Abstract
A deletion mutation (codons 595 to 603) in the cytomegalovirus (CMV) UL97 gene was recently reported after sequence analysis of leukocyte DNA from a patient receiving ganciclovir. The corresponding viral phenotype was examined by transfer of this mutation to a laboratory CMV strain (strain Towne). The recombinant virus was resistant to ganciclovir (8.4-fold increase in the 50% inhibitory concentration), was sensitive to foscarnet, and replicated normally in cell culture.
Drug resistance that develops after prolonged ganciclovir therapy for human cytomegalovirus (CMV) infection usually involves a mutation in the viral UL97 phosphotransferase gene (3). UL97 is responsible for the initial phosphorylation of the drug, which is necessary for its antiviral activity (15). Mutations in the CMV DNA polymerase gene may also contribute to ganciclovir resistance (6, 7, 9).
Previous studies have documented that mutations at UL97 codons 460, 520, and 591 to 607 are associated with ganciclovir resistance in clinical CMV isolates (1–6, 10), with amino acid substitutions at codons 460, 594, and 595 being the most common (4, 5). Deletion mutations conferring ganciclovir resistance are relatively unusual. Reported cases include a one-codon deletion at codon 595 (2) and a four-codon deletion (codons 590 to 593 or 591 to 594) (5, 15). Recently, a nine-codon deletion (UL97 codons 595 to 603) was detected in DNA extracted from leukocytes and colon tissue of a renal transplant recipient after prolonged exposure to ganciclovir (12). Although this mutation was associated with treatment-resistant CMV disease, no viral isolate was available from this patient to confirm the drug resistance phenotype that was suspected to be conferred by this mutation. Additionally, because this was the largest UL97 deletion that had been reported in a clinical specimen, the question of its replication competence arose, since UL97 activity appears to be necessary for normal CMV replication and laboratory mutants containing large deletions in UL97 are either difficult to make or are very much attenuated (13, 14). We therefore performed experiments to transfer the deletion from codons 595 to 603 into a laboratory CMV strain (strain Towne) in order to examine its associated phenotype.
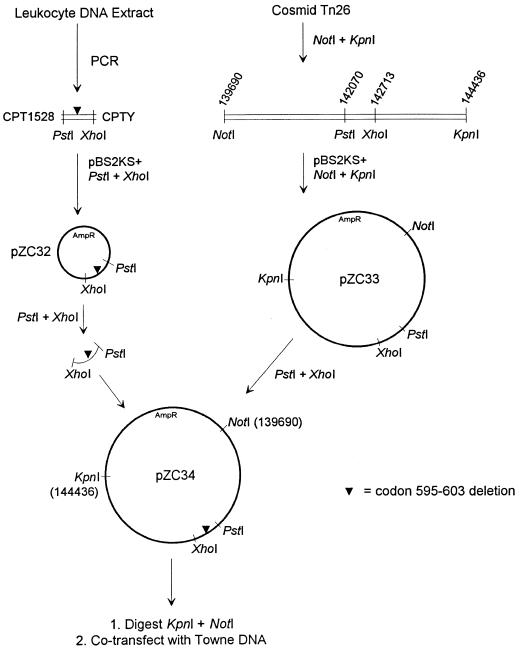
The transfer of the deletion into CMV Towne (VR-977; American Type Culture Collection, Manassas, Va.) was performed by homologous recombination as published previously (4). The virus was propagated in human fibroblasts, and Towne DNA was extracted from culture supernatants by ultracentrifugation and lysis of the viral pellet with sodium dodecyl sulfate and proteinase K, followed by phenol-chloroform extraction and ethanol precipitation (4). A PCR product (PCR41999) containing a segment of the CMV UL97 gene (from codon 510 onward) was amplified from a leukocyte extract of the patient previously reported to have the UL97 deletion from codons 595 to 603 (12). The PCR primers were CPT1528 (5′-AACTGCTCGCACCGTCTGCGC-3′) and CPTY (5′-GGTGAGCTCCTCATCGTCGTCG-3′). PCR41999 was digested with restriction enzymes PstI and XhoI, taking advantage of naturally occurring restriction sites within PCR41999 corresponding to nucleotides 142070 and 142713, respectively, of the published complete CMV sequence (GenBank accession no. X17403). The resulting 0.64-kb fragment was isolated by agarose gel electrophoresis and was cloned into Bluescript vector pBS2KS+ (Stratagene, La Jolla, Calif.), forming plasmid pZC32 (Fig. 1). A 4.7-kb segment of the Towne genome, corresponding to nucleotides 139690 to 144436 of the complete CMV sequence (Genbank accession no. X17403) and incorporating the UL97-coding sequence, was subcloned from cosmid Tn26 (11) by digestion with restriction enzymes NotI and KpnI, followed by preparative agarose gel electrophoresis and ligation into vector pBS2KS+, forming plasmid pZC33 (Fig. 1). The UL97 segment of pZC32 containing the deletion from codons 595 to 603 was then swapped into pZC33 (Fig. 1), forming pZC34. Homologous recombination was achieved by cotransfection of 20 μg of Towne DNA and 2 μg of pZC34 digested with NotI and KpnI into subconfluent human foreskin fibroblast monolayers by the calcium phosphate precipitation method (4). A viral cytopathic effect was noted at 9 days posttransfection, and the resulting extracellular virus was propagated twice in the presence of 20 μM ganciclovir and was then plaque purified once in the presence of 20 μM ganciclovir and twice in the presence of no drug.
