound and then declines. In fact, as Table 1 confirms, the same pattern is observed for any diameter selected for comparison. Therefore, of the five compounds analyzed, tri-n-propyltin chloride is the strongest PDR5 substrate, while the pentyl and methyl compounds are very weak.
FIG. 1.
Relationship between the ratio of zones of inhibition and the number of carbons in the tri-n-alkyltin chlorides and the concentrations of tri-n-alkyltin chlorides. The ratio is defined as the zone of inhibition with JW436 divided by the zone of inhibition with RW2802. The solid curve (○) is for zones of inhibition at the lowest concentration and the broken curve (●) is for zones of inhibition at the highest concentration of tri-n-alkyltin chlorides.
Comparison of results from zone-of-inhibition assays and fixed-concentration assays.
The quantitative comparisons made in the zone-of-inhibition assays with tri-n-alkyltin chlorides are valid provided that the diffusion of the compound from the disk is not rate limiting. Because we could not eliminate this possibility entirely, a second approach was devised. For each inhibitor, a series of media in which the concentrations of the tri-n-alkyltin compounds varied by twofold were made. The mutant (JG436) and the wild type (RW2802) were spotted in duplicate on each petri dish containing a specific drug concentration. The highest concentration of inhibitor permitting growth of the strains is shown in Table 2 for tri-n-ethyltin, tri-n-propyltin, tri-n-butyltin, and tri-n-pentyltin chlorides. Significantly, the results derived from this experiment mirror those found with the zone-of-inhibition assays. Thus, the greatest difference between the strains is the ability to grow on tri-n-propyltin chloride. In contrast, no distinction can be made with tri-n-pentyltin chloride, suggesting that the difference is less than twofold and that the zone-of-inhibition assay is more sensitive for this inhibitor.
TABLE 2.
Fixed-concentration assays of tri-n-alkyltin chlorides with RW2802 (PDR5) and JG436 (pdr5)
| Chloride compound | Highest concn permitting growth of:
|
Difference (fold) | |
|---|---|---|---|
| RW2802 | JG436 | ||
| Triethyltin | 1.8 μM | 0.9 μM | 2 |
| Tripropyltin | 2.4 nM | 0.24 nM | 10 |
| Tributyltin | 0.32 nM | 0.08 nM | 4 |
| Tripentyltin | 1 nM | 1 nM | 1 |
PDR5 substrate specificity as a function of anion composition.
The ability of PDR5 to mediate resistance is not strongly influenced by anion composition, as indicated by the data shown in Table 3. The relatively strong PDR5 substrate specificity of tri-n-butyltin chloride is also observed with the bromide and acetate compounds. In contrast, we also observed that tri-n-phenyltin chloride, tri-n-phenyltin acetate, and tri-n-phenyltin hydroxide are all relatively weak substrates, with ratios of about 1.1 to 1.2 (Table 3).
TABLE 3.
Effect of a counterion on zones of inhibition with tributyltin and triphenyltin compounds for RW2802 (PDR5) and JG436 (pdr5)a
| Compound | Concn (mmol) | Zone of inhibition (cm) for:
|
Ratio of zone for JG436 to zone for RW2802 | ClogP | |
|---|---|---|---|---|---|
| RW2802 | JG436 | ||||
| Tributyltin chloride | 0.36 | 2.0 ± 0.1 | 2.8 ± 0.1 | 1.4 ± 0.1 | 4.25 |
| Tributyltin bromide | 3.5 | 2.0 ± 0.1 | 2.9 ± 0.1 | 1.5 ± 0.1 | 4.45 |
| Tributyltin acetate | 0.02 | 1.9 ± 0.1 | 3.0 ± 0.2 | 1.6 ± 0.1 | 3.26 |
| Triphenyltin chloride | 0.08 | 2.2 ± 0.1 | 2.6 ± 0.1 | 1.2 ± 0.1 | 3.56 |
| Triphenyltin acetate | 0.02 | 2.2 ± 0.1 | 2.5 ± 0.1 | 1.1 ± 0.1 | 1.26 |
| Triphenyltin hydroxide | 0.01 | 2.1 ± 0.1 | 2.3 ± 0.1 | 1.1 ± 0.1 | |
Zones and ratios are given as means ± standard errors of the means.
PDR5 substrate specificity as a function of Kd.
The ability of the tri-n-alkyltin chlorides and other inhibitors to dissociate was determined by measuring their conductances in 95% ethanol as described in Materials and Methods. The Kd values for the five tri-n-alkyltin compounds are shown in Table 4. Unlike hydrophobicity, which increases with an increase in the hydrocarbon chain length, Kd and thus ionization capability decrease. Taken by themselves, these data can be interpreted in one of two ways. The ionization capability of a PDR5 substrate might be relatively unimportant. Alternately, ionization might be necessary but insufficient.
