Abstract
Objective
To develop a fully automated full-thickness cartilage segmentation and mapping of T1, T1ρ, and T2*, as well as macromolecular fraction (MMF) by combining a series of quantitative 3D ultrashort echo time (UTE) Cones MR imaging with a transfer learning-based U-Net convolutional neural networks (CNN) model.
Methods
65 participants (20 normal, 29 doubtful-minimal osteoarthritis (OA), and 16 moderate-severe OA) were scanned using 3D UTE Cones T1 (Cones-T1), adiabatic T1ρ (Cones-AdiabT1ρ), T2* (Cones-T2*), and magnetization transfer (Cones-MT) sequences at 3T. Manual segmentation was performed by two experienced radiologists, and automatic segmentation was completed using the proposed U-Net CNN model. The accuracy of cartilage segmentation was evaluated using Dice score and volumetric overlap error (VOE). Pearson correlation coefficient and intraclass correlation coefficient (ICC) were calculated to evaluate the consistency of quantitative MR parameters extracted from automatic and manual segmentations. UTE biomarkers were compared among different subject groups using one-way ANOVA.
Results
The U-Net CNN model provided reliable cartilage segmentation with a mean Dice score of 0.82 and a mean VOE of 29.86%. The consistency of Cones-T1, Cones-AdiabT1ρ, Cones-T2*, and MMF calculated using automatic and manual segmentations ranged from 0.91 to 0.99 for Pearson correlation coefficients, and from 0.91 to 0.96 for ICCs, respectively. Significant increases in Cones-T1, Cones-AdiabT1ρ, and Cones-T2* (P<0.05), and a decrease in MMF (P<0.001) were observed in doubtful-minimal OA and/or moderate-severe OA over normal controls.
Conclusion
Quantitative 3D UTE Cones MR imaging combined with the proposed U-Net CNN model allows fully automated comprehensive assessment of articular cartilage.
Keywords: deep learning, cartilage, osteoarthritis, biomarkers
Introduction
Though conventional MRI may detect the morphological changes in articular cartilage associated with osteoarthritis (OA), these changes typically occur during the late stages of OA when treatment is much less effective. The development of novel MRI techniques capable of detecting changes that occur in the early stages of OA, alterations in proteoglycan (PG) and collagen which may even be present while cartilage is still considered intact, will be critical to improving disease diagnosis and treatment opportunity [1]. A large number of studies suggest that quantitative measurement of T1, T2, T2*, and T1ρ, as well as magnetization transfer (MT) effects, may provide a portfolio of sensitive biomarkers that would facilitate effective and comprehensive monitoring of the progression of OA and would guide better assessment of disease treatment in vivo. For example, T2 and T2* have been shown to be sensitive to collagen degradation [2, 3], T1ρ has been shown to be sensitive to PG depletion [4, 5], and T1 has been shown to be sensitive to water content in cartilage [6]. MT ratio (MTR) and, in particular, MT modeling of macromolecular fraction (MMF) have been shown to be sensitive to changes in macromolecules (e.g., PG and collagen) [7].
It is well-known that cartilage contains both long and short T2 tissue components. These include the more superficial layers which have long T2s and can be imaged with conventional MRI sequences, and the deep radial and calcified layers which have very short T2s and are “invisible” with conventional MRI [8]. Fortunately, ultrashort echo time (UTE) MR sequences with echo times (TEs) ~100 times shorter than the TEs of conventional sequences are able to directly image these short T2 tissues whose alterations may contribute to OA and its symptoms [9–12]. Furthermore, with the widening acceptance of OA as a whole-organ disease, the development and validation of novel sequences which can provide quantitative assessment of all the disease’s major joint components are necessary for a truly comprehensive evaluation of OA [13, 14]. A series of 3D UTE Cones sequences to measure T1 (Cones-T1), adiabatic T1ρ (Cones-AdiabT1ρ), T2* (Cones-T2*), and MMF (by modeling Cones-MT data) of various musculoskeletal (MSK) tissues had been proposed [15–18].
Typically, in order to acquire the quantitative MR parameters for assessment of cartilage, the bone-cartilage interface and the boundary between cartilage and its adjacent structures must first be determined over the entire articulating surface. This segmentation process is time-consuming and prone to inter-observer variability and bias by manual methods [19, 20]. There has been great interest in developing an accurate and fully automated method to segment cartilage which would allow a more seamless workflow to obtain quantitative biomarkers.
Recently, deep learning has surfaced as a powerful machine learning approach. Unlike traditional machine learning techniques, which require a considerable amount of expertise and experience in data engineering and computer science to design and operate the networks, deep learning is a simple and versatile method based on a purely data-driven approach that can be easily adapted to a diverse array of datasets and problems [21, 22]. One such deep learning technique, U-Net, is based on deep convolutional neural networks (CNN), effective networks that can process images and which are based on a fully convolutional network comprised of two main paths: an encoder (or contracting) path and a decoder (or expansive) path. Moreover, shortcut connections are commonly added between the layers to improve object localization [23]. The purpose of this study was to develop a comprehensive approach for fully automated knee cartilage segmentation and extraction of quantitative parameters, including Cones-T1, Cones-AdiabT1ρ, and Cones-T2* relaxometry, and MMF, by combining a series of quantitative 3D UTE Cones MR imaging with a transfer learning-based U-Net CNN model.
Methods
Subjects
The study was approved by the local Institutional Review Board. A total of 65 human subjects (54.8±16.9 years; 33 females) was included in the study, and informed consent was obtained from all subjects. According to condensed Kellgren–Lawrence (KL) grade [24], subjects were categorized into three groups: normal controls (20, KL=0), doubtful-minimal OA (29, KL=1–2), and moderate-severe OA (16, KL=3–4). The whole knee joint (29 left, 36 right) was scanned using a transmit/receive 8-channel knee coil on a 3T clinical MR system (MR750, GE Healthcare Technologies).
MRI protocol
The protocol included a series of 3D UTE Cones sequences to measure Cones-T1, Cones-AdiabT1ρ, Cones-T2*, and MMF. Cones-T1 was performed based on actual flip angle imaging followed by variable flip angle (AFI-VFA) imaging, where four different flip angles (5°, 10°, 20°, and 30°) were applied [15]. Cones-AdiabT1ρ was performed using paired adiabatic inversion pulse (Silver-Hoult pulse, duration=6 ms, bandwidth=1500 Hz) for magic angle-insensitive spin-locking [16, 25]. The degree of spin-locking was controlled by changing the number of adiabatic inversion pulses (NIR) between 0, 2, 4, 6, 8, 12, and 16. Cones-T2* was performed using fat-suppressed multi-echo UTE imaging (TEs=0.032, 4.4, 8.8, 13.2, 17.6, and 22 ms). MMF was calculated from 3D UTE Cones-MT data with three radiofrequency (RF) powers (500°, 1000°, and 1500°) and five frequency offsets (2, 5, 10, 20, and 50 kHz) using a two-pool UTE-MT model [18, 26]. Other parameters included sagittal plane, field of view (FOV)=15×15×10.8 cm3, receiver bandwidth=166 kHz, and matrix=256×256×36 (except for AFI, where a smaller matrix of 128×128×18 was used). The total scan time for all four UTE Cones sequences was approximately 45 minutes.
