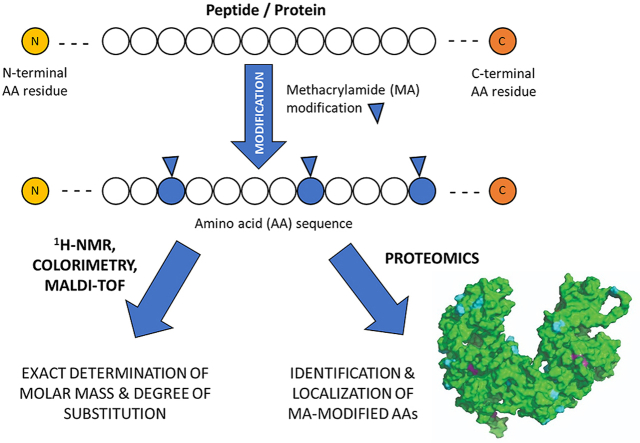
Abstract
The distribution of photo-crosslinkable moieties onto a protein backbone can affect a biomaterial's crosslinking behavior, and therefore also its mechanical and biological properties. A profound insight in this respect is essential for biomaterials exploited in tissue engineering and regenerative medicine. In the present work, photo-crosslinkable moieties have been introduced on the primary amine groups of: (i) a recombinant collagen peptide (RCPhC1) with a known amino acid (AA) sequence, and (ii) bovine skin collagen (COL BS) with an unknown AA sequence. The degree of substitution (DS) was quantified with two conventional techniques: an ortho-phthalic dialdehyde (OPA) assay and 1H NMR spectroscopy. However, neither of both provides information on the exact type and location of the modified AAs. Therefore, for the first time, proteomic analysis was evaluated herein as a tool to identify functionalized AAs as well as the exact position of photo-crosslinkable moieties along the AA sequence, thereby enabling an in-depth, unprecedented characterization of functionalized photo-crosslinkable biopolymers. Moreover, our strategy enabled to visualize the spatial distribution of the modifications within the overall structure of the protein. Proteomics has proven to provide unprecedented insight in the distribution of photo-crosslinkable moieties along the protein backbone, undoubtedly contributing to superior functional biomaterial design to serve regenerative medicine.
Keywords: (Photo-crosslinkable) biomaterials, Chemical modifications, Biomaterial characterization, Proteomics, Localization of functionalizations
Graphical abstract
Highlights
-
•
Photo-crosslinkable functionalized biomaterials for TE applications.
-
•
A toolkit to identify and localize functional groups on the AA sequence.
-
•
Proteomic analysis enables unprecedented characterization of functional biopolymers.
1. Introduction
Proteins are large, complex biomolecules composed of amino acid residues linked together into one or more chains. Depending on the amino acids present, combinations of hydrophobic, hydrophilic, polar and apolar regions can be present within a protein chain. Proteins differ from one another primarily in their sequence and composition of amino acids. Depending on this amino acid sequence, the protein usually folds into a specific three-dimensional (3D) structure. The structure and possible conformations of a protein affect a protein's function and bioactivity. For example, structural proteins like collagen, elastin, keratin, etc. maintain tissue shape and constitute structural elements in connective tissue like cartilage and bone [1].
Collagen is the most abundant structural protein, both in animals and humans. In the human body, collagen accounts for one third of the total protein content. Moreover, it forms the main component of the extracellular matrix (ECM) in various connective tissues in the body [2]. Collagen provides structural support and strength, and mediates local biological responses [3]. It is composed of three α-chains (i.e. two α1 and one α2) that are arranged in a triple helix. The sequence of this protein is characterized by the regular occurrence of glycine, proline and hydroxyproline [4].
In tissue engineering (TE) and regenerative medicine (RM), the aim is to regenerate, reconstruct or repair native tissue. Because connective tissue is mainly composed of fibrous ECM components, researchers have been exploiting ECM components as materials for TERM applications [5]. Due to the abundance of collagen in the ECM of human tissue, collagen has been studied frequently as a biomaterial for tissue engineering applications [6]. The main advantages of collagen include low antigenicity, biocompatibility, bioactivity, biodegradability and the capability to promote cell adhesion through cell receptors that recognize a specific peptide sequence (such as RGD (R: arginine; G: glycine; D: aspartic acid)) within collagen [[7], [8], [9]].
One of the most important limitations related to the use of extracted collagen for TERM applications is its mechanical properties, mainly at the viscoelastic level, such as the insufficient burst strength of collagen-based materials when exposed to the high stresses and pressures encountered in vascular tissue engineering (i.e. compliance of 4.5–6.2%/100 mm Hg, a burst pressure of 2031–4225 mm Hg, a maximum stress of 1.44 ± 0.87 MPa, a maximum strain of 0.54 ± 0.25 MPa and a physiological elastic modulus of 1.48 ± 0.24 MPa) [7,[10], [11], [12], [13], [14], [15], [16]]. Therefore, research has focused on various approaches to control the polymerization and the stability in solution and to reduce enzymatic sensitivity, in an attempt to improve the mechanical strength. Another approach involves chemical, physical or enzymatic crosslinking of the individual collagen chains [1,[17], [18], [19]]. Therefore, crosslinkable moieties (introduced upon reaction with e.g. methacrylic anhydride, 4-vinylbenzyl chloride, glycidyl methacrylate, 2-iminothiolane, maleic anhydride, itaconic anhydride, etc.) have already been introduced on the protein backbone to obtain a stable, crosslinked network [7]. A frequently applied approach in this respect is to exploit the primary amines present in the biopolymer backbone (i.e. hydroxylysine, lysine and ornithine) for functionalization purposes. Moreover, the type of modification, the corresponding crosslinking mechanism along with the crosslinking kinetics influence the processability of biomaterials into scaffolds along with their mechanical and biological properties (including the material's bioactivity) [20,21].
In order to engineer the structure reasonably allowing to achieve the targeted mechanical properties - and thus to mimic those of native tissue - one should obtain information on the molar mass (MM), the degree of functionalization, the location of the modified amino acids as well as the location of these functionalities within a protein's 3D structure (i.e. towards inner or outer side of the protein's 3D structure).
Conventional characterization techniques applied as quantitative tool to evaluate the degree of substitution (DS) of functionalized proteins include an ortho-phthalic dialdehyde (OPA) assay and proton nuclear magnetic resonance (1H NMR) spectroscopy [22,23]. However, these techniques only provide a quantitative evaluation of the amount of introduced functional groups but do not provide further insight in the distribution and the location of these modifications along the protein backbone.
In order to gain additional insight in biopolymer functionalization, proteomics can be a valuable tool. Proteomics involves the systematic, large-scale analysis of proteins. It is based on the study of the proteome, which is defined as a complete set of proteins produced by a given cell or organism under a defined set of conditions [24]. Proteomics is currently used to quantify and identify naturally occurring modifications present on a peptide/protein but more importantly, it enables to localize these modifications along the protein sequence [25,26].
During the past decade, mass spectrometry has become the method of choice in proteomics for the identification, quantification and study of post-translational modifications (PTMs) on proteins. It is a very sensitive (i.e. fmol∙μL−1 peptide), accurate and efficient method for sequencing proteins. Shotgun proteomics refers to the use of bottom-up proteomics techniques to study complex protein mixtures [26]. It utilizes the technology of high-performance liquid chromatography (HPLC) hyphenated with mass spectrometry (MS). The most distinct feature of shotgun proteomics is that it enables the identification and comparative quantification of a wide range of proteins at the same time while only requiring minimal separation between the LC peaks of the peptides. This technique is based on the extraction of proteins followed by their denaturation, reduction and alkylation. Next, the proteins are digested by an enzyme like trypsin. The cleaved, released peptides are separated with HPLC, followed by tandem MS/MS analysis to identify the amino acid sequence of each peptide. The identified peptide mass sequences are then compared with a protein database such as Swiss-Prot, which enables the identification of the proteins [[26], [27], [28]].
In other words, proteomics can show exactly where functional groups are located on the amino acid sequence, which has already been studied for naturally occurring modifications (e.g. proline hydroxylation, phosphorylation, etc.) in a biopolymer, and in the field of modifications with drug conjugates [29] but has not yet been investigated for introduced chemical modifications aiming at developing photo-crosslinkable biomaterials for TE purposes (e.g. methacrylamide modification). The latter could provide unprecedented insight in the distribution of introduced photo-crosslinkable functionalities along the chain. Moreover, proteomics can enable further unravelling of the efficiency of a biopolymer modification along with its potency to create a crosslinked network. This know-how is crucial to enable translation of novel, functionalized biomaterials from bench to bedside, given regulatory constraints and the need for perfectly defined and reproducible biomaterials.
In the present work, proteomic analysis was used to determine the position and to quantify the number of photo-crosslinkable groups (i.e. introduced methacrylamide groups) in comparison with conventional characterization techniques (i.e. OPA and 1H NMR spectroscopy). To the best of our knowledge, this is the first time that photo-crosslinkable biomaterials serving TE applications are fully characterized with respect to their MM, DS, location of functionalities along the protein backbone as well as their accessibility within the protein's primary, secondary and tertiary structure.
Herein, proteomics was assessed for its potential to elucidate a known peptide sequence, i.e. a recombinant peptide based on collagen type I (RCPhC1) as well as its derivatives, containing photo-crosslinkable methacrylamides (MA). Afterwards, the proteomics technique was applied on a material with an unknown amino acid sequence, i.e. bovine skin collagen (COL BS).
2. Experimental section/materials and methods
2.1. Materials
Recombinant peptide based on collagen I, commercially available as Cellnest™, was kindly provided by Fujifilm Manufacturing (Europe B·V.). Collagen type I, extracted from bovine skin [30], was supplied by the Department of Collagen Research (National Research & Development Institute for Textiles and Leather, Romania). Methacrylic anhydride (MeAnH), sodium hydroxide (NaOH) and potassium chloride (KCl) were obtained from Sigma-Aldrich (Diegem, Belgium). Potassium phosphate monobasic (KH2PO4) and sodium phosphate dibasic (Na2HPO4) were obtained from Acros Organics (Geel, Belgium). All 1H NMR spectra were recorded in deuterium oxide (D2O) provided by Euriso-top (Saint-Aubin Cedex, France). Spectra/Por7 dialysis membranes (MWCO of 12,000–14000 kDa) were obtained from Polylab (Antwerp, Belgium). UF filters were obtained from Amicon® units (10 kDa cutoff limit; Millipore, Billerica, MA) and the chemical products for proteomics were obtained from Sigma-Aldrich.
2.2. Derivatization of biopolymers
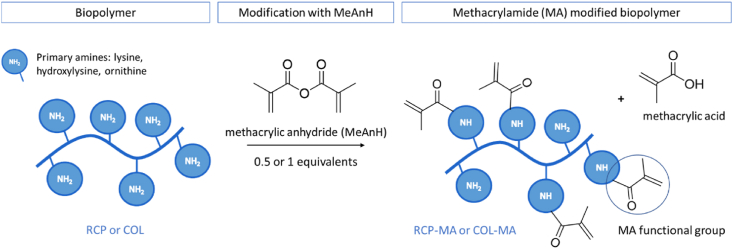
The methacrylation of biopolymers (Fig. 1) was performed according to the protocol of Tytgat et al. [31] In brief, methacrylamide-modified recombinant collagen (RCPhC1-MA) and methacrylamide-modified bovine skin collagen (COL-MA) were prepared through reaction of the primary amines with methacrylic anhydride (MeAnH). First, the biopolymer (10 w/v%) was dissolved in a phosphate buffer (pH = 7.8) at 37 °C. Next, 0.5 or 1 equivalents MeAnH with respect to the primary amines were calculated and added, followed by stirring during 1 h. Next, the reaction mixture was dialyzed (MWCO 12–14 kDa) against distilled water (37 °C, 24 h), followed by freeze-drying.
Fig. 1.
Development of methacrylamide-modified RCPhC1 (RCPhC1-MA) and methacrylamide-modified collagen (COL-MA) by introduction of methacrylamide moieties on the primary amines of the biopolymer (i.e. lysine, hydroxylysine and ornithine).
2.3. 1H NMR spectroscopy
The degree of substitution of the RCPhC1 and collagen derivatives was quantified via 1H NMR spectroscopy (Bruker WH 500 MHz) using D2O as solvent at elevated temperature (40 °C). The calculation of the DS was performed following the procedure as described earlier by Van Vlierberghe et al. [32] and using the MestReNova software. A Whittaker Smoother baseline correction was performed before analyzing the obtained spectra and integrating the peaks of interest. The 1H NMR spectra of the modified proteins show characteristic peaks at 5.75 and 5.55 ppm which correspond to the vinyl protons of the introduced MA functional groups (Fig. S1). The DS was quantified by comparing the integration of these characteristic peaks (i.e. 5.75 and 5.55 ppm) to the integration of the signal corresponding with the methyl protons present in Val, Leu and Ile (i.e. at 1.01 ppm) which are inert during the modification [32].
2.4. Ortho-phthalic dialdehyde assay
An OPA assay was applied as a quantitative tool to evaluate the DS of the functionalized polymers. To this end, 20 mg OPA was dissolved in 10 mL ethanol. Next, the mixture was diluted to 50 mL with double distilled water (deionized water). A second stock solution containing 25 μL 2-mercaptoethanol in a 50 mL borate buffer (pH = 10) was prepared. For 50 μL of heated (T = 37 °C) collagen solution (1 g/40 mL deionized water), 950 μL deionized water, 1500 μL 2-mercaptoethanol solution and 500 μL of the OPA stock solution were added, followed by vigorously mixing. Finally, the absorbance (Uvikon XL, BioTek Instruments) at 335 nm was measured compared to a blank (i.e. mixture with deionized water instead of collagen) at 37 °C. All measurements were performed in triplicate. Analogous measurements were performed with n-butylamine (0.002 M–0.01 M) standards to obtain a calibration curve. Calculation of the amount of unreacted amine groups remaining after the modification, enabled the determination of the DS.
