Abstract
Background
We aimed to analyze the prognostic power of CT-based radiomics models using data of 14,339 COVID-19 patients.
Methods
Whole lung segmentations were performed automatically using a deep learning-based model to extract 107 intensity and texture radiomics features. We used four feature selection algorithms and seven classifiers. We evaluated the models using ten different splitting and cross-validation strategies, including non-harmonized and ComBat-harmonized datasets. The sensitivity, specificity, and area under the receiver operating characteristic curve (AUC) were reported.
Results
In the test dataset (4,301) consisting of CT and/or RT-PCR positive cases, AUC, sensitivity, and specificity of 0.83 ± 0.01 (CI95%: 0.81–0.85), 0.81, and 0.72, respectively, were obtained by ANOVA feature selector + Random Forest (RF) classifier. Similar results were achieved in RT-PCR-only positive test sets (3,644). In ComBat harmonized dataset, Relief feature selector + RF classifier resulted in the highest performance of AUC, reaching 0.83 ± 0.01 (CI95%: 0.81–0.85), with a sensitivity and specificity of 0.77 and 0.74, respectively. ComBat harmonization did not depict statistically significant improvement compared to a non-harmonized dataset. In leave-one-center-out, the combination of ANOVA feature selector and RF classifier resulted in the highest performance.
Conclusion
Lung CT radiomics features can be used for robust prognostic modeling of COVID-19. The predictive power of the proposed CT radiomics model is more reliable when using a large multicentric heterogeneous dataset, and may be used prospectively in clinical setting to manage COVID-19 patients.
Keywords: X-ray CT, COVID-19, Radiomics, Prognosis, Machine learning
Graphical abstract
1. Introduction
The novel coronavirus disease, which emerged in 2019 (COVID-19), is now a major cause of death worldwide [1]. This highly contagious virus can cause a spectrum of pulmonary, hematological, neurological, and systemic complications, making it a highly lethal pathogen [2]. As of March 21st, 2022, there have been >400 million globally confirmed cases of COVID-19, including >6 million deaths and >10 billion vaccinations reported to the world health organization (WHO) [https://covid19.who.int/]. There remains an urgent need for addressing issues such as diagnosis, prognosis, and treatment options [3].
Diagnostic tools for COVID-19, such as reverse transcription-polymerase chain reaction (RT-PCR), aid in distinguishing between negative and positive cases [4]. Conversely, prognostic tools provide clinicians with insights to optimize treatment strategies, manage the hospitalization of patients both in the wards and intensive care units (ICU), and better handle patient follow-up plans [5]. Different studies have evaluated clinical and/or non-clinical features for determining the diagnosis and prognosis of patients with COVID-19. Yan et al. [6] used only clinical features to classify patients into different categories, ranging from mild to critical conditions. Zhou et al. [7] also aimed at establishing a prognostic model for outcome prediction of patients with COVID-19, utilizing their clinical data.
Computed Tomography (CT) plays a pivotal role in the management of a wide variety of diseases as a fast and non-invasive imaging modality. In the case of COVID-19, CT is used for both diagnostic (e.g., in case of limited access to RT-PCR) and prognostic purposes [8]. The clinical value of CT imaging relies mainly on the early detection of lung infections and high accuracy in quantifying the disease progression and severity [9]. Francone et al. [10] assessed the correlation of CT scores with COVID-19 pneumonia severity and outcome. In another study, Zhao et al. [11] specified pulmonary involvement severity by measuring the extent of pneumonia and consolidation that appear on CT images. Li et al. [12] concluded that high scores of CT images are associated with severe COVID-19 pneumonia.
Despite previously conducted research, there is still a need for studies with more accurate and comprehensive analyses [13]. In conventional analyses, CT features are visually and subjectively defined, while machine learning (ML) and/or deep learning (DL) models have the potential to provide a more comprehensive and objective assessment of images. Towards modeling of outcomes in COVID-19 patients, several ML and DL algorithms have been utilized to assess the severity and to predict the outcome of patients using CT imaging [[14], [15], [16], [17]]. To evaluate the sensitivity and specificity of CT for COVID-19 diagnostic purposes, Harmon et al. [14] achieved a sensitivity of 84% and specificity of 93% in an independent test set of 1337 patients applying DL on CT images. In another machine learning study, Mei et al. [15] reported a sensitivity of 84.3% and specificity of 82.8% based on combined CT and clinical data. Cai et al. [16] utilized a random forest model to assess the severity of COVID-19 disease in 99 patients and their need for a more extended hospital or ICU stay. Another study by Lessman et al. [17] reported the model performance of three DL models for severity assessment in COVID-19. In another multi-center study, Meng et al. [18] differentiated patients with high-risk of mortality versus low-risk ones using a convolutional neural network named De-COVID-Net. Ning et al. [19] used their pre-trained DL model on a dataset consisting of 351 patients that was capable of distinguishing between non-coronavirus pneumonia, mild coronavirus pneumonia, and severe forms of COVID-19 disease.
Various studies also reported remarkable prediction accuracies utilizing radiomics approaches using CT and chest x-ray imaging modalities [[20], [21], [22], [23]]. Medical images could be converted into high-dimensional data through radiomics, wherein radiomics features are selected from the images and combined using machine learning algorithms to arrive at radiomics signatures as biomarkers of disease. In addition to wide usage in several oncologic [[24], [25], [26]] as well as non-oncologic diseases [[27], [28], [29]], radiomics studies have indicated that imaging features extracted from CT or chest X-ray images could be used as parameters for outcome prediction of patients with COVID-19 pneumonia. Radiomics analyses have been applied to different aspects of COVID-19, including diagnosis, severity scoring, prognosis, hospital/ICU stay prediction, and survival analysis [[20], [21], [22]]. In a retrospective study, Fu et al. [30] constructed a predictive model based on CT radiomics, clinical and laboratory features. This signature could classify COVID-19 patients into stable and unstable (i.e., progressive phenotype). Homayounieh et al. [31] aimed to predict the severity of pneumonia in patients with COVID-19 using a radiomics model that outperformed models consisting of clinical-only features. Another study by Li et al. [32] analyzed a radiomics/DL model that distinguished severe from critical COVID-19 pneumonia patients. Cai et al. [33] developed a model combining CT radiomic features and clinical data to predict RT-PCR negativity during admission. Yue et al. [34] conducted a multicentric radiomics study on 52 patients to differentiate whether an individual needs a short-term or long-term hospital stay. Another study by Bae et al. [35] predicted the mortality of patients with COVID-19 using chest x-ray radiomics. Their model could identify whether a patient needs mechanical ventilation or not.
Artificial intelligence (AI) has been widely used in radiology to provide diagnostic and prognostic tools to help clinicians during the pandemic. However, owing to the lack of standardization in AI studies in terms of data collection, methodology, and evaluation, most of these studies were not pragmatic when it comes to clinical adoption [13,36,37]. In a recent study by Roberts et al. [13], possible sources of bias in more than 2000 AI articles in COVID-19 were evaluated in both deep learning and traditional machine learning-based studies. This review showed that bias do exist in most, if not all, of the studies in different domains, including dataset and methodology. In the dataset domain, several articles used public datasets which can contain duplicates, low-quality images, false demographics, or unknown clinical/lab data of patients. These public datasets can also induce bias in the outcome domain as they may fail to supply sufficient information about how they exactly proved a patient is COVID-19 positive or how imaging data were acquired in terms of image acquisition and reconstruction. They also mentioned using small datasets, Frankenstein datasets [13], and Toy datasets [36] in several articles. Most of the studies did not provide all details about methodological aspects or did not perform a standard AI analysis based on guidelines [38,39].
Overall, Roberts et al. [13] reviewed 69 traditional machine learning/radiomics studies and reported that 44 were excluded because of Radiomics Quality Score (RQS) [38] of less than six or not describing the datasets appropriately. From the remaining 25, six articles performed model evaluation using external validation sets, and only four papers reported the significance of their model along with the statistical parameters (agreement level). They also assessed bias in the prognostication studies in the four areas of the prediction model risk of bias assessment tool (PROBAST) guide and reported high bias in participants, predictors, outcomes, and analysis areas. Overall, there are several radiomics studies targeting improved COVID-19 diagnosis or prognosis. However, the models tend to overfit due to the limited sample size, the single-centered nature of most of the databases, and variability in data acquisition and image reconstruction protocols [13]. Providing a generalizable model which is reproducible on unseen datasets of other centers is highly desired. In this context, we designed a large multi-institutional study to build and evaluate a radiomics model based on a large-scale CT imaging dataset aimed at the prediction of survival (alive or deceased) in COVID-19 infected patients. We built and evaluated our model based on different guidelines and tested different machine learning algorithms in different strategies to assess model reproducibility and repeatability in a large dataset.
To summarize, CT images of 14,339 COVID-19 patients with overall survival outcomes were collected from 19 medical centers. Whole lung segmentations were performed automatically using a deep learning-based model, and regions of interest were further evaluated and modified by a human observer. All images were resampled to an isotropic voxel size, intensities were discretized, and 107 radiomic features were extracted from the lung mask. Radiomic features were normalized using Z-score normalization, and highly-correlated features were eliminated. We applied the Synthetic Minority Oversampling Technique (SMOT) algorithm to only the training set for different models to overcome unbalanced classes. By cross-combination of four feature selectors and seven classifiers, twenty-eight different combinations of machine learning models were built. The models were built and evaluated using training and testing sets, respectively. Specifically, we evaluated the models using 10 different splitting and cross-validation strategies, including different types of test datasets (e.g., non-harmonized vs. ComBat-harmonized datasets), and different metrics were reported for models evaluation.