Input data processing
Motion registration was applied to all 3D UTE Cones images using Elastix software [27], where a rigid affine transform was followed by a non-rigid b-spline registration [28]. Cones-AdiabT1ρ-weighted images with an NIR of 2, which provided the best cartilage contrast, were manually segmented by two MSK radiologists (Rad1 and Rad2) with 18 and 13 years of experience, respectively, and were input to the CNN model as ground truth. The same manual segmentation masks were applied to registered Cones-T1, Cones-T2*, and Cones-MT data for subsequent processing. The input images were pre-processed using a local contrast normalization [29] before inputting to the CNN model along with the corresponding manual labels of cartilage. Then, the 3D images were broken into 2D images (24 central slices were selected from each subject), which were input to the CNN model. All 65 subjects were divided into 40, 10, and 15 subjects for training, validation, and testing. Note that a validation dataset is commonly required to ensure an unbiased evaluation of model fit while tuning hyper-parameters in training. The training data were augmented by image rotation (5, 0, or −5 degree) with or without horizontal flipping to produce six times larger training data.
The images were analyzed using MATLAB 2017b (The MathWorks Inc.). A single-component model was applied to fit Cones-T1, Cones-AdiabT1ρ, and Cones-T2* using non-linear optimization based on a Levenberg–Marquardt algorithm, as described in earlier publications [15–17]. A modified rectangular pulse (RP) approximation model was applied to the Cones-MT data to extract MMF [30]. Morphological 3D UTE Cones images as well as quantitative maps were then input to our CNN model for further processing.
CNN model architecture
In this study, we used a modified attention U-Net CNN model for cartilage segmentation. The U-Net architecture for this model has achieved excellent segmentation performance in the past [23, 31]. In a standard U-Net CNN model, feature maps from the encoder path are directly transferred to the decoder path. Our network, on the other hand, includes attention gates which process feature maps propagated through skip connections from the encoder path in order to suppress irrelevant regions of the transferred feature maps and improve the model’s learning process. As shown in Figure 1, the attention gates of our model account for output of decoder convolutional blocks in order to determine what the important regions of the image are and consequently filter feature maps accordingly from the encoder path. Networks equipped with attention gates are usually able to focus better on important image structures and therefore provide better segmentation performance [32]. Since automatic cartilage segmentation is considered a technical challenge owing to the tissue’s irregular shape and the fact that it constitutes only a small portion of the entire image, our network’s inclusion of the attention mechanism is a significant improvement over the standard model’s ability to effectively account for imaging variations in cartilage appearance.
Figure 1.
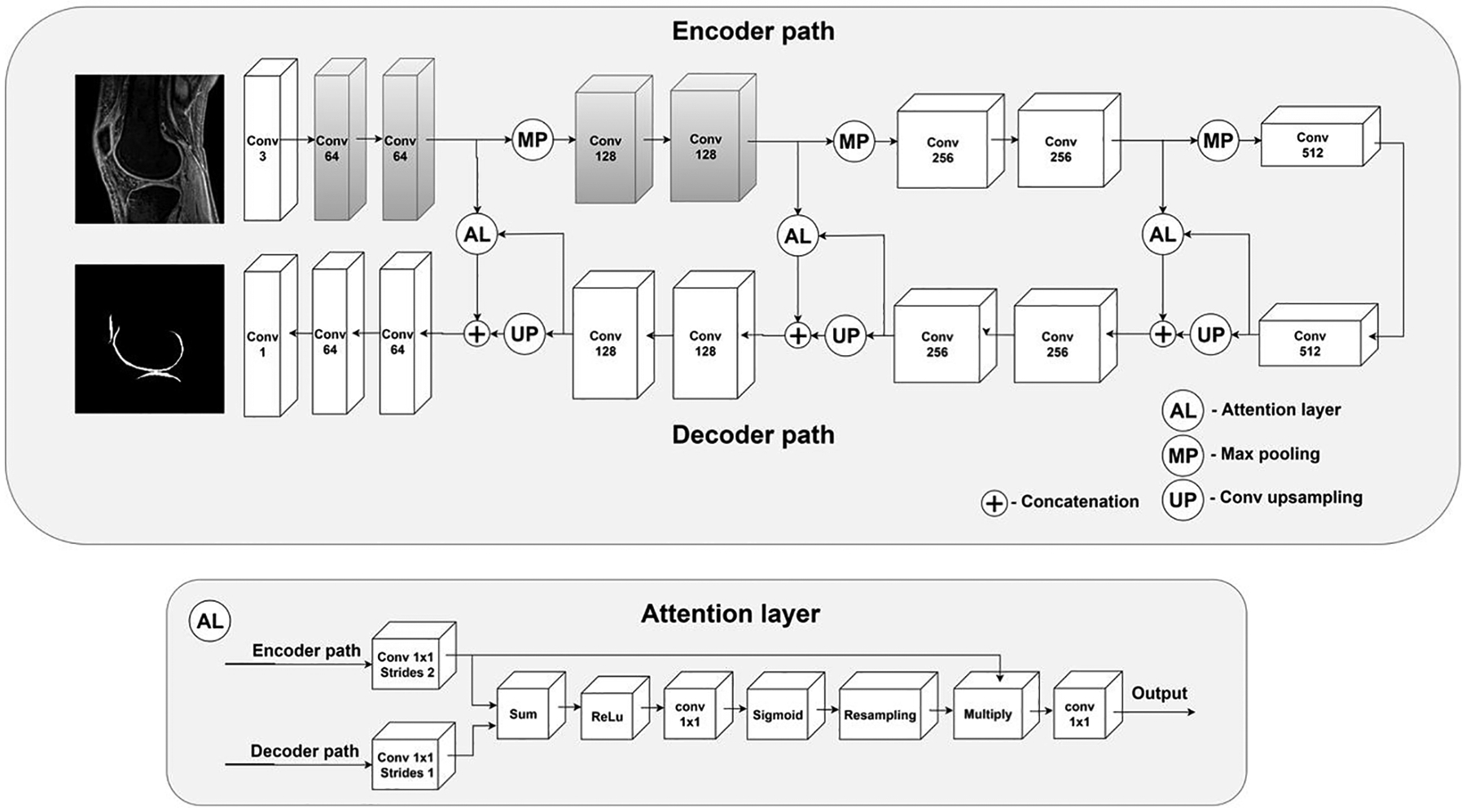
The architecture of the proposed 2D attention U-Net CNN model for automatic cartilage segmentation. Note that the blocks shaded in gray indicate the weights initialized from the VGG19 network for transfer learning.
Deep learning models in medical image analysis are commonly developed using transfer learning methods to allow robust training despite a limited number of input data, with the aim to leverage deep CNNs pre-trained on non-medical image datasets [33]. The first convolutional blocks in pre-trained deep models can be used to extract generic features from input images [34]. Building on this, we applied a new transfer learning method in this study to improve the network’s learning process and its object recognition capabilities. The first two convolutional blocks were replaced with the corresponding blocks from the VGG19 CNN pre-trained on the ImageNet dataset [35]. As a result, our network could extract generic features from input MR images and use them to develop more task-specific high-level features in deeper encoder convolutional blocks [36]. To further improve performance, we applied the matching layer technique: we added a 1D convolutional block to the front of our network in order to automatically convert input MR images from grayscale to Red Green Blue (RGB) since the VGG19 CNN was originally developed using RGB images, but MR images are grayscale [37].
Implementation of CNN
Our U-Net CNN model was trained to maximize the Dice coefficient-based loss function:
| (1) |
where X and Y represent manual and automatic segmentation, respectively. The Dice score-based cost function is a good choice for imbalanced data (i.e., when the objects to be segmented, namely cartilage in this case, vary significantly in shape [38].
We developed two segmentation networks using ROIs provided by Rad1 (i.e., CNN1) and Rad2 (i.e., CNN2), respectively, as ground truth. Each U-Net was trained using the backpropagation method with Adam optimizer to maximize the Dice score on the validation set. The initial learning rate was equal to 0.001 and exponentially decreased five epochs each time the Dice score on the validation set reached plateau. After training, the models which performed best on the validation set were selected for further evaluation on the test set. Networks were implemented and trained in Keras with Tensorflow backend [39].