FIG. 1.
Strategy for construction of CMV recombinant containing the deletion from codons 595 to 603.
The entire 2.1-kb UL97-coding sequence and the 3.7-kb DNA polymerase-coding sequence of plaque-purified (three times) virus T963-5-6-2 were sequenced by PCR amplification and multiple fluorescent dideoxy sequencing reactions (6). The DNA polymerase-coding sequence was identical to that of the Towne strain (GenBank accession no. D14980), and the UL97-coding sequence had no amino acid changes compared with the sequence of the Towne strain (GenBank accession no. U07355) other than the deletion from codons 595 to 603. The presence of the same deletion as in pZC34, along with absence of any other mutations in UL97 or DNA polymerase, the only known CMV genes that affect ganciclovir susceptibility, was interpreted as evidence that the deletion confers ganciclovir resistance, similar to conclusions drawn from results of studies of other UL97 marker transfers (1–4, 6, 10).
Cell culture susceptibility testing by plaque reduction (8) of recombinant virus T963-5-6-2, simultaneously performed three times with a wild-type Towne strain as a control, gave a mean ganciclovir 50% inhibitory concentration (IC50) of 24.4 μM (standard deviation [SD], 2.9 μM) and a mean foscarnet IC50 of 119 μM (SD, 31.5 μM). The mean ganciclovir IC50 for the Towne control virus was 3.1 μM (SD, 1.2 μM), and the mean foscarnet IC50 was 91 μM (SD, 9.6 μM). The ganciclovir IC50 for the recombinant virus was 8.4-fold (SD, 2.7-fold) increased over that for the Towne control strain, whereas the foscarnet IC50 was 1.3-fold that for the Towne control strain; the difference was considered to be not statistically significant. The degree of ganciclovir resistance is similar to the 7- to 10-fold increases reported previously for UL97 mutants (4, 6) and higher than the 3.5- to 5.3-fold increases reported for DNA polymerase mutants (6, 7).
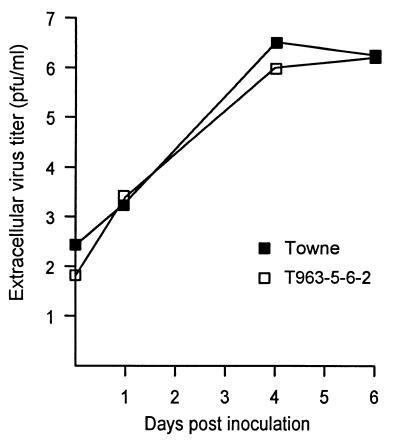
The recombinant virus T963-5-6-2 was observed to form normal-sized plaques and a cytopathic effect in fibroblast culture; the plaque sizes and cytopathic effects are comparable to those of the Towne strain at the same intervals postinfection. After inoculation of 7 × 105 fibroblasts with the same number of PFU of T963-5-6-2 or the Towne strain control, the virus titers in the supernatant were comparable at several time points afterward, as assayed by plaque titration on fibroblast monolayers (Fig. 2). We therefore conclude that the recombinant virus retains full replication competence.
FIG. 2.
Comparative growth curves of recombinant virus and Towne strain control. The day 0 value was determined 4 h after removal of the original inoculum.
Results of this study strengthen the current impression (3) that mutations at codons 460, 520, and 591 to 607 (or possibly slightly beyond codon 607) confer ganciclovir resistance and that genotypic screening of UL97 for ganciclovir resistance should focus on this relatively limited range of codons. Furthermore, since CMV strains with deletions of either codons 591 to 594 (15) or codons 595 to 603 appear to be fully replication competent, the range of codons from codons 591 to 607 seems to be dispensable for the normal kinase function of UL97, while it strongly affects ganciclovir phosphorylation. We may expect the continued occasional appearance of clinical isolates with various deletions over the range from codons 591 to 607. The frequency of such mutations cannot be high given the number of resistant isolates that have been analyzed (3), but their existence should be taken into account in the design of genotypic assays.
