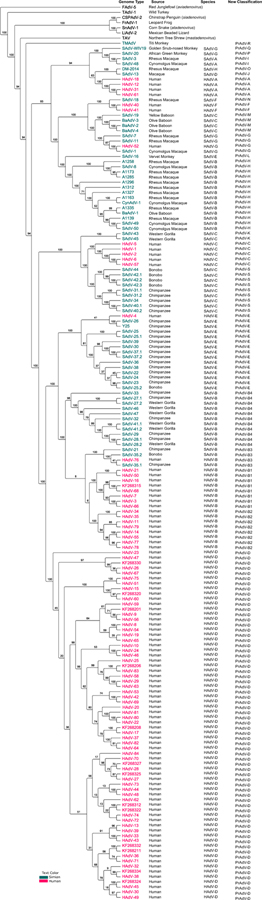
Fig. 1.
Heuristically parsimonious phylogenetic relationships of Mastadenovirus viruses. Whole genome sequences from 174 human, simian, and outgroup adenoviruses were aligned using MAFFT version 7 software, with default parameters (https://mafft.cbrc.jp/alignment/server/), and analyzed using the heuristically parsimonious phylogenetic tree algorithm embedded in the TNT software (http://www.lillo.org.ar/phylogeny/tnt/). Sequences from our recent analysis are indicated with GenBank no’s (REF: PMID: 29410402). The iterations were run with default parameters and 1000 bootstrap replicates. The current species designations of the human and simian adenovirus species are noted, followed by the proposed new designation “primate adenovirus species” (PrAdV). This classification recognizes the intertwined genomes and close phylogenetic distances. Bootstrap values are shown, with 80% noted as robust.