Abstract
With the advent of high-resolution and cost-effective genomics and bioinformatics tools and methods contributing to a large database of both human (HAdV) and simian (SAdV) adenoviruses, a genomics-based re-evaluation of their taxonomy is warranted. Interest in these particular adenoviruses is growing in part due to the applications of both in gene transfer protocols, including gene therapy and vaccines, as well in oncolytic protocols. In particular, the re-evaluation of SAdVs as appropriate vectors in humans is important as zoonosis precludes the assumption that human immune system may be naïve to these vectors. Additionally, as important pathogens, adenoviruses are a model organism system for understanding viral pathogen emergence through zoonosis and anthroponosis, particularly among the primate species, along with recombination, host adaptation, and selection, as evidenced by one long-standing human respiratory pathogen HAdV-4 and a recent re-evaluation of another, HAdV-76. The latter reflects the insights on amphizoonosis, defined as infections in both directions among host species including “other than human”, that are possible with the growing database of nonhuman adenovirus genomes. HAdV-76 is a recombinant that has been isolated from human, chimpanzee, and bonobo hosts. On-going and potential impacts of adenoviruses on public health and translational medicine drive this evaluation of 174 whole genome sequences from HAdVs and SAdVs archived in GenBank. The conclusion is that rather than separate HAdV and SAdV phylogenetic lineages, a single, intertwined tree is observed with all HAdVs and SAdVs forming mixed clades. Therefore, a single designation of “primate adenovirus” (PrAdV) superseding either HAdV and SAdV is proposed, or alternatively, keeping HAdV for human adenovirus but expanding the SAdV nomenclature officially to include host species identification as in ChAdV for chimpanzee adenovirus, GoAdV for gorilla adenovirus, BoAdV for bonobo adenovirus, and ad libitum.
Introduction
Adenoviruses are non-enveloped viruses with linear double-stranded DNA genomes of approximately 36 000 base pairs. The major capsid proteins of the adenovirus virion include the hexon, penton, and fibre proteins, which constitute the icosahedral outer shell. Each face of the icosahedron is composed of three identical copies of the hexon protein, with 12 pentons at the vertices of the icosahedron. Each penton unit comprises five identical penton proteins, with a fibre protein fixed to each vertex. This fibre is a trimer of three identical monomers. Sequence variation in the hexon and fibre proteins constitutes the major difference between adenoviral types (Harrach et al., 2011).
These viruses are found in all vertebrates examined to date, spanning fishes to amphibians to reptiles to birds to mammals (Harrach et al., 2011). The family Adenoviridae is subdivided into five genera into which human adenoviruses (HAdV) and simian adenoviruses (SAdV) comprise the genus Mastadenovirus (Harrach et al., 2011). Within the historical taxonomic scheme based on presumed hosts and biology (and speculation), arbitrarily separate HAdV and SAdV species were recognized, with the HAdV serotypes distributed across seven species, HAdV species A-G, and SAdV types distributed into at least three species, SAdV species A-C arbitrarily noted in the literature (Harrach et al., 2011; Chen et al., 2011b; Roy et al., 2012; Chiu et al., 2013; Malouli et al., 2014), or more, SAdV species A-G (Panto et al., 2015), all of which may or may not be recognized formally by the ICTV Adenovirus Working Group (Harrach et al., 2011). Recently, additional SAdV species D-I have been proposed to accommodate newly sequenced genomes (Dadakova et al., 2017). Oddly, a 2019 report of a global examination of selected adenoviruses noted the HAdV species but left SAdV entries as “unnamed” with respect to species (Harrach et al., 2019). Nevertheless, the shared, intertwined phylogenetic relationships between all SAdVs and HAdVs are noted in the speculation that several “members of three HAdV species most probably originated from [Old World Monkey] AdVs”, from one member of the ICTV (Panto et al., 2015). This reinforces the analyses presented in this report and supports the proposal that there should be no distinction between primate hosts amongst these clades or species, and, more importantly, the potential for zoonosis is substantial and worrisome (Bailey et al., 2018).
In this genomics era, HAdV identification and typing have evolved from the low-resolution, restrictive, and data-limited serological endeavours based solely on two antigenic epitopes (Aoki et al., 2011) to a broader, host-independent DNA sequence- and genome-based classification (Seto et al., 2011; Seto et al., 2013) that has allowed much more comprehensive and expansive views, with higher resolution understanding of their relationships providing important implications for their taxonomy. These implications include recognizing the branching of HAdV and SAdV genomes from a single phylogenetic tree with viruses of both groups forming subclades comprising the seven ICTV-recognized “HAdV” species and several novel but phylogenetically related species that could potentially include both SAdVs and HAdVs. That is, unlike the past taxonomic scheme of separating HAdV and SAdV species, the genomes of both “simian” and “human” AdV genotypes comingle, intertwine, and form clades and subclades together in one phylogenetic tree. Therefore, rather than segregating arbitrarily into HAdV species and SAdV species, we propose the designation of “primate adenovirus” (PrAdV) to recognize that these viruses encode highly similar genomes that share common ancestry and that apparently may interchange hosts. As the term “simian” in the past referred to only “nonhuman” SAdVs, for consistency with the historical literature, “simian” AdVs will refer to “nonhuman” simian AdVs in this report.
The timely re-evaluation of HAdV and SAdV taxonomy and phylogeny is of academic as well as clinical, epidemiological, and public health importance. HAdVs were among the first human viral respiratory pathogens to be isolated and characterized (Rowe et al., 1953; Hilleman and Werner, 1954). They are linked to a range of diseases in the respiratory, ocular, renal, hepatic, cardiac, and gastrointestinal systems, and are implicated in a metabolic disease, obesity (Arnold et al., 2010; Yamada et al., 2012; Lion, 2014). There are currently 103 GenBank-recognized genotypes of HAdVs (http://hadvwg.gmu.edu/), evaluated by a Human Adenovirus Working Group. Rather than being simply a basic biological research interest, SAdVs are increasingly recognized as important to human health from both an epidemiological perspective, e.g., zoonosis (Purkayastha et al., 2005a; Purkayastha et al., 2005b; Dehghan et al., 2013b), and a clinical applications perspective, e.g., gene delivery vectors (Graham and Prevec, 1992; Roy et al., 2004; Roy et al., 2006) and as oncolytic agents (Doronin and Shayakhmetov, 2012; Larson et al., 2015). Sequence recombination within the separate groups of HAdV and SAdV genomes has been reported as an important mechanism of adenovirus evolution, leading to novel and emergent viruses and pathogens (Walsh et al., 2009; Walsh et al., 2010; Robinson et al., 2011; Robinson et al., 2013; Hoppe et al., 2015b). Surprisingly, genome recombination between HAdVs and SAdVs has also been reported, suggesting zoonosis and anthroponosis (Dehghan et al., 2013a; Dehghan et al., 2019), along with complementing reports of cross-species viral transmissions between humans and other simians that elicited neutralizing antibodies in both (Xiang et al., 2006; Roy et al., 2009; Ersching et al., 2010; Wevers et al., 2011; Chen et al., 2011b; Chiu et al., 2013; Pauly et al., 2015; Hoppe et al., 2015a). To provide a high-resolution and more comprehensive view of the phylogenetic relationships and taxonomy of HAdVs and SAdVs, this report uses a broader examination of all published HAdV and SAdV genomes to yield heuristically parsimonious phylogenomic trees. This analysis yields a revised, broad taxonomic view of an intertwined phylogenetic tree with implications for human health, not only because these viruses are putative emergent and re-emergent pathogens (Xiang et al., 2006; Roy et al., 2009; Ersching et al., 2010; Wevers et al., 2011; Chen et al., 2011b; Chiu et al., 2013; Pauly et al., 2015; Hoppe et al., 2015a), but also because of their use as potential gene delivery and gene therapy vectors for medical therapies (Graham and Prevec, 1992; Roy et al., 2004; Roy et al., 2006). The latter is important as SAdVs have been presumed to be attractive, alternative vectors that bypass pre-existing human prior exposure and resultant immunity. It should be noted that due to the lack of whole genome data for many nonhuman and non-simian adenoviruses, the exploration of cross-species transmission may be incomplete. Therefore, this report is focused on primate hosts. To date, partial hexon sequences suggest cross-species transmissions may have occurred between human and bat hosts, and human and cat hosts, as noted in a recent literature survey of zoonosis, anthroponosis, and amphizoonosis (Borkenhagen et al., 2019). The hexon gene encodes the major adenovirus capsid protein, and is the source for antigenic variation within the adenoviral population. It has been shown to be an active locus for recombination between adenoviruses (Singh et al., 2012; Seto et al., 2013; Singh et al., 2015). It is the target for vaccine development. Hexon epitopes have been used as a marker for adenoviral identification and phylogeny for many years, prior to the advent of whole adenoviral genome sequencing (Aoki et al., 2011; Seto et al., 2011).
Materials and methods
Genomes
One hundred and seventy-four adenovirus whole genome sequences were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/). Among these, 98 genomes were from viruses infecting human hosts, 69 were isolated from simian hosts, and seven were from non-human and non-simian hosts, selected as representing outgroups for phylogeny analysis. Note, some genomes were deposited by researchers for patent application purposes. One example is SAdV-6, which apparently is a modified vector (genomic regions deleted and/or laboratory-derived chimeric) and therefore excluded from this study. All viruses were collected and reported in the literature between 1951 and 2017, with sequence data deposits relatively recent (1984–2007). Additional information associated with these viruses is described in Table S1.