2.5. Sample preparation for proteomic analysis
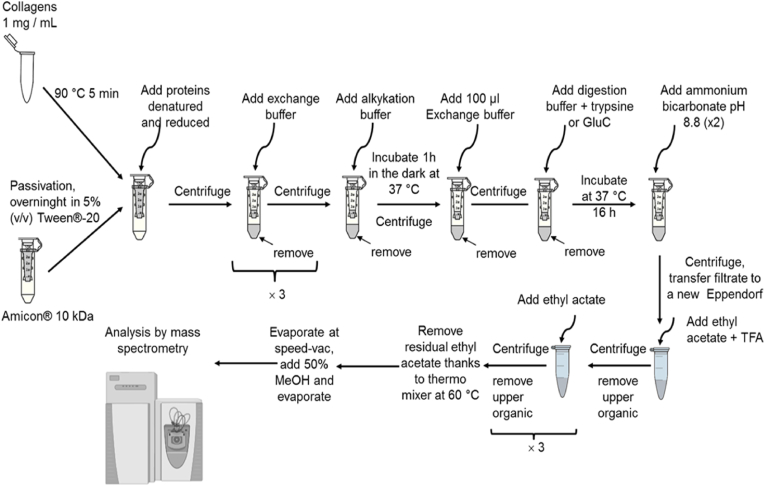
The sample preparation via the enhanced filter aided sample preparation (eFASP) tryptic digestion workflow is illustrated in Fig. 2. First, different collagens were dissolved in ultrapure water at 1 mg mL-1 in an Eppendorf® microtube (Eppendorf, Hamburg, Germany). The solutions were heated at 90 °C for 5 min to dissolve RCPhC1 or COL BS. The samples were prepared using a modified eFASP [33]. Before their use, 0.5 mL Amicon® ultra centrifugal filters equipped with a cut-off of 10 kDa (EMD Millipore, Darmstadt, Germany) were incubated overnight in a passivation solution containing 5% (v/v) Tween®-20 and then rinsed with ultrapure water. Next, 100 μg of protein was incubated in a 100 μL lysis buffer (8 M urea, 0.2% deoxycholic acid, 25 mM DTT, 100 mM ammonium bicarbonate pH 8.8). The solutions were transferred to an Amicon® filter and 100 μL of exchange buffer was added (8 M urea, 0.2% deoxycholic acid, 100 mM ammonium bicarbonate pH 8.8). After a centrifugation step of 20 min at 20,817 g, the filtrates were removed and 200 μL of exchange buffer was added to the Amicon® filters, which were consecutively centrifuged. This operation was repeated twice. The proteins were alkylated for 1 h at room temperature (20 °C) in the dark using 100 μL of alkylation buffer (8 M urea, 50 mM iodoacetamide and 100 mM ammonium bicarbonate, pH 8.8). The Amicon® filters were centrifuged again for 20 min at 20,817 g and the filtrates were discarded. After this alkylation step, 200 μL of exchange buffer was added to the Amicon® filters, which were again centrifuged for 20 min at 20,817 g and the filtrates were discarded. An aliquot (200 μL) of digestion buffer (0.2% deoxycholic acid, 50 mM ammonium bicarbonate, pH 8.8) was added to the Amicon® filters, prior to another centrifugation step (20 min at 20,817 g). This operation was repeated twice, with the filtrate being removed and discarded. The Amicon® filters were transferred to a new 2 mL concentrator collection tube. Next, 100 μL of digestion buffer containing 40 μL of endoproteinases Trypsin/LysC or GluC (Promega, Madison, WI) was added at 1/50 ratio enzyme/protein (w/w) and incubated in the Amicon® filters under continuous shaking in a heating block tube (MHR23, Hettich, Netherlands) during 16 h at 37 °C.
Fig. 2.
Workflow of the sample preparation (for proteomic analysis) by enhanced Filter Aided Sample Preparation (eFASP) digestion.
Thereafter, the peptides present in the Amicon® filters were recovered in the tube by centrifugation for 15 min at 20,817 g. To maximize the peptide recovery, two washing steps were implemented with 50 μL of ammonium bicarbonate solution (50 mM pH 8.8). The filtrates containing all peptides were transferred to a 1.5 mL Eppendorf® microtube. Next, 200 μL of ethyl acetate with 2.5 μL of TFA was added, to precipitate the peptide (white color). Again, 800 μL of ethyl acetate was added, the resulting solutions were centrifuged for 10 min at 10,681 g and the organic phases were eliminated. This operation was repeated twice. The Eppendorf® microtubes were placed for 5 min at 60 °C in a heating block (SBH130, Stuart, Staffordshire, UK) to enable evaporation of the remaining ethyl acetate. The samples were dried at room temperature in a SpeedVac™ Concentrator (EppendorfTM Concentrator Plus, Eppendorf). Next, 100 μL of a methanol/water (50/50) mixture was added to the resulting solid phase and let to evaporate.
For MS analysis, the samples were dissolved in 10 μL of ultrapure water supplemented with 0.1% of formic acid. The sample concentration was estimated by measuring the optical density (OD) at 215 nm of 1 μL of the solution using a droplet UV spectrometer (DS-11+, Denovix, Wilmington, USA). Finally, the concentration of the samples was adjusted to 1 μg∙μL-1 by dilution with ultrapure water containing 0.1% formic acid (FA) before analysis. Each sample was analyzed in triplicate.
2.6. Proteomic analysis using LC-MS/MS Orbitrap
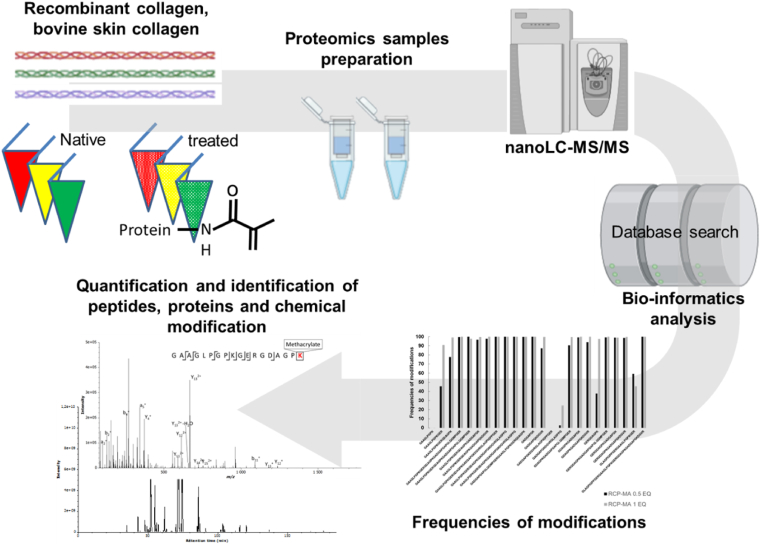
Liquid chromatography tandem mass spectrometry (LC-MS/MS) protein analyses were performed on an Orbitrap Q Exactive plus Mass Spectrometer hyphenated to a U3000 RSLC Microfluidic high-performance liquid chromatography (HPLC) System (ThermoFisher Scientific, Waltham, MA). An aliquot of the peptide mixture (1 μL) at a concentration of 1 μg∙μL-1 was injected with a solution A (5% acetonitrile, 94.9% H2O and 0.1% FA) for 3 min at an isocratic flow rate of 5 μL min-1 of solution A on an Acclaim PepMap100C18 pre-column (5 μm, 300 μm i.d. × 5 mm) (ThermoFisher Scientific). Next, the peptides were separated on a C18 Acclaim PepMap100C18 reversed phase column (3 μm, 75 mm i.d. × 500 mm) (ThermoFisher Scientific), using a linear gradient (5–40%) from solution A to solution B (75% ACN, 24.9% H2O and 0.1% FA) and a flow rate of 250 nL min-1 in 160 min followed by 100% solution B for 5 min, to clean the column and then re-equilibrated with solution A during 10 min. The column and the pre-column were placed in a column oven at a temperature of 45 °C. The total duration of the analysis of one sample was 180 min. The LC runs were acquired in positive ion mode. MS scans for DDA (data dependent acquisition) were acquired from m/z 350 to 1500 in the Orbitrap mass analyzer with a 70,000 resolution with maximum injection time of 80 ms and AGC target of 1 × 106. MS/MS scans were sequentially acquired in the high-energy collision dissociation cell for the 10 most-intense ions detected in the full MS survey scan. For MS/MS, the resolution was set to 35,000 with maximum injection time of 120 ms and AGC target of 5 × 105 and the normalized collision energy was set to 28 eV. Dynamic exclusion was set at 90 s and ions with 1 and more than 8 charges were excluded. The workflow of the proteomics approach for the identification and quantification is shown in Fig. 3.
Fig. 3.
Workflow of proteomics approach for identification and quantification of (modified) biopolymers.
2.7. Label-free quantification of modified peptides
The raw data from LC-MS/MS were processed by Proteome Discoverer version 2.2 (ThermoFisher Scientific) with a SEQUEST search engine against all entries of Swiss-Prot database (563,552 sequences, version UniProtKB 2020_05) or Bos taurus database from Swiss-Prot (37,512 sequences, version June 29, 2020) or home-made collagen sequences containing 3 sequences (i.e. recombinant collagen, collagen α1(I) chain P02453, collagen α2(I) chain P02465). The quantification was done with Minora node. The MS error was set to 10 ppm and 0.05 Da for MS/MS mass tolerance error. Trypsin with specific cleavage site (K, R) and GluC with specific cleavage site (E, D) were selected, together with variable modifications of the MA functionalities on lysine with a specific delta mass (+68.026 Da), oxidation of methionine (+15.994 Da), and/or deamidation of glutamine and asparagine (+0.984 Da). Oxidation of proline (+15.994 Da) was added for collagen. The fixed modification of carbamidomethyl cysteine (+57.021 Da) was also selected. The minimal peptide length of amino acids and the maximum number of missed cleavages were both set to six. The false discovery rate (FDR) threshold was set to 0.05 using the Percolator node. Relative abundances of peptides were calculated by integration of the area under the curve of the MS1 peaks using Minora LFQ node. The peptides were filtered on posterior error probability (PEP) score, q-value, Xcorr score. The PEP score and q-value threshold was set to less than 0.05 and Xcorr score greater than 1.
The percentage of the MA modified peptides (i.e. the degree of modification/substitution) is calculated using Equation (1). An example of the calculation of the degree of modification using proteomics can be found in Supplementary Information, on page 1.
| (1) |
With; DS = degree of substitution.
IntensityMA modified peptide = intensity of all MA modified peptides in the MA modified sample.
Intensitycontrol peptide = intensity of the MA modifiable peptides without modification in the MA modified sample.
2.8. Intact mass analysis using MALDI-TOF
The RCPhC1 and its derivatives were analyzed by matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF) (MALDI TOF/TOF) 4800+ (Sciex, Framingham, MA). Five μg of the RCPhC1 collagens were mixed with 1 μL of CHCA (α-cyano-4-hydroxycinnamic acid) matrix (70% ACN, 30% H2O, 0.1% TFA). The RCPhC1 collagens were acquired in linear positive ion mode. The mass range was set to 40,000 until 60,000 m/z and 2000 spectra were accumulated.
2.9. SDS-PAGE analysis on RCPhC1 and COL BS
Different collagens were dissolved in water at 5 mg mL-1 in Eppendorf. The solutions were heated at 90 °C during 5 min to dissolve the collagen. 50 μg of collagens samples were dissolved in 30 μl of laemmli buffer (4% SDS, 20% glycerol, 50 mM DTT, 0.004% bromophenol blue and 0.125 M Tris HCl, pH approx. 6.8) and heated 5 min at 90 °C. Collagens were separated on one-dimensional SDS-polyacrylamide gel electrophoresis. SDS-polyacrylamide gel was performed using standard methods on the Invitrogen SureCast™ system (10 cm × 10.5 cm minigels). The SDS–PAGE was carried out with 4% stacking gel and 12% resolving gel for the separation of recombinant collagen and the 10% polyacrylamide gels was used for the separation of bovine skin collagen. Dextran 500 kDa was incorporated in SDS–PAGE to improve the separation [34]. The voltage of power supply was set at 200 V for 60 min. The gel was stained with Coomassie Brilliant Blue R-250 for 120 min, then the stained gel was destained using a destaining solution (water/ethanol/acetic acid, 7:2:1, v/v/v). PageRuler Plus prestained protein ladder (Themoscientific, Baltics, UAB) was used to determinate the molar mass of proteins.
2.10. In gel tryptic digestion
The protein bands were excised from the gel and the gel slices were rinsed with a mixture of acetonitrile/ammonium bicarbonate (50 mM, pH 8.8) (50/50, v/v), then dehydrated with 100% acetonitrile. The gel slices were subjected to reduction of disulfide bonds by 10 mM DTT, 50 mM ammonium bicarbonate (pH 8.8) at 56 °C at 45 °C for 1 h. Alkylation step was then performed with 55 mM iodoacetamide, 50 mM ammonium bicarbonate (pH 8.8) at room temperature for 1 h in the dark. Before trypsin digestion, the gel slices were washed tree times with a mixture of acetonitrile: 50 mM ammonium bicarbonate (v/v) and dehydrated with acetonitrile. The gel slices were dried for 30 min at room temperature. Trypsin digestion was finally performed by incubating the gel slices with 25 μL of trypsin solution (20 μg of Trypsin Gold; Promega with 1 mL of 50 mM ammonium bicarbonate pH 8.8) during 15 min at 4 °C. The excess of solution was discarded and 50 μL of 50 mM ammonium bicarbonate pH 8.8 were added on gel slices and incubated 16 h with shaking in a heating block tube (MHR23, Hettich, Netherlands) overnight at 37 °C. Following digestion, the tryptic digested fragments present in the supernatant were collected. The gel slices were dehydrated with 100% acetonitrile containing 0.1% trifluoroacetic acid (TFA) for 30 min and the solution was added in the supernatant. The extracts were finally dried in a SpeedVac™ Concentrator (EppendorfTM Concentrator Plus, Eppendorf) and dissolved in a solution of 0.1% formic acid for mass spectrometric analysis.