2. Materials and methods
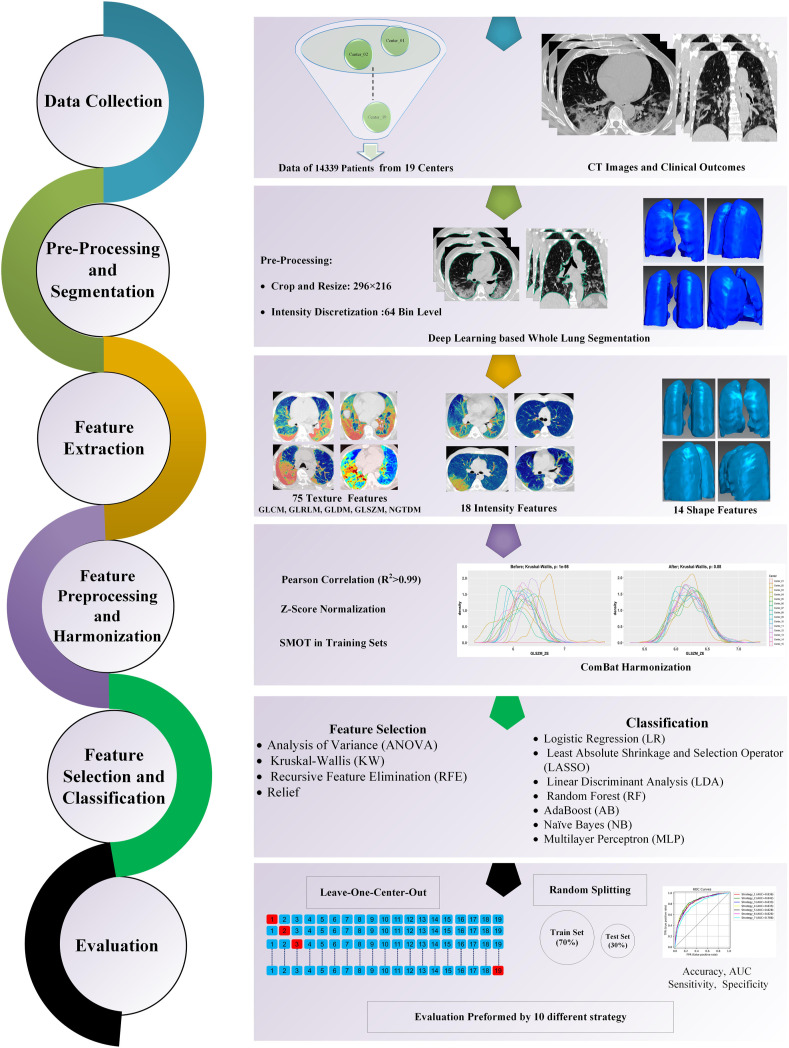
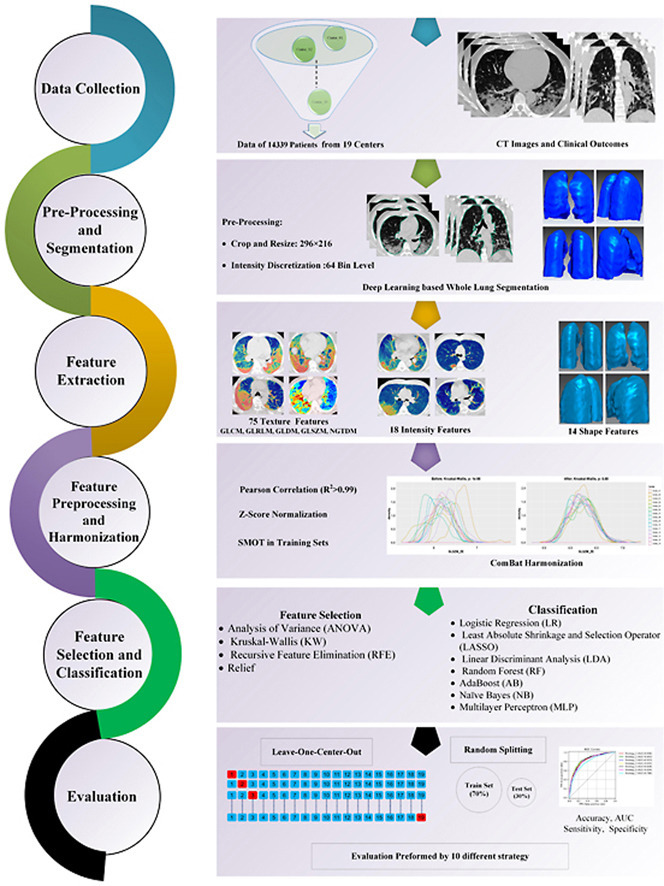
Fig. 1 summarizes the different steps adopted in this study. We completed different checklists/guidelines concerning predictive modeling, radiomics studies, and artificial intelligence studies to provide a standard and reproducible study. The Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) [40] checklist is provided in the supplemental material. We also reported the Radiomics Quality Score (RQS) based on Lambin et al. [38] and the Checklist for Artificial Intelligence in Medical imaging (CLAIM) [39] in supplemental material. These checklists were filled out by two individuals (with consensus) who are experts in the field of radiomics and not co-authors in this study.
Fig. 1.
The flowchart of our study represents the different radiomic steps. CT images of 14,339 COVID-19 patients with overall survival outcome were collected from 19 medical centers. Whole lung segmentations were performed automatically using a deep learning-based model. The models were evaluated using 10 different splitting and cross-validation strategies, including different types of test datasets and the different metrics reported for models evaluation.”
2.1. Patient population
This study was approved by institutional review boards (IRBs) of Shahid Beheshti University of Medical Sciences, Alborz University of Medical Sciences and Tehran University of Medical Sciences, Iran, and written informed consent of patients was waived by the ethics committees as anonymized data were used without any interventional effect on diagnosis, treatment, or management of COVID-19 patients.
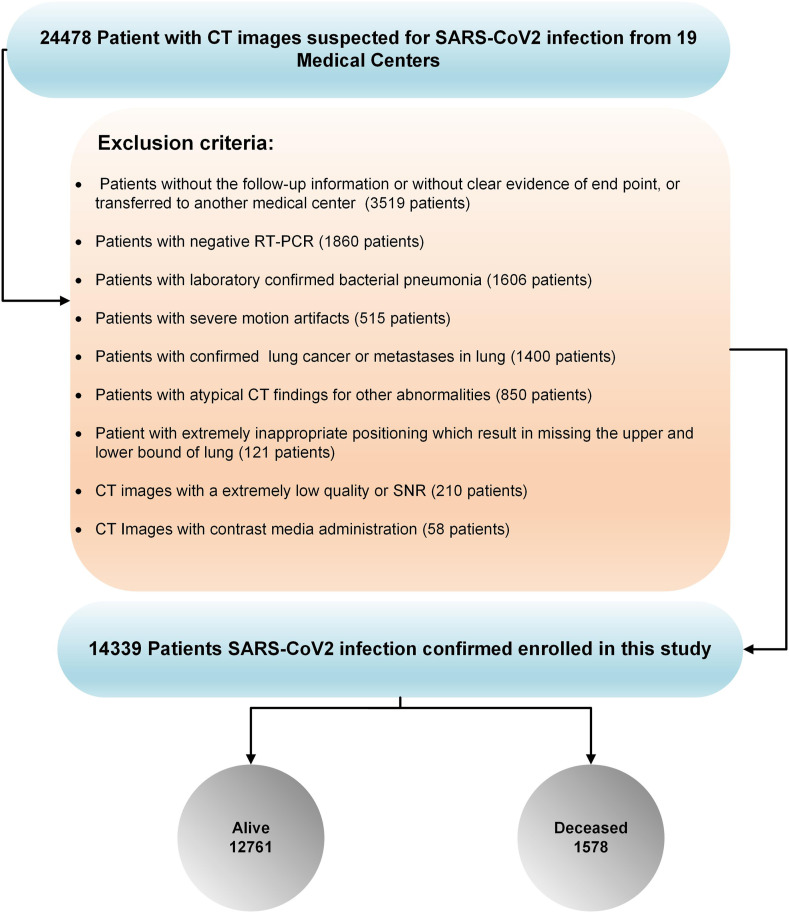
In the first step, 24,478 patients from 19 medical centers (18 centers from Iran and one from China) suspected of COVID-19 and with acquired chest CT images were included. Different exclusion criteria were applied to provide a reliable dataset. We excluded: (i) patients without follow-up information or clear evidence of clinical endpoint, or if they were transferred to another medical center (3519 patients), or patients with (ii) negative RT-PCR (1860 patients), (iii) laboratory-confirmed pneumonia of other types (1606 patients), (iv) confirmed lung cancer or metastases from other origins to the lungs (1400 patients), (v) atypical CT findings for other abnormalities (850 patients), (vi) CT images with contrast media administration (58 patients), (vii) severe motion or bulk motion artifacts in CT images were carefully checked (515 patients), (viii) extremely inappropriate positioning which resulted in missing the upper and lower bounds of the lungs (121 patients), or (ix) CT images with extremely low quality or SNR (210 patients).
Considering these criteria, we excluded 10,139 patients from further analysis (Fig. 2 ). Hence, 14,339 chest CT scans (with one scan per patient whose COVID-19 was confirmed either by RT-PCR or chest CT imaging) were included in this study (in case of negative RT-PCR test, they were excluded from the study). Common symptoms of COVID-19, including fever, respiratory symptoms, shortness of breath, dry cough and tiredness, were recorded and contact history with COVID-19 patients was also assessed. In each center, CT images were evaluated at least by two radiologists, and in case of discrepancy, a third radiologist was involved in settling the disagreement. As defined in the COVID-19 Reporting and Data System (CO-RADS) [40], typical manifestations of COVID-19, such as ground-glass opacity, consolidation, crazy-paving pattern, or dominant peripheral distribution of parenchymal abnormalities were considered diagnostic for COVID-19 in CT images.
Fig. 2.
Inclusion and exclusion criteria adopted in this study. Overall, 24,478 patients with CT images suspected of SAR-CoV2 infection from 19 centers were enrolled in this study and 10,139 patients were excluded based on exclusion criteria. Among the 14,339 patients with SARS-CoV2 infection confirmation used for further analysis, 12,761 and 1,578 cases were alive and deceased, respectively. The fraction of deceased patients is overrepresented in our data due to our exclusion criteria.
Among these studies, 13,731 CT images were collected from 18 centers in Iran (1,560 deceased, 12,171 alive; a fraction of deceased cases are significantly overrepresented due to our exclusion 1 images were gathered from online open-access databases from China (Center 9: 18 deceased, 590 alive) [41]. All patients from Iran received standard treatment regimens according to the interim national COVID-19 treatment guideline [corona.behdasht.gov.ir]. Only one center (Center 10) included outpatient studies, and the rest were inpatient-only studies from hospitalized patients. Follow-up was performed 3–4 months after the initial CT scan in the outpatient cases. For admitted patients (inpatient), follow-up was performed until discharge from the hospital, which was considered after careful evaluation of patients by the attending physician based on several criteria, including stable hemodynamics state (BP > 90/60, HR < 120), absence of fever for > 2 days, absence of any respiratory distress, blood oxygen saturation >93% in ambient oxygen without the need for supplementary oxygen, and no need for hospitalization for any other pathology.
