Summary
Background
Although sepsis accounts for 1 in 5 deaths globally, few molecular therapies exist for this condition. The development of effective biomarkers and treatments for sepsis requires a more complete understanding of host responses and pathogenic mechanisms at early stages of disease to minimize host-driven pathology.
Methods
An alternative to the current symptom-based approach used to diagnose sepsis is a precise assessment of blood proteomic changes during the onset and progression of Salmonella Typhimurium (ST) murine sepsis.
Findings
A distinct pattern of coagulation factor protein abundance was identified in the pre-septic state– prior to overt disease symptoms or bacteremia– that was predictive of the dysregulation of fibrinolytic and anti-coagulant activities and resultant consumptive coagulopathy during ST murine sepsis. Moreover, the changes in protein abundance observed generally have the same directionality (increased or decreased abundance) reported for human sepsis. Significant overlap of ST coagulopathic activities was observed in Gram-negative Escherichia coli– but not in Gram-positive staphylococcal or pneumococcal murine sepsis models. Treatment with matrix metalloprotease inhibitors prevented aberrant inflammatory and coagulopathic activities post-ST infection and increased survival. Antibiotic treatment regimens initiated after specific changes arise in the plasma proteome post-ST infection were predictive of an increase in disease relapse and death after cessation of antibiotic treatment.
Interpretation
Altered blood proteomics provides a platform to develop rapid and easy-to-perform tests to predict sepsis for early intervention via biomarker incorporation into existing blood tests prompted by patient presentation with general malaise, and to stratify Gram-negative and Gram-positive infections for appropriate treatment. Antibiotics are less effective in microbial clearance when initiated after the onset of altered blood proteomics as evidenced by increased disease relapse and death after termination of antibiotic therapy. Treatment failure is potentially due to altered bacterial / host-responses and associated increased host-driven pathology, providing insight into why delays in antibiotic administration in human sepsis are associated with increased risk for death. Delayed treatment may thus require prolonged therapy for microbial clearance despite the prevailing notion of antibiotic de-escalation and shortened courses of antibiotics to improve drug stewardship.
Funding
National Institutes of Health, U.S. Army.
Keywords: Sepsis, Sepsis diagnosis, Sepsis treatment, Factor XI, Coagulopathy, Matrix metalloprotease inhibitor
Research in context.
Evidence before this study
Patients with sepsis continue to experience significant morbidity and mortality despite coordinated efforts by the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) to align current understanding of research and patient management. In the clinical setting, sepsis is diagnosed by a symptom-based approach that may include decreased oxygen saturation, altered mental status, decreased systolic blood pressure, acidosis, abnormal kidney or liver function, decreased platelet counts and thrombosis or bleeding. A suspected infection may be confirmed by positive body fluid culture (blood, urine, cerebrospinal fluid etc.), but this may only occur at late stages of disease or not at all in a substantial fraction of cases. Biomarkers of inflammation and coagulopathy remain exceedingly rare and those identified have somewhat limited clinical utility (e.g., C-reactive protein, procalcitonin etc.). The development of effective biomarkers and treatments for sepsis thus requires a more complete understanding of host responses and pathogenic mechanisms at early stages of disease to minimize host-driven injury.
Added value of this study
An alternative to the current symptom-based approach for sepsis diagnosis is a precise assessment of blood proteomic changes that occur at early versus late stages of disease. We found that blood proteomic changes occurred in a closely-timed catastrophic fashion prior to overt disease symptoms or bacteremia post-Salmonella infection. The marked similarities of coagulopathic activities in Gram-negative– but not in Gram-positive– sepsis suggests that sepsis can be potentially stratified by different disease mechanisms elicited by discrete pathogens, dispelling the prevailing notion that sepsis manifests as a stereotypical process spanning inflammation and coagulopathy. Antibiotic treatment regimens initiated after specific changes arise in the plasma proteome were predictive of increased disease relapse and death after completion of antibiotic therapy.