2.11. LC-MS/MS of in gel tryptic digestion
LC-MS/MS protein analysis was performed on an Orbitrap Q Exactive plus Mass Spectrometer hyphenated to a U3000 RSLC Microfluidic HPLC System (ThermoFisher Scientific). 1 μL of the peptide mixture was injected with a solution A (5% acetonitrile, 94.9% H2O and 0.1% FA) for 3 min at a flow rate of 5 μL min-1 on an Acclaim PepMap100C18 pre-column (5 μm, 300 μm i.d. × 5 mm) (ThermoFisher Scientific). The peptides were next separated on a C18 Acclaim PepMap100C18 reversed phase column (3 μm, 75 mm i.d. × 500 mm) (ThermoFisher Scientific), using a linear gradient (5–40%) from solution A to solution B (75% ACN and 0.1% formic acid) using a flow rate of 250 mL min-1 in 50 min followed by 100% solution B for 5 min and then re-equilibrated with solution A during 10 min. The column and the pre-column were placed in an oven at a temperature of 45 °C. The total duration of the analysis was 70 min. The LC (liquid chromatography) runs were acquired in positive ion mode. MS scans for DDA were acquired from m/z 350 to 1500 in the Orbitrap mass analyzer with a 70,000 resolution with maximum injection time of 100 ms and AGC target of 1 × 106. MS/MS scans were sequentially acquired in the high-energy collision dissociation cell for the 15 most-intense ions detected in the full MS survey scan. For MSMS the resolution was set to 35,000 with maximum injection time of 140 ms and AGC target of 5 × 105 and the normalized collision energy was set to 28 eV. Dynamic exclusion was set at 30 s and ions with 1 and more than 8 charges were excluded.
Identification of protein in SDS-PAGE. The raw data from LC-MS/MS were processed by Proteome Discoverer version 2.2 (Thermo Scientific) with a SEQUEST search engine against Bos taurus database from Swiss-Prot (37,512 sequences, version june 29, 2020) and the sequence of the recombinant collagen. Proteins and peptides were identified and quantified with the same parameters than section 2.7.
2.12. Structural 3D-prediction of proteins via I-TASSER
For the 3D protein structure prediction of the proteins, the online I-TASSER platform was used (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). The I-TASSER procedure follows the sequence-to-structure-to-function paradigm and involves four steps: (1) retrieving template proteins of similar folds from the protein data bank (PDB) library by LOMETS, (2) fragment structure reassembly by replica-exchange Monte Carlo simulations, (3) atomic level structure refinement using REMO and FG-MD, and 4) structure-based function interpretations using COFACTOR [[35], [36], [37]].
The primary amino acid sequences of our proteins RCPhC1, COL1α1 and COL1α2 were submitted to the I-TASSER server, which each took 2–4 months to model. The sequences were submitted as such and thus no advanced options were used like specifying distance constraints nor structure templates to assist in modeling, excluding some templates from the I-TASSER template library and specifying secondary structure for specific residues.
The results from the I-TASSER simulation that are relevant to us in this paper include:
-
-
Up to five full-length atomic predicted models ranked based on cluster density via SPICKER. The confidence of each model is quantitatively measured by a C-score, a confidence score for estimating the quality of the predicted models by I-TASSER, in the range of 2 to −5, with higher values signifying high model confidence. It is calculated based on the significance of threading template alignments and the convergence parameters of the structure assembly simulations.
-
-
Predicted secondary structures
-
-
Predicted solvent accessibility
-
-
Top 10 threading templates from LOMETS
-
-
Top 10 proteins in PDB which are structurally the closest to the predicted models with a TM-score for the first model. The TM-score is a scale for measuring the structural similarity between two structures [38] with a value > 0.5 indicating a model of correct topology and a value of <0.17 meaning a random similarity.
Our simulation data have been uploaded and can be accessed through the RSCB PDB protein bank (https://www.rcsb.org/#Category-deposit).
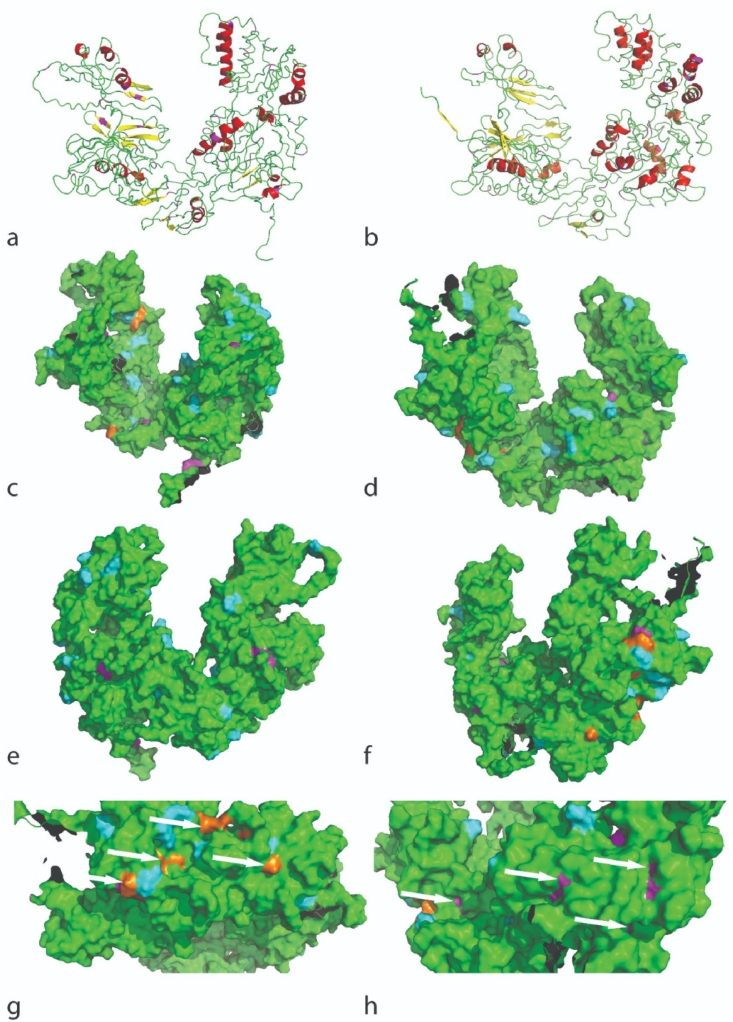
The generated PDB models from each protein were then visualized with Pymol 2.4 (Version 2.4.1). Each model is presented in cartoon and solvent surface accessibility view, and a consistent color code is implemented on all 3D-protein representations showing helices (red), sheets (yellow), loops (green (cartoon view)), RGD sequences at the surface (orange), protein surface (green (surface view)), lysine residues in the structure (magenta) and modified lysines (cyan). RCPhC1 is considered not to possess immunogenicity [39], and the 3D structural conformation of RCPhC1 as presented by this 3D model does not raise antibodies.
3. Results and discussion
In a first part, RCPhC1 and its derivative (i.e. methacrylamide-modified RCPhC1, RCPhC1-MA) were investigated. In proteomic analysis, the identification of the proteins is based on a partial sequence analysis with the aid of databank matching tools, or with the aid of an exactly known amino acid sequence as is the case for RCPhC1 [25]. Therefore, RCPhC1(-MA) acted as a proof-of-concept for evaluating proteomic analysis as characterization tool to study functionalized biopolymers. In a next step, the same protocol was used to analyze COL BS and its derivative (i.e. methacrylamide-modified COL BS, COL-MA BS) to prove that the proteomic analysis is also applicable to more complex samples with an unknown amino acid sequence.
3.1. Recombinant collagen (RCPhC1)
The potential of RCPhC1 as a source of hydrogel-based material for tissue engineering and regenerative medicine applications has already been showed by Tytgat et al. [31] and Fushimi et al. [39]. In this study, RCPhC1 was functionalized with methacrylamide moieties which enable UV-induced crosslinking to form networks in the presence of a photo-initiator. Two RCPhC1 derivatives were targeted namely with a lower and a higher degree of substitution (DS), based on literature [31]. In this work, 0.5 and 1 equivalents of MeAnH were added with respect to the number of lysine groups present. Because there are no available hydroxylysine nor ornithine groups in RCPhC1, only lysine groups were taken into consideration for functionalization (Table 1). A total of 33 lysine groups are present in RCPhC1 and could thus potentially be modified.
Table 1.
Overview of the amino acid composition of RCPhC1.
| AA | Number | % | mmol g−1 |
|---|---|---|---|
| Ala | 88 | 15.4 | 1.72 |
| Cys | 0 | 0.0 | 0.00 |
| Asp | 33 | 5.8 | 0.64 |
| Glu | 24 | 4.2 | 0.47 |
| Phe | 0 | 0.0 | 0.00 |
| Gly | 191 | 33.5 | 3.73 |
| His | 0 | 0.0 | 0.00 |
| Ile | 6 | 1.1 | 0.12 |
| Lys | 33 | 5.8 | 0.64 |
| Leu | 33 | 5.8 | 0.64 |
| Met | 9 | 1.6 | 0.18 |
| Asn | 0 | 0.0 | 0.00 |
| Pro | 100 | 17.5 | 1.95 |
| Gln | 12 | 2.1 | 0.23 |
| Arg | 33 | 5.8 | 0.64 |
| Ser | 0 | 0.0 | 0.00 |
| Thr | 0 | 0.0 | 0.00 |
| Val | 9 | 1.6 | 0.18 |
| Trp | 0 | 0.0 | 0.00 |
| Tyr | 0 | 0.00 | 0.00 |
| hydroxyLys | 0 | 0.00 | 0.00 |
| Ornithine | 0 | 0.00 | 0.00 |
3.1.1. Determination of the molar mass (MM) of RCPhC1 and its derivative
1H NMR spectroscopy and OPA do not allow the determination of the MM of RCPhC1. For polymers with a MM > 25 kDa [40], the determination of the MM by 1H NMR spectroscopy can be intractable because the resolution is diminished and the NMR spectra are too complex for natural biopolymers [40,41]. The OPA assay is used to measure the amount of primary amines in a sample, and is thus not appropriate for the determination of the MM.
Based on the known amino acid sequence of RCPhC1 (Table 1), the theoretical MM could be calculated. The unmodified RCPhC1 shows a MM of 51,185 Da (Table 2).
Table 2.
Theoretical and measured molar mass of RCPhC1 obtained by MALDI-TOF. The theoretical molar mass of RCPhC1 is calculated from the sequence of its amino acids (https://web.expasy.org/protparam/).
| Theoretical MM [Da] | Measured MM [Da] | Number of modified primary amines [n] | |
|---|---|---|---|
| Unmodified RCPhC1 | 51,185 | 51,188 | 0 |
| RCPhC1-MA 0.5 EQ | 52,205 | 52,280 | 16 |
| RCPhC1-MA 1 EQ | 53,429 | 53,380 | 32 |
MALDI-TOF analysis was used in order to confirm the theoretical MM and (the type of) PTMs (possibly) present in the analyzed samples (e.g. phosphorylations, glycolysations, etc.). This technique can be used on RCPhC1 because the MM of RCPhC1 is less than 150 kDa [42]. The intact protein analysis of the recombinant collagen showed a measured MM of 51,188 Da (Fig. S2), closely matching the theoretical MM of 51,185 Da. The number of modified lysines is zero for the unmodified RCPhC1. Additionally, the analysis showed that the recombinant protein does not have any naturally occurring PTMs (e.g. oxidations, deaminations). Analysis of the RCPhC1 protein modified with MA photo-crosslinkable groups (RCPhC1-MA) targeting a low and a high DS (i.e. by addition of 0.5 and 1 EQ MeAnH) showed higher MM of 52,280 Da and 53,380 Da, respectively. One MA modification increases the mass of the protein with +68.0 Da, corresponding to the incorporation of one C4H4O1 moiety on one lysine entity. RCPhC1 (unmodified) has 33 lysines in its amino acid sequence, whereas the intact mass analysis of the RCPhC1 derivatives (RCPhC1-MA) showed the presence of 16 and 32 modified lysines upon adding respectively 0.5 and 1 EQ MeAnH. The measured and theoretical MM of RCPhC1 are shown in Table 2.
Polyacrylamide SDS-PAGE gel analysis showed a MM for the RCPhC1 protein of roughly 50 kDa (Fig. S3). This is in agreement with literature [39]. The MM of the protein increases with an increase in the number of methacrylamides incorporated. These SDS-PAGE data are thus in correlation with the data obtained by MALDI-TOF. The digestion of the gel bands allowed the identification of the RCPhC1 protein in the gel bands (Supplementary Data Table 1). Peptides carrying a methacrylamide were identified in the gel bands corresponding to RCPhC1-MA 0.5EQ and RCPhC1-MA 1EQ.