Statistical analysis
Dice score was used to evaluate the accuracy of segmentation. Dice score was defined as
| (2) |
where X and Y represent manual and automatic segmentation, respectively, of the same cartilage. The Dice score was between 0 and 1, with a value of 1 indicating perfect segmentation and a value of 0 indicating no overlap whatsoever [23, 40]. Volumetric overlap error (VOE) was also calculated to evaluate the accuracy of segmentation. VOE was defined as
| (3) |
with a smaller VOE value indicating more accurate segmentation [19, 40].
Pearson correlation coefficient and intraclass correlation coefficient (ICC) were used to assess the consistency of relaxometry and MMF between the radiologists and CNN models. The pixel-wise parameters were estimated based on cartilage segmentation and compared using one-way ANOVA among different subject groups. All statistical analyses were performed with SPSS 26.0 (IBM). P < 0.05 indicated a significant difference.
Results
Segmentation performance
As shown in Figure 2, the CNN models provided reliable cartilage segmentation that was similar to the results provided by manual segmentation. Three representative subjects (A, 24-year-old female, KL=0; B, 43-year-old male, KL=1; and C, 74-year-old female, KL=3) were chosen to depict the input 3D UTE Cones images, corresponding manual labels drawn by Rad1 and Rad2, and resultant labels produced by CNN1 and CNN2. Strong inter-observer agreement was shown between Rad1 and CNN1 or between Rad2 and CNN2, where the morphology of labels produced by each CNN model showed high similarity with labels produced by the corresponding radiologist.
Figure 2.
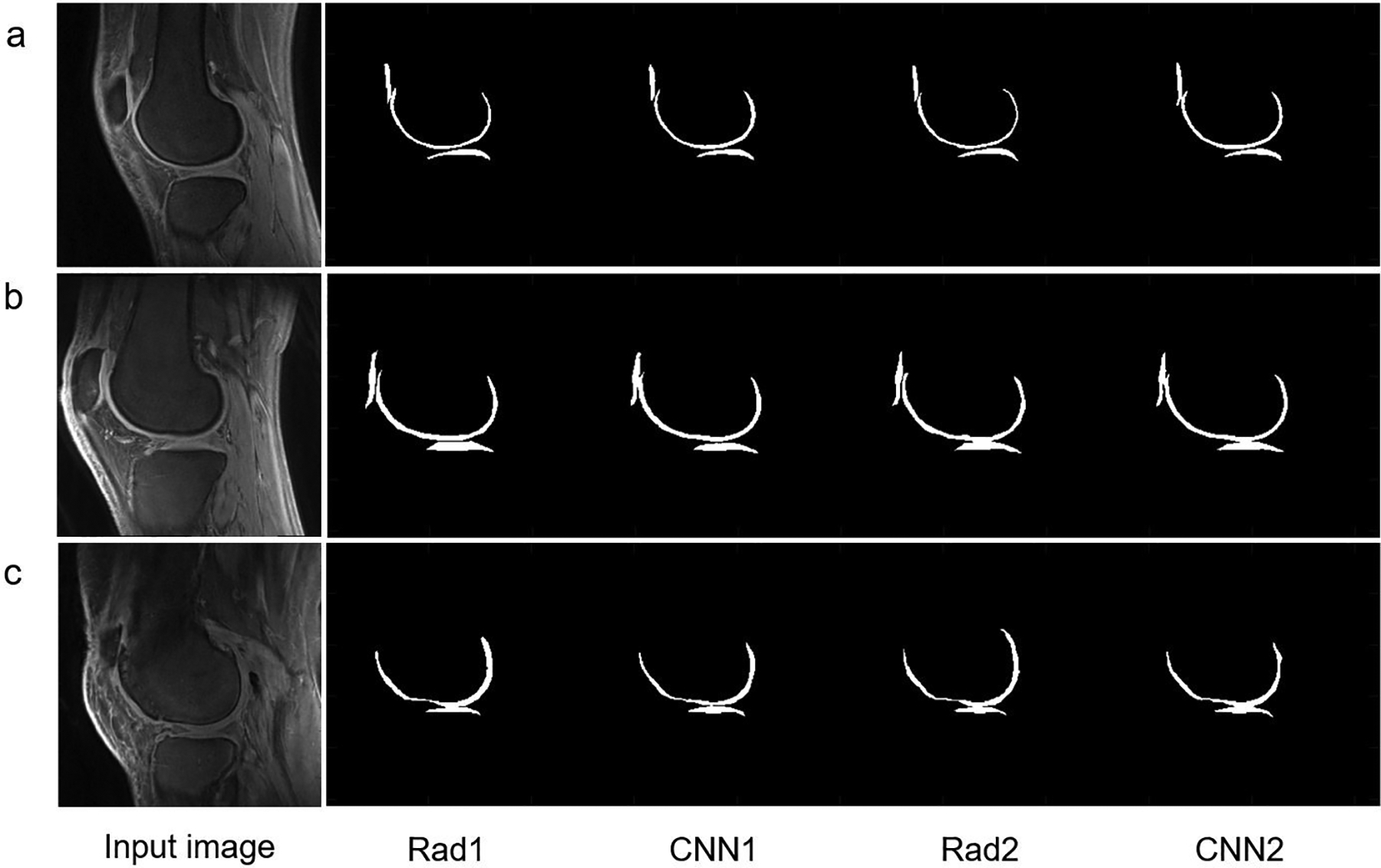
The segmentation performance of the CNN model in three representative subjects.
A: In a 24-year-old female, KL=0: Dice score=0.89, VOE=20.06% for Rad1 vs. CNN1 and Dice score=0.87, VOE=22.38% for Rad2 vs. CNN2, respectively. Compared to coefficients between Rad1 and Rad2 (Dice score=0.85, VOE=25.76%), CNN1 and CNN2 yielded a higher Dice score of 0.93 and a lower VOE of 13.20%.
B: In a 43-year-old male, KL=1: Dice score =0.88, VOE= 21.96% for Rad1 vs. CNN1 and Dice score=0.91, VOE=16.73% for Rad2 vs. CNN2, respectively. Compared to coefficients between Rad1 and Rad2 (Dice score=0.84, VOE=27.97%), CNN1 and CNN2 yielded a higher Dice score of 0.92 and a lower VOE of 15.08%.
C: In a 74-year-old female, KL=3: Dice score=0.84, VOE=28.00% for Rad1 vs. CNN1 and Dice score=0.86, VOE=24.74% for Rad2 vs. CNN2, respectively. Compared to coefficients between Rad1 and Rad2 (Dice score=0.87, VOE=22.41), CNN1 and CNN2 yielded a higher Dice score of 0.92 and a lower VOE of 14.94%.
Mean Dice scores were 0.81±0.11 for Rad1 vs. CNN1 and 0.82±0.08 for Rad2 vs. CNN2, respectively (see Table 1). Dice score was 0.87 for CNN1 and CNN2, which was higher than the Dice score between Rad1 and Rad2 (0.83), indicating improved inter-observer agreement in automatic segmentation over manual segmentation. The mean values of VOE between manual and automatic segmentations were 28.43±7.31%, 30.43±13.03%, 29.28±10.83%, and 22.15±10.51% for Rad1 vs. Rad2, Rad1 vs. CNN1, Rad2 vs. CNN2, and CNN1 vs. CNN2, respectively.
Table 1.