Implications of all the available evidence
Altered blood proteomics provides a platform to develop rapid and easy-to-perform tests to forecast sepsis for early intervention via biomarker incorporation into existing blood tests prompted by patient presentation with general malaise versus overt disease symptoms, and to stratify pathogens for appropriate treatment. Clinical benefit is manifested by the acquisition of risk profile data well-before overt disease symptoms necessary for Sequential Organ Failure Assessment (SOFA) and quick SOFA (qSOFA) scores used to evaluate intensive care unit (ICU) and non-ICU patients, respectively. Time to antibiotic treatment is the principal tenet for improved patient outcome during emergency care for sepsis and is consistent with the “Surviving Sepsis Campaign” recommendations for early and aggressive antibiotics for all patients with possible sepsis or septic shock. This study demonstrates that antibiotics are less effective in microbial clearance after the onset of altered blood proteomics as evidenced by increased disease relapse and death after termination of antibiotic therapy. Treatment failure may be due to altered bacterial / host responses and associated host-driven injury, providing insight into why delays in antibiotic administration in human sepsis are associated with increased risk for death. Delayed treatment may thus require extended therapy for microbial clearance despite established physician guidance of antibiotic de-escalation and shortened courses of antibiotics to improve drug stewardship.
Alt-text: Unlabelled box
Introduction
Sepsis is a life-threatening disease due to a dysregulated host response to infection, triggering severe inflammation, tissue coagulopathy, and organ dysfunction.1 Sepsis accounts for 1 in 5 deaths globally, is the most common cause of deaths in U.S. hospitals, and is more prevalent worldwide than cancer, with an estimated global annual burden of nearly 50 million cases and 11 million deaths.2 Even with adherence to rapid diagnosis and treatment protocols, mortality averages 25% following severe sepsis or septic shock, with many survivors experiencing lifelong physiological debilitation with cognitive decline reflecting vascular and organ damage.3, 4, 5 The costs of sepsis treatment are escalating and estimated to exceed $24 billion annually in the U.S.6 Despite this urgent medical need, few molecular therapies exist for this condition.7,8
Dysregulation of inflammation and coagulation during sepsis are the primary sources of host-driven pathology resulting in disability and death.9 The excessive presence of inflammatory cytokines in circulation and tissues during sepsis and septic shock– termed the “cytokine storm”– triggers severe systemic inflammation and resultant pathologic injury to vascular and organ systems.10,11 Although multiple cytokines that drive inflammatory tissue and vascular injury have been implicated, their therapeutic modulation has yet to lead to useful therapies and, in some cases, have increased patient mortality.12, 13, 14 Dysregulation of blood coagulation with pathologic thrombosis and hemorrhage– termed disseminated intravascular coagulation (DIC)– is the result of inflammatory cytokine-initiated coagulation coupled with dysregulation of anti-coagulant and fibrinolytic activities that lead to endothelial cell dysfunction and microvascular thrombosis.15,16 Once the DIC syndrome is triggered, these unrestrained activities can cause stroke, myocardial infarction, embolism, hemorrhage, and organ failure.
In the clinical setting, sepsis diagnosis is defined by the appearance of multiple physiologic and biochemical abnormalities that may include decreased oxygen saturation, altered mental status, decreased systolic blood pressure, abnormal serum creatinine, lactate, C-reactive protein (CRP) and bilirubin, decreased platelet counts and coagulopathic changes such as increased D-dimer, thrombosis, or bleeding.17 Biomarkers of inflammation and coagulopathy remain exceedingly rare and those identified have somewhat limited clinical utility (e.g., CRP, procalcitonin, IL-6, presepsin, CD64, lipopolysaccharide binding protein (LBP)).7,8,18 Currently, automated blood culture systems are the gold-standard to detect bacteremia, which are coupled to diagnostic techniques for pathogen identification.19 Examples include multiplex PCR (BioFire, bioMerieux), real time multiplex PCR (Xpert, Cepheid), DNA microarray (Verigene, Luminex), and fluorescent in situ hybridization (PNA Fish, AdvanDx). However, these techniques require positive blood cultures, detect only targeted pathogens, and are influenced by prior antibiotic exposure. Thus, the development of effective biomarkers and treatments for sepsis requires a more complete understanding of host responses and pathogenic mechanisms at early stages of disease to minimize host-driven injury.20