In conclusion, MALDI-TOF analysis is the only suitable technique (of the techniques discussed in this work: 1H NMR, OPA, MALDI-TOF and SDS PAGE) for the accurate determination of the MM of (functionalized) RCPhC1. Other suitable techniques that would allow the determination of the MM include (1) gel permeation chromatography (GPC), which are used rather for qualitative than for quantitative analysis [43,44], (2) membrane osmometry [44,45], (3) intrinsic viscosimetry in combination with static/dynamic light scattering (can be further combined with ultracentrifugation) [[45], [46], [47], [48], [49], [50], [51], [52]], and (4) a top-down method in proteomics using high resolution mass spectrometry [53,54]. Although all these techniques would allow the determination of the MM, herein, we have selected the most commonly applied methods in the state-of-the-art [20,21].
3.1.2. Identification and localization of the modified groups in the amino acid sequence of RCPhC1-MA
The RCPhC1 was digested by two different enzymes (that cut collagen at different sites) to improve the coverage, identification of the sequence and quantification of the modification [55]. Trypsin cuts after lysine (K) and arginine (R), while endoproteinases GluC cut after glutamic (E) and aspartic acid (D). The shotgun proteomic analysis identified 100% of the RCPhC1 sequence that contained 571 amino acids and provided a theoretical MM of 51,185 Da. The coverage of the sequence of RCPhC1-MA was found to be 100% for both types of digestions (Table 3). However, the proteomics approach identified more peptides in unmodified RCPhC1 compared to RCPhC1-MA 0.5 EQ and RCPhC1-MA 1 EQ, for both types of digestion. This difference may be explained by steric hindrance due to the introduced MA groups or unreachable sites in the protein structure (see section 3.1.4). The number of identified peptides (Table 3) corresponds to the number of digested peptides with and without chemically introduced modifications, showing a high coverage percentage.
Table 3.
Number of identified peptides, peptide spectrum matches (PSMs) and percentage of coverage for RCPhC1 from trypsin and GluC digestion by LC-MS/MS.
| RCPhC1 | RCPhC1-MA 0.5 EQ | RCPhC1-MA 1 EQ | ||
|---|---|---|---|---|
| Trypsin digestion | Peptides | 40 | 30 | 26 |
| Coverage (%) | 100 | 100 | 100 | |
| GluC digestion | Peptides | 56 | 37 | 39 |
| Coverage (%) | 100 | 100 | 100 | |
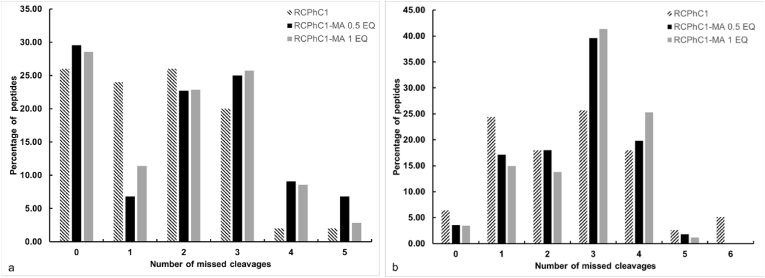
The number of missed cleavages observed is between 0 and 5 sites for trypsin and 0 to 6 for GluC (Fig. 4). The average size of the peptides after digestion with trypsin is 2340 Da and 355a0 Da for GluC. This result was anticipated because the number of lysine and arginine is higher than the number of glutamic acids, which induces a longer peptide size for GluC.
Fig. 4.
Histogram of the percentage of missed cleavages of peptides for RCPhC1 and its derivatives by trypsin digestion (a) and GluC digestion (b). The percentage of missed cleavages is obtained based on the ratio of the peptides with a missed cleavage identified by LC-MS/MS over the total number of identified peptides.
Fig. 4 shows a slight increase in the percentage of missed cleavages (3, 4, 5) for trypsin digestion, and (3,4) for GluC, when comparing the MA-modified RCPhC1 to unmodified RCPhC1 protein. This could be explained by the steric hindrance of the MA groups present near the trypsin cleavage site (after the lysine groups), and the fact that the size of the MA group can also hinder the cleavage by GluC.
The analysis indicated the presence of peptides containing 1 up to 3 MA moieties. An increase in missed cleavages makes the peptides (to be identified) larger in size and therefore these larger peptides can contain more than one lysine. Moreover, a trend was seen between the number of missed cleavage sites and the number of MAs introduced onto lysines (Fig. S4). When more MA groups were present in the sequence, an increase in the number of missed cleavages was detected. Both trypsin and Glu-C digestion showed a clear linear correlation with a higher missed cleavage with increasing modification, most likely due to steric hindrance of the enzyme.
The LC-MS/MS analysis confirmed that there are no identified MA sites in the unmodified RCPhC1. The analysis of RCPhC1-MA 0.5 EQ and RCPhC1-MA 1 EQ indicated that all lysine groups were MA modified lysines for both digestions, but at a different modification frequency (vide infra, section 3.1.3). Correlating these results with MALDI TOF indicates that on average 16 lysines are modified in case of RCPhC1-MA 0.5 EQ. This means that there are isoforms possible of RCPhC1-MA 0.5 EQ with 16 modified lysines, but at variable positions (see Fig. 8, c-d; section 3.1.4). In case of RCPhC1-MA 1 EQ, LC-MS/MS showed that all lysines groups were modified.
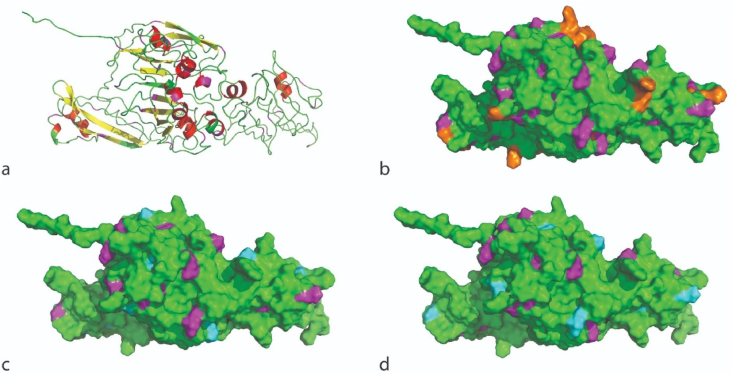
Fig. 8.
The protein 3D-conformation of the sequence of RCPhC1 modelled with I-TASSER; (a) in the cartoon view, showing helices (red), sheets (yellow), loops (green) and lysine residues in the structure (magenta), (b) in the solvent surface accessibility view, showing the protein accessibility surface (green), all lysines on the surface (magenta) and the RGD sequences at the surface (orange). The model in b is also the same as for RCPhC1-MA 1 EQ of which all lysines are modified (magenta = cyan) (Fig. S7). (c–d) 3D-conformation showing the modified lysines in two RCPhC1-MA 0.5 EQ isomorphs, and the corresponding RCP sequences in Supplementary Information: (c) the first isomorph (Fig. S8, blue) and (d) the second isomorph (Fig. S8, yellow), showing the unmodified lysines (magenta) and the modified lysines (cyan). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
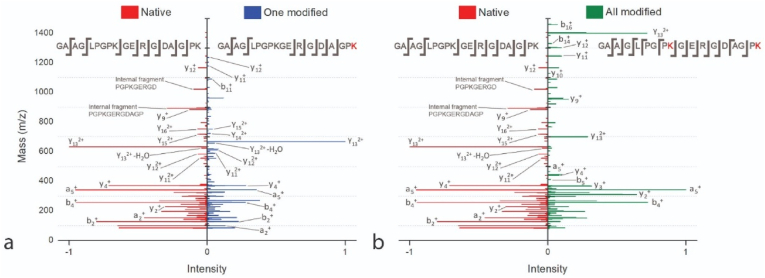
LC-MS/MS analysis also enables to determine the position of the MA functional groups. Fig. 5 shows the fragmentation of the peptide GAAGLPGPKGERGDAGPK. It is possible to localize the modification by observing a specific fragment obtained by MS/MS corresponding to the amino acid carrying the modification. In this work, HCD fragmentation (see section 2.6) leads to the formation of b (N-terminal) and y (C-terminal) charged fragment ions. The MS/MS spectrum from the peptide GAAGLPGPKGERGDAGPK with one or two MAs can be compared with the MS/MS spectrum of the unmodified peptide (Fig. 5, red spectrum). The MS/MS spectrum with one MA in Fig. 5 (a, blue spectrum) allowed the localization of the modifications at the C-terminal due to the series of b and y ions, because there is no fragment corresponding to lysine at position 9 with a MA group. The b11 and y11 ions made it possible to show that the lysine at position 9 did not carry a MA group. The MS/MS spectrum in Fig. 5 (b, green spectrum), showed that all available lysines in the peptide were modified. The spectrum allowed the identification of the MA on lysine in positions 9 and 18 on the sequence peptide GAAGLPGPKGERGDAGPK because the y11 ion at m/z 1150.5847 corresponds to the MA group on the lysines.
Fig. 5.
MS/MS of peptide GAAGLPGPKGERGDAGPK. Panel (a) shows the fragmentation of native peptide (red) and the modified peptide with 1 methacrylate on lysine in C-terminal position (blue). Panel (b) shows the fragmentation of native peptides (red) and peptides with all methacrylated lysines (green). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
On the one hand, the shotgun proteomic analysis highlights that there are isoforms of the RCPhC1 with different positions of the MA groups. On the other hand, the results showed that there are redundant (or repetitive) peptide sequences in the complete RCPhC1 sequence. This presents an additional challenge when identifying the exact position of the MA groups in the sequence. For example, the LC-MS/MS analysis identified the peptide GAAGLPGPKGERGDAGPK with a modified lysine (after trypsin digestion) but this peptide was identified at different positions in the RCPhC1 sequence, namely positions [[22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39]]; [[58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75]]; [100−117]; [157−174]; [211−228]; [247−264]; [289−306]; [346−363]; [400−417]; [436−453]; [478−495]; [535−552].
Furthermore, the incorporated MA groups might influence the ionization of the peptides, that can in turn lead to a decrease in sensitivity. Nevertheless, even though the number of identified peptides decreased (upon increasing the number of incorporated MA groups), the sensitivity remained sufficiently high to reach a sequence coverage of 100%, for all analyzed samples (i.e. RCPhC1, RCPhC1-MA 0.5 EQ and RCPhC1-MA 1 EQ) [56].
In conclusion, the shotgun proteomic analysis is the only technique that enables: (1) the identification of the fragmented peptides, and (2) the determination of the exact location of the MA-modified amino acids in the amino acid sequence. In case of RCPhC1 however, one major challenge was identified using the shotgun proteomic analysis, namely the occurrence of redundant peptides.
3.1.3. Determination of the degree of substitution (DS) of modified RCPhC1
The DS of the developed RCPhC1 derivatives can be determined via 1H NMR spectroscopy [31]. The 1H NMR spectra of modified RCPhC1 showed characteristic peaks at 5.75 and 5.55 ppm which correspond to the vinyl protons of the introduced MA functional groups. The DS was quantified by comparing the integration of these characteristic peaks (i.e. 5.75 and 5.55 ppm) to the integration of the methyl protons present in Val, Leu and Ile, (i.e. at 1.01 ppm) which are chemically inert during modification (Fig. S1, a-b) [32]. Based on 1H NMR spectroscopy, the addition of 0.5 and 1 equivalents MeAnH (i.e. RCPhC1-MA 0.5 EQ and RCPhC1-MA 1 EQ) resulted in a DS of 53.7% [0.346 mmol g−1 of available photo-crosslinkable MA groups] and 93.3% [0.597 mmol g−1], respectively.
In addition to 1H NMR spectroscopy, an OPA amine quantification assay was performed on RCPhC1 and its derivatives to determine the number of primary amines available prior to and after functionalization. By comparing those two values, the DS and thus the number of introduced crosslinkable groups can be determined. Based on the OPA assay, the DS values of RCPhC1-MA 0.5 EQ and RCPhC1-MA 1 EQ were 51.9 ± 1.6% and 99.2 ± 0.2%, respectively. The result of RCPhC1-MA 0.5 EQ are comparable to those obtained via NMR (i.e. 53.7%). For the 1 EQ derivative, there is a minor discrepancy between both techniques. Compared to 1H NMR spectroscopy, the OPA assay is based on the difference of the number of primary amines available before and after functionalization, and may therefore be seen as the most accurate technique for the determination of the DS [57,58].
Based on MALDI-TOF analysis, the measured MM could be determined for the original (unmodified) RCPhC1 showing 33 unmodified lysines, versus the modified RCPhC1 (Table 2) showing 16 and 32 modified lysines for RCPhC1-MA 0.5 EQ and 1 EQ respectively. The latter corresponds to a DS of 50.0% and 97.0% for RCPhC1-MA 0.5 EQ and 1 EQ, respectively.
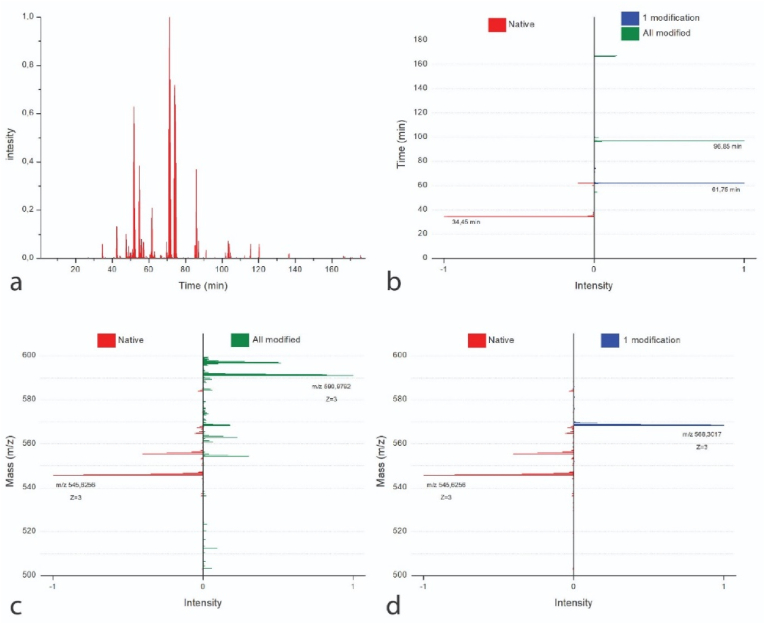
When performing the proteomic analysis, the quantification method (LC-MS/MS) is based on a label-free method, that aims at determining the relative amount of proteins and peptides in two or more biological samples. In other words, the quantification by LC-MS/MS was performed by carrying out the ion extraction of the modified and unmodified peptides in the samples (Fig. 6).