Average Dice scores and VOE (mean ± SD) calculated using manual and automatic segmentations.
| Groups | Rad1 | Rad2 | CNN1 | CNN2 | |
|---|---|---|---|---|---|
| Rad1 | Dice score | 1 | |||
| VOE(%) | |||||
| Rad2 | Dice score | 0.83±0.05 | 1 | ||
| VOE(%) | 28.43±7.31 | ||||
| CNN1 | Dice score | 0.81±0.11 | 0.80±0.10 | 1 | |
| VOE(%) | 30.43±13.03 | 32.07±11.85 | |||
| CNN2 | Dice score | 0.81±0.10 | 0.82±0.08 | 0.87±0.08 | 1 |
| VOE(%) | 30.57±12.00 | 29.28±10.83 | 22.15±10.51 | ||
Note: VOE= volumetric overlap error; CNN= convolutional neural networks; Rad= radiologist.
Quantitative MR parameters
The average values and ICCs of Cones-T1, Cones-AdiabT1ρ, Cones-T2*, and MMF calculated based on pixel-wise parameter map from manual and automatic segmentations, respectively, are listed in Table 2. The ICCs of these parameters were highly consistent between the two radiologists and the CNN models. Figures 3–6 show scatter plots for these parameters between Rad1 vs. CNN1, Rad2 vs. CNN2, Rad1 vs. Rad2, and CNN1 vs. CNN2, with corresponding Pearson correlation coefficients ranging from 0.91 to 0.99, respectively.
Table 2.
Values of Cones-T1, Cones-AdiabT1ρ, Cones-T2*, and MMF (mean ± SD) calculated using manual and automatic segmentations, and the consistency evaluation using ICC between radiologists and CNN models
| Groups | Cones-T1 (ms) | Cones-AdiabT1ρ (ms) | Cones-T2* (ms) | MMF (%) |
|---|---|---|---|---|
| Rad1 | 975.24 ± 127.94 | 41.45±4.59 | 30.67 ± 7.54 | 10.66 ± 1.00 |
| Rad2 | 971.46 ± 120.21 | 41.45± 4.34 | 30.39 ± 7.39 | 10.65 ± 1.00 |
| CNN1 | 973.57 ± 122.75 | 41.51± 4.41 | 30.95 ± 7.69 | 10.71 ± 1.00 |
| CNN2 | 971.11 ± 119.62 | 41.61 ± 4.41 | 30.48 ± 7.32 | 10.67 ± 1.04 |
| ICC1 | 0.93*** | 0.91*** | 0.94*** | 0.91*** |
| ICC2 | 0.93*** | 0.93*** | 0.94*** | 0.92*** |
| ICC3 | 0.94*** | 0.93*** | 0.96*** | 0.95*** |
| ICC4 | 0.95*** | 0.95*** | 0.95*** | 0.95*** |
Note:
P < 0.001;
CNN= convolutional neural networks; Rad= radiologist; MMF = macromolecular fraction;
ICC = intraclass correlation coefficient;
ICC1 = consistency for CNN1 vs. Rad1; ICC2 = consistency for CNN2 vs. Rad2;
ICC3 = consistency for Rad1 vs. Rad2; ICC4 = consistency for CNN1 vs. CNN2.
Figure 3.
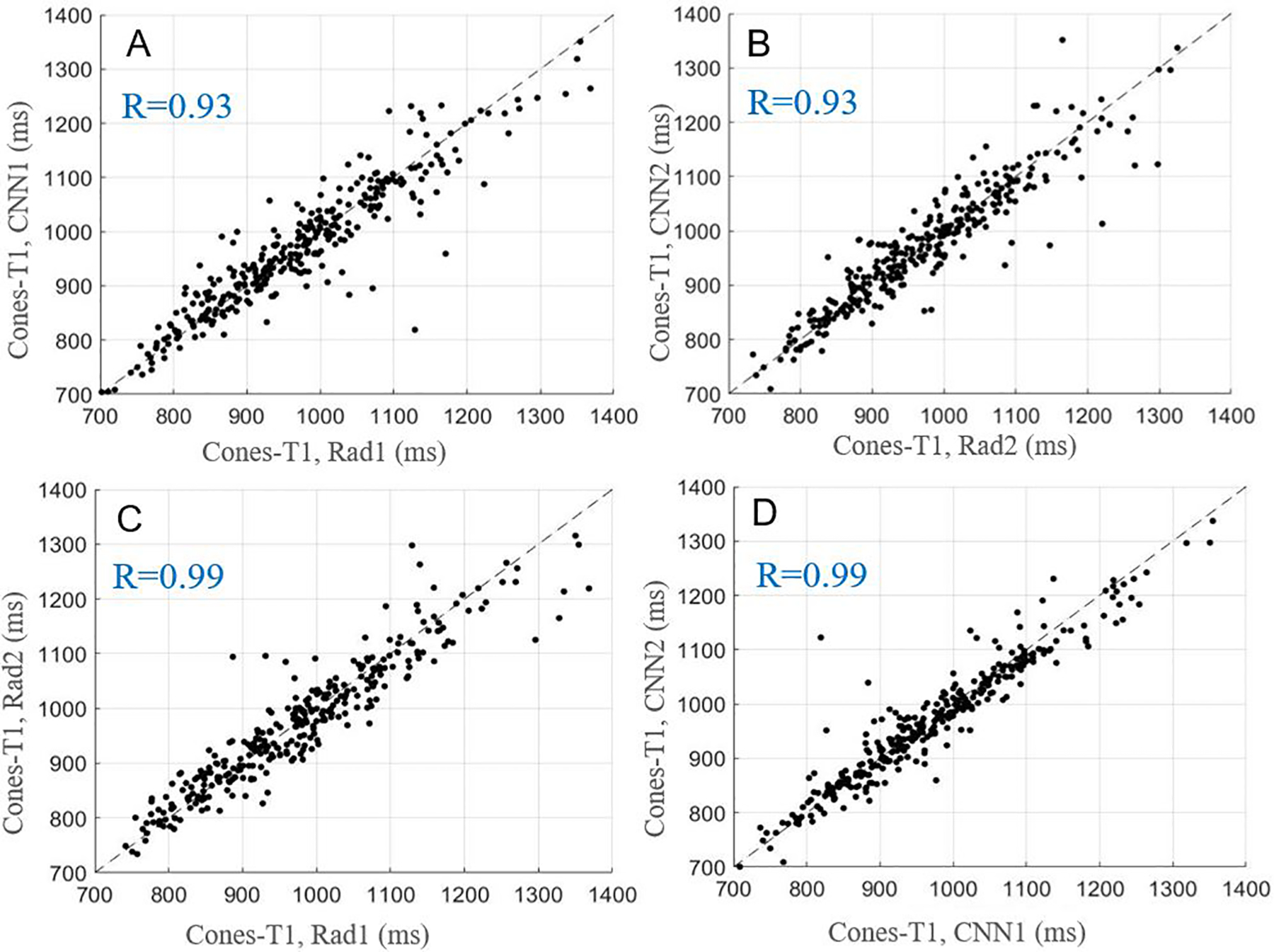
Scatter plots for average Cones-T1 values obtained with manual and automatic segmentations provided by the radiologists and CNN models, respectively. A-D show the relationships of Cones-T1 values for Rad1 vs. CNN1, Rad2 vs. CNN2, Rad1 vs. Rad2, and CNN1 vs. CNN2.
Figure 6.