Recent approaches are beginning to reveal important insights into the role of host responses and their impact on sepsis. The Plasma Proteome Signature of Sepsis (PPSS) is a functionally-connected network initially identified as being common to five Gram-negative and Gram-positive murine models of late-stage sepsis, with similar changes in the expression of these proteins observed in human sepsis.21 Additionally, serum analysis of sepsis patients revealed pathogenic-proteomic-signatures for prediction of Staphylococcus aureus patient mortality risk profiling.22 However, an early detection system that does not depend on blood culture or pathogen identification, and can stratify Gram-negative vs. Gram-positive sepsis, would be immensely important to patient care due to their potential vast differences in antibiotic susceptibilities. Herein, we applied temporal investigations of blood proteome remodeling in ST sepsis to provide a potential means for early sepsis diagnosis before overt symptoms or bacteremia, which is not available to late-stage sepsis studies alone, wherein host responses and pathogenic mechanisms may consolidate into a stereotypical process of aberrant inflammatory and coagulopathic activities.20
Methods
Bacterial strains and culture conditions
Salmonella enterica serovar Typhimurium (ST) reference strain ATCC 14028 (CDC 6516-60), Escherichia coli (EC) strain ATCC 25922 (clinical isolate, FDA strain Seattle 1946), methicillin-sensitive Staphylococcus aureus (MSSA) strain Newman, methicillin-resistant Staphylococcus aureus (MRSA) strain CA-MRSA USA300, and Streptococcus pneumoniae (SPN) serotype 2 strain D39 were grown as previously described.20
Bacterial infections
ST was administered via gastric intubation at a dose of 1 × 107 colony forming units (cfu) (20X LD50); EC and SPN were administered i.p.; EC at a dose of 1 × 107 cfu (10X LD50) and SPN at a dose of 1 × 104 cfu (10X LD50); MRSA and MSSA were administered i.v.; MRSA at a dose of 1 × 108 cfu (20X LD50) and MSSA at a dose of 5 × 108 cfu (20X LD50).20 Animal experiments were carried out with 8–12-week-old C57BL/6J mice and used equal numbers of males and females. All mice in the study were provided sterile pellet food and water ad libitum.
Sepsis time points
ST: t = 0 (pre-infection), t = 5 days (early sepsis), and t = 8 days (late sepsis); EC: t = 0 (pre-infection), t = 24 h (early sepsis), and t = 48 h (late sepsis); MSSA: t = 0 (pre-infection), t = 24 h (early sepsis), and t = 96 h (late sepsis); MRSA: t = 0 (pre-infection), t = 24 h (early sepsis), and t = 48 h (late sepsis); SPN: t = 0 (pre-infection), t = 24 h (early sepsis), and t = 48 h (late sepsis).20 For all five bacterial strains, disease severity and mortality are directly proportional to increased bacterial cfu in the bloodstream and not the bacterial dose or route of administration.20
Proteomics plasma sample preparation
Proteomics samples were prepared as described previously,21 with the exception of the depletion step, which was optimized as described below. Albumin and immunoglobulin depletion were performed by precipitating 10 μL of plasma with pre-chilled acetone (20 °C), air-dried and incubated for an hour at 4 °C with a mix of antibodies specific to albumin (SigmaAldrich LSKMAGL10) and IgA/G (ThermoFisherScientific 88802), crosslinked to magnetic 92 beads. The eluate was collected and incubated once more with magnetic beads. Protein eluates were cysteine-reduced and alkylated with DTT and iodoacetamide, respectively, followed by overnight digestion with mass spectrometry grade trypsin protease (PIERCETM 90057). Equal amounts of protein digests from six time points (days 0, 2, 3, 4, 5, 8 post-infection) were labeled with specific isotopically-labeled tandem mass tags (TMT). Each time point included plasma from two biological replicates (two mice).
Proteomics data collection, analysis and interpretation
TMT-labeled sample from each biological replicate was injected three times in a nano-Liquid Chromatography system (nLC) connected inline to a LUMOS-Trybrid Orbitrap tandem mass spectrometer (MS/MS). To increase the number of peptides targeted for sequencing by MS/MS fragmentation, a two-dimensional (2D) nLC prefractionation setup was used. The first dimension was a low resolution C18 reverse-phase separation at high pH that produced 12 fractions. Each fraction was in turn separated at low pH in a high resolution C8 reverse phase and ionized for MS/MS fragmentation and MS determination. The separation and data collection methods were as described previously.21 Data protein identification assignments and TMT ratio calculations were performed using the latest version of MaxQuant (1.6.8).23 Statistical calculations and plots were done in RStudio (1.1.463). To identify and quantify bacterial and host protein abundance simultaneously, we combined the most updated proteome databases from UniProtKB for Salmonella enterica serovar Typhimurium 14028 and the mouse strain C57BL/6J. Proteomics data is provided in Supplementary Table 1: a, network annotation; b, protein expression ratios; c, temporal analysis; d, STRING network coordinates; e, network calculations.