2.2. CT image acquisition
All chest CT images from the Iranian centers were acquired according to an institutional variation of the national society of radiology COVID-19 imaging guidelines [42]. Image acquisition was performed during breath-hold to reduce motion artifacts. Variations in CT imaging protocols among centers were observed, which led to considerable variability in image quality and radiation dose. Volumetric CT Dose Index (CTDIvol), as a parameter representing vendor-free information on radiation exposure, was reported to better reflect the intra/inter-institutional variability of our dataset. Table 1 summarizes the image acquisition characteristics of each center, including the number of images, acquisition parameters (slice thickness, tube current), and CTDIvol.
Table 1.
Demographics and data acquisition parameters across the different centers.
| Center |
Number (Deceased %) |
PCR |
Gender |
CTDIvol |
Tube current (mAs) |
Age |
Slice thickness |
|
|---|---|---|---|---|---|---|---|---|
| % of patients with available PCR result | Male | Female | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | ||
| Center_01 | 152 (26.3%) | 100% | 87 | 65 | 1.57 ± 0.32 | 45 ± 47.8 | 62.1 ± 17.0 | 1.50 ± 0.50 |
| Center_02 | 264 (41.6%) | 99% | 120 | 144 | 8.47 ± 0.53 | 186.2 ± 34.0 | 71.8 ± 12.4 | 3.44 ± 0.95 |
| Center_03 | 319 (1.5%) | 100% | 168 | 151 | 8.55 ± 4.61 | 185 ± 65.3 | 51.6 ± 17.9 | 2.82 ± 0.99 |
| Center_04 | 373 (22.5%) | 77% | 157 | 216 | 7.52 ± 1.23 | 147.1 ± 43.2 | 54.1 ± 19.3 | 5.22 ± 0.46 |
| Center_05 | 382 (19.1%) | 100% | 215 | 167 | 6.37 ± 0.67 | 128.3 ± 4.5 | 58.2 ± 18.25 | 7.67 ± 1.7 |
| Center_06 | 394 (38.6%) | 100% | 257 | 137 | 5.8 ± 2.52 | 149 ± 48.3 | 56.8 ± 16.8 | 2.34 ± 0.24 |
| Center_07 | 492 (5.2%) | 100% | 276 | 216 | 9.68 ± 2.91 | 349 ± 181 | 33.0 ± 5.66 | 2.57 ± 1.36 |
| Center_08 | 539 (19.1%) | 94% | 287 | 252 | 4.10 ± 4.57 | 167 ± 39.1 | 54.1 ± 17.4 | 7.71 ± 0.77 |
| Center_09 | 608 (3.0%) | 100% | 274 | 334 | 6.61 ± 4.87 | 164 ± 128 | 55.2 + 15.99 | 2.10 ± 1.21 |
| Center_10 | 685 (14.8%) | 89% | 348 | 337 | 5.01 ± 3.57 | 171 ± 33.3 | 56.4 ± 18.7 | 7.16 ± 0.81 |
| Center_11 | 866 (13.2%) | 100% | 520 | 346 | 3.40 ± 0.42 | 123.6 ± 9.6 | 55.4 ± 17.9 | 2.04 ± 0.23 |
| Center_12 | 883 (11.2%) | 62% | 448 | 435 | 4.30 ± 1.54 | 166.2 ± 51.3 | 55.4 ± 15.6 | 4.99 ± 0.16 |
| Center_13 | 949 (10.9%) | 76% | 568 | 381 | 5.57 ± 1.21 | 210.7 ± 39.6 | 52.6 ± 19.3 | 2.17 ± 1.51 |
| Center_14 | 1022 (9.7%) | 100% | 526 | 496 | 5.01 ± 3.19 | 91.6 ± 48.6 | 56.2 ± 18.8 | 4.93 ± 0.49 |
| Center_15 | 1024 (1.0%) | 29% | 542 | 482 | 6.82 ± 1.02 | 228.4 ± 27.2 | 48.3 ± 14.5 | 2.48 ± 1.96 |
| Center_16 | 1180 (12.7%) | 84% | 578 | 602 | 5.63 ± 2.98 | 172 ± 34.1 | 55.4 ± 16.7 | 6.38 ± 0.82 |
| Center_17 | 1323 (12.7%) | 100% | 680 | 643 | 6.14 ± 1.41 | 175 ± 38.2 | 57.4 ± 18.7 | 5.66 ± 0.82 |
| Center_18 | 1355 (2.5%) | 70% | 752 | 603 | 4.21 ± 3.18 | 98.4 ± 41.1 | 55.1 ± 19.1 | 4.75 ± 0.69 |
| Center_19 | 1529 (5.6%) | 93% | 814 | 715 | 8.92 ± 1.40 | 220.7 ± 27.9 | 54.5 ± 19.0 | 6.96 ± 0.34 |
2.3. Image segmentation and image preprocessing
The lungs were automatically segmented using our previously developed DL-based algorithm named COLI-Net [43]. For efficient radiomics feature extraction (feature extraction time), all images were first cropped to the lung region and then resized to 296 × 216 to obtain a computationally efficient feature extraction. After reviewing the segmentations, the image voxel was resized to an isotropic voxel size of 1 × 1 × 1 mm3, and the intensity discretized to 64-binning size [44].
2.4. Radiomics feature extraction and harmonization
After image preprocessing, radiomics feature extraction was performed using ITK and PyRadiomics [45] Python library, respectively. Radiomics features, including morphological (n = 14), intensity (n = 18), and texture features including second-order features, such as Gray Level Co-occurrence Matrix (GLCM, n = 24), higher-order features, namely Gray Level Size Zone Matrix (GLSZM, n = 16), Neighboring Gray Tone Difference Matrix (NGTDM, n = 5), Gray Level Run Length Matrix (GLRLM, n = 16), and Gray Level Dependence Matrix (GLDM, n = 14) were extracted in compliance with the Image Biomarker Standardization Initiative (IBSI) guidelines [44]. Details about the extracted features were presented in supplemental section.
2.5. Feature preprocessing
For each feature vector, the mean and standard deviation were calculated (in training sets) and then normalized using Z-Score normalization, which consists of subtracting each feature vector from the mean followed by division by the standard deviation. For Z-score normalization, the mean and standard deviation were calculated for the training set and then applied to the test set. Features’ correlation was evaluated using Pearson correlation, and features with high correlation (R2 > 0.99) were eliminated. Owing to unbalanced datasets in the training and test set, we applied Synthetic Minority Oversampling Technique (SMOT) algorithm to only the training set for the different models.
2.6. Feature selection and classification
In this study, we used four feature selection algorithms, including Analysis of Variance (ANOVA), Kruskal-Wallis (KW), Recursive Feature Elimination (RFE), and Relief. Feature preprocessing and selection were performed on training sets and then applied on test sets. F-value was calculated for radiomic features and outcome relationship in ANOVA and Kruskal-Wallis. Subsequently, the features were sorted according to F-values and finally selected the top n features according to validation performance. We automatically selected the features based on RFE by recursively considering a smaller set of the features. We selected sub-radiomic features set recursively by Relief algorithm according to the outcome. We selected the features for each feature selection separately, where the number of features were chosen automatically based on validation set for each model and strategy. All test and external validation sets were unseen to feature processing and the selection and model building process. For the classification task, we used seven classifiers, including Logistic Regression (LR), Least Absolute Shrinkage and Selection Operator (LASSO), Linear Discriminant Analysis (LDA), Random Forest (RF), AdaBoost (AB), Naïve Bayes (NB) and Multilayer Perceptron (MLP). We tested twenty-eight different combinations by cross-combination of four feature selectors and seven classifiers. Classification algorithms were optimized during training using grid search algorithms. The best models were chosen by one standard deviation rule in 10-fold cross-validation and then evaluated on the test or external validation sets. All feature selection and classification algorithms were implemented in Scikit-Learn Python library [5].
2.7. Evaluation
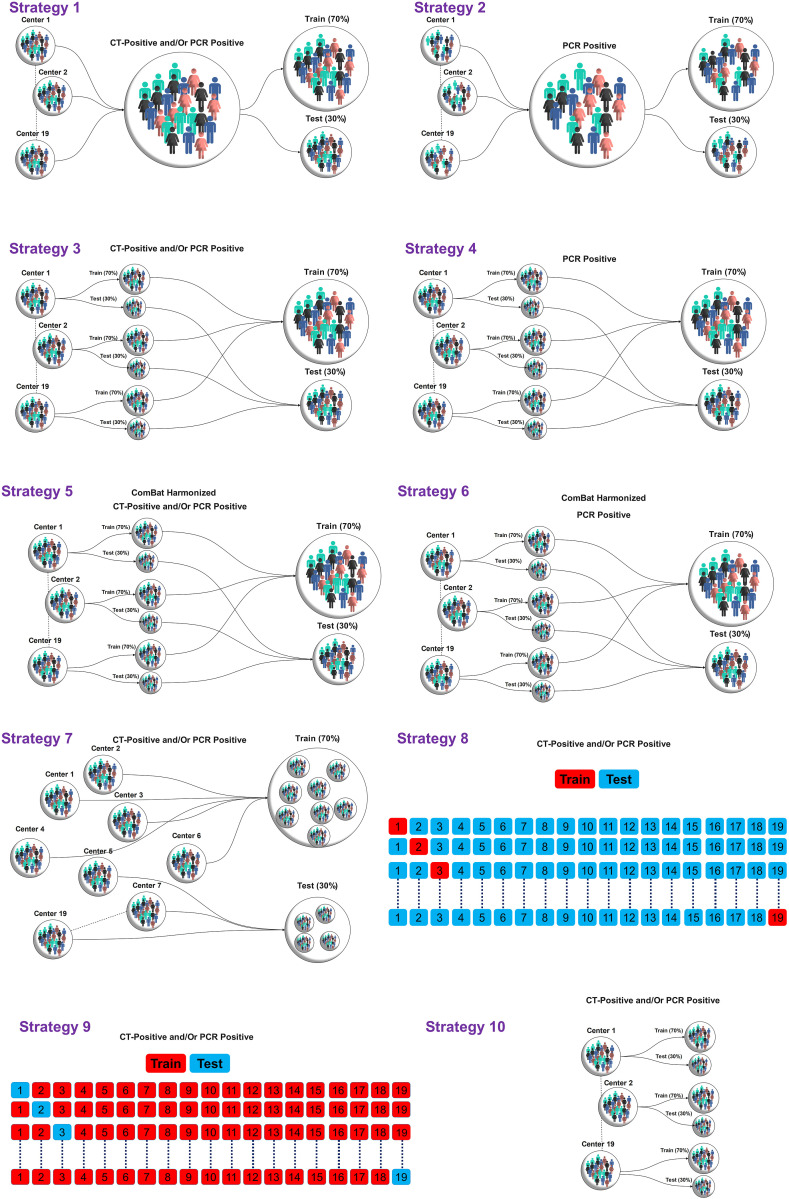
We trained and evaluated our models for thorough assessment using ten different strategies as summarized in Fig. 3 . To evaluate the models on whole datasets without considering data variability in each center, we divided the dataset of each center into 70% training and 30% tests sets resulting in the following two strategies (1 and 2):
-
-
Random Splitting method #1: Non-harmonized datasets were randomly split into 70% (10,038 patients) and 30% (4,301 patients) for training and test sets, respectively, without considering centers. The data included patients whose COVID-19 was confirmed using RT-PCR and patients confirmed only by imaging. This test dataset included both populations.