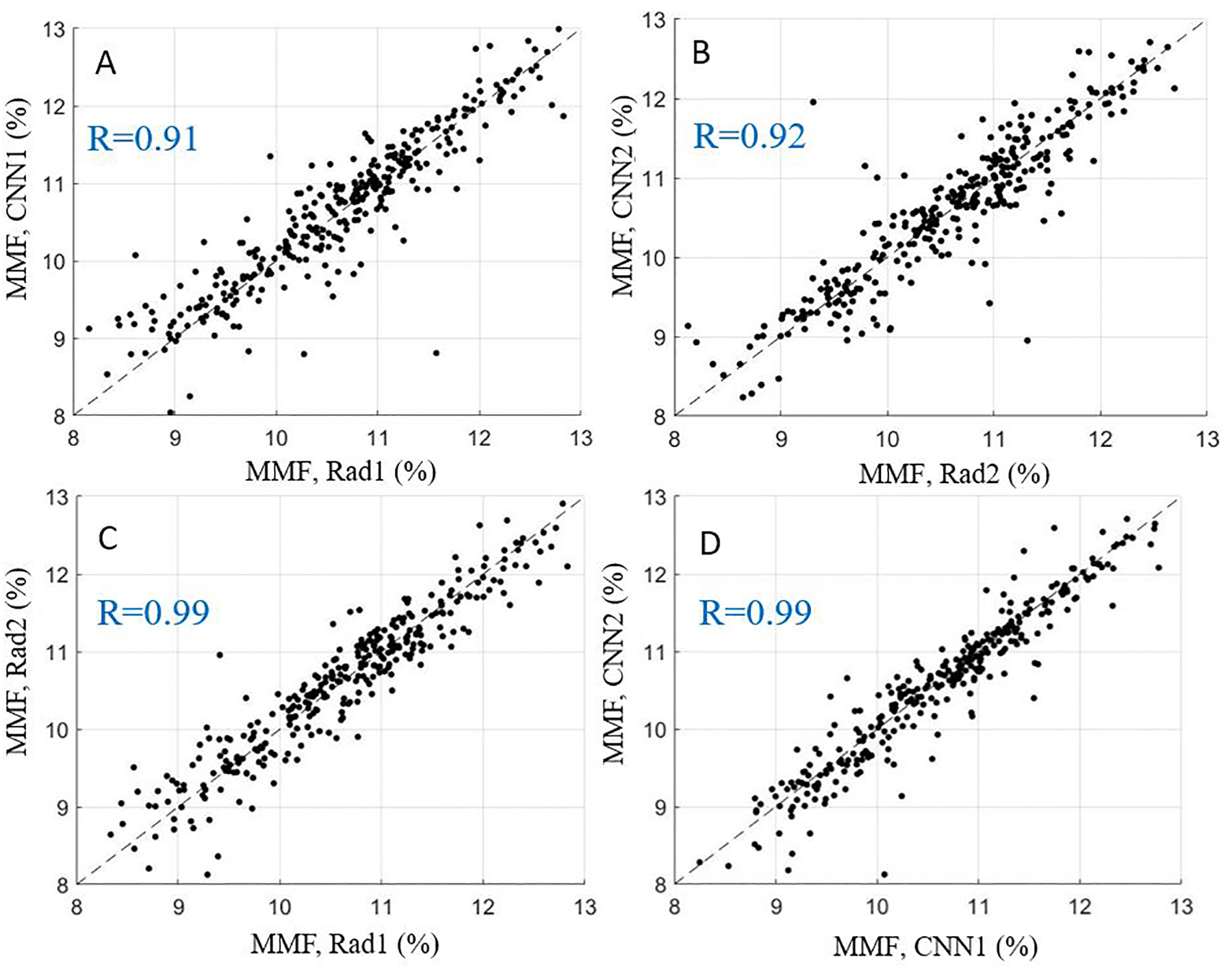
Scatter plots for average MMF values obtained with manual and automatic segmentations provided by the radiologists and CNN models, respectively. A-D show relationships of MMF for Rad1 vs. CNN1, Rad2 vs. CNN2, Rad1 vs. Rad2, and CNN1 vs. CNN2.
Performance in OA evaluation
The average values of Cones-T1, Cones-AdiabT1ρ, Cones-T2*, and MMF for normal controls and OA patients calculated based on manual and automatic segmentations, respectively, are presented in Table 3. High consistency was observed between the two radiologists and the CNN models. Significant increases in T1, T1ρ, and T2* values (P < 0.05), as well as a decrease in MMF (P < 0.001) were observed in doubtful-minimal OA and/or moderate-severe OA compared with normal controls, suggesting that fully automated quantitative UTE biomarkers may be used for evaluation of OA.
Table 3.
Global values (mean ± SD) and comparison of the quantitative MR parameters in different KL groups
| Groups | KL=0 | KL=1–2 | KL=3–4 |
|---|---|---|---|
| Cones-T1 (ms) | |||
| Rad1 | 932.01±112.62 | 978.92±110.30** | 1006.14±150.70** |
| CNN1 | 929.35±112.73 | 977.11±99.29** | 1005.49±146.40*** |
| Rad2 | 932.63±113.35 | 968.63±99.16* | 1007.05±139.82*** |
| CNN2 | 922.64±107.42 | 970.94±92.27** | 1011.25±144.33***# |
| Cones-T1ρ (ms) | |||
| Rad1 | 39.08±4.29 | 43.72±3.64*** | 41.54±4.80**## |
| CNN1 | 38.97±4.18 | 43.86±3.51*** | 41.68±4.32***### |
| Rad2 | 39.07±4.20 | 43.44±3.36*** | 41.84±4.51***## |
| CNN2 | 38.90±4.15 | 43.92±3.35*** | 42.02±4.43***## |
| Cones-T2*(ms) | |||
| Rad1 | 29.16±6.94 | 32.60±8.37** | 31.61±7.44 |
| CNN1 | 29.43±6.99 | 32.69±8.35** | 31.86±8.00 |
| Rad2 | 28.88±6.87 | 31.80±8.15* | 31.65±7.35* |
| CNN2 | 28.53±6.68 | 32.27±7.85** | 31.65±7.35* |
| MMF (%) | |||
| Rad1 | 11.33±0.87 | 10.63±0.88*** | 10.02±0.86***### |
| CNN1 | 11.41±0.89 | 10.66±0.88*** | 10.06±0.79***### |
| Rad2 | 11.29±0.88 | 10.68±0.83*** | 9.98±0.92***### |
| CNN2 | 11.38±0.92 | 10.61±0.88*** | 9.98±0.91***### |
Note: CNN = convolutional neural networks; Rad = radiologist; MMF = macromolecular fraction;
KL = Kellgren–Lawrence grade; OA = osteoarthritis;
P < 0.05,
P < 0.01,
P < 0.001, compared with normal controls (KL=0);
P < 0.05,
P < 0.01,
P < 0.001, compared with doubtful-minimal OA (KL=1–2).
Discussion
In this study, we evaluated a transfer learning-based U-Net CNN model to automatically segment whole-knee full-thickness cartilage and to extract quantitative parameters in 3D UTE Cones MR imaging in vivo. Maps of Cones-T1, Cones-AdiabT1ρ, and Cones-T2* relaxometry, as well as MMF, were automatically generated for whole-knee cartilage. The robustness of this novel approach was validated by high consistency in global mean values of Cones-T1, Cones-AdiabT1ρ, Cones-T2*, and MMF between manually and automatically segmented cartilage. Significant increases in T1, T1ρ, and T2* values, as well as a decrease in MMF, were observed in doubtful-minimal OA and/or moderate-severe OA compared with normal controls. In particular, a significant difference was seen between doubtful-minimal OA and moderate-severe OA for Cones-AdiabT1ρ and MMF. Our preliminary results suggest the potential of fully automated panels of quantitative UTE biomarkers for comprehensive evaluation of OA. To the best of our knowledge, this is the first time that a CNN model with transfer learning has been used to segment whole-knee full-thickness cartilage in 3D UTE Cones MR imaging for accurate assessment of cartilage relaxometry and MT effect in normal volunteers and OA patients.