TABLE 4.
Kds of tri-n-alkyltin chlorides in 95% ethanol at 25°C
| Chloride compound | Kd (M)a |
|---|---|
| Trimethyltin | (2.38 ± 0.48) × 10−3 |
| Triethyltin | (8.77 ± 0.32) × 10−4 |
| Tripropyltin | (5.18 ± 1.28) × 10−4 |
| Tributyltin | (4.59 ± 0.94) × 10−4 |
| Tripentyltin | (2.14 ± 0.32) × 10−4 |
Values in parentheses are means ± standard errors of the means.
PDR5 substrate specificity as a function of hydrophobicity.
It has been proposed that hydrophobicity is a central feature of MDR1 substrate recognition (5). The data shown in Table 5 indicate that, as expected, logP increases markedly as the length of the hydrocarbon chain increases in the tri-n-alkyltin chlorides. In contrast, as noted above, the ability of the PDR5 gene product to mediate resistance rises and then falls as a function of chain length. Thus, there is no obvious direct relationship between hydrophobicity and the ability of a tri-n-alkyltin compound to undergo efflux mediated by the PDR5 gene product. This conclusion also can be drawn from the data shown in Table 3. Although tri-n-butyltin chloride and tri-n-butyltin acetate have P values that differ by 10-fold, they are equal within experimental error as PDR5 substrates. One could argue for a model of recognition based upon a range of hydrophobicity values. This does not appear to be the case for the PDR5 gene product, as indicated by the data shown in Table 5, which also shows logP values and zones of inhibition for additional substrates of fairly diverse structures. As an example, tri-n-phenyltin chloride is a weak substrate, yet it has a logP value between those of the very strong substrates tri-n-propyl chloride and tri-n-butyl chloride. Cycloheximide and lincomycin, which are medium to strong PDR5 substrates, like tri-n-ethyl chloride and tri-n-butyl chloride, have logP values that are negative. Chloramphenicol, a strong PDR5 substrate (15), is only modestly hydrophobic, with a logP estimated at 0.69. The multivariate analysis shows that for the coefficient on ClogP, the Student t test value of −1.298 indicates that the coefficient is not statistically significant. Accordingly, the null hypothesis that ClogP has no influence on ratios cannot be rejected. The results also show that the ratios are directly related to the number of alkyl carbons and TSA and inversely related to the MV and Kd.
Effect of mutations in SNQ2 and SIN4.
It is now established that the SNQ2 and SIN4 proteins mediate resistance to some of the same inhibitors as the PDR5 protein (4, 16). The SNQ2 protein is an ATP-binding cassette transporter with high homology to the PDR5 protein, while SIN4 encodes a global transcriptional regulator (7). Interpretation of our data might be confounded if SNQ2 and SIN4 mediated resistance to some but not all of the tin compounds. The data in Table 6 demonstrate that mutations in either of these genes fail to alter inhibition by the tin compounds in zone-of-inhibition assays.
TABLE 6.
Effect of snq2 and sin4 mutations on tri-n-alkyltin chloride hypersensitivity
| Strain | Genotype | Zone of inhibition (cm)a with:
|
||||
|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | C5 | ||
| RW2802 | Wild type | 3.0 ± 0.2 | 2.9 ± 0.1 | 2.6 ± 0.2 | 3.5 ± 0.1 | 2.2 ± 0.2 |
| JG436 | pdr5 | 2.9 ± 0.2 | 3.9 ± 0.2 | 3.9 ± 0.2 | 4.9 ± 0.1 | 2.5 ± 0.1 |
| JG545 | snq2 | 2.9 ± 0.1 | 3.0 ± 0.1 | 2.3 ± 0.1 | 3.2 ± 0.2 | 2.0 ± 0.1 |
| JG546 | pdr5 snq2 | 3.0 ± 0.2 | ND | 3.9 ± 0.2 | 4.7 ± 0.1 | 2.6 ± 0.2 |
| DY150 | Wild type | 2.9 ± 0.1 | 3.3 ± 0.2 | 3.7 ± 0.1 | 2.6 ± 0.2 | 2.1 ± 0.1 |
| DY1704 | sin4::URA3 | 2.6 ± 0.2 | 3.1 ± 0.2 | 3.6 ± 0.2 | 2.5 ± 0.3 | 2.1 ± 0.1 |
C1, 10 mmol of trimethyltin chloride; C2, 4 mmol of triethyltin chloride; C3, 0.02 mmol of tripropyltin chloride; C4, 3.6 mmol of tributyltin chloride; C5, 0.34 mmol of tripentyltin chloride. ND, not determined.