-
-
Random Splitting method #2: Non-harmonized datasets were randomly split into 70% (8,503 patients) and 30% (3,644 patients) for the training and test sets, respectively, without considering centers. The train and test sets consisted of only patients with positive RT-PCR.-To evaluate the models on whole datasets considering data variability in each center, we divided the dataset of each center into 70% training and 30% test sets resulting in the following two strategies (3 and 4):
-
-
Random Splitting method #3: Data from each center (non-harmonized) were randomly split into 70% (10,048 patients) and 30% (4,291 patients) for the training and test sets, respectively. As our data included patients whose COVID-19 was confirmed using RT-PCR and patients confirmed only by imaging, this test dataset included both populations.
-
-
Random Splitting method #4: Data from each center were randomly split into 70% (8,512 patients) and 30% (3,635 patients) for the training and test sets, respectively. The train and test sets consist of only patients with positive RT-PCR.
Fig. 3.
Different strategies implemented in this study for model evaluation. In strategies 1–6, the data were split into 70%/30% train/test sets. In strategy 7, the centers were split into 70%/30% train/test sets. In strategy 8, the model was trained on one center and evaluated on 18 centers and repeated for all centers. In strategy 9, on each of the 19 iterations, 18 centers were used as the training set, and one as the external validation set. In strategy 10, the models were built and evaluated on each center separately using data splitting of 70%/30% train/test sets, respectively.
To evaluate the models in the whole dataset by removing data variability due to acquisition/reconstruction from different centers, ComBat harmonization proposed by Johnson et al. [46] was applied to the extracted features to tackle the effect of center-based imaging variability. The impact of ComBat harmonization on radiomics features was assessed by the Kruskal-Wallis test. After applying ComBat harmonization, we divided the datasets of each center into 70/30% train/test sets resulting in the following two strategies (5 and 6):
-
-
Random Splitting method #5: Data from each center (ComBat harmonization) were randomly split into 70% (10,048 patients) and 30% (4,291 patients) for the training and test sets, respectively. As our data included patients whose COVID-19 was confirmed using RT-PCR and patients confirmed only by imaging, this test dataset included both populations.
-
-
Random Splitting method #6: Data from each center (ComBat harmonization) were randomly split into 70% (8,512 patients) and 30% (3,635 patients) for the training and test sets, respectively. The train and test sets consisted of only patients with positive RT-PCR.
To evaluate model generalizability and sensitivity to datasets, we performed model assessment using the following strategies (7–9) on the external validation sets:
-
-
Random Splitting method #7: Data (non-harmonized) were randomly split into 70% (10,655 patients) and 30% (3,684 patients) for the training and external validation sets, respectively. The center's number in the test set appears in the test sets.
-
-
Center-based model evaluation #8: we built models on one center's dataset (non-harmonized) and then evaluated on 18 remaining centers (external validation set) and then repeated this process for all datasets.
-
-
Leave-one-center-out (LOCO) #9: On each of the 19 iterations, 18 centers were used as the training set, and one as the external validation set (unseen data during training). We repeated this process for all center datasets (non-harmonized).
To evaluate model sensitivity to each dataset, we trained and tested the models in each center separately on each center dataset using the following strategies:
-
-
Random Splitting method #10: Data from each center (non-harmonized) were randomly split into 70% and 30% for the training and test sets, respectively. The models were built and evaluated on each center separately.
All multivariate steps, including feature preprocessing, feature selection and classification were performed separately for each strategy. Classification algorithms were optimized during training using grid search algorithms. The best models were chosen by one standard deviation rule in 10-fold cross-validation and then evaluated on test or external validation sets. The accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve (AUC) were reported for the test or external validation sets (unseen during training). Statistical comparison of AUCs (by 10,000 bootstrapping) between models was performed using the DeLong test [47]. The significance level was considered at a level of 0.05. All multivariate analysis steps were performed using Python Scikit-Learn [48] open-source library.
3. Results
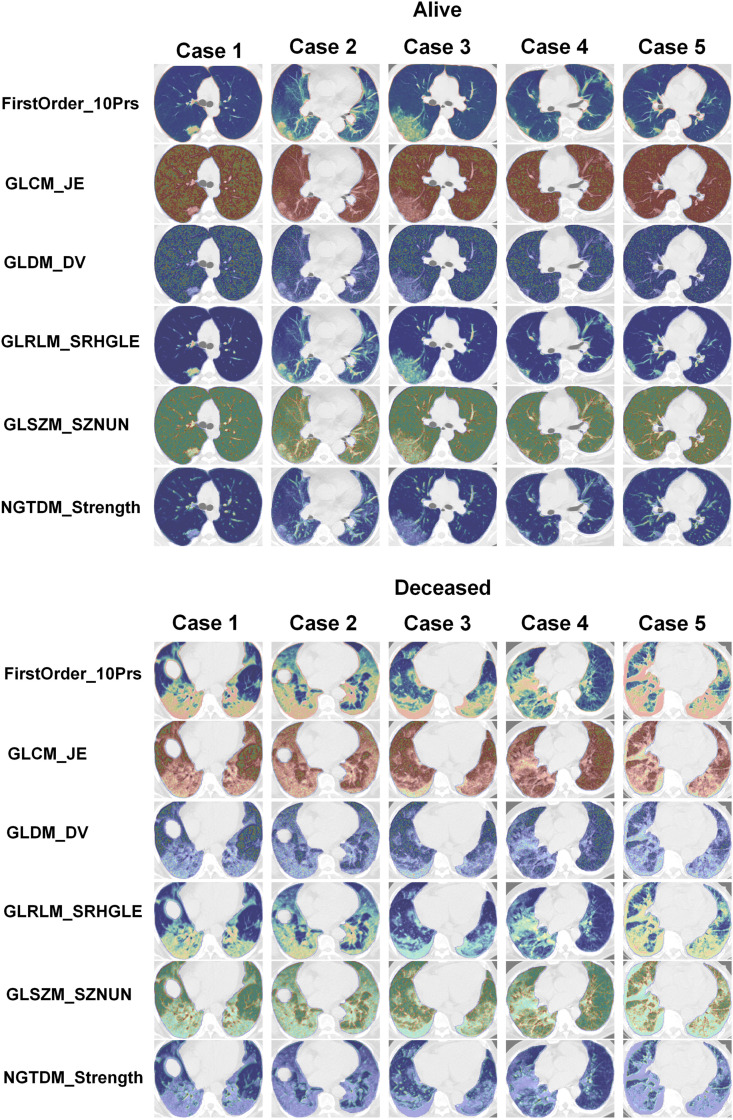
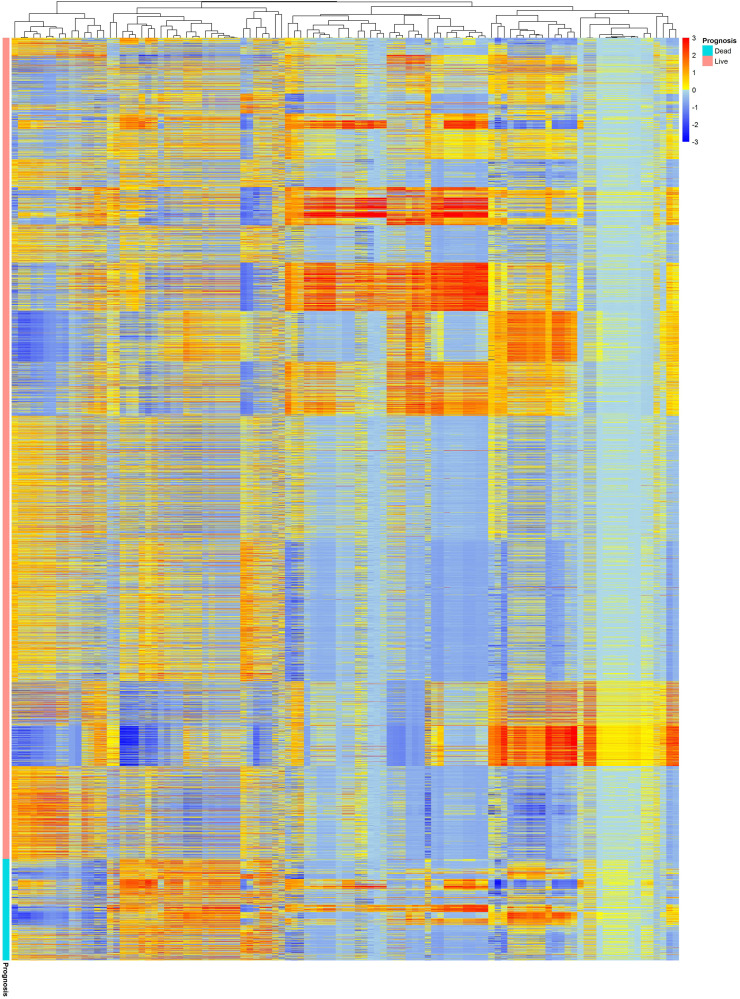
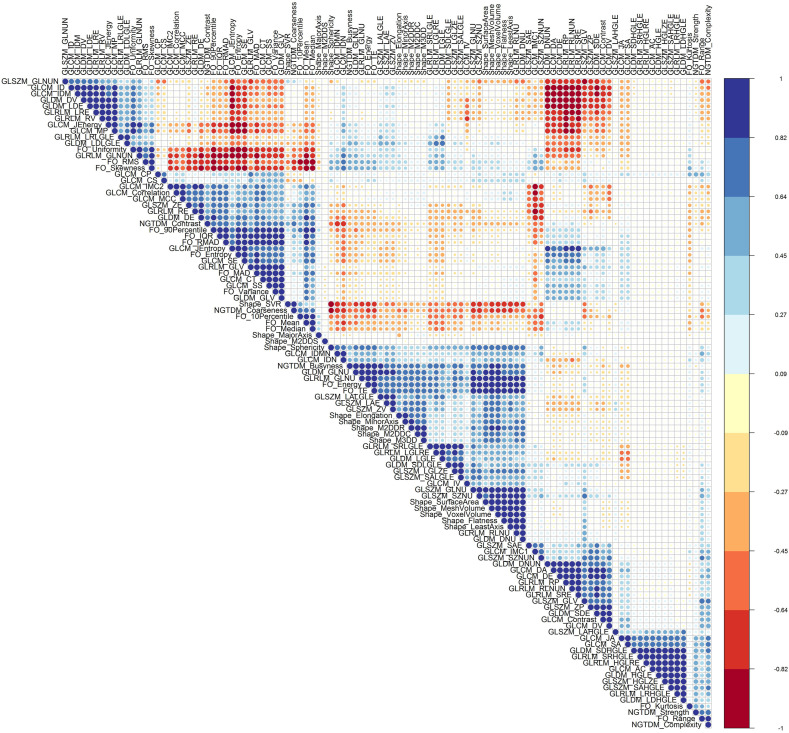
Fig. 4 depicts radiomic feature maps for ten different cases (five alive and five deceased). The rows represent the different radiomic feature maps whereas the columns represent different cases. We represented one feature from each feature set, including 10Percentile from the first-order, Joint Entropy from GLCM, Dependence Variance from GLDM, Short Run High Gray Level Emphasis from GLRLM, Size Zone Non-Uniformity Normalized from GLSZM, and Strength from NGTDM. These features highlighted visually the difference between alive and deceased cases. Fig. 5 depicts the hierarchical clustering heat map of the distribution of radiomic features in alive and deceased groups for the whole dataset prior to ComBat harmonization. It demonstrates that clustering does not show any significant correlation or cluster in alive and deceased cases. Supplemental Fig. 1 shows the cluster heat map of radiomic features in the non-harmonized data set. Fig. 6 shows the correlation of radiomic features in the whole dataset, whereas Supplemental Fig. 2 represents the same for ComBat harmonized features. Highly correlated features (both positive and negative) were eliminated from further analysis. The statistical differences calculated using the Kruskal-Wallis test are presented in Supplemental Table 1 before and after ComBat harmonization. The results of ComBat harmonization showed that the algorithm properly eliminated the center effect on radiomics features in most features. ComBat harmonization data only were used for strategies 5 and 6. Fig. 7, Fig. 8, Fig. 9 provide the classifications power indices of AUC, sensitivity and specificity, respectively, for splitting strategies 1–10. More detailed results were presented in Supplemental Tables 2–11 for the different strategies.