Fig. 6.
Ion extraction chromatogram of the peptide GAAGLPGPKGERGDAGPK (as an example) with and without MA groups in RCPhC1-MA 1 EQ. Panel (a) shows the total ion extraction chromatogram. Panel (b) shows the retention time (RT) and corresponding signal intensity of the unmodified peptide (red, RT 34.45 min) versus 1 lysine modification (blue, 61.75 min) and all lysines modified (green, 96.85 min). Panel (c) and Panel (d) show mass (m/z) with z = 3 and the corresponding signal intensity of the unmodified peptide (red, m/z 545.6256) versus all lysines modified (c, green, m/z 590.9792) and versus 1 lysine modification (d, blue m/z 568.3017), respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6 shows the signal intensity of the unmodified peptide (red), the peptide functionalized with one MA (blue) and the peptide with all lysines functionalized with MAs extracted from the total ion extraction spectrum of the RCPhC1-MA 1 EQ sample. This analysis showed a different retention time depending on the peptide containing one MA, the maximum amount of MAs or no MA (i.e. unmodified). This difference was caused by the introduction of MA groups, modifying the physico-chemical properties of the peptide thereby leading to a shift in retention time (Fig. 6, a-d).
Using both the Proteome Discoverer 2.2 software and the Minora quantification node, the peptides of the samples were quantified for the RCPhC1, RCPhC1-MA 0.5 EQ and RCPhC1-MA 1 EQ samples. The software extracted the intensity of the peptides, allowing the comparison between samples. The number of quantified peptides with one or more MA modifications was determined to be 34 for the trypsin digestions and 66 for the GluC digestions. These numbers included the peptides with oxidations on methionine, deamination of asparagine and glutamine and peptides with missed cleavages. Supplementary Data Table 2 provides a list of all quantified peptides.
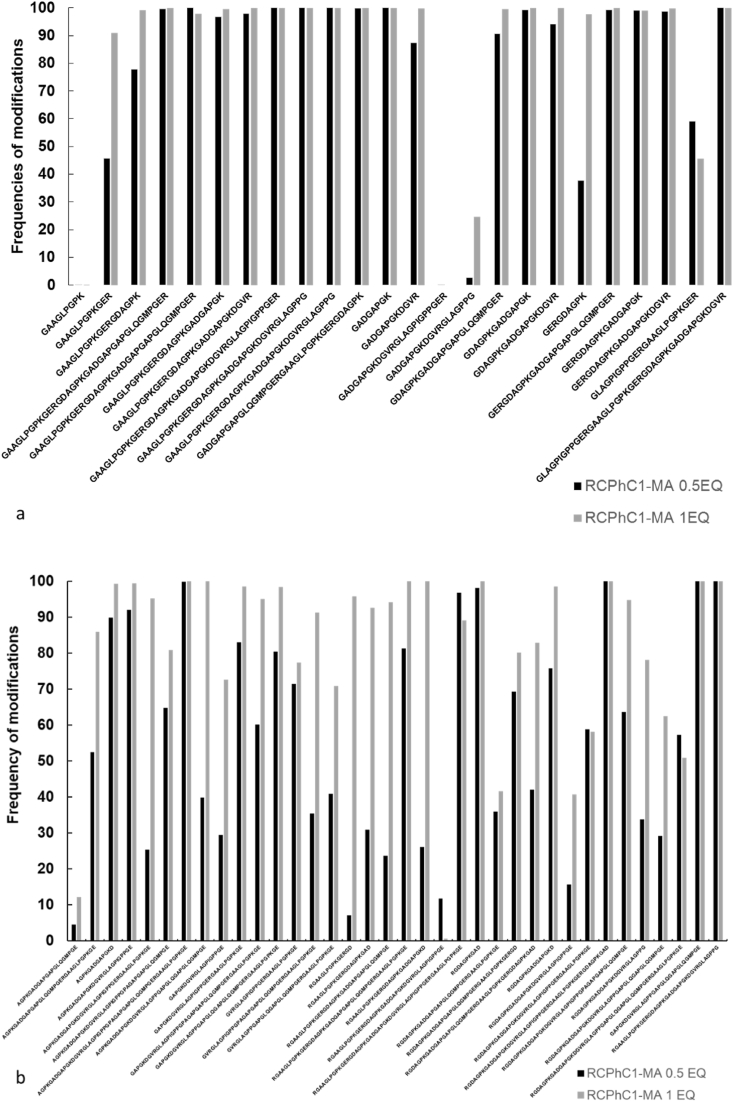
In addition, quantitative proteomic analysis provides information on the frequency of the modifications at one specific location (i.e. one specific AA in the AA sequence). The modification frequency can be defined as the probability that a certain peptide can be modified and is calculated with Equation (1) (section 2.7). In general, the quantification by LC-MS/MS shows a higher frequency of modification for RCPhC1-MA 1 EQ than for RCPhC1-MA 0.5 EQ. This can be deduced from the peptide frequency histogram (Fig. 7). The majority of peptides in RCPhC1-MA 1 EQ have shown a modification frequency close to 100%, indicating that all lysine groups present in RCPhC1-MA 1 EQ were almost completely modified (i.e. a DS close to 100%). The latter is in excellent agreement with the data obtained from MALDI-TOF (i.e. DS of 97.0%), OPA (i.e. DS of 99.2%) and NMR spectroscopy (i.e. DS of 93.3%). For RCPhC1-MA 0.5 EQ, the modification frequency of the modified lysines was lower. For example, the peptide GAAGLPGPKGER containing one MA identified on the lysine group after trypsin digestion, exhibited a modification frequency of 45% for RCPhC1-MA 0.5 EQ and 90% for the RCPhC1-MA 1 EQ (Fig. 7, a). The co-elution of the peptides can affect the quantification of the chemical modification independently of the quantification methods used, therefore, proteomics analysis as a stand-alone technique is not ideal for the exact quantification of the DS [59].
Fig. 7.
Histogram of frequency of MA modification (on lysines) for peptides resulting from cleavage with enzyme Trypsin (a) and GluC (b). Black bars correspond to RCPhC1-MA 0.5 EQ and grey bars to RCPhC1-MA 1 EQ.
On the one hand, some peptides contained certain lysine positions that had a 100% modification frequency in both the low and high DS RCPhC1-MA (i.e. RCPhC1-MA 0.5 and 1 EQ). On the other hand, some peptides were not modified in either of the RCPhC1-MA derivatives. An example is the peptide GAAGLPGPK, that did not constitute a modified lysine in either of the derivatives. An explanation could be that this peptide was frequently found to have 1 or 2 missed cleavages, indicating that either the MA modification or the position of the lysine in the overall structure of RCPhC1 (i.e. inner versus outer side of the protein's structural conformation, vide infra) interferes with the function of the enzyme. A similar result was observed for the frequency of MA upon GluC digestion (Fig. 7, b). The majority of peptides in RCPhC1-MA 1 EQ showed a modification frequency close to 100%, which shows that the lysine groups in RCPhC1-MA 1 EQ were almost completely modified.
Using shotgun proteomic analysis for the calculation of the DS of RCPhC1-MA is more challenging as the identification is complex due to the presence of redundant peptides (i.e. more challenging to identify them in the AA sequence because of repeating units) and because a higher DS results in more steric hindrance interfering with the enzyme function. Casey et al. [60] have already shown that chemical derivatization can modify the charge state distribution and the ionization. In one case, chemical derivatization was even shown to enhance the ionization of the peptide [61]. It can be concluded that for a protein of this size (i.e. RCPhC1, MM of 53 kDa), MALDI-TOF is the most sensitive and thus preferred technique to determine the DS.
As described above, proteomic analysis enabled to determine the modification frequency at one specific position in the sequence. Based on these modification frequencies, an estimation of the DS becomes possible. In order to increase the accuracy of the identification and quantification using proteomic analysis, the results of the analyses using two different types of digestion enzymes (trypsin and GluC) were combined. The average of the DS obtained by shotgun proteomics was 66.2% for RCPhC1-MA 0.5 EQ and 81.7% for RCPhC1-MA 1 EQ. Table 4 shows the DS based on the different techniques applied.
Table 4.
Overview of the obtained data for RCPhC1-MA 0.5 and 1 EQ: Determination of the molar mass using MALDI-TOF and determination of the degree of substitutions (DS) using MALDI-TOF, 1H NMR spectroscopy (based on the known amino acid composition), OPA assay and shotgun proteomic analysis.
| MM obtained by MALDI-TOF [kDa] | DS obtained by MALDI-TOF [%] | DS obtained via 1H NMR (based on known AA composition) [%] | DS obtained via OPA [%] | DS obtained by shotgun analysis [%] | |
|---|---|---|---|---|---|
| RCPhC1-MA 0.5 EQ | 52.28 | 50.0 | 53.7 | 51.9 ± 1.6 | 66.2 |
| RCPhC1-MA 1 EQ | 53.38 | 97.0 | 93.3 | 99.2 ± 0.2 | 81.7 |
In conclusion, proteomics is not an accurate technique to determine the exact DS, but only enables to determine an average DS, based on the modification frequencies. However, as stated in section 3.1.2, one of the aforementioned „conventional“ techniques (i.e. MALDI-TOF, OPA or 1H NMR) combined together with proteomic analysis makes it possible to fully characterize RCPhC1 and its derivatives.
3.1.4. Localization of modified amino acids in the 3D-structure of RCPhC1 modified biopolymers
To gain insight in the 3D-conformation of RCPhC1 and its derivatives, the I-TASSER server was used to model the 3D structure of RCPhC1 (see section 2.12). These 3D-models cannot be confirmed by X-ray studies and should be treated as possible folding configurations. However, the I-TASSER has a strong history of predicting simulation models close to X-ray confirmed models. The I-TASSER provides different models that are ranked according to a c-score (i.e. a confidence score for estimating the quality of the predicted models) with the first ranked model being the most reliable. The first model of RCPhC1 had a c-score of 0.44 and the estimated TM-score and root mean square deviation (RMSD) were 0.77 (±0.10) and 6.6 (±4.02 Å) respectively, signifying a high confidence in the model.
For RCPhC1, the obtained 3D-model is presented in Fig. 8 (a), showing the occurrence of helices (red) and sheets (yellow) in the main structure of the protein. In contrast to native COL1α1 (Fig. 8, a-b), the RCPhC1 derivative consists of considerably higher amounts of sheets and less helices. This is in accordance with the study of Fushimi et al. [39] in which the secondary structure of RCPhC1 was characterized by circular dichroism and based on a sequence-derived in silico model. This could be due to (i) the introduction of multiple RGD sequences in the sequence causing repetition (Fig. S5), (ii) its high content in alanine, glycine and proline, and/or (iii) the absence of hydroxyprolines in RCPhC1. This also explains why the second model that is predicted by I-TASSER has the same c-score as the first model and only contains β-sheets (Figure S6, a and b). This second model will not be discussed herein as it resembled to a lesser extent the structure of COL1α1.
The predicted model for RCPhC1 showed a good spatial distribution of the introduced RGD sequences in the protein, favoring optimal cell interaction [9,62], which is important for TE applications (Fig. 8, b, orange). More importantly, a good spatial distribution was observed for the lysines present in the amino acid sequence of the protein (Fig. 8, b, magenta), which is vital for the network formation of the functionalized proteins into crosslinkable materials after modification. This assumption is strengthened by the ability to form a solid and strong cross-linked biomaterial exploiting these protein derivatives as starting materials [31].
For RCPhC1-MA 1 EQ, it was shown that all lysines were modified (DS 100%), meaning that all magenta colored lysines can be colored in cyan in Fig. 8 (b). For RCPhC1 EQ (0.5), however, only 50% (based on MALDI-TOF, Table 4) of the lysines have been modified and obtaining a detailed insight into the modification is extremely challenging. In this respect, two possible isoforms of RCPhC1-MA 0.5 EQ are shown in Fig. 8 (c and d), as derived from the sequences shown in Fig. S8.
The two isoforms indicate that the spatial distribution of the modified lysines is somewhat reduced compared to RCPhC1-MA 1 EQ which might imply that the crosslinking efficiency may be reduced. Overall, it seems that all lysines are accessible in the solvent accessibility view in the protein's pristine condition, meaning that they have almost equal chances of being modified with MA or being cleaved by the enzymes. This also supports the observation that complete modification of the lysines was realized for RCPhC1-MA 1 EQ, along with a successful protein identification (i.e. 100 % coverage of identification).
3.2. Collagen bovine skin (COL BS)
Exploiting proteomic analysis to quantify and localize the chemical modifications introduced on a recombinant protein RCPhC1 with a known amino acid sequence, was shown to be a successful tool in the study of modified biopolymers. In a next step, the potential of the elaborated proteomics approach was evaluated on “more complex” protein-based biopolymers [6] with an unknown amino acid composition and/or sequence. The main difference between natural proteins and RCPhC1 is that PTMs and amino acid substitutions can occur in natural proteins. Herein, collagen from bovine skin origin (COL BS) was modified with MA-functional groups, resulting in different degrees of substitution (through addition of 0.5 and 1 EQ MeAnH).