The labels used as ground truth for evaluating the model’s performance were manually generated by two MSK radiologists. Dice score and VOE were calculated to evaluate the accuracy of cartilage segmentation. The proposed CNN model produced high Dice score and low VOE, meaning that our CNN model was able to accurately segment cartilage. Dice score and VOE of the CNN model were comparable to or outperformed those from other state-of-the-art techniques proposed for automatic cartilage segmentation in the literature. For example, Pang et al. reported a mean Dice score of 0.76 using their proposed pattern recognition-based approach [41]. A mean Dice score of 0.72 was reported by Lee et al. using their atlas-based method [42]. More recently, advanced methods based on CNN showed improved performance, where Liu et al. [43] reported a mean VOE of 33.1% with SegNet combined with deformable modeling and Raj et al. [44] reported a mean Dice score of 0.83 and a mean VOE of ~28–29% with U-Net. However, it is difficult to directly compare the different studies performed using different input datasets, and more systematic comparison will be required for this purpose.
Compared to coefficients between Rad1 and Rad2, CNN1 and CNN2 yielded a higher Dice score and a lower VOE, suggesting that the CNN model better compensated for inter-observer variability and bias. Pearson correlation coefficients and ICCs between MR parameters, namely Cones-T1, Cones-AdiabT1ρ, Cones-T2*, and MMF, estimated using manual and automatic segmentations reached a high level of consistency, indicating that the CNN model can precisely extract quantitative biomarkers from automatically segmented cartilage. Among the results, the ICC reached 0.94 between the radiologists and the CNN models. The ICC between the two CNN models was 0.95, similar to the ICC between the two radiologists (0.96), indicating that biomarkers extracted using the CNN model are at least as accurate, if not more accurate, than those extracted using manual methods.
In previous studies, Norman et al. [45] presented a similar U-net CNN model applied to two conventional MR sequences to perform cartilage segmentation and extraction of T1ρ and T2 relaxometry. Byra et al. [23] applied a similar U-Net CNN model to segment menisci and determine meniscal relaxometry. Our CNN model provides not only automatic segmentation of whole-knee full-thickness cartilage, but also mapping of Cones-T1, Cones-T2*, Cones-AdiabT1ρ, and MMF. Those UTE biomarkers may provide comprehensive quantitative assessment of macromolecules (e.g., PG depletion and collagen degradation), all of which may occur during the early stages of OA before the typically observed morphological changes occur.
While our initial results are promising, there are several limitations to our study. First, the size of our training dataset was limited to 40 subjects. While our CNN model greatly reduced the size of the dataset necessary for successful network training, there still needs to investigate the optimal number of training data points for more robust segmentation. Second, in order to achieve a high enough SNR for reliable quantification of parameters under a reasonable scan time, an anisotropic resolution was used for cartilage imaging, where the relatively poor slice resolution and the curved cartilage surface may have led to significant partial volume artifact, and therefore errors in deep learning-based segmentation. This is a big challenge in quantitative MR imaging which might be solved or alleviated through higher performance hardware and more advanced image reconstruction algorithms. Third, since simple global mean values were used to evaluate OA, it is likely that regional analysis would significantly improve the diagnosis. Panels of UTE biomarkers could also have been used for spatial pattern analysis [46] to further improve diagnosis of the varying degrees of OA. Finally, the comprehensive approach developed in this study needs to be applied to other MSK tissues in the knee joint, including the menisci, ligaments, tendons, and bone, in order to provide a truly whole-organ approach for assessment of knee joint degeneration [13, 47].
In conclusion, our automated knee cartilage segmentation technique combined with quantitative 3D UTE Cones MR imaging to map Cones-T1, Cones-AdiabT1ρ, Cones-T2*, and MMF achieved performance that was comparable with two experienced radiologists. The efficiency and accuracy of this technology make it suitable as a key tool for obtaining quantitative UTE biomarkers for assessment of OA in imaging studies and clinical trials.
Figure 4.
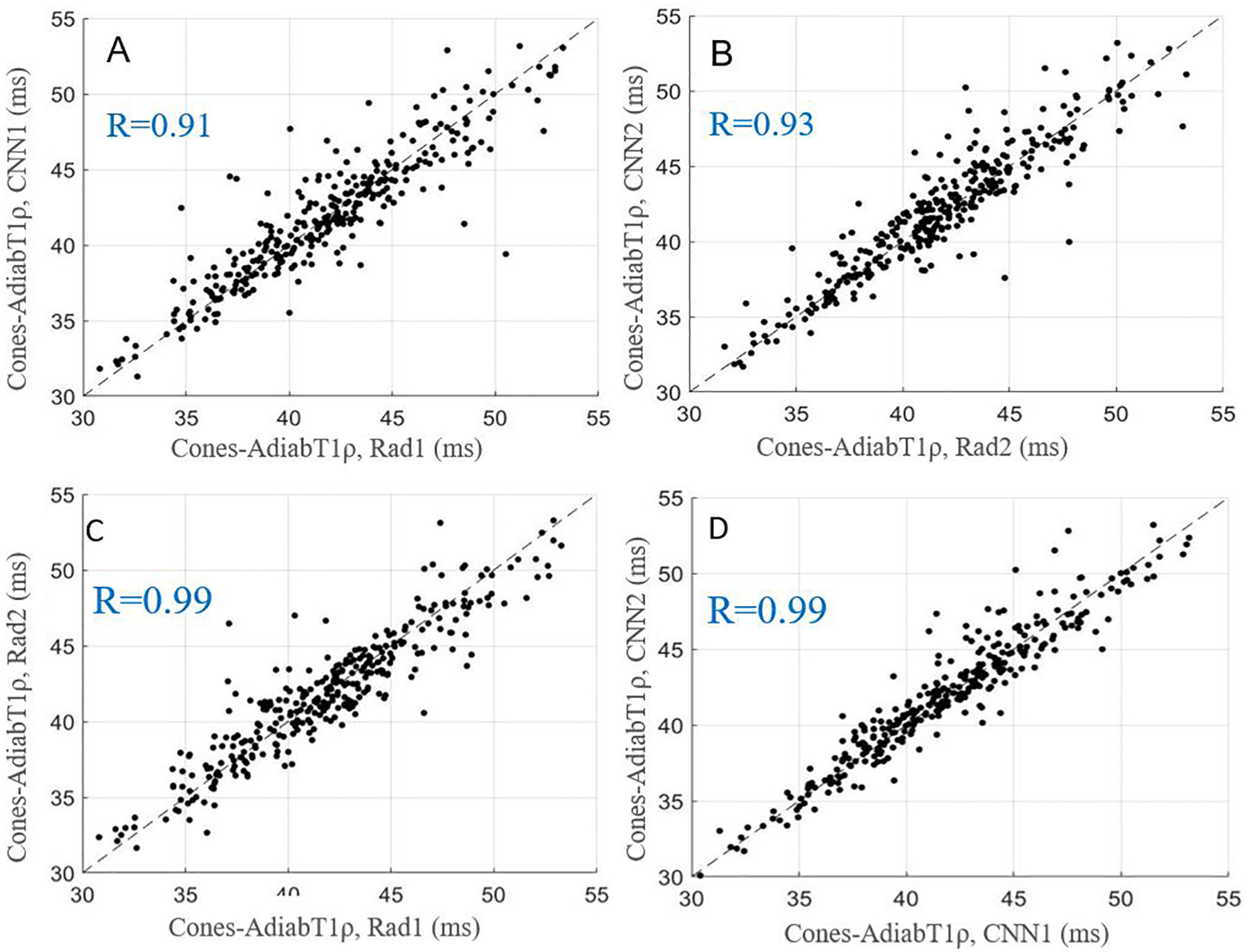
Scatter plots for average Cones-AdiabT1ρ values obtained with manual and automatic segmentations provided by the radiologists and CNN models, respectively. A-D show the relationships of Cones-AdiabT1ρ for Rad1 vs. CNN1, Rad2 vs. CNN2, Rad1 vs. Rad2, and CNN1 vs. CNN2.