ELISA assays
Cytokine enzyme-linked immunosorbent assay (ELISA) assays were performed and validated according to manufacturer's instructions, including IL-1β (BMS6002; Thermo Fisher Scientific), IL-6 (KMC0061; Thermo Fisher Scientific), IFNγ (KMC4021; Thermo Fisher Scientific) and TNFα (BMS607-3; Thermo Fisher Scientific).
Coagulation factor and platelet assays
Coagulation factor assays. All clot-based coagulation assays were performed on platelet-poor citrated plasma in duplicate using an ST-4 semi-automated coagulometer (DiagnosticaStago, NJ). Chromogenic substrate-based assays were performed with SpectraMax iD3 Multi-Mode Microplate Reader (Molecular Devices, CA). The methodology of all coagulation studies were performed according to established protocols.24,25Platelet Assays. Approximately 20 µL of whole blood collected from mice by cardiac puncture was added to microtainer tubes with EDTA (BD Biosciences) and analyzed within 2 h of collection using a Hemavet 950 FS instrument (Drew Scientific) programmed with mouse hematology settings.25
Alkaline phosphatase assay
Mice were infected via gastric intubation with ST (5 × 106 cfu) and blood was collected into microtainer serum separator tubes (BD Biosciences) with no anticoagulant and allowed to clot for 30 m at room temperature. Serum was collected after centrifugation at 13,000 rpm for 10 m. Serum chemistry analyses, including measurements of alkaline phosphatase (AP) activity, were acquired with a VetScan Comprehensive Diagnostic Profile reagent rotor using the VetScan Chemistry Analyzer as previously reported.20
Neuraminidase assay
Mice were infected via gastric intubation with ST (5 × 106 cfu) and blood was collected into K2 EDTA microtainer tubes (BD Biosciences) and mixed on a rotator for 10 m at room temperature. Plasma was collected after centrifugation at 13,000 rpm for 10 m. Neuraminidase (Neu) activity was detected via Amplex Red (Thermo Fisher Scientific) using 25 µL plasma as per manufacturer recommendations. Control Neu activity was determined using a neuraminidase standard (Arthrobacter ureafaciens; EY Laboratories) serially diluted 2-fold in PBS starting at 500 U/L.
Matrix metalloproteinase inhibitor treatment
Mice were infected via gastric intubation with ST (5 × 106 cfu). At 2 h post-infection, mice were treated with MMP inhibitor, GM6001 (inhibits MMP 1/2/3/8/9; CC1100, Millipore Sigma Aldrich),26 administered i.p. at a dose of 200 µg per mouse per day for 8 days; vehicle control mice were administered GM6001 diluent on the same treatment regimen.
Antibiotic treatment
Mice infected via gastric intubation with ST (107 cfu) were treated with the following dosing regimens: ceftriaxone (50 mg/kg/day) or ciprofloxacin (30 mg/kg/day) were administered every 12 h via the intraperitoneal route of administration on the days indicated.
Ethics
All animal experimentation was conducted following the National Institutes of Health guidelines for housing and care of laboratory animals and performed in accordance with Institutional regulations after pertinent review and approval by the Institutional Animal Care and Use Committees at the University of California, Santa Barbara (#445) and Sanford Burnham Prebys Medical Discovery Institute (#A3053) (La Jolla, CA).