Fig. 4.
Radiomics feature maps for ten different cases (5 alive and 5 deceased). Rows represent different radiomics feature map and columns represent different cases. We represented one feature from each feature sets including, 10Percentile from first order, Joint Entropy from GLCM, Dependence Variance from GLDM, Short Run High Gray Level Emphasis from GLRLM, Size Zone Non-Uniformity Normalized from GLSZM, and Strength from NGTDM.
Fig. 5.
Cluster heat map of radiomic features in non-harmonized data set. This illustration demonstrates that clustering cannot depict significant correlation or cluster in alive and deceased cases.
Fig. 6.
Radiomic features correlation using Pearson correlation in non-harmonized data set. Highly correlated features (both positive and negative) were eliminated from further analysis.
Fig. 7.
Heat map of AUC for cross combination of feature selectors and classifiers for the defined ten strategies. The heat map rows represent twenty-eighth cross combination of feature selectors and classifiers, whereas the columns depict strategies 1–10.
Fig. 8.
Heat map of sensitivity for cross combination of feature selectors and classifiers for the defined ten strategies. Heat map rows represent twenty-eighth cross combination of feature selectors and classifiers, whereas the columns depict strategies 1–10.
Fig. 9.
Heat map of specificity for cross combination of feature selectors and classifiers for the defined ten strategies. Heat map rows represent twenty-eighth cross combination of feature selectors and classifiers, whereas the columns depict strategies 1–10.
In strategy 1 where the data were randomly split into train and test sets (without considering centers), RFE feature selection and RF classifier results in the highest performance of AUC 0.84 ± 0.01 (CI95%: 0.82–0.85) with sensitivity and specificity of 0.78 and 0.76, respectively. In strategy 2 where only PCR positive studies were randomly split into train and test sets (without considering centers), KW feature selection and RF classifier combination resulted in the highest performance with an AUC of 0.84 ± 0.01 (CI95%: 0.82–0.86) and sensitivity and specificity of 0.81 and 0.76, respectively. There was no statistically significant difference (DeLong test, p-value>0.05) between Strategies 1 and 2, the main difference being the inclusion of CT and PCR positive studies in strategy one and only PCR positive studies in strategy 2.
In strategy 3, where whole data splitting was performed in each center separately for train and test sets, ANOVA feature selector and RF classifier combination resulted in the highest performance with AUC of 0.83 ± 0.01 (CI95%: 0.81–0.85), sensitivity and specificity of 0.81 and 0.72, respectively. Similar results as above were achieved for strategy 4 where data splitting was performed in each center separately to train and test set for PCR positive dataset. There was no statistically significant difference (DeLong test, p-value>0.05) between strategies 3 and 4, where the main difference was including CT and PCR positive in strategy 3 and only PCR positive in strategy 4.
In strategy 5, where Combat harmonized whole data splitting was performed in each center separately to the train, and test sets, Relief feature selector and RF classifier combination resulted in the highest performance with an AUC of 0.83 ± 0.01 (CI95%: 0.81–0.85), sensitivity and specificity of 0.77 and 0.74, respectively. In strategy 6, where Combat harmonized data splitting was performed in each center separately to the train, and test sets for PCR positive studies, Relief feature selector and RF classifier combination resulted in the highest performance with an AUC 0.83 ± 0.01 (CI95%: 0.81–0.84), sensitivity and specificity of 0.79 and 0.72, respectively.There were no statistically significant differences between strategies 5 and 6. The statistical comparison of AUCs between ComBat harmonization strategies 5 and 6 and the same splitting in strategies 3 and 4 using the DeLong test didn't reveal any statistically significant differences (p-value>0.05).
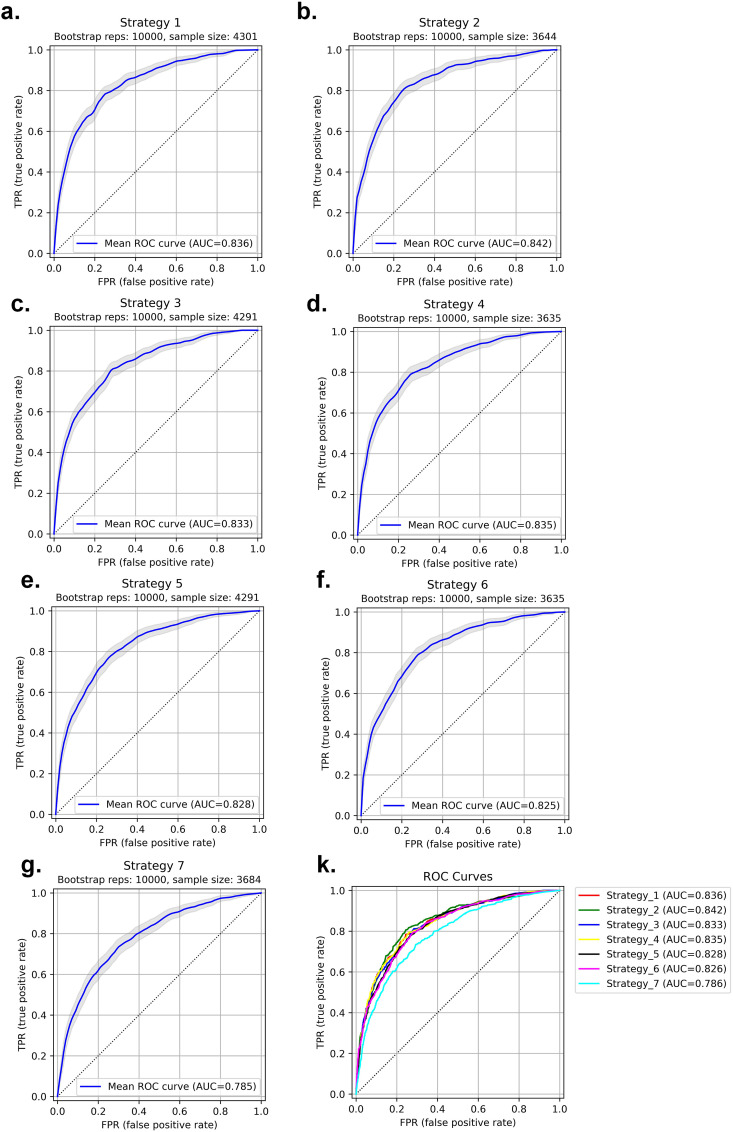
In strategy 7, where the splitting into train and test sets was performed based on centers (centers appear in training and test sets only once), the RFE selector and RF classifier combination resulted in the highest performance with an AUC of 0.79 ± 0.01 (CI95%: 0.76–0.81), sensitivity and specificity of 0.73 and 0.71, respectively. In Fig. 10 , the ROC curves of the test set for strategies 1–7, as well as the comparison of the different strategies are depicted. In strategy 8, where the model is built based on one center dataset (non-harmonized) and then evaluated on the 18 remaining centers (external validation set), ANOVA feature selector and NB classifier combination resulted in the highest performance with an AUC of 0.74 ± 0.034, sensitivity and specificity of 0.71 ± 0.026 and 0.69 ± 0.033, respectively. The results of each center were presented in Supplemental Table 12.
Fig. 10.
ROC curve for test sets in strategies 1–7. Strategy 1: AUC 0.84 ± 0.01 (a), Strategy 2: AUC 0.84 ± 0.01 (b), Strategy 3: AUC 0.83 ± 0.01 (c), Strategy 4: AUC 0.83 ± 0.01 (d), Strategy 5: AUC 0.83 ± 0.01 (e), Strategy 6: AUC 0.83 ± 0.01 (f), Strategy 7: AUC 0.79 ± 0.01 (g) and different strategies comparison (k). P-values for all ROCs were significant (p-value<0.05).
To evaluate the model on external validation sets, we reported the results of each center in the LOCO strategy 9. ANOVA feature selector and LR classifier combination resulted in the highest performance with an AUC of 0.80 ± 0.084, sensitivity and specificity of 0.77 ± 0.11 and 0.76 ± 0.075, respectively. In strategy 10, the data from each center (non-harmonized) were randomly split into 70% and 30% for training and test sets, respectively, and the models were built and evaluated on each center separately. ANOVA feature selector and LR classifier combination resulted in the highest performance with an AUC of 0.82 ± 0.10, sensitivity and specificity of 0.84 ± 0.12 and 0.77 ± 0.09, respectively. The results of each center were presented in Supplemental Tables 13 and 14 for strategies 9 and 10, respectively.