Genome sequence processing
Genome sequence alignments of the 174 strains were generated using tools in the software Multiple Alignment using Fast Fourier Transform package (MAFFT version 7), with default parameters (https://mafft.cbrc.jp/alignment/server/). MAFFT was selected for all alignments, including gene sequences, due to its speed and its capacity to handle a large amount of data. The whole genome alignment was additionally, manually inspected for accuracy using BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html). To provide a more detailed analysis, eight genes and DNA sequences were selected, extracted, and analyzed in addition to the whole genome. These include the major capsid proteins used for recognizing and characterizing HAdV genotypes: penton base, hexon, and fibre. Additionally, the E1A, DNA polymerase, and E4ORF6 coding regions were sampled. The variable regions, loops 1 and 2, comprising one antigenic epitope, “epsilon”, are contained in the hexon gene, while the second antigenic epitope, “gamma”, is contained in the fibre gene. For individual gene processing, the genome coordinates of the start and end of each gene were mapped through GenBank records. Genes of interest were extracted from the whole genome alignment based its genome position in BioEdit. The sequences of each set of genes were aligned in MAFFT using default parameters. From this, both fasta and Nexus output alignment formats were saved. The fasta sequence alignment was visualized and inspected in BioEdit, and the Nexus alignment was used in PAUP to generate the heuristically parsimonious tree. Other alignment software, such as ClustalW2, were also used to confirm alignments produced by MAFFT, which was used for all of the trees. Trees generated from either MAFFT and ClustalW2 alignments were not significantly distinct.
Outgroup choices
Representative genomes were chosen as outgroups from the other genera in Adenoviridae for context, as the focus is on primate AdVs. Two AdVs were picked from each of the non-Mastadenovirus genera: Aviadenovirus, Atadenovirus, and Siaadenovirus. These were isolates from mammalian-distinct species including turkey, junglefowl, lizard, snake, frog, and penguin, respectively. A fourth genus, Ichtadenovirus, is not represented as the prototype (white sturgeon) genome only contains three coding sequences. As an additional outgroup reference, a closely related mammalian but nonprimate-hosted adenovirus, the northern treeshrew adenovirus (TAV), was used in the analyses. It forms a clade within the Mastadenovirus genus.
Phylogenetic analysis
Phylogenetic relationships amongst the sequences were determined using a heuristically parsimonious tree algorithm for the whole genomes and select individual genes as implemented by “Phylogenetic Analysis Using Parsimony” (PAUP, version 4.0a build156; http://paup.sc.fsu.edu/) with 1000 bootstrap replicates. All bootstrap analyses first were resampled with a full heuristic search. A random stepwise addition with 10 replicates was applied. The whole genome phylogenetic tree was also reconfirmed using the software “Tree analysis using New Technology” (TNT, version 1.5; http://www.lillo.org.ar/phylogeny/tnt/) (Nixon, 1999). A full search with Ratchet, tree drifting, and tree fusing set in default was used until an arbitrary minimum length was discovered by TNT that was hit five times as a stopping rule. Phylogenetic trees were annotated and manipulated in “Treegraph2” (version 2.13.0–748 beta; http://treegraph.bioinfweb.info/). Nodes and tip labels were edited in “Inkscape” (version 0.91; https://inkscape.org/en/).
Phylogenetic network
A phylogenetic networks analysis based on default parameters was constructed using “SplitsTree4” (version 2.14.6; http://www.splitstree.org/) (Huson and Bryant, 2006). This was performed by submitting a “characters block” to NeighborNet and EqualAngle tools implementing a Neighbor-net network and equal angle algorithm. A model comparison test was performed in jModelTest-2.1.10 to select the best-fit model of nucleotide substitution. The likelihood score with the best base tree search was computed and the model with the lowest Bayesian Information Criterion score (delta = 0) was considered to best fit the nucleotide substitution pattern. Consequently, the GTR + I + G nucleotide substitution model was chosen for the character transformations. Phylogenetic network analysis was used to account for the horizontal gene transfer, hybridization, and recombination using the realistic model. Nodes and edges were reformatted to illustrate a more accurate visualization.
Percent identity clustering
Genome sequence identity matrices for each lineage were generated using BioEdit. To analyze the identity matrices data, an R script was compiled to evaluate the percent identity clustering using the package ‘gplots’ (R Core Team 2017; https://cran.r-project.org/web/packages/gplots/index.html). Heat maps, reflecting sequence identities, for each designated PrAdV species were created to visualize the lineages in phylogenetic trees. The lowest percent identity between two human adenovirus genomes was set as the cutoff threshold for species discrimination.
Comparative genome analysis
Detailed character optimization analyses of the sequences at the nucleotide (“character”) level were performed. In the context of the very large dataset generated, to define lineages and gene changes that vary between subgroups of the phylogenetic tree of PrAdV that have CI = 1 were examined in more detail (Table S2). Conserved single nucleotide polymorphisms (SNP) across the genomes within the designated PrAdV-A, B, C, D, E, and H phylogenetic lineages (“species”) were interpreted.
Non-gap phylogenetic character changes were manually identified using the alignments in BioEdit. The Expasy translate tool allowed each nucleotide character identified to be translated into its corresponding protein sequence (http://web.expasy.org/translate/), which enabled the identification of point mutations based on nucleotide changes. Nucleotide and amino acid were then realigned in Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) to locate their positions and to confirm the changes in the genes.
Results
Phylogenetic analysis of the whole genomes
A heuristically parsimonious phylogenetic analysis was performed on available whole genome sequences as well as on select gene/coding sequences of 174 HAdV and SAdV isolates collected and reported between 1951 and the present. These genome sequences include representatives of all HAdV and SAdV genotypes, along with seven outgroup representatives for reference. In toto, these genomes range in size from 25 000 to 45 000 nucleotides, with the average genome length amongst Mastadenovirus viruses being approximately 35 000 nucleotides, reconfirming earlier analyses with an incomplete set of whole genome data (Seto et al., 2010). The average G + C content is about 56%, which is distinctive for the Mastadenovirus genus, with characteristic percentages reflecting each of the seven HAdV species, and again reconfirming earlier observations (Seto et al., 2010).
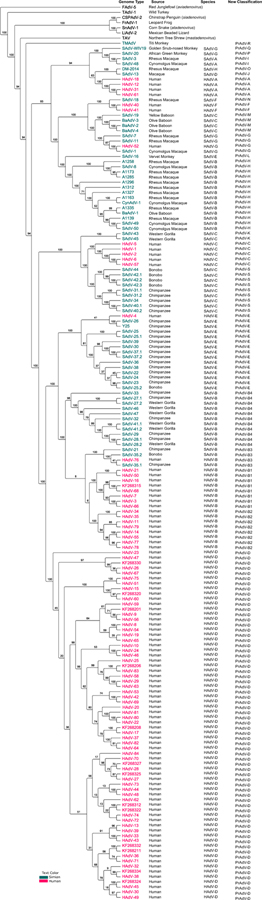
In this parsimony search, 68 414 characters were resampled within each replicate. Within this parsimony analysis, all characters have equal weight and are of the type ‘unord’. In summary, of the 68 414 total characters, 20 493 characters are constant, 9598 variable characters are parsimony-uninformative, and 38 323 characters are parsimony-informative. The latter class of characters was then used to generate the lineages in the heuristically parsimonious phylogenetic tree using TNT, shown in Fig. 1. Detailed information of the genomes is presented in Table S1, including GenBank accession numbers. Within this single phylogenetic tree, it should be noted that among clades comprising solely HAdV genotypes or SAdV genotypes, there are clades containing both. All clades branch closely together and given additional novel genomes, further iterations may show even closer phylogenetic relationships, as reported in the past, for example the inclusion of SAdV-21 into HAdV species B.
Fig. 1.
Heuristically parsimonious phylogenetic relationships of Mastadenovirus viruses. Whole genome sequences from 174 human, simian, and outgroup adenoviruses were aligned using MAFFT version 7 software, with default parameters (https://mafft.cbrc.jp/alignment/server/), and analyzed using the heuristically parsimonious phylogenetic tree algorithm embedded in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/). Sequences from our recent analysis are indicated with GenBank no’s (REF: PMID: 29410402). The iterations were run with default parameters and 1000 bootstrap replicates. The current species designations of the human and simian adenovirus species are noted, followed by the proposed new designation “primate adenovirus species” (PrAdV). This classification recognizes the intertwined genomes and close phylogenetic distances. Bootstrap values are shown, with 80% noted as robust.
A comparison of two phylogenetic analyses using PAUP and TNT algorithms reconfirmed the heuristically parsimonious tree results, as shown in Supplemental Figs. S1 and S2, respectively. The analyses provided similar Consistency Index (CI) and Retention Index (RI) values, and resulted in highly similar lineages in both trees. A RI value of 0.782 was generated from TNT, versus 0.7809 for PAUP, with a reference of “1” representing the scenario in which the characters fit the tree perfectly, in a range from 0 to 1 (Farris, 1989). For trees generated in TNT with equal weight, this indicates a moderate “fit for character” measure. TNT analysis yielded a CI of 0.289, with “1” representing no homoplasy, in a range from 0 to 1 (Kluge and Farris, 1969), which is consistent with the PAUP analysis at 0.2884. The CI value may indicate homoplasy in alignments, as the MAFFT tool generated reproducible alignments, which were additionally re-evaluated and confirmed manually, and were supported by the Mauve results. The total branch lengths generated using TNT and PAUP were also similar. The total length reported in TNT is 348 219, which is less than the 348 940, generated in PAUP. TNT showed a superior heuristic search than PAUP. Total branch length differences suggest fluctuations in evolutionary time estimation between nodes.