Statistics
Student's unpaired t-test was used to compare the means of two groups for cytokine and platelet analyses; GraphPad Prism software version 8.0 was used to determine statistical significance determined by unpaired 2-tailed t-test. Log-rank (Mantel-Cox) test was used to compare differences in survival between groups for Kaplan-Meier survival curves; significance was determined using GraphPad Prism version 9.0. P values of less than 0.05 were considered significant. The exact value of n, representing the number of mice, was indicated in the figure legends. Proteomic responses to infection were screened according to the ratio of the amount of each protein following infection relative to pre-infection (Supplementary Table 1b) as described previously.21 A two-sample t-test was calculated, based on three technical replicates (injections) per sample in control and sepsis samples. Inflammatory and coagulopathic proteins with changes in the ratio of sepsis/control (SE/CTL) of >0.6 (log2)/1.5-fold, (natural number), with P values < 0.1 selected to be included in the subsequent assessment of activity experiment. The statistical analysis of Tables 1 and 2 as well as the protein expression ratios in Supplementary Table 1b was conducted utilizing RStudio.27 As the data was not normally distributed, the distribution of the data was analyzed using Permutational Multivariate Analysis of Variance utilizing the vegan package.28 Table 1: Pathogen and time were included as independent variables and the results of each analyte as dependent variables. Time, pathogen and the interaction between time and pathogen were significant (P < 0.05). Pairwise comparisons were performed between uninfected mice (n = 60) and early (n = 12) and late (n = 12) septic mice for each analyte and pathogen. Benjamini-Hochberg was utilized to adjust P values for multiple comparisons. Table 2: Time and treatment were included as independent variables and the results of each analyte as dependent variables. Time, treatment and the interaction between time and treatment were significant (P < 0.05). Pairwise comparisons were performed between the untreated and the corresponding treated group for each analyte (early sepsis: untreated vs. treated; late sepsis: untreated vs. treated). Benjamini-Hochberg was utilized to adjust P values for multiple comparisons. n = 15 mice per group.
Table 1.
Temporal analysis of coagulation factor activities in mice infected with Gram-negative and Gram-positive pathogens.
| Not infected |
S. Typhimurium |
E. coli |
MSSA |
MRSA |
S. pneumoniae |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Analyte (% NMP) | Early sepsis (Day 5) | Late sepsis (Day 8) | Early sepsis (24 h) | Late sepsis (48 h) | Early sepsis (24 h) | Late sepsis (96 h) | Early sepsis (24 h) | Late sepsis (48 h) | Early sepsis (24 h) | Late sepsis (48 h) | |
| Antithrombin | 142 | 134 | 108* | 108* | 91* | 104* | 123 | 120 | 109* | 143 | 79* |
| Protein C | 106 | 90 | 87 | 107 | 100 | 85 | 172* | 135* | 131 | 82* | 76* |
| Protein S | 97 | 84 | 131* | 210* | 171* | 181* | 131* | 193* | 221* | 93 | 151* |
| α-2 antiplasmin | 165 | 167 | 159 | 170 | 146 | 158 | 246* | 200 | 224* | 173 | 153 |
| Fibrinogen (mg/dL) | 218 | 292* | 383* | 577* | 429* | 675* | 839* | 720* | 964* | 325* | 399* |
| Factor II | 87 | 97* | 99* | 123* | 98* | 118* | 179* | 129* | 131* | 79* | 65* |
| Factor V | 75 | 82 | 107* | 151* | 127* | 109* | 189* | 129* | 128* | 68 | 60* |
| Factor VII | 73 | 78 | 92* | 102* | 79 | 94* | 157* | 118* | 114* | 60* | 61* |
| Factor VIII | 76 | 78 | 64 | 72 | 63 | 89 | 190* | 100* | 119* | 63 | 49* |
| Factor IX | 82 | 76 | 84 | 79 | 69 | 74 | 166* | 101* | 120* | 67* | 54* |
| Factor X | 91 | 89 | 74* | 107* | 91 | 87 | 117* | 111* | 118* | 77* | 58* |
| Factor XI | 66 | 82* | 190* | 78* | 86* | 91* | 180* | 89* | 110* | 51* | 81* |
| Factor XII | 95 | 81* | 77* | 88 | 88 | 54* | 64* | 74* | 66* | 103* | 92 |
| vWF | 100 | 82 | 121 | 230* | 255* | 239* | 389* | 220* | 328* | 109 | 108 |
Mice were infected with S. Typhimurium, E. coli, MSSA, MRSA or S. pneumoniae as described in the methods and plasma was collected at the indicated times and analyzed for coagulation activities. Mean % values are relative to mouse normal pooled plasma (NMP). Pathogen and time were included as independent variables and the results of each analyte as dependent variables. Time, pathogen and the interaction between time and pathogen were significant (*P < 0.05). Pairwise comparisons were performed between uninfected mice (n = 60) and early (n = 12) and late (n = 12) septic mice for each analyte and pathogen. Benjamini-Hochberg was utilized to adjust P values for multiple comparisons.