4. Discussion
In this multi-centric study, we conducted a CT-based radiomics analysis to assess the ability of our model in predicting the overall survival of patients with COVID-19 using a large multi-institutional dataset. We included 14,339 patients along with their CT images, segmented the lungs, and extracted distinct radiomics features. As there is no “one fits all” machine learning approaches for radiomic studies, given that their performance is task-dependent and there is large variability across models [[49], [50], [51], [52], [53], [54], [55]], we tested the cross-combination of four feature selectors and seven classifiers, which resulted in twenty-eight different combinations of algorithms to find the best performing model. Since the dataset was gathered from different centers, we applied the ComBat Harmonization algorithm that has been successfully applied in radiomics studies over the extracted features [25,55,56]. As our dataset consisted of imbalanced classes, we first used the SMOT algorithm in the training sets. Our model was trained, and the results of 3 different testing methods were reported.
Prognostic modeling can be regarded as an important framework towards a better understanding of the disease, its management, monitoring, and identification of the best treatment options. A number of reports have shown the effectiveness of image-based, laboratory-based, or combined models in outcome prediction of COVID-19 infected patients [57,58]. Qiu et al. [59] constructed a radiomics model trained to classify the severity of COVID-19 lesions (mild vs severe) using CT images. Their study included a medium-to-large number of patients (n = 1,160) and achieved an AUC of 0.87 in the test dataset. They showed that the radiomics signature is potent in aiding physicians to manage patients in a more precise way. Fu et al. [30] conducted a similar experiment with a radiomics-based model using CT images and applied it to data from 64 patients to classify them into progressive and stable groups. Their model could accurately perform the given task (AUC = 0.83). While the results were promising, their study did not include a large cohort.
A study by Chao et al. [58] included different types of information, such as CT-based radiomics features clinical, and demographic data, to employ a holistic prognostic model. Their model could predict whether the patients will demand an ICU admission or not with an AUC of 0.88. Tang et al. [60] also assessed a random forest model for classifying patients into categories of severe and non-severe based on CT imaging radiomics features along with laboratory test results. The model performed well (AUC = 0.98) on their dataset consisting of 118 patients. In a study by Wu et al. [61], the authors assessed the predictive power of a radiomic signature for showing poor patient outcomes defined as ICU admission, need for mechanical ventilation, or death. Their model could reach an AUC of 0.97 in the prediction of 28-day outcomes after CT images were taken. This highly promising result was achieved with the help of clinical data and the harmonization of the features. At the same time, in our study, ComBat harmonization did not appear to impact outcome prediction.
One should note that both clinical-only and radiomics-only survival prediction models have advantages. However, studies have shown that radiomics features yield superior accuracy in most cases. In a study by Homayounieh et al. [62], the authors developed a radiomics-based signature and compared it with a clinical-only signature in terms of mortality prediction. They concluded that the radiomics-based model can outperform the clinical-only model with a wide margin (AUC of 0.81 versus 0.68). Their study included 315 adults and was applied to other clinical outcomes as well, such as the prediction of outpatient/inpatient care and ICU admission. In addition, other reports indicated that adding clinical features to the radiomics model only slightly improved the results [63]. In a recent study, Shiri et al. [63] performed a radiomics study for prognostication purpose (alive or deceased) of COVID-19 patients using clinical (demographic, laboratory, and radiological scoring), COVID-19 pneumonia lesion radiomics features and whole lung radiomics features, separately and in combination. They trained a machine learning algorithm, Maximum Relevance Minimum Redundancy (MRMR) as the feature selector and XGBoost as the classifier, on 106 patients and evaluated and reported results on 46 test sets. They reported an AUC of 0.87 ± 0.04 for clinical-only, 0.92 ± 0.03 for whole lung radiomics, 0.92 ± 0.03 for lesion radiomics, 0.91 ± 0.04 for lung + lesion radiomics, 0.92 ± 0.03 for lung radiomics + clinical data, 0.94 ± 0.03 for lesion radiomics + clinical data and 0.95 ± 0.03 for lung + lesion radiomics + clinical data. The lung and lesion radiomics-only models showed similar performance, while the integration of features resulted in the highest accuracy.
Lassau et al. [64] combined CT-based DL models biological and clinical features for severity prediction in 1,003 COVID-19 patients, confirmed by either CT or RT-PCR. They showed clinical and biological features correlation with CT markers. Zhang et al. [57] conducted a diagnostic and prognostic study using 3,777 COVID-19 patients. They reported a high positive and negative correlation of lung-lesion CT manifestations with a number of clinical and laboratory tests. They also reported that their diagnostic model (COVID-19 from common pneumonia and normal control) can improve radiologists’ performance from junior to senior level (AUC = 0.98) for progression to severe/critical disease in their prognostic model. They reported an AUC of 0.90 with sensitivity and specificity of 0.80 and 0.86, respectively. Feng et al. [65] built a machine learning prognostic model using a multicenter COVID-19 dataset. They reported a high correlation of CT features with clinical findings, also utilizing a multivariable model in the validation set consisting of 106 patients. The AUC was 0.89 (95% CI: 0.81–0.98). Recently, Xu et al. [66] conducted a multicentric study for the prediction of ICU admission, mechanical ventilation, and mortality of hospitalized patients with COVID-19. CT radiomics features were integrated with demographic and laboratory tests. The evaluation was performed in 1362 patients from nine hospitals reporting an AUC of 0.916, 0.919 and 0.853 for ICU admission, mechanical ventilation, and mortality of hospitalized patients, respectively. For the radiomics-only model, they reached an AUC of 0.86, 0.80, and 0.66 for the above three mentioned outcomes, respectively.
Most previous studies suffered from a common limitation of COVID-19 RT-PCR not being available for the entire dataset when using multicentric data. In our study, COVID-19 positivity was confirmed by either RT-PCR or CT images, and different strategies were adopted to evaluate the models, including random splits and leave-one-center-out. We randomly split the data to train and test sets containing both CT positive and RT-PCR positive patients. Furthermore, to ensure the reproducibility of our results on RT-PCR positive patients, we split the dataset in a way that the test set consisted only of RT-PCR positive patients. To maximize the generalizability of the model and avoid overfitting on training sets, owing to variability in acquisition and reconstruction protocols, our model was developed on multicentric datasets with a wide variety of acquisition and reconstruction parameters. To test the generalizability of our model, we repeated the evaluation of our model using leave-one-center-out cross-validation. The results were reported for ten different strategies of splitting and cross-validation scenarios.
Several studies reported on the use of CT radiomics or DL algorithms for diagnostic and prognostic purposes in patients with COVID-19 [57,58]. However, most studies were performed using a small sample size. Overall, establishing evidence that radiomics features can help prioritize patients based on the severity of their disease and/or predicting their survival requires assessment using larger cohorts for a more generalizable model because of the wide variability in COVID-19 manifestations in different patients. In this study, we provided a large multinational multicentric dataset and evaluated our model in different scenarios to ensure model reproducibility, robustness and generalizability.
While attempting to address bias and limitations to create a generalizable model, the results should be interpreted considering some issues. First, motion artifacts were unavoidable in some COVID-19 patient scans which resulted in overlapping pneumonia regions. We removed patients with severe motion artifacts to omit this effect on model generalization. Second, we enrolled patients with common symptoms of COVID-19 whose infection was confirmed by either RT-PCR or CT imaging (typical manifestation of COVID-19 defined by interim guidelines). We handled this issue by testing different scenarios, including training a model using RT-PCR or CT positive patients and held out only RT-PCR patients in the test set and reported reproducible and repeatable results. Third, we did not include comorbidities (increased risk of adverse outcome) clinical or laboratory data during modeling. However, previous studies showed a high correlation of lung features with these findings [57,64,65]. Future studies combining various information to build a holistic model using a large dataset could improve the model's performance. Forth, we built a prognostic model based on all lung radiomics features. However, COVID-19 can result in imaging manifestations in other organs, such as the heart. Including features from different organs has the potential of improving prognostic performance [67]. Fifth, therapeutic regimens for different patients were not considered during modeling, although providing this information may help improving the accuracy of the model. Sixth, only binary classification was considered for the prognostic model in this study. Future studies should perform survival analysis using time-to-event models to account for the time of the adverse event. Lastly, we did not evaluate the impact of image acquisition or reconstruction parameters on radiomics features at the same time. We applied the ComBat harmonization algorithm to eliminate center-specific parameter effects on CT radiomics features.
5. Conclusion
A very large heterogeneous COVID-19 database was gathered from multiple centers, and a predictive model of survival outcome was derived and extensively tested to evaluate its reproducibility and generalizability. We demonstrated that lung CT radiomics features could be used as biomarkers for prognostic modeling in COVID-19. Through the use of a large imaging dataset, the predictive power of the proposed CT radiomics model is more reliable and may be prospectively used in a clinical setting to manage COVID-19 patients.