Nineteen distinct OTUs (Operational Taxonomic Units), clades, or lineages (to be referred to as “species” in accordance with the adenovirus literature) are classified from the whole genome heuristically parsimonious phylogenetic tree analysis. These species are proposed to be renamed “Primate Adenovirus” species (PrAdV-A to S) to acknowledge their close relationships branching from a single tree and to reflect the intermingling of viral lineages regardless of host. This tree includes the established and formally-recognized human adenovirus (HAdV-A to G) species along with previously proposed and/or informally recognized various SAdV species that are noted in the literature (Harrach et al., 2011; Chen et al., 2011b; Roy et al., 2012; Chiu et al., 2013; Malouli et al., 2014; Panto et al., 2015; Dadakova et al., 2017), to be renamed “primate adenoviruses” with PrAdV-A to –G for the former and PrAdV-H to -S for the latter. Again, as noted in Fig. 1, each of these species designations may include both HAdVs and SAdVs within the same clade, rather than the previous arbitrary separation and designation of species for each (Harrach et al., 2011; Chen et al., 2011b; Roy et al., 2012; Chiu et al., 2013; Malouli et al., 2014; Panto et al., 2015; Dadakova et al., 2017). This observation is consistent with recent reports of and analyses incorporating additional SAdV genomes (Roy et al., 2009; Wevers et al., 2010; Wevers et al., 2011; Pauly et al., 2015; Hoppe et al., 2015a; Hoppe et al., 2015b).
Several major clades in the whole genome tree showed a more significant lineage. For example, species PrAdV-D is distinct and separated from all other AdV species. PrAdV-B clade comprises HAdV-B and SAdV-B isolates. This supports the hypothesis that the distinction between the HAdVs and SAdVs are trivial. The PrAdV-B clade is further divided into four subclades: PrAdV-B1, PrAdV-B2, PrAdV-B3 and PrAdV-B4. The lineages of these heuristically parsimonious phylogenetic trees are consistent with previous analyses of smaller whole genome data sets and for individual HAdV genes.
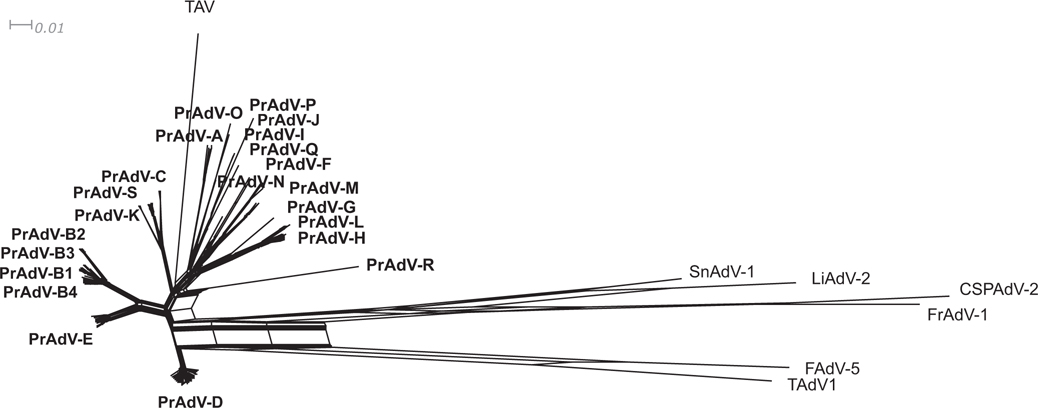
PrAdV-E comprises HAdV-4 and all SAdVs within the current HAdV-E species clade. PrAdV-C contains HAdV-C and a part of the SAdV-C subclade. PrAdV-G contains HAdV-52 and several SAdV-G strains. SAdV-43 and SAdV-45 form a separate clade, Finally, PrAdV-K is a clade of SAdV-43 and SAdV-45, leaving the following that do not cluster with any other genotypes and are separated into individual clades: SAdV-16, SAdV-18, SAdV-13, DM-2014, SAdV-20, SAdV-WIV19, and TMAdV. Note, changes to the existing parameters in PAUP generate the same results. The topology of the heuristically parsimonious phylogenetic tree computed from the whole genome data was similar to the topologies of heuristically parsimonious trees obtained for each individual gene and DNA sequence sets (hexon, penton base, fibre, DNA polymerase, E4ORF6, E1A, and hexon loops 1 and 2 (antigenic epitopes). That is, no significant changes in the clades were evident. Phylogenetic network analysis (Neighbor-net algorithm; SplitsTree4, v2.14.6) illustrated a similar relationship with multiple reticulate events at the outgroup regions and PrAdV-R. In general, both the phylogenetic network and the heuristically parsimonious tree produce similar clades and node connections, as shown in Fig. 2. This supports the parsing of species within human and simian adenoviruses.
Fig. 2.
Phylogenetic network analysis of Mastadenovirus genomes. Whole genome sequences (174 total) that have been archived in GenBank were collected and analyzed. A phylogenetic network was constructed using SplitsTree software with default parameters applied to character blocks using the GTR + I + G algorithm model (http://www.splitstree.org/). These genomes represented all human and simian adenoviruses contained in the genus Mastadenovirus, along with six non-Mastadenovirus outgroup viruses and one closely related nonprimate adenovirus (treeshrew, TAV). As shown in this network, the proposed primate adenovirus species (PrAdV) segregate into 19 species or clades, with one comprising subclades (PRAdV-B1 to B4). The ingroup for the adenovirus clades is in bold for each cluster. Provided is a scale bar that represents the number of weighted splits (“bipartisan”).
Percent identity matrix analysis
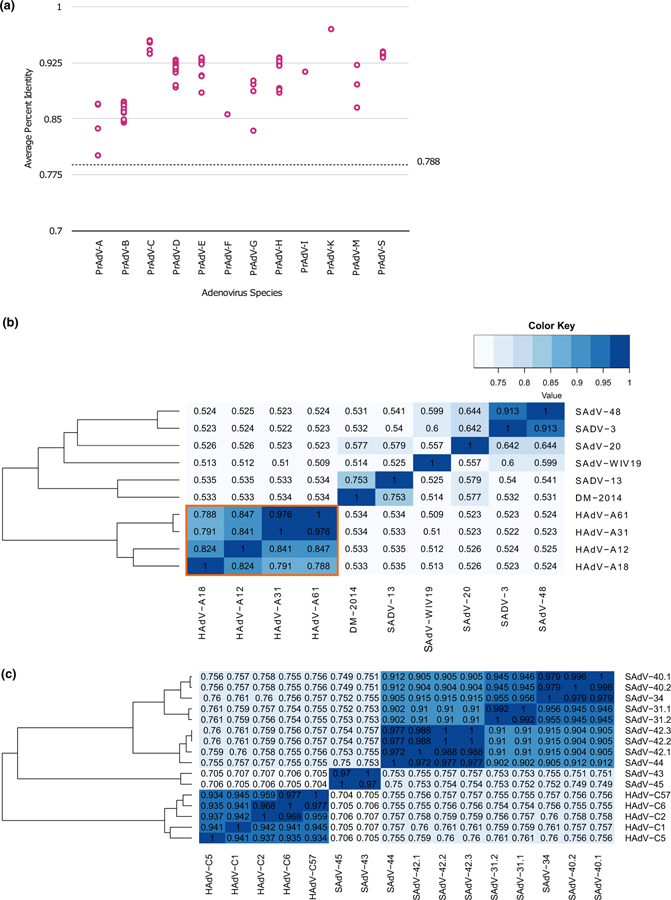
Percent identity matrices of whole genome sequences were generated to determine the level of sequence similarities supporting the phylogenetic clades. To be consistent with the literature, pairs of HAdV genomes were compared to yield the minimum threshold observed for the separation of these HAdVs. An arbitrary threshold cutoff of 78.8% was determined and designated to be the minimum for two HAdVs belonging within a species clade, as shown for PrAdV-A in Table 1; the next minimum value observed was 79.1%, for a pair of HAdVs in the PRAdV-B clade. This designated threshold value parses the whole genome phylogenetic tree into 19 distinct clades or species. As shown in Fig. 3A, the average percent identity separating each AdV from its nearest neighbor within each cluster is above the threshold value.
Table 1.
Genome percent identity matrix summary table for 19 distinct primate adenovirus species noting the number of samples. The grey highlights the cutoff value for speciation amongst these genomes. The minimum for each primate adenovirus species is above the cutoff.
| Type | # Isolates | Maximum | Minimum | Average | SD |
|---|---|---|---|---|---|
| PrAdV-A | 4 | 0.976 | 0.788 | 0.883 | 0.089 |
| PrAdV-B | 31 | 0.997 | 0.791 | 0.866 | 0.059 |
| PrAdV-C | 5 | 0.977 | 0.934 | 0.958 | 0.025 |
| PrAdV-D | 68 | 0.983 | 0.875 | 0.924 | 0.016 |
| PrAdV-E | 15 | 0.999 | 0.869 | 0.928 | 0.032 |
| PrAdV-F | 2 | 0.856 | 0.856 | 0.928 | 0.083 |
| PrAdV-G | 4 | 0.954 | 0.826 | 0.91 | 0.069 |
| PrAdV-H | 14 | 0.989 | 0.854 | 0.922 | 0.05 |
| PrAdV-I | 2 | 0.913 | 0.913 | 0.9565 | 0.05 |
| PrAdV-J | 1 | – | – | – | – |
| PrAdV-K | 2 | 0.97 | 0.97 | 0.986 | 0.017 |
| PrAdV-L | 1 | – | – | – | – |
| PrAdV-M | 4 | 1 | 0.86 | 0.926 | 0.061 |
| PrAdV-N | 1 | – | – | – | – |
| PrAdV-O | 1 | – | – | – | – |
| PrAdV-P | 1 | – | – | – | – |
| PrAdV-Q | 1 | – | – | – | – |
| PrAdV-R | 1 | – | – | – | – |
| PrAdV-S | 9 | 0.996 | 0.902 | 0.943 | 0.038 |
Fig. 3.