3.2.1. Determination of the molar mass (MM) of COL BS and its derivative
The determination of the MM of COL BS by the three aforementioned techniques for RCPhC1 is not possible for several reasons:
-
-
As described in section 3.1.1 for RCPhC1, and keeping in mind that the size of collagen is approx. 6 times larger than RCPhC1, 1H NMR spectroscopy and OPA do not enable determining the MM of collagen.
-
-
MALDI-TOF is not able to determine the MM of collagen because the MM of collagen is around 300 kDa and the practical limit of conventional MALDI-TOF is around 80 kDa upon ensuring excellent resolution (theoretically to 150 kDa, yet resulting in low resolution). [42]. The MALDI-TOF used in this study only allowed screening of masses of modified proteins in the range of 2 up to 100 kDa approximately [31,63]. It would however be possible to determine the mass of large proteins more precisely with the use of very high resolution mass spectrometers such as FT-ICR [64,65] and the use of fragmentation techniques, making it possible to identify the protein sequence as well as the PTMs [66], by a technique called Top Down [67]. In case of complex proteins, this technique only gives the stoichiometry but not the individual MM.
Polyacrylamide SDS-PAGE gel analysis revealed the typical pattern for type I collagen, with bands corresponding to the MM for COL BS pointing to β (215 kDa), α1 (130 kDa), and α2 (115 kDa) chains [68,69]. (Fig. S9). The collagen contains numerous PTMs such as oxidations and deamidations that increase the MM. Three bands were observed in COL BS which correspond to COL1α1 and COL1α2 while the bands corresponding with a higher MM indicated a mixture of two collagens. The MM of the collagen protein increases with an increase in MeAnH equivalents. The digestion of the gel bands allowed identification of COL1α1, COL1α2, COL3α1, COL2α1. The mass spectrometry results showed that COL1α2 was the most abundant protein in the band corresponding with the lowest MM (Supplementary Data Table 1). COL1α1 was the most abundant protein in the next band. A band of a high MM compound (±200 kDa) was observed on the SDS-PAGE gel. The LC-MS/MS analysis of the digested bands corresponding with higher MM showed the presence of COL1α1, COL1α2, COL3α1, COL2α1. The COL1α1 and the COL1α2 were the most abundant proteins. It is hypothesized that this identification showed the crosslinks between collagens. Previous research showed the presence of the crosslinks on the collagen protein and were mainly derived from the allysine route [70,71]. Mass spectrometry analysis showed the presence of peptides with methacrylamides in the gel bands corresponding to COL-MA BS 0.5EQ and COL-MA BS 1EQ.
The shotgun proteomic analysis does not allow the determination of the MM of COL BS directly. However, the theoretical MM of COL BS can be estimated based on the identified proteins (vide infra, cfr. 3.2.2) using the Swiss-Prot database (COL1α1 and COL1α2), resulting in an estimated MM of 94673.0 Da for the COL1α1 chain precursor (amino acid 162 until 1217) and 93415.3 Da for the COL1α1 chain precursor (amino acid 80 until 1117). Collagen is constituted of two α1 chains and one α2 chain, thus resulting in an approximate theoretical MM of 282761.3 Da.
3.2.2. Identification and localization of the modified groups in the amino acid sequence of COL-MA BS
The shotgun proteomic analysis enabled the study of more complex samples such as COL BS. By using LC, it became possible to separate the peptides resulting from the tryptic or GluC digestion of the protein. After digestion, a larger number of peptides was cleaved compared to the digestion of RCPhC1 because of the higher MM of COL BS (Fig. S10). The identification of the most abundant proteins present in COL BS was carried out by querying the total Swiss-Prot database containing 563,082 sequences and 13,936 taxons. The first identified protein is the collagen α1(I) chain from Bos taurus, conforming with the origin of the sample. Then, the identification of the most abundant proteins in COL BS was carried out by querying the Bos taurus database of Swiss-Prot. The results indicated that the most abundant proteins are the COL1α1 and COL1α2, amounting for 57% and 28% respectively (Supplementary Data Table 3). The sample also contained low quantities of other proteins like collagen α1(III) chain, collagen α1(II) chain and keratin. LC-MS/MS analysis showed that the N- and C-terminal pro-peptides are not present in COL1α1 and COL1α2. This indicated that it was the matured protein of COL1α1 and COL1α2 without its pro-peptides ends. Therefore, the LC-MS/MS analysis was carried out on the sequences of the proteins COL1α1 and COL1α2 of Bos taurus without the pro-peptides. Table 5 shows the number of identified peptides, the percentage of coverage and the PSMs for the COL BS samples from COL1α1 and COL1α2 (Bos taurus) for both types of digestion (trypsin and GluC). The number of identified peptides decreased with an increasing number of introduced MA moieties, as was also observed for RCPhC1 (Fig. S10).
Table 5.
Overview of the number of identified peptides, the percentage of coverage and the PSMs for trypsin and Gluc digestion from the bovine skin collagen samples. The information is given for the 2 major proteins which are COL1α1 and COL1α2.
| Peptides [n] |
Coverage [%] |
||||
|---|---|---|---|---|---|
| COL1α1 | COL1α2 | COL1α1 | COL1α2 | ||
| Trypsin digestion | COL BS | 127 | 103 | 91 | 92 |
| COL-MA BS 0.5EQ | 119 | 89 | 88 | 81 | |
| COL-MA BS 1EQ | 79 | 75 | 75 | 76 | |
| GluC digestion | COL BS | 92 | 59 | 65 | 54 |
| COL MA BS 0.5EQ | 77 | 49 | 60 | 48 | |
| COL MA BS 1EQ | 73 | 49 | 56 | 48 | |
The shotgun proteomics approach allows identifying 91 and 92% of the sequence of COL1α1 and COL1α2 for the unmodified COL BS by trypsin digestion (Fig. 9) thereby providing very good coverage [72]. However, the coverage decreased upon higher degrees of modification (i.e. COL MA BS 0.5EQ and COL MA BS 1EQ). A similar trend was observed for the GluC digested proteins with an overall coverage (i.e. 65 and 54%) that is lower than for trypsin. However, by combining the two digestions it is possible to obtain more than 90% of coverage for COL BS with and without MA functional groups [73].
Fig. 9.
Coverage of the identified and quantified peptides on the sequence of COL1α1 and COL1α2 for the samples of COL BS from both digestions. Green areas show the protein sequence coverage. Yellow areas show the non-covered parts of the protein. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The identified peptides (green) were distributed homogeneously along the sequence as shown in Fig. 9. The number of identified peptides decreases with increasing equivalents of MeAnH addition, thus increasing protein modification (i.e. higher DS). This was also observed for RCPhC1 for which we hypothesized that this was due to steric hindrance of the enzyme and a decrease in ionization. Therefore, we can assume that the MA modifications influence the identification of all types of collagen-based biomaterials. For collagen with trypsin or GluC digestion, the number of missed cleavages varied between 1 and 6 (Figs. S11–S14). The data showed that the longer the peptides were, the larger the number of MAs on the peptide.
This is also visualized in Fig. S4 where a clear positive trend is visible between the number of missed cleavages and the number of MA's for the three peptides RCPhC1, COL1α1 and COL1α2. This was logical because increasing the number of missed cleavages increased the number of lysines present in the peptide.
Moreover, we have observed a slight increase in the percentage of peptides with 1 and 2 missed cleavages for COL1α1 in the modified collagens (i.e. COL-MA BS 0.5EQ and COL-MA BS 1EQ) compared to COL BS (Figs. S11 and S13), for both digestions. For COL1α2 with trypsin digestion (Fig. S12), a slight increase in the percentage of peptides with 1, 2, 3 missed cleavages was observed. But for the digestion with GluC (Fig. S14), a slight increase in the percentage of peptides with 0 missed cleavages has been noticed. We also saw a high percentage of peptides with 0 and 1 missed cleavages.
The results were not identical to those from the analysis of the RCPhC1 protein. COL1α1 (94673.0 Da) and COL1α2 (93415.3 Da) were composed of 38 and 31 lysines respectively, compared to the RCPhC1 protein (51,185 Da) which has 33 lysines. The RCPhC1 protein had more lysines compared to collagen COL1α1 or COL1α2 considering the size, which may allow a better action of the enzymes on COL1α1 and COL1α2.
In addition to MA modification, PTMs were observed such as the oxidation of methionine and the deamidation of asparagine and glutamine. These modifications are common in shotgun proteomics approaches on “complex” natural biopolymers [27]. As an example, hydroxyproline was also observed in the analysis of COL BS. These PTMs are common for collagen because they are involved in the structure of the protein, and thus inherently present [4].
Next, the sequence of the different α strands (i.e. COL1α1 and COL1α2) in COL BS was studied, together with the number of modified lysines. The COL1α1 sequence contains a total of 38 lysines, whereas COL1α2 contains a total of 31 lysines (based on Swiss-Prot databases). Table 6 shows the numbers of MA-modified lysines identified by LC-MS/MS with trypsin and Gluc digestion.
Table 6.
The numbers of MA-modified lysines identified by LC-MS/MS with trypsin and Gluc digestion. The MA sites are identified on COL1α1 and COL1α2 from Bos taurus. Based on the AA sequences from the Swiss-Prot database, the total number of lysines for COL1α1 and COL1α2 were 38 and 31, respectively.
| Trypsin digestion | GluC digestion | ||
|---|---|---|---|
| COL-MA BS 0.5 EQ | COL1α1 | 30 | 17 |
| COL1α2 | 23 | 16 | |
| COL-MA BS 1 EQ | COL1α1 | 26 | 19 |
| COL1α2 | 21 | 15 |
This enabled the identification of the MA sites on COL1α1 and COL1α2. The sequences of COL1α1 and COL1α2 were reconstructed from the digested peptides and the MA modifications were localized in these sequences. The sequences of COL1α1 and COL1α2 together with the modified lysines for the trypsin and GluC digestion can be found in Supplementary Info (Figs. S15 and S16, Figs. S17 and S18, respectively). The digestion with trypsin and GluC makes it possible to identify a total of 4 unmodified lysines and 34 modified lysines (with MA) in the COL1α1 sequence. For the COL1α2 sequence, digestion with trypsin and GluC resulted in an identification of a total of 6 unmodified lysines and 25 modified lysines. These modifications will be further discussed and shown in the next section 3.2.4, “Localization of modified amino acids in the 3D-structure of the modified biopolymer”.
To conclude, proteomic analysis showed that MA modifications were present over the entire sequence of COL BS, determined by using both types of digestion. Moreover, the proteomic analysis identified that the modifications were present on lysine groups, and that the MA modified lysines were distributed homogeneously over the sequence. This is key in the development of biomaterials with an intentional use for TERM applications, because the distribution of the modified, photo-crosslinkable MA groups affects the crosslinking behavior of the biomaterial. A homogeneously spreading of the modified groups will result in a better crosslinked network (due to the homogeneously spread crosslinks), which will influence the mechanical properties of the biomaterial. In turn, these mechanical properties influence the biological properties (i.e. cell-biomaterial interaction). This highlights the importance of the insights given by proteomic analysis. However, although homogeneous spreading of the lysines over the primary amino acid composition is positive for their distribution, it does not ensure that they will be homogeneously spread in their 3D conformation, hence the study in section 3.2.4.
3.2.3. Determination of the degree of substitution (DS) of modified COL BS
Similar to the analysis of RCPhC1, the COL BS and its derivatives (i.e. COL-MA BS) were analyzed using 1H NMR spectroscopy (Fig. S1, c-d). Based on 1H NMR data, the MA-modification of collagen led to a DS of 76.6 and 103.7% for the COL-MA BS 0.5 EQ and 1 EQ, respectively. The degree of substitution higher than 100% (for the higher DS) can be explained by the fact that the calculations are based on the amino acid composition as found in the Swiss-Prot and NCBi database (which do not give the exact amino acid composition as the COL BS used in this work) and the fact that the NMR data might overestimate the DS due to hydroxyl groups that could be modified as well and/or integration errors. Instead of using a database, the amino acid composition can be analyzed in order to know the exact AA composition [74]. Calculating the DS of the COL-MA derivatives using the results from the AA analysis, a DS of 75.9% and 99.6% were obtained for COL-MA 0.5 EQ and COL-MA 1 EQ, respectively.
Based on the OPA assay, the amount of primary amines available for functionalization was found to be 0.252 mmol g−1 in COL BS. For the COL-MA BS 0.5 and 1 EQ, a DS of 74.1% and 95.8% was obtained. An overview of the obtained DS values via the different methods is provided in Table 8.
Table 8.
Determination of the degree of substitution (DS) of COL-MA BS 0.5 and 1 EQ using 1H NMR spectroscopy (based on the amino acid sequence from the Swiss-Prot database, and the analyzed amino acid composition), OPA assay and shotgun proteomic analysis.
| DS obtained via 1H NMR (based on database Swiss-Prot) [%] | DS obtained via 1H NMR (based on known AA composition) [%] | DS obtained via OPA [%] | DS obtained via shotgun analysis [%] | |
|---|---|---|---|---|
| COL-MA BS 0.5 EQ | 76.6 | 75.9 | 74.1 | 74.9 |
| COL-MA BS 1 EQ | 103.7 | 99.6 | 95.8 | 80.4 |
Because it was not possible to perform MALDI-TOF analysis on the COL BS and its derivatives, due to their high MM (>150 kDa), it was also not possible to determine the DS using our currently available MALDI-TOF.