Data and code availability
Radiomics features and code would be available upon request.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Swiss National Science Foundation under grant SNRF 320030_176052.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105467.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Woolf S.H., Chapman D.A., Lee J.H. COVID-19 as the leading cause of death in the United States. JAMA. 2021;325:123–124. doi: 10.1001/jama.2020.24865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. 105924-105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai C.-C., Ko W.-C., Lee P.-I., Jean S.-S., Hsueh P.-R. Extra-respiratory manifestations of COVID-19. Int. J. Antimicrob. Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.106024. 106024-106024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J. Adv. Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill T.M. The central role of prognosis in clinical decision making. JAMA. 2012;307:199–200. doi: 10.1001/jama.2011.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan X., Han X., Peng D., Fan Y., Fang Z., Long D., Xie Y., Zhu S., Chen F., Lin W., Zhu Y. Clinical characteristics and prognosis of 218 patients with COVID-19: a retrospective study based on clinical classification. Front. Med. 2020;7 doi: 10.3389/fmed.2020.00485. 485-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou W., Qin X., Hu X., Lu Y., Pan J. Prognosis models for severe and critical COVID-19 based on the Charlson and Elixhauser comorbidity indices. Int. J. Med. Sci. 2020;17:2257–2263. doi: 10.7150/ijms.50007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pontone G., Scafuri S., Mancini M.E., Agalbato C., Guglielmo M., Baggiano A., Muscogiuri G., Fusini L., Andreini D., Mushtaq S. J Cardiovasc Comput Tomog; 2020. Role of Computed Tomography in COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang R., Li X., Liu H., Zhen Y., Zhang X., Xiong Q., Luo Y., Gao C., Zeng W. Chest CT severity score: an imaging tool for assessing severe COVID-19. Radiology: Cardioth. Imag. 2020;2 doi: 10.1148/ryct.2020200047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francone M., Iafrate F., Masci G.M., Coco S., Cilia F., Manganaro L., Panebianco V., Andreoli C., Colaiacomo M.C., Zingaropoli M.A., Ciardi M.R., Mastroianni C.M., Pugliese F., Alessandri F., Turriziani O., Ricci P., Catalano C. Chest CT score in COVID-19 patients: correlation with disease severity and short-term prognosis. Eur. Radiol. 2020;30:6808–6817. doi: 10.1007/s00330-020-07033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao W., Zhong Z., Xie X., Yu Q., Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. AJR Am. J. Roentgenol. 2020;214:1072–1077. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 12.Li K., Wu J., Wu F., Guo D., Chen L., Fang Z., Li C. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Invest. Radiol. 2020;55:327–331. doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts M., Driggs D., Thorpe M., Gilbey J., Yeung M., Ursprung S., Aviles-Rivero A.I., Etmann C., McCague C., Beer L., Weir-McCall J.R., Teng Z., Gkrania-Klotsas E., Ruggiero A., Korhonen A., Jefferson E., Ako E., Langs G., Gozaliasl G., Yang G., Prosch H., Preller J., Stanczuk J., Tang J., Hofmanninger J., Babar J., Sánchez L.E., Thillai M., Gonzalez P.M., Teare P., Zhu X., Patel M., Cafolla C., Azadbakht H., Jacob J., Lowe J., Zhang K., Bradley K., Wassin M., Holzer M., Ji K., Ortet M.D., Ai T., Walton N., Lio P., Stranks S., Shadbahr T., Lin W., Zha Y., Niu Z., Rudd J.H.F., Sala E., Schönlieb C.-B., Aix C. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat. Mach. Intell. 2021;3:199–217. [Google Scholar]
- 14.Harmon S.A., Sanford T.H., Xu S., Turkbey E.B., Roth H., Xu Z., Yang D., Myronenko A., Anderson V., Amalou A., Blain M., Kassin M., Long D., Varble N., Walker S.M., Bagci U., Ierardi A.M., Stellato E., Plensich G.G., Franceschelli G., Girlando C., Irmici G., Labella D., Hammoud D., Malayeri A., Jones E., Summers R.M., Choyke P.L., Xu D., Flores M., Tamura K., Obinata H., Mori H., Patella F., Cariati M., Carrafiello G., An P., Wood B.J., Turkbey B. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat. Commun. 2020;11:4080. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mei X., Lee H.C., Diao K.Y., Huang M., Lin B., Liu C., Xie Z., Ma Y., Robson P.M., Chung M., Bernheim A., Mani V., Calcagno C., Li K., Li S., Shan H., Lv J., Zhao T., Xia J., Long Q., Steinberger S., Jacobi A., Deyer T., Luksza M., Liu F., Little B.P., Fayad Z.A., Yang Y. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26:1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai W., Liu T., Xue X., Luo G., Wang X., Shen Y., Fang Q., Sheng J., Chen F., Liang T. CT quantification and machine-learning models for assessment of disease severity and prognosis of COVID-19 patients. Acad. Radiol. 2020;27:1665–1678. doi: 10.1016/j.acra.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lessmann N., Sánchez C.I., Beenen L., Boulogne L.H., Brink M., Calli E., Charbonnier J.P., Dofferhoff T., van Everdingen W.M., Gerke P.K., Geurts B., Gietema H.A., Groeneveld M., van Harten L., Hendrix N., Hendrix W., Huisman H.J., Išgum I., Jacobs C., Kluge R., Kok M., Krdzalic J., Lassen-Schmidt B., van Leeuwen K., Meakin J., Overkamp M., van Rees Vellinga T., van Rikxoort E.M., Samperna R., Schaefer-Prokop C., Schalekamp S., Scholten E.T., Sital C., Stöger L., Teuwen J., Vaidhya Venkadesh K., de Vente C., Vermaat M., Xie W., de Wilde B., Prokop M., van Ginneken B. 2020. Automated Assessment of CO-RADS and Chest CT Severity Scores in Patients with Suspected COVID-19 Using Artificial Intelligence, Radiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng L., Dong D., Li L., Niu M., Bai Y., Wang M., Qiu X., Zha Y., Tian J. IEEE J Biomed Health Inform; 2020. A Deep Learning Prognosis Model Help Alert for COVID-19 Patients at High-Risk of Death: A Multi-Center Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ning W., Lei S., Yang J., Cao Y., Jiang P., Yang Q., Zhang J., Wang X., Chen F., Geng Z., Xiong L., Zhou H., Guo Y., Zeng Y., Shi H., Wang L., Xue Y., Wang Z. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nat. Biomed. Eng. 2020 doi: 10.1038/s41551-020-00633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang M., He B., Li L., Dong D., Yang X., Li C., Meng L., Zhong L., Li H., Li H. CT radiomics can help screen the coronavirus disease 2019 (COVID-19): a preliminary study. Sci. China Life Sci. 2020;63:1–8. [Google Scholar]
- 21.Homayounieh F., Ebrahimian S., Babaei R., Karimi Mobin H., Zhang E., Bizzo B.C., Mohseni I., Digumarthy S.R., Kalra M.K. CT radiomics, radiologists and clinical information in predicting outcome of patients with COVID-19 pneumonia. Radiology: Cardioth. Imag. 2020;2 doi: 10.1148/ryct.2020200322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H., Wang L., Lee E.H., Zheng J., Zhang W., Halabi S., Liu C., Deng K., Song J., Yeom K.W. Decoding COVID-19 pneumonia: comparison of deep learning and radiomics CT image signatures. Eur. J. Nucl. Med. Mol. Imag. 2020:1–9. doi: 10.1007/s00259-020-05075-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiri I., Salimi Y., Saberi A., Pakbin M., Hajianfar G., Avval A.H., Sanaat A., Akhavanallaf A., Mostafaei S., Mansouri Z. medRxiv; 2021. Diagnosis of COVID-19 Using CT Image Radiomics Features: A Comprehensive Machine Learning Study Involving 26,307 Patients. [Google Scholar]
- 24.Rahmim A., Toosi A., Salmanpour M.R., Dubljevic N., Janzen I., Shiri I., Ramezani M.A., Yuan R., Ho C., Zaidi H. 2022. Tensor Radiomics: Paradigm for Systematic Incorporation of Multi-Flavoured Radiomics Features. arXiv preprint arXiv:2203.06314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiri I., Amini M., Nazari M., Hajianfar G., Haddadi Avval A., Abdollahi H., Oveisi M., Arabi H., Rahmim A., Zaidi H. Impact of feature harmonization on radiogenomics analysis: prediction of EGFR and KRAS mutations from non-small cell lung cancer PET/CT images. Comput. Biol. Med. 2022;142:105230. doi: 10.1016/j.compbiomed.2022.105230. [DOI] [PubMed] [Google Scholar]
- 26.Amini M., Nazari M., Shiri I., Hajianfar G., Deevband M.R., Abdollahi H., Arabi H., Rahmim A., Zaidi H. Multi-level multi-modality (PET and CT) fusion radiomics: prognostic modeling for non-small cell lung carcinoma. Phys. Med. Biol. 2021;66 doi: 10.1088/1361-6560/ac287d. [DOI] [PubMed] [Google Scholar]
- 27.Edalat-Javid M., Shiri I., Hajianfar G., Abdollahi H., Arabi H., Oveisi N., Javadian M., Shamsaei Zafarghandi M., Malek H., Bitarafan-Rajabi A., Oveisi M., Zaidi H. J Nucl Cardiol; 2020. Cardiac SPECT Radiomic Features Repeatability and Reproducibility: A Multi-Scanner Phantom Study. [DOI] [PubMed] [Google Scholar]
- 28.Bouchareb Y., Moradi Khaniabadi P., Al Kindi F., Al Dhuhli H., Shiri I., Zaidi H., Rahmim A. Artificial intelligence-driven assessment of radiological images for COVID-19. Comput. Biol. Med. 2021;136:104665. doi: 10.1016/j.compbiomed.2021.104665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdollahi H., Shiri I., Heydari M. Medical imaging technologists in radiomics era: an alice in wonderland problem. Iran. J. Public Health. 2019;48:184–186. [PMC free article] [PubMed] [Google Scholar]
- 30.Fu L., Li Y., Cheng A., Pang P., Shu Z. A novel machine learning-derived radiomic signature of the whole lung differentiates stable from progressive COVID-19 infection: a retrospective cohort study. J. Thorac. Imag. 2020 doi: 10.1097/RTI.0000000000000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Homayounieh F., Babaei R., Karimi Mobin H., Arru C.D., Sharifian M., Mohseni I., Zhang E., Digumarthy S.R., Kalra M.K. Computed tomography radiomics can predict disease severity and outcome in coronavirus disease 2019 pneumonia. J. Comput. Assist. Tomogr. 2020;44:640–646. doi: 10.1097/RCT.0000000000001094. [DOI] [PubMed] [Google Scholar]