Figure 5.
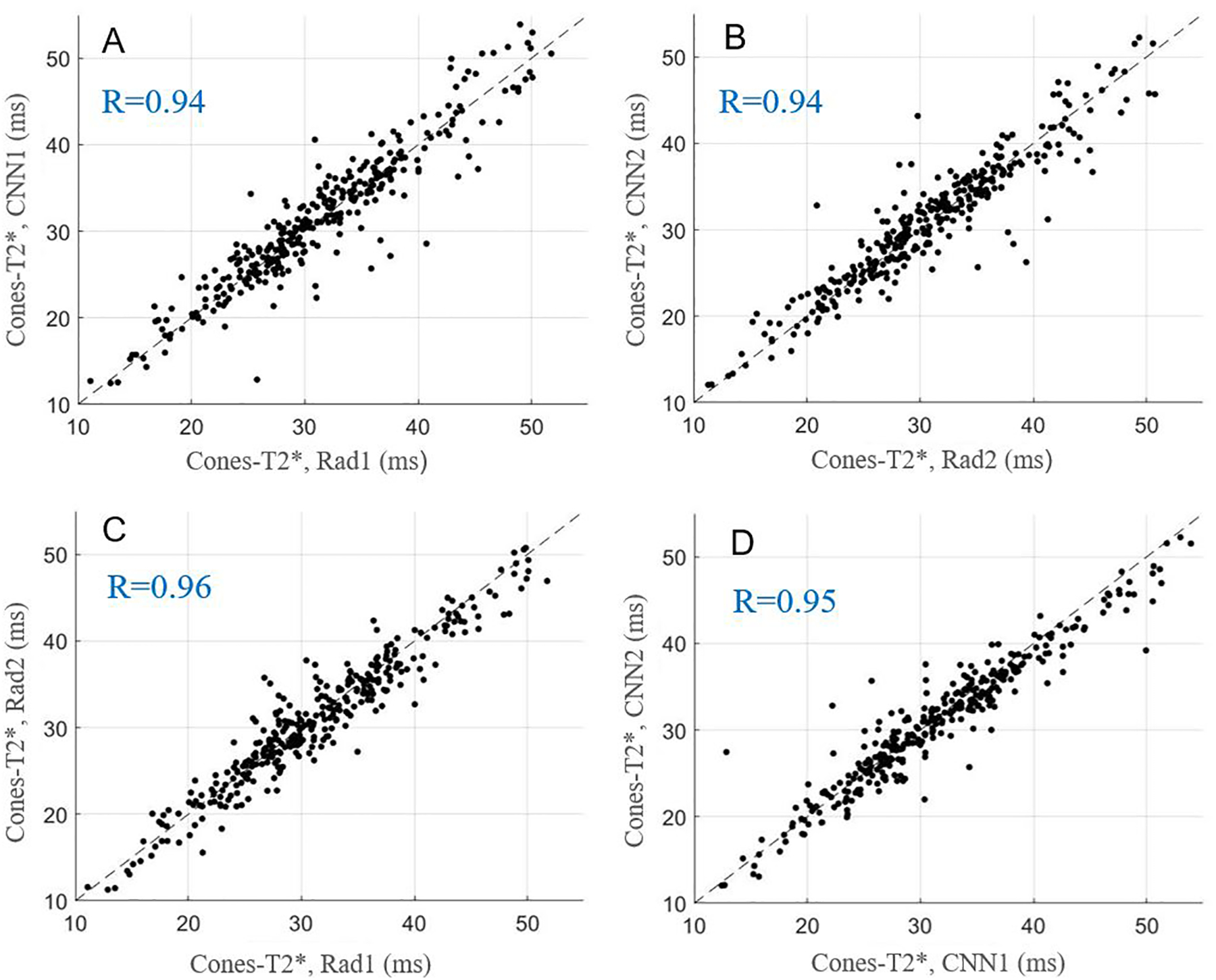
Scatter plots for average Cones-T2* values obtained with manual and automatic segmentations provided by the radiologists and CNN models, respectively. A-D show the relationships of Cones-T2* for Rad1 vs. CNN1, Rad2 vs. CNN2, Rad1 vs. Rad2, and CNN1 vs. CNN2.
Key Points.
3D UTE Cones imaging combined with U-Net CNN model was able to provide fully automated cartilage segmentation.
UTE parameters obtained from automatic segmentation were able to reliably provide quantitative assessment of cartilage.
Acknowledgements
The authors acknowledge grant support from GE Healthcare, NIH (1R01 AR062581 and 1R01 AR068987), and the VA Clinical Science Research & Development Service (1I01CX001388 and I21RX002367).
Abbreviations
- UTE
ultrashort echo time
- FOV
field of view
- TR
repetition time
- TE
echo time
- MT
magnetization transfer
- MMF
macromolecular fraction
- AFI
actual flip angle imaging
- VFA
variable flip angle
- OA
osteoarthritis
- Cones-T1
UTE Cones T1
- Cones-AdiabT1ρ
UTE Cones Adiabatic T1ρ
- Cones-T2*
UTE Cones T2*
- UTE Cones Magnetization Transfer
Cones-MT
- MSK
musculoskeletal
- RF
radiofrequency
- NIR
number of adiabatic inversion pulses
- RP
rectangular pulse
- SD
standard deviation
- CNN
convolutional neural networks.