In addition to the conventional characterization techniques, proteomic analysis was performed on COL BS and its derivatives. The percentage of coverage for COL BS by trypsin digestion was found to be very high (i.e. 91 % and 92%), which is important for enabling superior identification and quantification of the modified groups in the AA sequence (Table 5, cfr. 3.2.2). Moreover, this method makes it possible to identify almost the entire sequence [75,76].
The numbers of quantified peptides after trypsin digestion of COL-MA BS was 195 for COL1α1 and 106 for COL1α2. LC-MS/MS analysis quantified 32 lysines with a MA on COL1α1 and 24 lysines with a MA on COL1α2. The numbers of quantified peptides after GluC digestion of COL-MA BS was 115 for COL1α1 and 68 for COL1α2. LC-MS/MS analysis quantified 20 lysines with a MA, on COL1α1 and 18 lysines with a MA on COL1α2 (Table 7).
Table 7.
The numbers of MA-modified lysines quantified by LC-MS/MS with trypsin and Gluc digestion. The MA sites are identified on COL1α1 and COL1α2.
| Trypsin digestion |
GluC digestion |
|||
|---|---|---|---|---|
| Quantified peptides | Quantified modified lysines | Quantified peptides | Quantified modified lysines | |
| COL1α1 | 195 | 32 | 115 | 20 |
| COL1α2 | 106 | 24 | 62 | 18 |
For the COL1α1, the first and last lysines were not modified with MA. However, for the COL1α2, the last lysine (Fig. S16) was modified. The position of this lysine within the collagen 3D structure could have an influence on the accessibility of the lysine and thus on the potential modification. Also, as shown in Fig. 9, the coverage of the identified and quantified peptides from both digestions showed non-covered parts of the protein at the end of the sequence, implying that the last lysine might not be detected using proteomics analysis (which can be due to the length of the last peptide in this sequence).
The LC-MS/MS analysis identified a few lysines without modification but the NMR and OPA showed a modification percentage close to 100% for COL-MA BS 1 EQ. Proteomic analysis indicated that the two chains α1 and α2 were modified and it was possible to locate the preferential positions of these modifications. The quantitative analysis of the modifications showed a higher MA modification frequency in the COL-MA BS 1 EQ than in the COL-MA BS 0.5 EQ samples. The results using the trypsin digestion indicated that COL-MA BS 1 EQ had an overall frequency of MA modification higher than COL-MA BS 0.5 EQ (Figs. S19 and S20), for both sequences of COL1α1 and COL1α2. The results were similar for GluC digestion (Figs. S21 and S22). Based on the modification frequency, an average DS could be calculated of 74.9 and 80.4% for COL-MA BS 0.5 and 1 EQ respectively. This trend (increase in DS when increasing eq MeAnH) correlates with the OPA and NMR data on the COL BS derivatives. An overview of the data is shown in Table 8.
To conclude, unlike 1H NMR and OPA, proteomic analysis does not allow to calculate the exact DS of a modified protein. Proteomic analysis gives information on the modification frequency (in %) of a specific amino acid, meaning that an average can be calculated to get an estimation on the DS. It is thus possible to identify which positions are easily modified (i.e. modification frequency of 100% in both COL-MA BS 0.5 EQ and 1 EQ) and thus to determine the preferential (lysine) sites for MA modification. It can be hypothesized that the location of the lysine groups in the 3D-structure of COL BS (i.e. inner or outer side of the 3D conformation, easy or difficult to access) is (partly) responsible for the ease of modification of specific positions in the sequence (section 3.2.4).
3.2.4. Localization of modified amino acids in the 3D-structure of COL BS modified biopolymers
Similarly to RCPhC1, the more complex sequences of COL1α1 and COL1α2 (P02453 and P02465 in Swiss-Prot DB respectively) were modelled using the I-TASSER server to obtain an idea of the 3D-conformation of the protein. The two COL BS sequences were a lot longer (i.e. 1463 and 1364 AA for the COL1α1 and COL1α2 respectively) than the RCPhC1 sequence (i.e. 571 AA), taking about 2–3 months per sequence to calculate a model (based on the protein data bank). Again, these 3D-models could not be confirmed by X-ray studies (not possible yet for such large proteins) and should be treated as possible folding configurations of the proteins studied. Moreover, the longer the protein sequence being modelled, the more possible degrees of freedom resulting in a decreasing confidence in the final protein model. However, even though the two sequences, COL1α1 and COL1α2, were separately submitted and ran on the I-TASSER, they resulted in remarkably similar models. Both models exhibited the same U-shape morphology and had similar amounts and positions of helices and sheets. Moreover, they resembled the model for RCPhC1, a derivative of collagen type I. These observations suggest that these models could be very close to the native protein configuration.
The best predicted model with I-TASSER for COL1α1 had a c-score of 0.22, which is excellent for such a large protein sequence. The estimated TM-score and RMSD were 0.74 (±0.11) and 9.3 (±4.6 Å) respectively, signifying a high confidence in the model. The best predicted model with I-TASSER for COL1α2 had a c-score of 0.63, which is extremely high for such a large protein sequence. The estimated TM-score and RMSD were 0.8 (±0.09 Å) and 8.2 (±4.4 Å) respectively, signifying the highest confidence in the model of all three modelled protein sequences in this paper.
For COL1α1 and COL1α2, the obtained 3D-models are shown in Fig. 10 in a and b respectively, showing the occurrence of the helices (red), sheets (yellow), lysines (magenta) in the main structure of the protein.
Fig. 10.
The protein 3D-conformation of the COL1α1 (a) and COL1α2 (b) proteins modelled with I-TASSER in the cartoon view showing helices (red), sheets (yellow), loops (green), and lysine residues in the structure (magenta). The same models shown in the solvent surface accessibility view for COL1α1for one side (c) and 180° turn around Y-axis site (e), and COL1α2 for one side (d) and 180° turn around Y-axis site (f), showing the protein accessibility surface (green), unmodified lysines on the surface (magenta), modified lysine (cyan) and the RGD sequences at the surface (orange). Close-ups of the protein COL1α2 showing (g) the grouped RGD sequences (orange, indicated with arrow) and proximity of the modified lysines (cyan) and (h) the buried unmodified lysines (magenta, indicated with arrow). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The predicted models for COL1α1 and COL1α2 show that they only have 2 and 4 RGD sequences, respectively, in contrast to RCPhC1 that has 12 RGD sites for a shorter overall sequence. Moreover, unlike RCPhC1, these RGD sites were not homogeneously distributed over both proteins COL1α1 and COL1α2 and even were aggregated into one location for COL1α2 (Fig. 10, c-f, orange). Moreover, these RGD sites were very closely positioned to modified lysines (cyan), meaning that cell accessibility may be limited once crosslinked into networks (Fig. 10, c-g, orange-cyan). This should be tested and confirmed in future work.
The spatial distribution of modified lysines in COL1α1 (Fig. 10, c and e) occurred to be better than in COL1α2 (Fig. 10, d and f), with the latter having large areas on the protein surface without any modified lysines present. As discussed above, the homogeneous distribution of the modified lysines is vital for the network formation of the proteins into useable biomaterials, important for TERM applications.
Although many lysines were modified (cyan), some were not modified (magenta) (Fig. 10, c-f). Whereas most modified lysines seem to be very accessible in the solvent accessibility view, some of the non-modified lysines occurred to be less accessible/more buried in the protein structure (Fig. 10, h (arrows)). This was certainly true for COL1α2, where some residues even barely reached the surface, possibly influencing their modification efficiency (Fig. 10, h) This also holds true for enzymes, making it more difficult to cut them due to steric hindrance caused by the natural protein shape and induced hindrance due to lysine modifications.
4. Conclusion
In this work, photo-crosslinkable moieties have been introduced onto RCPhC1 and COL BS, followed by applying different characterization techniques to determine the most important properties: the MM, the DS, the location of the modifications introduced in the AA sequence and the location of these modifications in the protein's 3D structure (exposed at the surface or less exposed and deeper within the protein's conformation). An overview of these properties in relation to the characterization techniques can be found in Table 9, and will also be used as a guideline in this conclusions.
Table 9.
Summary of the characterization techniques discussed in this work, and what information these techniques can reveal on modified biopolymers.
| Molar mass (MM) | Localization of modified group | Degree of substitution (DS) | Localization in 3D model possible | |
|---|---|---|---|---|
| MALDI-TOF | +* | – | +* | – |
| 1H NMR | – | – | + | – |
| OPA | – | – | + | – |
| PROTEOMICS | ± | + | ± | + |
(*) MALDI-TOF: only possible if MM < 150 kDa.
In general, the molar mass could not be determined by 1H NMR spectroscopy, nor by OPA. In case of low MM proteins (<150 kDa) such as RCPhC1, MALDI-TOF is very useful for the determination of the MM. However, in case of high MM proteins (>150 kDa), the MM could only be estimated using the sequences (found in the Swiss-Prot database) of the abundant proteins as determined by proteomic analysis.
Besides the molar mass, the identification and localization of the modified peptides is important information to optimize the biopolymer modification procedure and/or to control the resulting physico-chemical properties. Neither MALDI-TOF, 1H NMR spectroscopy, nor OPA enabled the identification and the localization of the introduced functionalities. Conversely, proteomic analysis enabled: (i) the identification of the peptides and modified AAs, and (ii) the localization of the introduced photo-crosslinkable groups. This is vital information when pursuing the development of a biomaterial for TERM applications because it provides insight in the distribution of these photo-crosslinkable groups throughout a biopolymer chain. It is hypothesized that this distribution directly affects the crosslinking behavior of a biopolymer and hence the mechanical properties, that in turn influence the biological properties (i.e. cell-biomaterial interactions and bioactivity).
Whereas the identification and localization of the modified peptides is important, the quantification of modifications present along the protein backbone is as essential. The degree of substitution/functionalization (DS) could be determined by MALDI-TOF (for MM < 150 kDa), 1H NMR spectroscopy and OPA. However, with proteomic analysis, some challenges were encountered when quantifying the photo-crosslinkable moieties: (i) the presence of redundant peptide sequences, and (ii) a higher DS (corresponding with more photo-crosslinkable moieties) resulting in more steric hindrance. Despite these challenges, proteomic analysis was able to provide insight in the modification frequency of specific modification sites, enabling the calculation of an average DS. Obtaining modification frequencies is exciting as they also indicated that some positions were more accessible towards modification compared to others, resulting in additional information which was absent upon applying the other techniques.
Based on the proteomic analysis and the obtained information on the localization of the modified groups, it was possible to 3D model the biopolymers. Moreover, it permitted the identification, localization and distribution of each unmodified and modified AA in its 3D structural conformation, providing crucial insight in the overall distribution of the modified sites along the protein backbone.
In conclusion, proteomic analysis cannot (yet) be used as a stand-alone technique to fully characterize a modified (photo-crosslinkable) biopolymer because it only provides an average DS based on the modification frequencies. However, it is the only technique that enables the identification and localization of the functionalized AA along with supplying the required information for establishing the 3D model. This enabled to gain unprecedented insight in the distribution of the introduced functionalities along the protein backbone which is crucial with respect to reproducibility and regulatory aspects for its use as a biomaterial for TERM applications, and further unravelling of the efficiency of the biopolymer modification process and the effect on the crosslinked network.
This paper is a first step towards understanding protein modification in relation to biomaterial properties. It can be anticipated that in the near future with the continuously emerging technologies, it will also become possible to gain in-depth information regarding non-covalent modification sites on proteins and their influence on the final biomaterial properties (e.g. protein-protein and protein-ligand interactions).
Funding
The authors would like to thank Fujifilm Manufacturing (Europe B·V.) and the Department of Collagen Research (M. Albu, National Research & Development Institute for Textiles and Leather, Romania) for kindly provinding RCPhC1 and bovine skin collagen, respectively. The authors acknowledge Prof. J. Martins and Tim Courtin of the NMR department for helping with the NMR measurements, and Dr. Francesco Copes for his valuable input and feedback.
P. Dubruel and S. Van Vlierberghe would like to acknowledge the financial support of the Research Foundation Flanders (FWO) under the form of research grants and the financial support of the Special Research Fund (BOF, Ghent University). The authors would like to acknowledge funding from the Interreg 2Seas 3DMed. The work of N. Pien was supported by a Vanier Canada Graduate Scholarship.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Nele Pien: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing, Visualization, Project administration, Funding acquisition. Fabrice Bray: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization, Project administration, Funding acquisition. Tom Gheysens: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization, Project administration, Funding acquisition. Liesbeth Tytgat: Methodology, Resources. Christian Rolando: Supervision, Writing – review & editing, Visualization, Project administration, Funding acquisition. Diego Mantovani: Supervision, Writing – review & editing, Visualization, Project administration, Funding acquisition. Peter Dubruel: Supervision, Writing – review & editing, Visualization, Project administration, Funding acquisition. Sandra Van Vlierberghe: Supervision, Writing – review & editing, Visualization, Project administration, Funding acquisition.
Acknowledgements
The authors acknowledge the IBiSA network for financial support of the USR 3290 (MSAP) proteomics facility TOP_OMICS. The mass spectrometers were funded by the University of Lille, the CNRS, the Région Hauts-de-France and the European Regional Development Fund (ERDF).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.01.023.
Contributor Information
Peter Dubruel, Email: peter.dubruel@ugent.be.