Percent identity analysis of human and simian adenovirus genomes. (a) Average percent identity analysis is plotted for individual pairs of adenovirus genomes within each primate adenovirus species. The cutoff threshold for separating clades is indicated, at 0.788. (b) Genome percent identity matrix is presented as a heatmap for the PrAdV-A, PrAdV-O, PrAdV-J, PrAdV-Q, PrAdV-P, and PrAdV-I pairs. (c) Genome percent identity matrix heatmap for the PrAdv-C, PrAdV-K, and PrAdV-S pairs. Heatmap values are presented such that the darker colour reflects higher similarity (“Colour Key”).
Genomes within each individual clade identify with themselves and are highly correlated, i.e., they have high sequence similarities to each other. For PrAdV-D, a total of 68 genomes were used in the percent identity matrix analysis, resulting in a minimal percent identity of 87.5%. Analysis of the 15 PrAdV-E genomes reveals a minimal percent identity of 86.9%. Average percent identities for PrAdV-E and PrAdV-D are 92.2% ± 0.13% and 92.3% ± 0.6%, respectively. The PrAdV-B heat map showed a clear separation among the four groups which directly correlate with the B1, B2, B3, and B4 in the whole genome tree. As a control, genomes have a percent identity of 1 when run against themselves. The average for individual clades is not skewed by this control in the matrix. The standard deviation shows that the error is in the acceptable range.
Character optimization of genome-wide substitution analysis
The character changes for whole genome parsimony tree lineages identified 119 important amino acid changes that may define the six commonly seen PrAdV species A total of 18 character informative silent mutations were observed throughout the genome. The genetic variations in all the genes define each PrAdV clade. Single nucleotide polymorphisms (SNPs) are presented in all gene regions except the whole L5 and part of E3 and E4 gene regions due to higher base variations. The most character changes observed are in pIVa2, polymerase, L1 52K, and L4 100K. Hexon sequences also contained a higher number of SNPs at the conserved area C1 and C4 epitope. A base change of G to A at the polymerase and pTP region resulted in two different amino acid change R to H (position 45) and V to I (position 618), respectively (Table 2).
Table 2.
Nonsynonymous SNPs in select genes that may define human adenoviruses comprising distinct PrAdV species. Twenty-one select genes spanning the adenovirus genome were selected and scored for conserved SNPs that are nonsynonymous. Those SNPs that are conserved amongst members of a species indicate a common ancestry and may be indicative of the species. For reference, human adenoviruses traditionally are classified across seven species (HAdV-A to G) and are proposed to be renamed as seven primate species (PrAdV-A to G), along with letters (H-S) to designate additional clades. Font colours green, blue, and orange indicate the polar, hydrophobic, and charged R-groups, respectively. Amino acids with dark font colour represent synonymous mutations
| E1A 28K | E1B 19K | E1B 55K | IX | IVa2 | pol | pTP | LI 52K | Ilia | Penton base | VII | V | X | VI | Hexon | Protease | DBP | L4 100K | L4 22K | L4 33K | VIII | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PrAdV-A | D256D | T107S | F370C |
S350T G286S E133Q L118L |
L1089I L1044S D953N S907S C758A A336K R45H* |
V618I* V524I H476L M447L R173Q |
G390A | A153T | R15R A34S |
R16R |
T99S R524R S579N |
H188N |
E539T K594R |
R141R | H178L |
Y13F S147T |
|||||
| PrAdV-B |
L232Y R321R |
M368L |
T65M Y100F |
||||||||||||||||||
| PrAdV-C | P19S |
E395Q T252I |
L94L |
R114K
T211M |
Q311L | K200R | Y14F |
G43N F702Y S729A |
I83V | K676R |
M31I Q53R |
||||||||||
| PrAdV-D | A74Q |
P225A T255S V271A K303Q Q358N |
V358A | G327A |
A44S S147A S224S E293R |
N357T Q425L |
K52R N218H |
R471K |
T97S
K110Q |
S311S |
A259I P261A G273A M302L Y357C Y602F E723V |
K172K |
R165N S174S |
||||||||
| PrAdV-E | C275N | P228I |
Y264F R275R |
S293N | T888N | T368I | M101L | ||||||||||||||
| PrAdV-H | E47A |
K554Q F338Y |
I587V |
M86L V127L K148Q P185A |
P29A S293S |
C358G | G20A |
S5S R46R |
P441T F347Y D303N |
C362A K363Q G495A L600A |
G133A | R51R |
Same base in an overlapping region.
Discussion
The goals of this study are to understand the taxonomic relationships between HAdVs and SAdVs based on whole genome, gene, and DNA sequences; to identify micro-evolutionary differences that may link to the causation of adenovirus variations; and to reaffirm AdVs as an exceptional model system in the genomics era, one in which the origins of emergent human pathogens from zoonotic and anthroponotic sources and their routes in adaptation to a new host can be elucidated. To that end, we present the largest, comprehensive, phylogenetic analysis performed to date of HAdVs and SAdVs circulating worldwide. The trees in this study are very similar to those constructed previously both as preliminary trials and in other studies reported, albeit in small-scale datasets and recovered using different phylogenetic methods (Prado-Irwin et al., 2018; Harrach et al., 2019), with some minor changes in tree topology that do not affect our major conclusions. This larger phylogenomic study of the primate adenoviruses, particularly of 174 whole genomes, allows a more precise and comprehensive understanding of the evolutionary relationship of human and simian adenoviruses.
Recognition and elucidation of virus-host interactions, particularly viral-host adaptive evolution, have been enhanced greatly by genomics technology and databases. Many viruses undergo cross-species transmission (Parrish et al., 2008), resulting in emergent pathogens and novel human diseases. Human Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) is an example of an emergent cross-species zoonotic pathogen first identified and characterized using DNA probe hybridization and array technology, coupled with the then-expanding genome and DNA sequence databases (Chen et al., 2011a). Subsequent and continuing analyses have identified bats as a reservoir or natural host (Li et al., 2005; Hu et al., 2017). A second example is the recent emergence of the Middle East Respiratory Syndrome-related Coronavirus (MERS-CoV) virus, from which genomic evidence has suggested origins or non-human reservoirs of bats and camels (Anthony et al., 2017; Chu et al., 2018).
HIV is yet another example of a zoonotically derived emergent human viral pathogen that has been thoroughly documented and analyzed using genomics and phylogeny analysis tools. The origins of HIV appear to lie in multiple cross-species infections from chimpanzees to human over a period of time, but with only one of these cases leading to the establishment of HIV in humans. This host adaptation was likely the result of genome changes that were selected for optimal virus fitness in the new host. The related human disease, AIDS, and epidemic were apparently driven and amplified by the confluence of social and technological events in 1920 Kinshasa, the Congo (Keele et al., 2006; Faria et al., 2014).
AdVs provide an ideal model system for applying genomics technology, bioinformatics methodology, and resultant sequence databases in studying and understanding virus-host interactions and host adaptation and virus evolution in the context of zoonosis and anthroponosis. HAdVs have been the subject of countless research inquiries across multiple disciplines and have been well-studied since the first dual simultaneous isolations in 1952 (Rowe et al., 1953; Hilleman and Werner, 1954). Through the years, a vast number of HAdVs, SAdVs, and other AdVs have been isolated, characterized, and genome-sequenced, with the most recent version of the GenBank database yielding 730 complete genomes across a myriad of vertebrate hosts (November 19, 2018). These AdV genomes are represented in the largest number with isolates from human and simian hosts. In particular, the diversity of the SAdVs provide multiple routes of zoonotic entry into new hosts and opportunities for host adaptation, particularly if the resultant human disease is benign or non-life threatening, as observed for most HAdV infections.
Zoonotic transmission of a TMAdV from titi monkeys to a human at the California National Primate Research Center, with subsequent infections and human familial transmission has been reported (Chen et al., 2011b). Another example of primate to human zoonosis was reported in an acute respiratory disease (ARD) outbreak involving baboon AdVs (BaAdV-3), in an olive baboon colony in 1997 at the Texas Biomedical Research Institute (Chiu et al., 2013). Following that event, neutralizing antibody titres for BaAdV-3 were detected in both baboons and humans, with the additional discovery of neutralizing antibodies amongst the healthy baboons from 1996 to 2003, as well as in staff personnel from 1997 (Chiu et al., 2013). In non-captive environments, a survey of human sera that tested positive for antibodies against SAdVs may be paired with a sub-Saharan Africa dataset that reported a PCR-based survey of nonhuman primates to support the hypothesis that cross-species transmissions occur between humans and non-human primates (Xiang et al., 2006; Wevers et al., 2011). Additional reports of cross-species transmission of AdVs include sequence data and/or sero-neutralization data (Ersching et al., 2010; Pauly et al., 2015; Hoppe et al., 2015a; Wang et al., 2018). Again, the importance of these reports is two-fold. First, the bidirectional transmissions and the genome similarities may limit the advantage of using SAdV genomes as vectors for gene therapy and vaccine development. Second, significant, novel and emergent human pathogens may arise.