References
- 1.Saarakkala S, Julkunen P, Kiviranta P, Mäkitalo J, Jurvelin JS, Korhonen RK (2010) Depth-wise progression of osteoarthritis in human articular cartilage: investigation of composition, structure, and biomechanics. Osteoarthritis Cartilage 18:73–81 [DOI] [PubMed] [Google Scholar]
- 2.Stehling C, Liebl H, Krug R, et al. (2010) Patellar cartilage: T2 values and morphologic abnormalities at 3.0-T MR imaging in relation to physical activity in asymptomatic subjects from the osteoarthritis initiative. Radiology 254:509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu J, Zhang Y, Duan C, Peng X, Hu P, Lu H (2017) Feasibility study for evaluating early lumbar facet joint degeneration using axial T1ρ, T2, and T2* mapping in cartilage. J Magn Reson Imaging 46: 468–475 [DOI] [PubMed] [Google Scholar]
- 4.Su F, Hilton JF, Nardo L, et al. (2013) Cartilage morphology and T1ρ and T2 quantification in ACL reconstructed knees: A 2-year follow-up. Osteoarthritis Cartilage 21:1058–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Cheng J, Lin K, et al. (2011) Quantitative MRI using T1ρ and T2 in human osteoarthritic cartilage specimens: correlation with biochemical measurements and histology. Magn Reson Imaging 29:324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berberat JE, Nissi MJ, Jurvelin JS, Nieminen MT (2009) Assessment of interstitial water content of articular cartilage with T1 relaxation. Magn Reson Imaging 27:727–732 [DOI] [PubMed] [Google Scholar]
- 7.Lattanzio PJ, Marshall KW, Damyanovich AZ, Peemoeller H (2000) Macromolecule and water magnetization exchange modeling in articular cartilage. Magn Reson Med 44:840–851 [DOI] [PubMed] [Google Scholar]
- 8.Ma YJ, Jerban S, Carl M, et al. (2019) Imaging of the region of the osteochondral junction (OCJ) using a 3D adiabatic inversion recovery prepared ultrashort echo time cones (3D IR-UTE-cones) sequence at 3 T. NMR Biomed. DOI: 10.1002/nbm.4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oei EH, van Tiel J, Robinson WH, Gold GE (2014) Quantitative radiologic imaging techniques for articular cartilage composition: toward early diagnosis and development of disease-modifying therapeutics for osteoarthritis. 66:1129–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du J, Carl M, Diaz E, et al. (2010) Ultrashort TE T1rho (UTE T1rho) imaging of the Achilles tendon and meniscus. Magn Reson Med 64:834–842 [DOI] [PubMed] [Google Scholar]
- 11.Du J, Carl M, Bae WC, et al. (2013) Dual inversion recovery ultrashort echo time (DIR-UTE) imaging and quantification of the zone of calcified cartilage (ZCC). Osteoarthritis Cartilage 21:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang EY, Du J, Chung CB (2015) UTE imaging in the musculoskeletal system. J Magn Reson Imaging 41:870–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandt KD, Radin EL, Dieppe PA, van de Putte L (2006) Yet more evidence that osteoarthritis is not a cartilage disease. Ann Rheum Dis 65:1261–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter DJ, Zhang YQ, Niu JB, et al. (2006) The association of meniscal pathologic changes with cartilage loss in symptomatic knee osteoarthritis. Arthritis Rheum 54:795–801 [DOI] [PubMed] [Google Scholar]
- 15.Ma YJ, Zhao W, Wan LD, et al. (2019) Whole knee joint T1 values measured in vivo at 3T by combined 3D ultrashort echo time cones actual flip angle and variable flip angle methods. Magn Reson Med 81:1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma YJ, Carl M, Searleman A, Lu X, Chang EY, Du J (2018) 3D adiabatic T1ρ prepared ultrashort echo time cones sequence for whole knee imaging. Magn Reson Med 80:1429–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Drygalski A, Barnes RFW, Jang H, et al. (2019) Advanced magnetic resonance imaging of cartilage components in hemophilic joints reveals that cartilage hemosiderin correlates with joint deterioration. Haemophilia 25:851–858 [DOI] [PubMed] [Google Scholar]
- 18.Jerban S, Ma YJ, Wan L, et al. (2019) Collagen proton fraction from ultrashort echo time magnetization transfer (UTE‐MT) MRI modelling correlates significantly with cortical bone porosity measured with microcomputed tomography (μCT). NMR Biomed. DOI: 10.1002/nbm.4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R (2018) Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 79:2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhari A, Fang Z, Hyung Lee J, Gold G, Hargreaves B (2018) Deep Learning Super-Resolution Enables Rapid Simultaneous Morphological and Quantitative Magnetic Resonance Imaging. MLMIR. DOI: 10.1007/978-3-030-00129-2_1 [DOI] [Google Scholar]
- 21.Hinton G, Deng L, Yu D, et al. (2012). Deep neural networks for acoustic modeling in speech recognition: the shared views of four research groups. Signal Processing Magazine IEEE 29:82–97 [Google Scholar]
- 22.LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444 [DOI] [PubMed] [Google Scholar]
- 23.Byra M, Wu M, Zhang X, et al. (2020) Knee menisci segmentation and relaxometry of 3D ultrashort echo time cones MR imaging using attention U-Net with transfer learning. Magn Reson Med 83:1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Chang G, Xu J, et al. (2012) T1rho MRI of menisci and cartilage in patients with osteoarthritis at 3T. Eur J Radiol 81:2329–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu M, Ma YJ, Kasibhatla A, et al. (2020) Convincing evidence for magic angle less-sensitive quantitative T1ρ imaging of articular cartilage using the 3D ultrashort echo time cones adiabatic T1ρ (3D UTE cones-AdiabT1ρ) sequence. Magn Reson Med. DOI: 10.1002/mrm.28317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma YJ, Shao H, Du J, Chang EY (2016) Ultrashort echo time magnetization transfer (UTE-MT) imaging and modeling: magic angle independent biomarkers of tissue properties. NMR Biomed 29:1546–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein S, Staring M, Murphy K, Viergever MA, Pluim JP (2010) Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 29:196–205 [DOI] [PubMed] [Google Scholar]
- 28.Wu M, Zhao W, Wan L, et al. (2020) Quantitative three‐dimensional ultrashort echo time cones imaging of the knee joint with motion correction. NMR Biomed. DOI: 10.1002/nbm.4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paris S, Hasinoff SW, Kautz J (2015) Local Laplacian filters: edge-aware image processing with a Laplacian Pyramid. Commun ACM 58:81–91 [Google Scholar]
- 30.Ma YJ, Chang EY, Carl M, Du J (2018) Quantitative magnetization transfer ultrashort echo time imaging using a time-efficient 3D multispoke Cones sequence. Magn Reson Med 79: 692–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oktay O, Schlemper J, Folgoc LL, et al. (2018) Attention U-Net: learning where to look for the pancreas. Available via https://arxiv.org/abs/1804.03999. Accessed 11 Apr 2018
- 32.Schlemper J, Oktay O, Schaap M, et al. (2019) Attention gated networks: learning to leverage salient regions in medical images. Med Image Anal 53: 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng J, Dong W, Socher R, Li LJ, Li K, Li FF (2009) Imagenet: A large-scale hierarchical image database. 2009 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR 2009). DOI: 10.1109/CVPR.2009.5206848 [DOI] [Google Scholar]
- 34.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A (2014) Object detectors emerge in deep scene CNNs. Available via https://arxiv.org/abs/1412.6856. Accessed 22 Dec 2014
- 35.Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. Available via https://arxiv.org/abs/1409.1556. Accessed 4 Sep 2014 [Google Scholar]
- 36.Raghu M, Zhang CY, Kleinberg J, Bengio S (2019) Transfusion: Understanding transfer learning for medical imaging. Available via https://arxiv.org/abs/1902.07208v2. Accessed 30 May 2019 [Google Scholar]
- 37.Byra M, Galperin M, Ojeda-Fournier H, et al. (2019) Breast mass classification in sonography with transfer learning using a deep convolutional neural network and color conversion. Med Phys. 46:746–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sudre CH, Li W, Vercauteren T, Ourselin Sébastien, Cardoso MJ (2017) Generalised Dice overlap as a deep learning loss function for highly unbalanced segmentations. Available via https://arxiv.org/abs/1707.03237. Accessed 14 Jul 2017 [DOI] [PMC free article] [PubMed]
- 39.Abadi M, Barham P, Chen J, et al. (2016) TensorFlow: A system for large-scale machine learning. Available via https://arxiv.org/abs/1605.08695v2. Accessed 31 May 2016 [Google Scholar]
- 40.Zhou ZY, Zhao GY, Kijowski R, Liu F (2018) Deep Convolutional Neural Network for Segmentation of Knee Joint Anatomy. Magn Reson Med 80:2759–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pang J, Li PY, Qiu M, Chen W, Qiao L (2015). Automatic Articular Cartilage Segmentation Based on Pattern Recognition from Knee MRI Images. J. Digit. Imaging 28:695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J-G, Gumus S, Moon CH, Kwoh CK, Bae KT (2014) Fully automated segmentation of cartilage from the MR images of knee using a multi-atlas and local structural analysis method. Med. Phys 41:092303 doi: 10.1118/1.4893533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R (2018) Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn. Reson. Med 79:2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raj A, Vishwanathan S, Ajani B, Krishnan K, Agarwal H (2018) Automatic knee cartilage segmentation using fully volumetric convolutional neural networks for evaluation of osteoarthritis. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018): 851–854. doi: 10.1109/ISBI.2018.8363705 [DOI] [Google Scholar]
- 45.Norman B, Pedoia V, Majumdar S (2018) Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology 288:177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eckstein F, Wirth W (2011) Quantitative Cartilage Imaging in Knee Osteoarthritis. Arthritis DOI: 10.1155/2011/475684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loeser RF, Goldring SR, Scanzello CR, Goldring MB (2012) Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum 64 :1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]