- 32.Li C., Dong D., Li L., Gong W., Li X., Bai Y., Wang M., Hu Z., Zha Y., Tian J. IEEE J Biomed Health Inform; 2020. Classification of Severe and Critical COVID-19 Using Deep Learning and Radiomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai Q., Du S.Y., Gao S., Huang G.L., Zhang Z., Li S., Wang X., Li P.L., Lv P., Hou G., Zhang L.N. A model based on CT radiomic features for predicting RT-PCR becoming negative in coronavirus disease 2019 (COVID-19) patients. BMC Med. Imag. 2020;20:118. doi: 10.1186/s12880-020-00521-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue H., Yu Q., Liu C., Huang Y., Jiang Z., Shao C., Zhang H., Ma B., Wang Y., Xie G., Zhang H., Li X., Kang N., Meng X., Huang S., Xu D., Lei J., Huang H., Yang J., Ji J., Pan H., Zou S., Ju S., Qi X. Machine learning-based CT radiomics method for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: a multicenter study. Ann. Transl. Med. 2020;8:859. doi: 10.21037/atm-20-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bae J., Kapse S., Singh G., Phatak T., Green J., Madan N., Prasanna P. ArXiv; 2020. Predicting Mechanical Ventilation Requirement and Mortality in COVID-19 Using Radiomics and Deep Learning on Chest Radiographs: A Multi-Institutional Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tizhoosh H.R., Fratesi J. COVID-19, AI enthusiasts, and toy datasets: radiology without radiologists. Eur. Radiol. 2021;31:3553–3554. doi: 10.1007/s00330-020-07453-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Summers R.M. Artificial intelligence of COVID-19 imaging: a hammer in search of a nail. Radiology. 2021;298:E162–e164. doi: 10.1148/radiol.2020204226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambin P., Leijenaar R.T.H., Deist T.M., Peerlings J., de Jong E.E.C., van Timmeren J., Sanduleanu S., Larue R., Even A.J.G., Jochems A., van Wijk Y., Woodruff H., van Soest J., Lustberg T., Roelofs E., van Elmpt W., Dekker A., Mottaghy F.M., Wildberger J.E., Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 39.Mongan J., Moy L., Kahn J. Charles E. Checklist for artificial intelligence in medical imaging (CLAIM): a guide for authors and reviewers. Radiology: Artif. Intell. 2020;2 doi: 10.1148/ryai.2020200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prokop M., van Everdingen W., van Rees Vellinga T., Quarles van Ufford H., Stöger L., Beenen L., Geurts B., Gietema H., Krdzalic J., Schaefer-Prokop C., van Ginneken B., Brink M., Co-Rads A categorical CT assessment scheme for patients suspected of having COVID-19-definition and evaluation. Radiology. 2020;296:E97–e104. doi: 10.1148/radiol.2020201473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ning W., Lei S., Yang J., Cao Y., Jiang P., Yang Q., Zhang J., Wang X., Chen F., Geng Z., Xiong L., Zhou H., Guo Y., Zeng Y., Shi H., Wang L., Xue Y., Wang Z. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nat. Biomed. Eng. 2020;4:1197–1207. doi: 10.1038/s41551-020-00633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radpour A., Bahrami-Motlagh H., Taaghi M.T., Sedaghat A., Karimi M.A., Hekmatnia A., Haghighatkhah H.-R., Sanei-Taheri M., Arab-Ahmadi M., Azhideh A. COVID-19 evaluation by low-dose high resolution CT scans protocol. Acad. Radiol. 2020;27:901. doi: 10.1016/j.acra.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiri I., Arabi H., Salimi Y., Sanaat A., Akhavanallaf A., Hajianfar G., Askari D., Moradi S., Mansouri Z., Pakbin M., Sandoughdaran S., Abdollahi H., Radmard A.R., Rezaei-Kalantari K., Ghelich Oghli M., Zaidi H., Coli-Net Deep learning-assisted fully automated COVID-19 lung and infection pneumonia lesion detection and segmentation from chest computed tomography images. Int. J. Imag. Syst. Technol. 2021 doi: 10.1002/ima.22672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zwanenburg A., Vallières M., Abdalah M.A., Aerts H.J., Andrearczyk V., Apte A., Ashrafinia S., Bakas S., Beukinga R.J., Boellaard R. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., Beets-Tan R.G.H., Fillion-Robin J.C., Pieper S., Aerts H. Computational radiomics System to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2006;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 47.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12:1–8. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 49.Du D., Feng H., Lv W., Ashrafinia S., Yuan Q., Wang Q., Yang W., Feng Q., Chen W., Rahmim A. Machine learning methods for optimal radiomics-based differentiation between recurrence and inflammation: application to nasopharyngeal carcinoma post-therapy PET/CT images. Mol. Imag. Biol. 2019:1–9. doi: 10.1007/s11307-019-01411-9. [DOI] [PubMed] [Google Scholar]
- 50.Amini M., Hajianfar G., Hadadi Avval A., Nazari M., Deevband M.R., Oveisi M., Shiri I., Zaidi H. Overall survival prognostic modelling of non-small cell lung cancer patients using positron emission tomography/computed tomography harmonised radiomics features: the quest for the optimal machine learning algorithm. Clin. Oncol. 2022;34:114–127. doi: 10.1016/j.clon.2021.11.014. [DOI] [PubMed] [Google Scholar]
- 51.Shiri I., Maleki H., Hajianfar G., Abdollahi H., Ashrafinia S., Hatt M., Zaidi H., Oveisi M., Rahmim A. Next-generation radiogenomics sequencing for prediction of EGFR and KRAS mutation status in NSCLC patients using multimodal imaging and machine learning algorithms. Mol. Imag. Biol. 2020;22:1132–1148. doi: 10.1007/s11307-020-01487-8. [DOI] [PubMed] [Google Scholar]
- 52.Avard E., Shiri I., Hajianfar G., Abdollahi H., Kalantari K.R., Houshmand G., Kasani K., Bitarafan-Rajabi A., Deevband M.R., Oveisi M., Zaidi H. Non-contrast Cine Cardiac Magnetic Resonance image radiomics features and machine learning algorithms for myocardial infarction detection. Comput. Biol. Med. 2022;141:105145. doi: 10.1016/j.compbiomed.2021.105145. [DOI] [PubMed] [Google Scholar]
- 53.Khodabakhshi Z., Amini M., Mostafaei S., Haddadi Avval A., Nazari M., Oveisi M., Shiri I., Zaidi H. Overall survival prediction in renal cell carcinoma patients using computed tomography radiomic and clinical information. J. Digit. Imag. 2021;34:1086–1098. doi: 10.1007/s10278-021-00500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khodabakhshi Z., Mostafaei S., Arabi H., Oveisi M., Shiri I., Zaidi H. Non-small cell lung carcinoma histopathological subtype phenotyping using high-dimensional multinomial multiclass CT radiomics signature. Comput. Biol. Med. 2021;136:104752. doi: 10.1016/j.compbiomed.2021.104752. [DOI] [PubMed] [Google Scholar]
- 55.Shayesteh S., Nazari M., Salahshour A., Sandoughdaran S., Hajianfar G., Khateri M., Yaghobi Joybari A., Jozian F., Fatehi Feyzabad S.H., Arabi H., Shiri I., Zaidi H. Treatment response prediction using MRI-based pre-, post-, and delta-radiomic features and machine learning algorithms in colorectal cancer. Med. Phys. 2021;48:3691–3701. doi: 10.1002/mp.14896. [DOI] [PubMed] [Google Scholar]
- 56.Da-Ano R., Visvikis D., Hatt M. Harmonization strategies for multicenter radiomics investigations. Phys. Med. Biol. 2020;65:24tr02. doi: 10.1088/1361-6560/aba798. [DOI] [PubMed] [Google Scholar]
- 57.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K., Ye L., Gao M., Zhou Z., Li L., Wang J., Yang Z., Cai H., Xu J., Yang L., Cai W., Xu W., Wu S., Zhang W., Jiang S., Zheng L., Zhang X., Wang L., Lu L., Li J., Yin H., Wang W., Li O., Zhang C., Liang L., Wu T., Deng R., Wei K., Zhou Y., Chen T., Lau J.Y., Fok M., He J., Lin T., Li W., Wang G. Clinically applicable AI System for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181:1423–1433. doi: 10.1016/j.cell.2020.04.045. e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chao H., Fang X., Zhang J., Homayounieh F., Arru C.D., Digumarthy S.R., Babaei R., Mobin H.K., Mohseni I., Saba L., Carriero A., Falaschi Z., Pasche A., Wang G., Kalra M.K., Yan P. Integrative analysis for COVID-19 patient outcome prediction. Med. Image Anal. 2020;67 doi: 10.1016/j.media.2020.101844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiu J., Peng S., Yin J., Wang J., Jiang J., Li Z., Song H., Zhang W. A radiomics signature to quantitatively analyze COVID-19-infected pulmonary lesions. Interdiscipl. Sci. Comput. Life Sci. 2021:1–12. doi: 10.1007/s12539-020-00410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang Z., Zhao W., Xie X., Zhong Z., Shi F., Liu J., Shen D. Severity assessment of COVID-19 using CT image features and laboratory indices. Phys. Med. Biol. 2020 doi: 10.1088/1361-6560/abbf9e. [DOI] [PubMed] [Google Scholar]
- 61.Wu Q., Wang S., Li L., Wu Q., Qian W., Hu Y., Li L., Zhou X., Ma H., Li H., Wang M., Qiu X., Zha Y., Tian J. Radiomics Analysis of Computed Tomography helps predict poor prognostic outcome in COVID-19. Theranostics. 2020;10:7231–7244. doi: 10.7150/thno.46428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Homayounieh F., Ebrahimian S., Babaei R., Mobin H.K., Zhang E., Bizzo B.C., Mohseni I., Digumarthy S.R., Kalra M.K. CT radiomics, radiologists, and clinical information in predicting outcome of patients with COVID-19 pneumonia. Radiology: Cardioth. Imag. 2020;2 doi: 10.1148/ryct.2020200322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shiri I., Sorouri M., Geramifar P., Nazari M., Abdollahi M., Salimi Y., Khosravi B., Askari D., Aghaghazvini L., Hajianfar G., Kasaeian A., Abdollahi H., Arabi H., Rahmim A., Radmard A.R., Zaidi H. Machine learning-based prognostic modeling using clinical data and quantitative radiomic features from chest CT images in COVID-19 patients. Comput. Biol. Med. 2021 doi: 10.1016/j.compbiomed.2021.104304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lassau N., Ammari S., Chouzenoux E., Gortais H., Herent P., Devilder M., Soliman S., Meyrignac O., Talabard M.-P., Lamarque J.-P. Integrating deep learning CT-scan model, biological and clinical variables to predict severity of COVID-19 patients. Nat. Commun. 2021;12:1–11. doi: 10.1038/s41467-020-20657-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng Z., Yu Q., Yao S., Luo L., Zhou W., Mao X., Li J., Duan J., Yan Z., Yang M., Tan H., Ma M., Li T., Yi D., Mi Z., Zhao H., Jiang Y., He Z., Li H., Nie W., Liu Y., Zhao J., Luo M., Liu X., Rong P., Wang W. Early prediction of disease progression in COVID-19 pneumonia patients with chest CT and clinical characteristics. Nat. Commun. 2020;11:4968. doi: 10.1038/s41467-020-18786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Q., Zhan X., Zhou Z., Li Y., Xie P., Zhang S., Li X., Yu Y., Zhou C., Zhang L.J. 2020. CT-based Rapid Triage of COVID-19 Patients: Risk Prediction and Progression Estimation of ICU Admission, Mechanical Ventilation, and Death of Hospitalized Patients, medRxiv : the Preprint Server for Health Sciences. [Google Scholar]
- 67.Chassagnon G., Vakalopoulou M., Battistella E., Christodoulidis S., Hoang-Thi T.-N., Dangeard S., Deutsch E., Andre F., Guillo E., Halm N. AI-driven quantification, staging and outcome prediction of COVID-19 pneumonia. Med. Image Anal. 2021;67 doi: 10.1016/j.media.2020.101860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Radiomics features and code would be available upon request.