DISCUSSION
The PDR5 protein causes the efflux of a very large array of cellular inhibitors (8, 14). To understand the chemical basis for PDR5 specificity, we compared the relative sensitivities of isogenic mutant and wild-type strains toward a group of simple, structurally related tri-n-alkyltin chloride inhibitors of mitochondrial ATPase. Using a quantitative zone-of-inhibition assay, we determined that tri-n-propyltin chloride was the strongest PDR5 substrate of the five compounds tested. In contrast, the methyl and pentyl compounds were very weak. Based on earlier work (11), it is assumed that the differences in the zones reflect differences in the ability of the strains to cause efflux of the inhibitor in question, because this is the only known difference between the otherwise isogenic strains.
The zone-of-inhibition assay is subject to the criticism that if inhibitor diffusion is the limiting step, the diameter of the corresponding zone might be artificially small and erroneous conclusions could be drawn in comparisons of different tri-n-alkyltin chlorides. A second approach in which two strains were spotted on a series of media containing fixed concentrations of each inhibitor was therefore used. Using this assay, we drew the same conclusions with respect to the relative PDR5 substrate strengths of the tri-n-alkyltin chlorides. Thus, the greatest difference between the strains was observed with tri-n-propyltin chloride. Modest differences were found with tri-n-butyl and tri-n-ethyltin chlorides (Table 2). It should be pointed out, however, that the zone-of-inhibition assay is more sensitive. Using this latter assay, we observed a small but significant difference between the PDR5 and the pdr5 strains when tri-n-pentyltin chloride was tested. No difference was found with the fixed-concentration approach.
The two methods do, however, yield different orders of relative toxicity. With the zone-of-inhibition assay, toxicity can be approximately determined by finding what concentration gives the same diameter in the wild-type strain. Based upon the concentration of tri-n-alkyltin chlorides giving a zone of about 1.7 cm, the order of increasing toxicity is methyl, ethyl, butyl, pentyl, and propyl. In the second approach, toxicity is determined from the lowest concentration preventing the growth of the wild-type strain. With this criterion, the order of increasing toxicity is methyl, ethyl, propyl, pentyl, and butyl. It is likely that the toxicity estimate from the fixed-concentration assay is more accurate.
Once the zone-of-inhibition data were collected, the hydrophobicity, anion composition, and dissociation properties of the inhibitors were analyzed. Our study with the tri-n-alkyltin chlorides and different anions rules out hydrophobicity and anion composition as the major parameters in the PDR5-mediated resistance of substrates from tri-n-alkyltin chlorides and from other inhibitors of diverse structures. For instance, cycloheximide, a strong PDR5 substrate, is quite hydrophilic, whereas the hydrophobic compounds triphenyltin chloride and tri-n-pentyltin chloride are relatively weak. The range in logP values (Table 3) for effective substrates is more than 5 orders of magnitude.
The zone-of-inhibition curve (Fig. 1) can be interpreted in two ways. The parabolic shape may indicate two interacting parameters with opposing effects on PDR5 substrate affinity. Alternatively, the plot may represent recognition based on a range values for some chemical property, for example, the size of the alkyl groups, as represented by the MV and the TSA. The multivariate analysis shows that the ratios are related directly to TSA and inversely to MV. Because most of the known PDR5 substrates have ionizable groups, it is not unreasonable to suggest that there is a relationship between substrate affinity and the proportion of the substrate in the ionic form. This suggestion is also confirmed by a significant relationship between ratio and Kd, as determined by the multivariate analysis.
It is important to compare the behavior of PDR5 with that of the well-studied mammalian MDR1 locus. There are two major studies on the chemical specificity of the latter (15, 18). Both were concerned primarily with compounds that bind to and modulate the multidrug transporter so that it is less active (as opposed to compounds whose efflux from cells is mediated by the MDR1 product). Both studies concluded that hydrophobicity was the principal chemical property shared by the modulators. Zamora et al. (18) also analyzed the chemical properties of anticancer agents known to be transported out of cells by the MDR1 P-glycoprotein and noted that most were hydrophobic and cationic. There were, however, several exceptions (for instance, doxorubicin, which is a strong MDR1 substrate but has a negative logP value). Our data suggest that PDR5-mediated efflux does not have the strict requirements of the flippase model proposed for the MDR1 P-glycoprotein (5). Thus, at least some of the PDR5 protein substrates are hydrophilic and probably do not have to be intercalated into the plasma membrane prior to interaction with the efflux apparatus. It is therefore not clear whether the PDR5 and MDR1 proteins are really different in their means of recognizing or causing efflux of their substrates.