As an example of a “significant, novel and emergent human pathogen” that has arisen, the highly contagious and “long-established” human respiratory pathogen HAdV-4 serves as a precedent, highlighting zoonosis and host adaptation through genome recombination. This genotype was one of the first HAdVs isolated and characterized as a human pathogen associated with ARD. Its pathology and epidemiology were of such importance that it is one of only two HAdVs for which vaccines have been developed, thrice (1956, 1971, and 2011; https://www.historyofvaccines.org/content/blog/adenovirus-vaccines-reinstated-after-long-absence) (Top et al., 1971; Lyons et al., 2008). Each vaccine pause resulted in the immediate re-emergence of HAdV-4 as a major pathogen (Barraza et al., 1999), as the prevalence of HAdV-4 returned to pre-vaccine levels upon discontinuation of the vaccine. Unexpectedly, this significant human pathogen was shown, using genomics and bioinformatics, to be a chimpanzee AdV (Purkayastha et al., 2005a; Purkayastha et al., 2005b; Dehghan et al., 2013b). An apparent recent genome recombination event involving a parental HAdV genome (Dehghan et al., 2013b) and its host protein-binding (NF1) motif that provides for optimized AdV replication in human cells (Nagata et al., 1982; Stillman et al., 1982; Kenny et al., 1988; Mul et al., 1990; Hatfield and Hearing, 1991). This may have facilitated its adaptation to humans (Dehghan et al., 2013b; Zhang et al., 2019; Coleman et al., 2020). Intriguingly, HAdV-4 is still the sole HAdV member of “HAdV” species E (Purkayastha et al., 2005a; Dehghan et al., 2013b), with chimpanzee AdVs comprising the rest of the clade. To re-emphasize, it is known as “HAdV-4” despite its high genome similarity to fourteen SAdVs, comprising a clade historically named “HAdV species E” or the proposed PrAdV-E, as shown in Figure S1. In more detail, HAdV-4 showed a ~100% genomic identity to SAdV-26, suggesting it is SAdV-26 (or vice versa as perhaps a simian host acquired HAdV-4 through anthroponosis, as it is difficult to ascertain with certainty the direction of gene/genome flow), which again highlights and supports the hypothesis that these HAdVs and SAdVs may be interchangeable with respect to these phylogenetically related hosts. As a model system, this raises the conundrum of whether nomenclature should be modified given high-resolution insights, that is, high-resolution data supersedes “historical entrenchment”, and provides a truer perspective for biological, clinical, and public health reality.
Chimpanzee AdVs, for example strain Y25, have been shown to be either antigenically related to HAdV-2 (Hillis et al., 1969) or distinguishable from HAdVs (Hillis et al., 1968). Other chimpanzee AdV isolates, including strains Pan5 and Pan 9, were not neutralized by antisera against HAdVs (Rogers et al., 1967; Basnight et al., 1971).
Complicating the cross-species transmissions are recent genome analysis reports on HAdVs and SAdVs that indicate recombination events may be more common than previously thought, both within AdVs of each species and also between AdVs of different host species. These are significant with respect to public health, as the recombination events do lead to novel, emergent human pathogens, e.g., epidemic keratoconjunctivitis (Walsh et al., 2009) and acute respiratory disease (Walsh et al., 2010). In humans, adenovirus genomes undergo relatively frequent genetic recombination, particularly in the hypervariable regions of the genome, with sequence parameters reported (Lee et al., 2018). Although there are not many reports to date of HAdVs and SAdVs cross-species recombinants in the literature, there is one compelling example. SAdV-35.1 and –35.2 have been isolated from captive populations of chimpanzees and bonobos at two different sites (Dehghan et al., 2013a). The genome sequences show a near 100% identity, hence the non-standard and confusing names. A third, archived genome from 1967, has been recently sequenced and reported to show near identity to the above two genomes as well (manuscript in preparation). As it was isolated from a human host and led to a respiratory illness and fatality, it has been renamed HAdV-76 (Dehghan et al., 2019), rather than “SAdV-35.3” or the original designation “21 + 16/16”. These three genomes contain recombination events from SAdV-21 (chimpanzee), HAdV-21 (human), SAdV-27 (chimpanzee), and HAdV-16 (human) (Dehghan et al., 2013a; Dehghan et al., 2019). This striking example underscores why AdVs may be one of the few, if not the only, model organisms to explore and understand the phenomenon of “amphizoonosis”, that is, cross-species transmissions occurring in both directions.
A further complication is anthroponosis, which is the transmission of microbe from human to other animal species (Yu et al., 2009). Of concern is transmission to nonprimate host species. Should subsequent amphizoonosis occur, that is, from nonhuman host species and back to humans, potential recombinant AdVs may result in novel, emergent human pathogens. Recent reports indicate that bat and cat harbour at least one AdV each that have gene sequences with significant sequence similarities to their HAdV counterparts (Ongradi, 1999; Phan et al., 2006; Baker et al., 2013; Ongradi et al., 2019). Phylogenetic analysis of the fruit bat AdV-1 hexon shows it forms a clade separate from previously sequenced bat AdVs, and clusters with the HAdV clade with a high bootstrap value of 93 (Baker et al., 2013). The bat hexon shared 77% and 90% amino acid similarities with the HAdV counterpart across 58 and 63 amino acids, respectively, of the 900 amino acid hexon protein. The hexon gene of an isolated cat AdV presents with high sequence similarity to HAdV-1 as well (Ongradi, 1999; Ongradi et al., 2019). Independently, and geographically separated, the isolation of a cat AdV in a one-year old child hospitalized for acute gastroenteritis was reported in 2006. Sequence analysis shows 100% amino acid sequence identity in the seven hypervariable regions of the hexon gene and 97% amino acid sequence identity in the fibre gene to the HAdV-1 counterparts (Phan et al., 2006).
The current nomenclature, formally ICTV-approved or informally proposed in reports, is highly confusing and is misleading. In the absence of guidance, suggestions for a universal set of rules go unheard as researchers assign apparently random names. The example alluded to in the previous discussion is indicative of many similar names for different AdVs, isolated from different hosts albeit with near-identical or highly similar genomes. Other examples include SAdV-35.1 (chimpanzee; New Iberia Research Center) and SAdV-35.2 (bonobo; San Diego Zoo); SAdV-37.1 (chimpanzee; New Iberia Research Center) and SAdV-37.2 (bonobo; San Diego Zoo); SAdV-25 and SAdV-25.1; SAdV-27.1 (chimpanzee; New Iberia Research Center); and SAdV-27.2 (gorilla; Atlanta Zoo). In contrast, SAdV-25 and SAdV-25.1 are both chimpanzee viruses. These isolates and the nomenclature confusion reflect the intriguing hypothesis that certain SAdVs and HAdVs are able to infect hosts across species lines, perhaps as multiple events across multiple species hosts over time, and recombine genomes. Although the directionality of transmission is difficult to determine, the result is a novel adenovirus that may be an emergent pathogen. In some, or perhaps most instances, the zoonosis or anthroponosis is short-lived as the virus does not have sufficient fitness to spread significantly in the new host, e.g., TMAdV and BaAdV-3. Perhaps in some instances, conditions allow for a suboptimal grasp on the new host, selecting for a mutation or recombinational event that allows the novel host-adapted virus to thrive in an immune-naïve population, e.g., HAdV-4.
It may be argued that all SAdVs isolated from the primates in captive settings actually originate from human caretakers, as primates in captivity are often bred for generations and may have never been in contact with wild primates. As a counterargument, it should also be noted that, with the exception of HAdV-4 and perhaps HAdV-76, these SAdVs have never been found in human populations, at least the past 67 years since the first recognition and isolation of HAdVs in 1952 (Rowe et al., 1953; Hilleman and Werner, 1954).
Examination of the sequences of 174 isolates suggested that adenoviruses may be parsed into 19 distinct clades. Of these, there may be something fundamentally different about PrAdV-D genomes. First, the PrAdV-D clade is found exclusively in human hosts. Also, PrAdV-D genotypes will only recombine with others within the PrAdV-D clade. This indicates that there might be a barrier between the PrAdV-D clade and other clades that prevents lateral genome transfer. More data are required for a better understanding of these distinctions of the PRAdV-D clade. The average sequence identities for PrAdV-E and PrAdV-D genomes are 0.928 ± 0.032 and 0.924 ± 0.016, respectively. The approximately 7% difference in genome percent identity are centered in the hypervariable regions that include the penton base, hexon, and fibre genes. Similar results have been observed in other studies with smaller datasets (Robinson et al., 2013). PrAdV-B clade is the second largest clade in the whole genome phylogenetic tree. It can be further divided into four subclades: PrAdV-B1, PrAdV-B2, PrAdV-B3, and PrAdV-B4. While PrAdV-B1, PrAdV-B2, and PrAdV-B4 each contain solely either HAdV or SAdV, PrAdV-B3 contains both, that is, the SAdV-21, SAdV-35.2, SAdV-35.1, and HAdV-76 genotypes. Specifically, SAdV-35.1, SAdV-35.2 and HAdV-76 shared a percent identity of > 0.996.
A total of 119 SNPs were identified for six PrAdV clades across four genes examined. These were found to have character informative mutations equal to or greater than the number of SNPs detected in DNA polymerase, which was used as a reference as it is presumably conserved. These four genes, IVa2, L1 52K, hexon C1 and C4 epitopes, and L4 100K may have conserved regions that are important for viral survival. All four genes are essential to produce mature virions, as IVa2 and L1 52K are required for DNA packaging (Zhang et al., 2001); L1 52K interacts with the IVa2 to aid the adenovirus assembly (Zhang et al., 2001; Ma and Hearing, 2011); and L4 100K is a chaperone protein for hexon and essential for cellular infectivity and adenovirus replication (Koyuncu and Dobner, 2009).
A difficulty and a caveat of this study is the potential quality issues in some of the sequences, as genomic sequences were performed by different laboratories, with a range of sequencing platforms and methodologies, and across three decades – some of the strains were sequenced when sequencing technology was still immature. As an example, the first HAdV genome sequences were composites generated in pieces from multiple research groups and included manual radioactive sequencing data. The first HAdV-17 genome data set has been discarded for being of poor quality; in fact, all of the original five HAdV genomes archived in GenBank have been resequenced. Nevertheless, inaccurate data would contribute to an imprecise result. However, since nearly all of the sequences were performed recently and as there are vast amounts of quality data involved in this study, it is likely that the effects of sequencing errors are insignificant.