Acknowledgments
We thank Gail Marousek, Wei Zhang, and Matthew Shiveley for technical assistance, Carlos Paya (12) for referring the clinical specimen, and George Kemble (11) for providing cosmid Tn26.
This work was supported by U.S. Department of Veterans Affairs Research Funds and by NIH grant AI39938.
REFERENCES
- 1.Baldanti F, Silini E, Sarasini A, Talarico C L, Stanat S C, Biron K K, Furione M, Bono F, Palu G, Gerna G. A three-nucleotide deletion in the UL97 open reading frame is responsible for the ganciclovir resistance of a human cytomegalovirus clinical isolate. J Virol. 1995;69:796–800. doi: 10.1128/jvi.69.2.796-800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldanti F, Underwood M R, Talarico C L, Simoncini L, Sarasini A, Biron K K, Gerna G. The Cys607→Tyr change in the UL97 phosphotransferase confers ganciclovir resistance to two human cytomegalovirus strains recovered from two immunocompromised patients. Antimicrob Agents Chemother. 1998;42:444–446. doi: 10.1128/aac.42.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou S. Antiviral drug resistance in human cytomegalovirus. Transplant Infect Dis. 1999;1:105–114. doi: 10.1034/j.1399-3062.1999.010204.x. [DOI] [PubMed] [Google Scholar]
- 4.Chou S, Erice A, Jordan M C, Vercellotti G M, Michels K R, Talarico C L, Stanat S C, Biron K K. Analysis of the UL97 phosphotransferase coding sequence in clinical cytomegalovirus isolates and identification of mutations conferring ganciclovir resistance. J Infect Dis. 1995;171:576–583. doi: 10.1093/infdis/171.3.576. [DOI] [PubMed] [Google Scholar]
- 5.Chou S, Guentzel S, Michels K R, Miner R C, Drew W L. Frequency of UL97 phosphotransferase mutations related to ganciclovir resistance in clinical cytomegalovirus isolates. J Infect Dis. 1995;172:239–242. doi: 10.1093/infdis/172.1.239. [DOI] [PubMed] [Google Scholar]
- 6.Chou S, Marousek G, Guentzel S, et al. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J Infect Dis. 1997;176:786–789. doi: 10.1086/517302. [DOI] [PubMed] [Google Scholar]
- 7.Cihlar T, Fuller M D, Cherrington J M. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J Virol. 1998;72:5927–5936. doi: 10.1128/jvi.72.7.5927-5936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drew W L, Miner R, Saleh E. Antiviral susceptibility testing of cytomegalovirus: criteria for detecting resistance to antivirals. Clin Diagn Virol. 1993;1:179–185. doi: 10.1016/0928-0197(93)90012-t. [DOI] [PubMed] [Google Scholar]
- 9.Erice A, Gil-Roda C, Perez J L, et al. Antiviral susceptibilities and analysis of UL97 and DNA polymerase sequences of clinical cytomegalovirus isolates from immunocompromised patients. J Infect Dis. 1997;175:1087–1092. doi: 10.1086/516446. [DOI] [PubMed] [Google Scholar]
- 10.Hanson M N, Preheim L C, Chou S, Talarico C L, Biron K K, Erice A. Novel mutation in the UL97 gene of a clinical cytomegalovirus strain conferring resistance to ganciclovir. Antimicrob Agents Chemother. 1995;39:1204–1205. doi: 10.1128/aac.39.5.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kemble G, Duke G, Winter R, Spaete R. Defined large scale alteration of the human cytomegalovirus genome constructed by cotransfection of overlapping cosmids. J Virol. 1996;70:2044–2048. doi: 10.1128/jvi.70.3.2044-2048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendez J C, Sia I G, Tau K R, Espy M J, Smith T F, Chou S, Paya C V. Novel mutation in the CMV UL97 gene associated with resistance to ganciclovir therapy. Transplantation. 1999;67:755–757. doi: 10.1097/00007890-199903150-00020. [DOI] [PubMed] [Google Scholar]
- 13.Michel D, Pavic I, Zimmermann A, Haupt E, Wunderlich K, Heuschmid M, Mertens T. The UL97 gene product of human cytomegalovirus is an early-late protein with a nuclear localization but is not a nucleoside kinase. J Virol. 1996;70:6340–6346. doi: 10.1128/jvi.70.9.6340-6346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prichard M N, Gao N, Jairath S, Mulamba G, Krosky P, Coen D M, Parker B O, Pari G S. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J Virol. 1999;73:5663–5670. doi: 10.1128/jvi.73.7.5663-5670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan V, Talarico C L, Stanat S C, Davis M, Coen D M, Biron K K. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature. 1992;358:162–164. doi: 10.1038/358162a0. [DOI] [PubMed] [Google Scholar]