ACKNOWLEDGMENTS
We appreciate the assistance of Kevin Forbes in interpreting and performing the multivarent analysis. We thank James Keeven for suggesting the experiment described in Table 2 and David Stillwell and Scott Moye-Rowley for strain and plasmid donations.
This work was supported by a grant from the National Science Foundation (MCB-9603553).
REFERENCES
- 1.Balzi E, Wang M, Leterme S, Van Dyck L, Goffeau A. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcriptional regulator PDR1. J Biol Chem. 1994;269:2206–2214. [PubMed] [Google Scholar]
- 2.Cain K, Griffiths D E. Studies of energy-linked reaction: localization of the site of action of trialkyltin in yeast mitochondria. Biochem J. 1977;162:575–580. doi: 10.1042/bj1620575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egner R, Rosenthal F, Kralli A, Sanglard D, Kuchler K. Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol Biol Cell. 1998;9:523–543. doi: 10.1091/mbc.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleckenstein A, Shallom J, Golin J. A PDR5-independent pathway of multi-drug resistance regulated by the SIN4 gene product. Yeast. 1999;15:133–137. doi: 10.1002/(SICI)1097-0061(19990130)15:2<133::AID-YEA354>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 5.Gottesmann M, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 6.Gros P, Dhir R, Croop J, Talbot F. A single amino acid substitution strongly modulates the activity and substrate specificity of mouse mdr1 and mdr3 drug efflux pumps. Proc Natl Acad Sci USA. 1991;88:7289–7293. doi: 10.1073/pnas.88.16.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Y W, Stillman D J. Regulation of HIS4 expression by the Saccharomyces cerevisiae SIN4 transcriptional regulator. Genetics. 1995;140:103–114. doi: 10.1093/genetics/140.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolaczkowski M, Vanderest M, Eybularz-Kolaczkowski A, Soumillion J, Konings W, Goffeau A. Anticancer drugs, ionophoric peptides and steroids as substrates of the yeast multidrug transporter Pdr5p. J Biol Chem. 1996;271:31543–31548. doi: 10.1074/jbc.271.49.31543. [DOI] [PubMed] [Google Scholar]
- 9.Kraus C A, Callis C C. Studies relating to the metallo-organic compounds. I. Introduction. II. The equivalent conductance of trimethylstannyl chloride in ethyl alcohol. J Am Chem Soc. 1923;45:2624–2632. [Google Scholar]
- 10.Leo A J. Calculating log POCT from structures. Chem Rev. 1993;93:1281–1306. [Google Scholar]
- 11.Leonard P J, Rathod P K, Golin J. Loss of function mutation in the yeast multiple drug resistance gene PDR5 causes a reduction in chloramphenicol efflux. Antimicrob Agents Chemother. 1994;38:2492–2494. doi: 10.1128/aac.38.10.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loo A J, Clark D M. Functional consequences of proline mutations in the predicted transmembrane domains of P-glycoprotein. J Biol Chem. 1993;268:3143–3149. [PubMed] [Google Scholar]
- 13.Luedke E, Lucero E, Eng G. Molecular volume as a predictor of organotin biotoxicity. Main Group Met Chem. 1991;14:59–66. [Google Scholar]
- 14.Meyers S, Schauer W, Balzi E, Wagner M, Goffeau A, Golin J. Interaction of the yeast pleiotropic drug resistance genes—PDR1 and PDR5. Curr Genet. 1992;21:431–436. doi: 10.1007/BF00351651. [DOI] [PubMed] [Google Scholar]
- 15.Nogac I, Kohno K, Kikuchi J, Kawano J, Akuyama M. Analysis of structural features of dihydropyridine analogs needed to reverse MDR and inhibit photo affinity labeling of P-glycoprotein. Biol Pharm. 1989;38:519–517. doi: 10.1016/0006-2952(89)90393-6. [DOI] [PubMed] [Google Scholar]
- 16.Servos J, Hasse E, Brendel M. Gene SNQ2 of Saccharomyces cerevisiae, which confers resistance to 4-nitroquinoline-N-oxide and other chemicals, encodes a 169 kDa protein homologous to ATP-dependent permeases. Mol Gen Genet. 1993;236:214–218. doi: 10.1007/BF00277115. [DOI] [PubMed] [Google Scholar]
- 17.Wulf R G, Byington K H. On the structure-activity relationships and mechanism of organotin induced nonenergy dependent swelling of liver mitochondria. Arch Biochem Biophys. 1975;167:176–185. doi: 10.1016/0003-9861(75)90454-3. [DOI] [PubMed] [Google Scholar]
- 18.Zamora J, Pearce H L, Beck W T. Physical-chemical properties shared by compounds that modulate MDR in human leukemic cells. Mol Pharmacol. 1988;33:454–462. [PubMed] [Google Scholar]