Sandra Van Vlierberghe, Email: sandra.vanvlierberghe@ugent.be.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Liu X., Zheng C., Luo X., Wang X., Jiang H. Mater. Sci. Eng. C. 2019;99:1509. doi: 10.1016/j.msec.2019.02.070. [DOI] [PubMed] [Google Scholar]
- 2.Wang T., Lew J., Premkumar J., Poh C.L., Win Naing M. Eng. Biol. 2017;1:18. [Google Scholar]
- 3.Miranda-Nieves D., Chaikof E.L. ACS biomater. Sci. Eng. 2017;3:694. doi: 10.1021/acsbiomaterials.6b00250. [DOI] [PubMed] [Google Scholar]
- 4.Shoulders M.D., Raines R.T. Annu. Rev. Biochem. 2009;78:929. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinderer S., Layland S.L., Schenke-Layland K. Adv. Drug Deliv. Rev. 2016;97:260. doi: 10.1016/j.addr.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Meyer M. Biomed. Eng. Online. 2019;18:1. doi: 10.1186/s12938-019-0647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Copes F., Pien N., Van Vlierberghe S., Boccafoschi F., Mantovani D. Front. Bioeng. Biotechnol. 2019;7:1. doi: 10.3389/fbioe.2019.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen F.M., Liu X. Prog. Polym. Sci. 2016;53:86. doi: 10.1016/j.progpolymsci.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hersel U., Dahmen C., Kessler H. Biomaterials. 2003;24:4385. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 10.Chen H., Kassab G.S. J. Biomech. 2016;49:2548. doi: 10.1016/j.jbiomech.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.L'Heureux N., Dusserre N., Konig G., Victor B., Keire P., Wight T.N., Chronos N.A.F., Kyles A.E., Gregory C.R., Hoyt G., et al. Nat. Med. 2006;12:361. doi: 10.1038/nm1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar S., Salacinski H.J., Hamilton G., Seifalian A.M. Eur. J. Vasc. Endovasc. Surg. 2006;31:627. doi: 10.1016/j.ejvs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Montini-Ballarin F., Calvo D., Caracciolo P.C., Rojo F., Frontini P.M., Abraham G.A., Guinea G.V. J. Mech. Behav. Biomed. Mater. 2016;60:220. doi: 10.1016/j.jmbbm.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Karimi A., Navidbakhsh M., Shojaei A., Faghihi S. Mater. Sci. Eng. C. 2013;33:2550. doi: 10.1016/j.msec.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 15.Goins A., Webb A.R., Allen J.B. Mater. Sci. Eng. C. 2019;97:896. doi: 10.1016/j.msec.2018.12.067. [DOI] [PubMed] [Google Scholar]
- 16.Tronci G., Russell S.J., Wood D.J. J. Mater. Chem. B. 2013;1:3705. doi: 10.1039/C3TB20720J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davidenko N., Schuster C.F., Bax D.V., Raynal N., Farndale R.W., Best S.M., Cameron R.E. Acta Biomater. 2015;25:131. doi: 10.1016/j.actbio.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallop P.M., aPaz M. Physiol. Rev. 1975;55:418. doi: 10.1152/physrev.1975.55.3.418. [DOI] [PubMed] [Google Scholar]
- 19.Rýglová Š., Braun M., Suchý T. Macromol. Mater. Eng. 2017;302:1. [Google Scholar]
- 20.Van Hoorick J., Dobos A., Markovic M., Gheysens T., Van Damme L., Gruber P., Tytgat L., Van Erps J., Thienpont H., Dubruel P., et al. Biofabrication. 2021;13(015017) doi: 10.1088/1758-5090/abc95f. [DOI] [PubMed] [Google Scholar]
- 21.Arslan A., Steiger W., Roose P., Van den Bergen H., Gruber P., Zerobin E., Gantner F., Guillaume O., Ovsianikov A., Van Vlierberghe S., et al. Mater. Today. 2021;44:25. [Google Scholar]
- 22.Tytgat L., Van Damme L., Van Hoorick J., Declercq H., Thienpont H., Ottevaere H., Blondeel P., Dubruel P., Van Vlierberghe S. Acta Biomater. 2019;94:340. doi: 10.1016/j.actbio.2019.05.062. [DOI] [PubMed] [Google Scholar]
- 23.Ravichandran R., Islam M.M., Alarcon E.I., Samanta A., Wang S., Lundström P., Hilborn J., Griffith M., Phopase J. J. Mater. Chem. B. 2016;4:318. doi: 10.1039/c5tb02035b. [DOI] [PubMed] [Google Scholar]
- 24.Twyman R.M. BIOS Scientific Publishers; 2004. Priciples of Proteomics. [Google Scholar]
- 25.Liebler D.C. Humana Press Inc; 2002. Introduction to Proteomics, Tools for the New Biology. [Google Scholar]
- 26.Aebersold R., Mann M. Nature. 2003;422 doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y., Fonslow B.R., Shan B., Baek M.C., Yates J.R. Chem. Rev. 2013;113:2343. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen M.R., Trelle M.B., Thingholm T.E., Jensen O.N. Biotechniques. 2006;40:790. doi: 10.2144/000112201. [DOI] [PubMed] [Google Scholar]
- 29.Gahoual R., Bolbach G., Ould-Melha I., Clodic G., François Y.N., Scherman D., Mignet N., Houzé P. J. Pharm. Biomed. Anal. 2020;185 doi: 10.1016/j.jpba.2020.113242. [DOI] [PubMed] [Google Scholar]
- 30.Albu M.G. Lambert Academic Publishing; 2011. Collagen Gels and Matrices for Biomedical Applications. ISBN: 9783844330571. [Google Scholar]
- 31.Tytgat L., Markovic M., Qazi T.H., Vagenende M., Bray F., Martins J.C., Rolando C., Thienpont H., Ottevaere H., Ovsianikov A., et al. J. Mater. Chem. B. 2019;7:3100. doi: 10.1039/c8tb03308k. [DOI] [PubMed] [Google Scholar]
- 32.Van Vlierberghe S., Fritzinger B., Martins J.C., Dubruel P. Appl. Spectrosc. 2010;64:1176. doi: 10.1366/000370210792973550. [DOI] [PubMed] [Google Scholar]
- 33.Helle S., Bray F., Verbeke J., Devassine S., Courseaux A., Facon M., Tokarski C., Rolando C., Szydlowski N. Front. Plant Sci. 2018;9:1. doi: 10.3389/fpls.2018.00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ainseba-Chirani N., Dembahri Z., Tokarski C., Rolando C., Benmouna M. Polym. Int. 2011;60:1024. [Google Scholar]
- 35.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. Nat. Publ. Gr. 2015;12:7. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy A., Kucukural A., Zhang Y. Nat. Protoc. 2010;5:725. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J., Zhang Y. Curr. Protoc. Bioinforma. 2015;52(5) doi: 10.1002/0471250953.bi0508s52. 8.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y., Skolnick J. Nucleic Acids Res. 2005;33:2302. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fushimi H., Hiratsuka T., Okamura A., Ono Y., Ogura I., Nishimura I. Commun. Mater. 2020;1:1. [Google Scholar]
- 40.Izunobi J.U., Higginbotham C.L. J. Chem. Educ. 2011;88:1098. [Google Scholar]
- 41.Arrabal-Campos F.M., Aguilera-Sáez L.M., Fernández I. Anal. Methods. 2019;11:142. [Google Scholar]
- 42.Signor L., Erba E.B. JoVE. 2013:1. [Google Scholar]
- 43.Martinsen A., Skjåk-Bræk G., Smidsrød O., Zanetti F., Paoletti S. Carbohydr. Polym. 1991;15 doi: 10.1016/0144-8617(91)90031-7. [DOI] [Google Scholar]
- 44.Mohammad R.K. J. Polym. Biopolym. Phys. Chem. 2018:6. [Google Scholar]
- 45.Kasaai M.R., Arul J., Charlet G. J. Polym. Sci., Part B: Polym. Phys. 2000:38. 10.1002/1099-0488(20001001)38:19<2591::AID-POLB110>3.0.CO;2–6 [Google Scholar]
- 46.Garth W., Hastings C. CRC Press; 2018. [Google Scholar]
- 47.Nobbmann U., Connah M., Fish B., Varley P., Gee C., Mulot S., Chen J., Zhou L., Lu Y., Sheng F., et al. Biotechnol. Genet. Eng. Rev. 2007;24 doi: 10.1080/02648725.2007.10648095. [DOI] [PubMed] [Google Scholar]
- 48.Burchard W. Static and dynamic light scattering from branched polymers and biopolymers. Light Scattering from Polymers. Advances in Polymer Science. 1983;48 doi: 10.1007/3-540-12030-0_1. ISBN 978-3-540-12030-8. [DOI] [Google Scholar]
- 49.Langer R.S., Peppas N.A. Biomaterials. 1981;2 doi: 10.1016/0142-9612(81)90059-4. [DOI] [PubMed] [Google Scholar]
- 50.Metters A.T., Lin C.-C. Biodegradable Hydrogels: Tailoring Properties and Function through Chemistry and Structure in “Biomaterials”. CRC Press. 2007 doi: 10.1201/9780849378898. ISBN: 9780429116520. [DOI] [Google Scholar]
- 51.Stetefeld J., McKenna S.A., Patel T.R. Biophys. Rev. 2016;8 doi: 10.1007/s12551-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wineinger H.B., Shamshina J.L., Kelly A., King C., Rogers R.D. Green Chem. 2020;22(12) doi: 10.1039/d0gc00753f. [DOI] [Google Scholar]
- 53.Kelleher N.L. Anal. Chem. 2004;76 doi: 10.1021/ac0415657. [DOI] [PubMed] [Google Scholar]
- 54.Cai W., Tucholski T., Chen B., Alpert A.J., McIlwain S., Kohmoto T., Jin S., Ge Y. Anal. Chem. 2017;89 doi: 10.1021/acs.analchem.7b00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giansanti P., Tsiatsiani L., Low T.Y., Heck A.J.R. Nat. Protoc. 2016;11:993. doi: 10.1038/nprot.2016.057. [DOI] [PubMed] [Google Scholar]
- 56.Olsen J.V., Mann M. Mol. Cell. Proteomics. 2013;12:3444. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bharti S.K., Roy R. TrAC - trends anal. Inside Chem. 2012;35:5. [Google Scholar]
- 58.Benson J.R., Hare P.E. Proc. Natl. Acad. Sci. U.S.A. 1975;72:619. doi: 10.1073/pnas.72.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim M.-S., Zhong J., Pandey A. Proteomics. 2016;16:700. doi: 10.1002/pmic.201500355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krusemark C.J., Frey B.L., Belshaw P.J., Smith L.M. J. Am. Soc. Mass Spectrom. 2009;20:1617. doi: 10.1016/j.jasms.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qi B.-L., Liu P., Wang Q.-Y., Cai W.-J., Yuan B.-F., Feng Y.-Q. TrAC trends anal. Inside Chem. 2014;59:121. [Google Scholar]
- 62.Bellis S.L. Biomaterials. 2011;32:4205. doi: 10.1016/j.biomaterials.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kubala M., Geleticova J., Huliciak M., Zatloukalova M., Vacek J., Sebela M. Biomed. Pap. 2014;158:194. doi: 10.5507/bp.2014.018. [DOI] [PubMed] [Google Scholar]
- 64.Tolmachev A.V., Robinson E.W., Wu S., Paša-Tolić L., Smith R.D. Int. J. Mass Spectrom. 2009;287:32. doi: 10.1016/j.ijms.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valeja S.G., Kaiser N.K., Xian F., Hendrickson C.L., Rouse J.C., Marshall A.G. Anal. Chem. 2011;83:8391. doi: 10.1021/ac202429c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bogdanov B., Smith R.D. Mass spectrom. Rev. 2005;24:168. doi: 10.1002/mas.20015. [DOI] [PubMed] [Google Scholar]
- 67.Catherman A.D., Skinner O.S., Kelleher N.L. Biochem. Biophys. Res. Commun. 2014;445:683. doi: 10.1016/j.bbrc.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Techatanawat S., Surarit R., Suddhasthira T., Khovidhunkit S.O.P. Asian Biomed. 2011;5:787. [Google Scholar]
- 69.Abraham L.C., Zuena E., Perez-Ramirez B., Kaplan D.L. J. Biomed. Mater. Res. B Appl. Biomater. 2008;87:264. doi: 10.1002/jbm.b.31078. [DOI] [PubMed] [Google Scholar]
- 70.Eyre D.R., Koob T.J., Van Ness K.P. Anal. Biochem. 1984;137:380. doi: 10.1016/0003-2697(84)90101-5. [DOI] [PubMed] [Google Scholar]
- 71.Van Der Slot-Verhoeven A.J., Van Dura E.A., Attema J., Blauw B., DeGroot J., Huizinga T.W.J., Zuurmond A.M., Bank R.A. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2005;1740:60. doi: 10.1016/j.bbadis.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 72.Li Q., Uygun B.E., Geerts S., Ozer S., Scalf M., Gilpin S.E., Ott H.C., Yarmush M.L., Smith L.M., Welham N.V., et al. Biomaterials. 2016;75:37. doi: 10.1016/j.biomaterials.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dau T., Bartolomucci G., Rappsilber J. Anal. Chem. 2020;92:9523. doi: 10.1021/acs.analchem.0c00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pien N., Pezzoli D., Van Hoorick J., Copes F., Vansteenland M., Albu M., De Meulenaer B., Mantovani D., Van Vlierberghe S., Dubruel P. Mater. Sci. Eng. C. 2021;130:112460. doi: 10.1016/j.msec.2021.112460. [DOI] [PubMed] [Google Scholar]
- 75.Buckley M. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang C.T., Ghosh D., Beaudry F. Food addit. Contam. - Part A Chem. Anal. Control. Expo. Risk Assess. 2018;35:599. doi: 10.1080/19440049.2017.1416680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.