A substantial amount of sampling bias is also observed in this study since HAdVs have been studied disproportionally more than other AdVs. Originally, SAdV sequences were few in number and some were poorly sequenced and annotated. Although HAdVs currently outnumber the SAdV sequences available in this study, there are now sufficient sequences to ensure an accurate analysis and conclusions. As more SAdVs are being isolated, even higher quality analyses can be performed. However, it is anticipated that these additional genomes will support and enhance the current findings.
Phylogenetic networks were computed to represent and quantify the uncertainty in the parsimony trees. The reticulate event in the neighbour-net may be due to genetic variations among distinct species. PrAdV-R stands out as an apparent anomaly among the various species comprising the primate adenoviruses as it seemingly separates like the outgroups, which are included for reference. As noted in Fig. 2, the network analysis displays lines for PrAdV-R and SnAdV-1, indicating past recombination events between the two. The overall layout of the tree and network displayed a high similarity that proves that sampling error and systematic error had little or no effect on the tree construction. Horizontal gene transfer and recombination were also taken into account in the phylogenetic network. This more realistic approach has proven the validity of using phylogenetic tree lineages for adenovirus speciation.
This study demonstrates that the differences between HAdVs and SAdVs are negligible in terms of phylogeny and taxonomy. Clades or “species” within this phylogenetic tree are intermixed with both AdVs as the relationships between HAdVs and SAdVs are complex and entangled, some of which have been revealed with high-resolution analyses. HAdVs and SAdVs do not appear to have evolved separately as would be implied by separate phylogenetic trees accepted by the ICTV Adenovirus Working Group. These AdVs are from the same origins and utilized cross-species recombination as an evolutionary mechanism to survive and prosper within hosts, presumably also in new hosts with immune-naïve surveillance. Again, the prospect and reality of zoonosis and anthroponosis of AdVs amongst human and other simian hosts is relevant in terms of public health and applications to medicine.
Supplementary Material
Fig. S1. Heuristically parsimonious phylogenetic tree of Mastadenovirus viruses. Whole genome sequences of 174 adenoviruses were aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the PAUP software (http://paup.sc.fsu.edu), with default parameters and 1000 bootstrap replicates. The scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as significant.
Fig. S2. Heuristically parsimonious phylogenetic tree of the Mastadenovirus viruses. Whole genome sequences of 174 adenoviruses were aligned using the “Multiple Alignment” software tool incorporating a” Fast Fourier Transform” (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using the heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/), with default parameters and 1000 bootstrap replicates. The scale bar indicates the branch length. Bootstrap values > 50% are displayed, with 80% noted as significant.
Fig. S3. Heuristically parsimonious phylogenetic tree of the adenoviral hexon gene with 1000 bootstrap replicates. Hexon gene sequences of 174 adenoviruses were extracted from the whole genome sequence and aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/), with default parameters and 1000 bootstrap replicates. Scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as significant.
Fig. S4. Heuristically parsimonious phylogenetic tree of the adenoviral penton base gene with 1000 bootstrap replicates. Penton base gene sequences of 174 adenoviruses were extracted from the whole genome sequence and aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/) with default parameters and 1000 bootstrap replicates. Scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as significant.
Fig. S5. Heuristically parsimonious phylogenetic tree of the adenoviral fibre gene with 1000 bootstrap replicates. Fibre gene sequences of 174 adenoviruses were extracted from the whole genome sequence and aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/) with default parameters and 1000 bootstrap replicates. Scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as significant.
Fig. S6. Heuristically parsimonious phylogenetic tree of the adenoviral DNA polymerase gene with 1000 bootstrap replicates. DNA polymerase gene sequences of 174 adenoviruses were extracted from the whole genome sequence and aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/) with default parameters and 1000 bootstrap replicates. Scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as significant.
Fig. S7. Heuristically parsimonious phylogenetic tree of the adenoviral E4ORF6 gene with 1000 bootstrap replicates. E4ORF6 gene sequences of 174 adenoviruses were extracted from the whole genome sequence and aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/) with default parameters and 1000 bootstrap replicates. Scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as significant.
Fig. S8. Heuristically parsimonious phylogenetic tree phylogeny of the adenoviral E1A gene with 1000 bootstrap replicates. E1A gene sequences of 174 adenoviruses were extracted from the whole genome sequence and aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/) with default parameters and 1000 bootstrap replicates. Scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as robust and significant.
Table S1. Inventory of adenoviruses contained within the genus Mastadenovirus used in this study. All genomes, available from GenBank, are noted along with their accession numbers, host from which the virus was isolated, year of isolation, and the country in which the virus was isolated. Simian adenoviruses, in many cases, were isolated from captive populations, including zoos and primate colonies established for medical and scientific research.
Table S2. Variable genes that may define select PrAdV clades, including PrAdV-A, B, C, D, E, and H. Each gene and protein with character informative changes are aligned to locate and confirm the SNPs for each clade. All synonymous and nonsynonymous SNPs are recorded based on the nucleotide and amino acid positions in the gene and gene product. These SNPs are conserved within individual clades which may serve as a potential indicator for a common ancestor.
Acknowledgments
In memoriam: Professor Clark Tibbetts (2019), who provided the impetus and inspiration for this work. D.S. also thanks Mr. Chuck Ellison for generously providing an exceptional, quiet writing retreat during Thanksgiving 2018 at “Maison Jaureguy” in Tardets-Sorholus, France, and Sr. Harold Suarez del Toro and the staff at the Biwa Hotel in Tulum, Mexico during Christmases 2018 and 2019 for the same, as well as for “bottomless” cups of coffee. We thank Dr. Patrick Gillevet for discussions and advice on the methods, analyses, and critical readings. This project was supported in part by U.S. Public Health Service grants EY013124, EY021558, and EY014104, and a Research to Prevent Blindness Senior Scientific Investigator Award (to J.C.).
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
Supporting Information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
References
- Anthony SJ, Gilardi K, Menachery VD, Goldstein T, Ssebide B, Mbabazi R, Navarrete-Macias I, Liang E, Wells H, Hicks A, Petrosov A, Byarugaba DK, Debbink K, Dinnon KH, Scobey T, Randell SH, Yount BL, Cranfield M, Johnson CK, Baric RS, Lipkin WI and Mazet JA, 2017. Further evidence for bats as the evolutionary source of middle east respiratory syndrome coronavirus. MBio 8, e00373–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Benko M, Davison AJ, Echavarria M, Erdman DD, Harrach B, Kajon AE, Schnurr D and Wadell G, 2011. Toward an integrated human adenovirus designation system that utilizes molecular and serological data and serves both clinical and fundamental virology. J. Virol. 85, 5703–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J, Janoska M, Kajon AE, Metzgar D, Hudson NR, Torres S, Harrach B, Seto D, Chodosh J and Jones MS, 2010. Genomic characterization of human adenovirus 36, a putative obesity agent. Virus Res. 149, 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey ES, Fieldhouse JK, Choi JY and Gray GC, 2018. A mini review of the zoonotic threat potential of influenza viruses, coronaviruses, adenoviruses, and enteroviruses. Front. Public Health 6, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KS, Leggett RM, Bexfield NH, Alston M, Daly G, Todd S, Tachedjian M, Holmes CE, Crameri S, Wang LF, Heeney JL, Suu-Ire R, Kellam P, Cunningham AA, Wood JL, Caccamo M and Murcia PR, 2013. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology 441, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraza EM, Ludwig SL, Gaydos JC and Brundage JF, 1999. Reemergence of adenovirus type 4 acute respiratory disease in military trainees: report of an outbreak during a lapse in vaccination. J. Infect. Dis. 179, 1531–1533. [DOI] [PubMed] [Google Scholar]
- Basnight M Jr, Rogers NG, Gibbs CJ Jr and Gajdusek DC, 1971. Characterization of four new adenovirus serotypes isolated from chimpanzee tissue explants. Am. J. Epidemiol. 94, 166–171. [DOI] [PubMed] [Google Scholar]
- Borkenhagen LK, Fieldhouse JK, Seto D and Gray GC, 2019. Are adenoviruses zoonotic? A systematic review of the evidence. Emerg. Microbes Infect. 8, 1679–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EC, Miller SA, DeRisi JL and Chiu CY, 2011a. Using a pan-viral microarray assay (Virochip) to screen clinical samples for viral pathogens. J. Vis. Exp. 2536–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EC, Yagi S, Kelly KR, Mendoza SP, Tarara RP, Canfield DR, Maninger N, Rosenthal A, Spinner A, Bales KL, Schnurr DP, Lerche NW and Chiu CY, 2011b. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a New World monkey colony. PLoS Pathog. 7, e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CY, Yagi S, Lu X, Yu G, Chen EC, Liu M, Dick EJ Jr, Carey KD, Erdman DD, Leland MM and Patterson JL, 2013. A novel adenovirus species associated with an acute respiratory outbreak in a baboon colony and evidence of coincident human infection. MBio 4, e00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DKW, Hui KPY, Perera R, Miguel E, Niemeyer D, Zhao J, Channappanavar R, Dudas G, Oladipo JO, Traore A, Fassi-Fihri O, Ali A, Demissie GF, Muth D, Chan MCW, Nicholls JM, Meyerholz DK, Kuranga SA, Mamo G, Zhou Z, So RTY, Hemida MG, Webby RJ, Roger F, Rambaut A, Poon LLM, Perlman S, Drosten C, Chevalier V and Peiris M, 2018. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc. Natl Acad. Sci. USA 115, 3144–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman KK, Wong CC, Jayakumar J, Nguyen TT, Wong AWL, Yadana S, Thoon KC, Chan KP, Low JG, Kalimuddin S, Dehghan S, Kang J, Shamsaddini A, Seto D, Su YCF and Gray GC, 2020. Adenoviral infections in Singapore: should new antiviral therapies and vaccines be adopted? J. Infect. Dis. 221, 566–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadakova E, Chrudimsky T, Brozova K, Modry D, Celer V and Hrazdilova K, 2017. New adenoviruses from new primate hosts – growing diversity reveals taxonomic weak points. Mol. Phylogenet. Evol. 107, 305–307. [DOI] [PubMed] [Google Scholar]
- Dehghan S, Seto J, Jones MS, Dyer DW, Chodosh J and Seto D, 2013a. Simian adenovirus type 35 has a recombinant genome comprising human and simian adenovirus sequences, which predicts its potential emergence as a human respiratory pathogen. Virology 447, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghan S, Seto J, Liu EB, Walsh MP, Dyer DW, Chodosh J and Seto D, 2013b. Computational analysis of four human adenovirus type 4 genomes reveals molecular evolution through two interspecies recombination events. Virology 443, 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghan S, Seto J, Liu EB, Ismail AM, Madupu R, Heim A, Jones MS, Dyer DW, Chodosh J and Seto D, 2019. A zoonotic adenoviral human pathogen emerged through genomic recombination amongst human and nonhuman simian hosts. J. Virol. 93, e00564–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronin K and Shayakhmetov DM, 2012. Construction of targeted and armed oncolytic adenoviruses. Methods Mol. Biol. 797, 35–52. [DOI] [PubMed] [Google Scholar]
- Ersching J, Hernandez MI, Cezarotto FS, Ferreira JD, Martins AB, Switzer WM, Xiang Z, Ertl HC, Zanetti CR and Pinto AR, 2010. Neutralizing antibodies to human and simian adenoviruses in humans and New-World monkeys. Virology 407, 1–6. [DOI] [PubMed] [Google Scholar]
- Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pepin J, Posada D, Peeters M, Pybus OG and Lemey P, 2014. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science 346, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris JS, 1989. The retention index and the rescaled consistency index. Cladistics 5, 417–419. [DOI] [PubMed] [Google Scholar]
- Graham FL and Prevec L, 1992. Adenovirus-based expression vectors and recombinant vaccines. Biotechnology 20, 363–390. [DOI] [PubMed] [Google Scholar]
- Harrach B, Benkö M, Both GW, Brown M, Davison AJ, Echavarria M, Hess M, Jones MS, Kajon A, Lehmkuhl HD, Mautner V, Mittal SK and Wadell G, 2011. Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. In: King AMQ, Adams MJ, Carstens EB and Lefkowitz EJ (Eds.), Family Adenoviridae. Elsevier Press, San Diego, CA, pp. 125–141. [Google Scholar]
- Harrach B, Tarjan ZL and Benko M, 2019. Adenoviruses across the animal kingdom: a walk in the zoo. FEBS Lett. 593, 3660–3673. [DOI] [PubMed] [Google Scholar]
- Hatfield L and Hearing P, 1991. Redundant elements in the adenovirus type 5 inverted terminal repeat promote bidirectional transcription in vitro and are important for virus growth in vivo. Virology 184, 265–276. [DOI] [PubMed] [Google Scholar]
- Hilleman MR and Werner JH, 1954. Recovery of new agent from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med. 85, 183–188. [DOI] [PubMed] [Google Scholar]
- Hillis WD, Holmes AW and Davison V, 1968. Serologic characterization of adenoviruses isolated from chimpanzees associated with viral hepatitis. Proc. Soc. Exp. Biol. Med. 129, 366–369. [DOI] [PubMed] [Google Scholar]
- Hillis WD, Garner AC and Hillis AI, 1969. A new simian adenovirus serologically related to human adenovirus type 2 and a chimpanzee with “viral hepatitis”. Am. J. Epidemiol. 90, 344–353. [DOI] [PubMed] [Google Scholar]
- Hoppe E, Pauly M, Gillespie TR, Akoua-Koffi C, Hohmann G, Fruth B, Karhemere S, Madinda NF, Mugisha L, Muyembe JJ, Todd A, Petrzelkova KJ, Gray M, Robbins M, Bergl RA, Wittig RM, Zuberbuhler K, Boesch C, Schubert G, Leendertz FH, Ehlers B and Calvignac-Spencer S, 2015a. Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Mol. Biol. Evol. 32, 2072–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe E, Pauly M, Robbins M, Gray M, Kujirakwinja D, Nishuli R, Boji Mungu-Akonkwa DD, Leendertz FH and Ehlers B, 2015b. Phylogenomic evidence for recombination of adenoviruses in wild gorillas. J. Gen. Virol. 96, 3090–3098. [DOI] [PubMed] [Google Scholar]
- Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B, Xie JZ, Shen XR, Zhang YZ, Wang N, Luo DS, Zheng XS, Wang MN, Daszak P, Wang LF, Cui J and Shi ZL, 2017. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 13, e1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH and Bryant D, 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. [DOI] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M and Hahn BH, 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313, 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny MK, Balogh LA and Hurwitz J, 1988. Initiation of adenovirus DNA replication. I. Mechanism of action of a host protein required for replication of adenovirus DNA templates devoid of the terminal protein. J. Biol. Chem. 263, 9801–9808. [PubMed] [Google Scholar]
- Kluge AG and Farris JS, 1969. Quantitative phyletics and the evolution of anurans. Syst. Biol. 18, 1–32. [Google Scholar]
- Koyuncu OO and Dobner T, 2009. Arginine methylation of human adenovirus type 5 L4 100-kilodalton protein is required for efficient virus production. J. Virol. 83, 4778–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson C, Oronsky B, Scicinski J, Fanger GR, Stirn M, Oronsky A and Reid TR, 2015. Going viral: a review of replication-selective oncolytic adenoviruses. Oncotarget 6, 19976–19989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Lee JS, Materne EC, Rajala R, Ismail AM, Seto D, Dyer DW, Rajaiya J and Chodosh J, 2018. Bacterial RecA protein promotes adenoviral recombination during in vitro, infection. mSphere 3, e00105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S and Wang LF, 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–679. [DOI] [PubMed] [Google Scholar]
- Lion T, 2014. Adenovirus infections in immunocompetent and immunocompromised patients. Clin. Microbiol. Rev. 27, 441–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons A, Longfield J, Kuschner R, Straight T, Binn L, Seriwatana J, Reitstetter R, Froh IB, Craft D, McNabb K, Russell K, Metzgar D, Liss A, Sun X, Towle A and Sun W, 2008. A double-blind, placebo-controlled study of the safety and immunogenicity of live, oral type 4 and type 7 adenovirus vaccines in adults. Vaccine, 26, 2890–2898. [DOI] [PubMed] [Google Scholar]
- Ma HC and Hearing P, 2011. Adenovirus structural protein IIIa is involved in the serotype specificity of viral DNA packaging. J. Virol. 85, 7849–7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malouli D, Howell GL, Legasse AW, Kahl C, Axthelm MK, Hansen SG and Fruh K, 2014. Full genome sequence analysis of a novel adenovirus of rhesus macaque origin indicates a new simian adenovirus type and species. Virol. Rep. 3–4, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mul YM, Verrijzer CP and van der Vliet PC, 1990. Transcription factors NFI and NFIII/oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J. Virol. 64, 5510–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Guggenheimer RA, Enomoto T, Lichy JH and Hurwitz J, 1982. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc. Natl Acad. Sci. USA 79, 6438–6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon KC, 1999. The Parsimony Ratchet, a new method for rapid Parsimony analysis. Cladistics 15, 407–414. [DOI] [PubMed] [Google Scholar]
- Ongradi J, 1999. Identification of a feline adenovirus isolate that replicates in monkey and human cells in vitro. Am. J. Vet. Res. 60, 1463. [PubMed] [Google Scholar]
- Ongradi J, Chatlynne LG, Tarcsai KR, Stercz B, Lakatos B, Pring-Akerblom P, Gooss D Sr, Nagy K and Ablashi DV, 2019. Adenovirus isolated from a cat is related to human adenovirus 1. Front. Microbiol. 10, 1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panto L, Podgorski II, Janoska M, Marko O and Harrach B, 2015. Taxonomy proposal for Old World monkey adenoviruses: characterisation of several non-human, non-ape primate adenovirus lineages. Adv. Virol. 160, 3165–3177. [DOI] [PubMed] [Google Scholar]
- Parrish CR, Holmes EC, Morens DM, Park EC, Burke DS, Calisher CH, Laughlin CA, Saif LJ and Daszak P, 2008. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 72, 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Akoua-Koffi C, Buchwald N, Schubert G, Weiss S, Couacy-Hymann E, Anoh AE, Mossoun A, Calvignac-Spencer S, Leendertz SA, Leendertz FH and Ehlers B, 2015. Adenovirus in rural Cote D’Ivoire: high diversity and cross-species detection. EcoHealth 12, 441–452. [DOI] [PubMed] [Google Scholar]
- Phan TG, Shimizu H, Nishimura S, Okitsu S, Maneekarn N and Ushijima H, 2006. Human adenovirus type 1 related to feline adenovirus: evidence of interspecies transmission. Clin. Lab. 52, 515–518. [PubMed] [Google Scholar]
- Prado-Irwin SR, van de Schoot M and Geneva AJ, 2018. Detection and phylogenetic analysis of adenoviruses occurring in a single anole species. PeerJ 6, e5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A, Ditty SE, Su J, McGraw J, Hadfield TL, Tibbetts C and Seto D, 2005a. Genomic and bioinformatics analysis of HAdV-4, a human adenovirus causing acute respiratory disease: implications for gene therapy and vaccine vector development. J. Virol. 79, 2559–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A, Su J, McGraw J, Ditty SE, Hadfield TL, Seto J, Russell KL, Tibbetts C and Seto D, 2005b. Genomic and bioinformatics analyses of HAdV-4vac and HAdV-7vac, two human adenovirus (HAdV) strains that constituted original prophylaxis against HAdV-related acute respiratory disease, a reemerging epidemic disease. J. Clin. Microbiol. 43, 3083–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CM, Singh G, Henquell C, Walsh MP, Peigue-Lafeuille H, Seto D, Jones MS, Dyer DW and Chodosh J, 2011. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology 409, 141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CM, Zhou X, Rajaiya J, Yousuf MA, Singh G, Deserres JJ, Walsh MP, Wong S, Seto D, Dyer DW, Chodosh J and Jones MS, 2013. Predicting the next eye pathogen: analysis of a novel adenovirus. MBio 4, e00595–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers NG, Basnight M, Gibbs CJ and Gajdusek DC, 1967. Latent viruses in chimpanzees with experimental kuru. Nature 216, 446–449. [DOI] [PubMed] [Google Scholar]
- Rowe WP, Huebner RJ, Gilmore LK, Parrott RH and Ward TG, 1953. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Exp. Biol. Med. 84, 570–573. [DOI] [PubMed] [Google Scholar]
- Roy S, Gao G, Lu Y, Zhou X, Lock M, Calcedo R and Wilson JM, 2004. Characterization of a family of chimpanzee adenoviruses and development of molecular clones for gene transfer vectors. Hum. Gene Ther. 15, 519–530. [DOI] [PubMed] [Google Scholar]
- Roy S, Zhi Y, Kobinger GP, Figueredo J, Calcedo R, Miller JR, Feldmann H and Wilson JM, 2006. Generation of an adenoviral vaccine vector based on simian adenovirus 21. J. Gen. Virol. 87, 2477–2485. [DOI] [PubMed] [Google Scholar]
- Roy S, Vandenberghe LH, Kryazhimskiy S, Grant R, Calcedo R, Yuan X, Keough M, Sandhu A, Wang Q, Medina-Jaszek CA, Plotkin JB and Wilson JM, 2009. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 5, e1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Sandhu A, Medina A, Clawson DS and Wilson JM, 2012. Adenoviruses in fecal samples from asymptomatic rhesus macaques, United States. Emerg. Infect. Dis. 18, 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto J, Walsh MP, Mahadevan P, Zhang Q and Seto D, 2010. Applying genomic and bioinformatic resources to human adenovirus genomes for use in vaccine development and for applications in vector development for gene delivery. Viruses 2, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto D, Chodosh J, Brister JR and Jones MS, 2011. Using the whole-genome sequence to characterize and name human adenoviruses. J. Virol. 85, 5701–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto D, Jones MS, Dyer DW and Chodosh J, 2013. Characterizing, typing, and naming human adenovirus type 55 in the era of whole genome data. J. Clin. Virol. 58, 741–742. [DOI] [PubMed] [Google Scholar]
- Singh G, Robinson CM, Dehghan S, Schmidt T, Seto D, Jones MS, Dyer DW and Chodosh J, 2012. Overreliance on the hexon gene, leading to misclassification of human adenoviruses. J. Virol. 86, 4693–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Zhou X, Lee JY, Yousuf MA, Ramke M, Ismail AM, Lee JS, Robinson CM, Seto D, Dyer DW, Jones MS, Rajaiya J and Chodosh J, 2015. Recombination of the epsilon determinant and corneal tropism: Human adenovirus species D types 15, 29, 56, and 69. Virology 485, 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman BW, Topp WC and Engler JA, 1982. Conserved sequences at the origin of adenovirus DNA replication. J. Virol. 44, 530–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top FH Jr, Grossman RA, Bartelloni PJ, Segal HE, Dudding BA, Russell PK and Buescher EL, 1971. Immunization with live types 7 and 4 adenovirus vaccines. I. Safety, infectivity, antigenicity, and potency of adenovirus type 7 vaccine in humans. J. Infect. Dis. 124, 148–154. [DOI] [PubMed] [Google Scholar]
- Walsh MP, Chintakuntlawar A, Robinson CM, Madisch I, Harrach B, Hudson NR, Schnurr D, Heim A, Chodosh J, Seto D and Jones MS, 2009. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS ONE 4, e5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MP, Seto J, Jones MS, Chodosh J, Xu W and Seto D, 2010. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J. Clin. Microbiol. 48, 991–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Li J, Wu S, Chen Y, Zhang Z, Zhai Y, Guo Q, Zhang J, Song X, Zhao Z, Hou L and Chen W, 2018. Seroepidemiological investigation of HAdV-4 infection among healthy adults in China and in Sierra Leone, West Africa. Emerg. Microbes. Infect. 7, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevers D, Leendertz FH, Scuda N, Boesch C, Robbins MM, Head J, Ludwig C, Kuhn J and Ehlers B, 2010. A novel adenovirus of Western lowland gorillas (Gorilla gorilla gorilla). Virol. J. 7, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevers D, Metzger S, Babweteera F, Bieberbach M, Boesch C, Cameron K, Couacy-Hymann E, Cranfield M, Gray M, Harris LA, Head J, Jeffery K, Knauf S, Lankester F, Leendertz SA, Lonsdorf E, Mugisha L, Nitsche A, Reed P, Robbins M, Travis DA, Zommers Z, Leendertz FH and Ehlers B, 2011. Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions. J. Virol. 85, 10774–10784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Li Y, Cun A, Yang W, Ellenberg S, Switzer WM, Kalish ML and Ertl HC, 2006. Chimpanzee adenovirus antibodies in humans, sub-Saharan Africa. Emerg. Infect. Dis. 12, 1596–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Hara K and Kadowaki T, 2012. Association of adenovirus 36 infection with obesity and metabolic markers in humans: a meta-analysis of observational studies. PLoS ONE 7, e42031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Zhou YJ, Li GX, Zhang GH, Liu HL, Yan LP, Liao M and Tong GZ, 2009. Further evidence for infection of pigs with human-like H1N1 influenza viruses in China. Virus Res. 140, 85–90. [DOI] [PubMed] [Google Scholar]
- Zhang W, Low JA, Christensen JB and Imperiale MJ, 2001. Role for the adenovirus IVa2 protein in packaging of viral DNA. J. Virol. 75, 10446–10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kang J, Dehghan S, Sridhar S, Lau SKP, Ou J, Woo PCY, Zhang Q and Seto D, 2019. A survey of recent adenoviral respiratory pathogens in Hong Kong reveals emergent and recombinant human adenovirus type 4 (HAdV-E4) circulating in civilian populations. Viruses 11, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Heuristically parsimonious phylogenetic tree of Mastadenovirus viruses. Whole genome sequences of 174 adenoviruses were aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the PAUP software (http://paup.sc.fsu.edu), with default parameters and 1000 bootstrap replicates. The scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as significant.
Fig. S2. Heuristically parsimonious phylogenetic tree of the Mastadenovirus viruses. Whole genome sequences of 174 adenoviruses were aligned using the “Multiple Alignment” software tool incorporating a” Fast Fourier Transform” (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using the heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/), with default parameters and 1000 bootstrap replicates. The scale bar indicates the branch length. Bootstrap values > 50% are displayed, with 80% noted as significant.
Fig. S3. Heuristically parsimonious phylogenetic tree of the adenoviral hexon gene with 1000 bootstrap replicates. Hexon gene sequences of 174 adenoviruses were extracted from the whole genome sequence and aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/), with default parameters and 1000 bootstrap replicates. Scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as significant.
Fig. S4. Heuristically parsimonious phylogenetic tree of the adenoviral penton base gene with 1000 bootstrap replicates. Penton base gene sequences of 174 adenoviruses were extracted from the whole genome sequence and aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/) with default parameters and 1000 bootstrap replicates. Scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as significant.
Fig. S5. Heuristically parsimonious phylogenetic tree of the adenoviral fibre gene with 1000 bootstrap replicates. Fibre gene sequences of 174 adenoviruses were extracted from the whole genome sequence and aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/) with default parameters and 1000 bootstrap replicates. Scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as significant.
Fig. S6. Heuristically parsimonious phylogenetic tree of the adenoviral DNA polymerase gene with 1000 bootstrap replicates. DNA polymerase gene sequences of 174 adenoviruses were extracted from the whole genome sequence and aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/) with default parameters and 1000 bootstrap replicates. Scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as significant.
Fig. S7. Heuristically parsimonious phylogenetic tree of the adenoviral E4ORF6 gene with 1000 bootstrap replicates. E4ORF6 gene sequences of 174 adenoviruses were extracted from the whole genome sequence and aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/) with default parameters and 1000 bootstrap replicates. Scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as significant.
Fig. S8. Heuristically parsimonious phylogenetic tree phylogeny of the adenoviral E1A gene with 1000 bootstrap replicates. E1A gene sequences of 174 adenoviruses were extracted from the whole genome sequence and aligned using a Multiple Alignment software tool incorporating a Fast Fourier Transform (MAFFT version 7) with default parameters (https://mafft.cbrc.jp/alignment/server/). Post-alignment, the data were parsed using a heuristically parsimonious phylogenetic tree algorithm in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/) with default parameters and 1000 bootstrap replicates. Scale bar indicates the branch length. Bootstrap values > 50% are shown, with 80% noted as robust and significant.
Table S1. Inventory of adenoviruses contained within the genus Mastadenovirus used in this study. All genomes, available from GenBank, are noted along with their accession numbers, host from which the virus was isolated, year of isolation, and the country in which the virus was isolated. Simian adenoviruses, in many cases, were isolated from captive populations, including zoos and primate colonies established for medical and scientific research.
Table S2. Variable genes that may define select PrAdV clades, including PrAdV-A, B, C, D, E, and H. Each gene and protein with character informative changes are aligned to locate and confirm the SNPs for each clade. All synonymous and nonsynonymous SNPs are recorded based on the nucleotide and amino acid positions in the gene and gene product. These SNPs are conserved within individual clades which may serve as a potential indicator for a common ancestor.