Table 2.
Effect of MMP inhibitor GM6001 on coagulation factor activities during S. Typhimurium sepsis.
| S. Typhimurium | − GM6001 |
+ GM6001 |
||||
|---|---|---|---|---|---|---|
| Analyte (% NMP) | Not infected | Early sepsis (Day 5) | Late sepsis (Day 8) | Not infected | Early sepsis (Day 5) | Late sepsis (Day 8) |
| Fibrinogen (mg/dL) | 189 | 227 | 351 | 152 | 261 | 226* |
| Factor II | 76 | 67 | 46 | 73 | 73 | 76* |
| Factor V | 83 | 80 | 60 | 70 | 80 | 92* |
| Factor VII | 93 | 90 | 105 | 81 | 104 | 111 |
| Factor VIII | 72 | 74 | 53 | 58 | 94 | 72 |
| Factor IX | 70 | 64 | 90 | 52* | 60 | 60* |
| Factor X | 87 | 81 | 45 | 82 | 82 | 75* |
| Factor XI | 72 | 71 | 260 | 62 | 51* | 70* |
| Factor XII | 84 | 78 | 61 | 77 | 66 | 64 |
Mice were orally infected with S. Typhimurium (107 cfus) and treated in the presence/absence of MMP inhibitor, GM6001. Plasma was collected at the indicated times and analyzed for coagulation activities. % values are relative to mouse normal pooled plasma (NMP). Time and treatment were included as independent variables and the results of each analyte as dependent variables. Time, treatment and the interaction between time and treatment were significant (*P < 0.05). Pairwise comparisons were performed between the untreated and the corresponding treated group for each analyte (early sepsis: untreated vs. treated; late sepsis: untreated vs. treated). Benjamini-Hochberg was utilized to adjust P values for multiple comparisons. n = 15 mice per group.
Role of funders
Funders had no role in the study design, data collection, data analyses, interpretation, or writing of the report.
Results
A distinct pattern of coagulation factor protein abundance in the pre-septic state was predictive of coagulopathic activities during late-stage disease
Plasma proteomics was determined at five time points post-ST infection (day 0, 2, 3, 4, 5, 8). Early-onset remodeling of the plasma proteome occurred in a narrowly-timed, catastrophic fashion by day 4 post-infection–prior to onset of overt disease symptoms or bacteremia– and correlated with the onset of tissue colonization (Figure 1a–c). Examples include the early-onset detection of the acute-phase immune response proteins, CRP and LBP, which are benchmark biomarkers for human sepsis7,8,18; and increased abundance of immune modulator proteins CD14 and myeloperoxidase (MPO) that stimulate TLR4 and neutrophil activation29,30 (Figure 1a). As expected, such changes ultimately led to proinflammatory cytokine production during late-stage sepsis (IL-1β, IL-6, IFNγ, TNFα) (Figure 1d). Further, marked overlap was observed with recently-discovered proteomic-signatures of Gram-positive S. aureus patient mortality, sharing 14 of the top 25 biomarkers, including decreased expression of anticoagulant heparin cofactor 2 (HCF2) and increased expression of hemostasis proteins fibronectin and proteoglycan 4 (PRG4).22 However, immensely important to patient care are early-onset changes that are not available to late-sepsis studies alone wherein host responses and pathogenic mechanisms may consolidate into stereotypical processes of aberrant inflammatory and coagulopathic activities.
Figure 1.
Temporal changes to the plasma proteome, cfus in circulation, and innate immune cytokines following S. Typhimurium infection. Plasma proteomics was determined by MS analysis of samples collected from mice at five time points post-S. Typhimurium (ST) infection (day 0, 2, 3, 4, 5, 8). Heatmap representation of the fold-change (Log2) of (a) functional sub-networks and (b) coagulation pathways upon unsupervised hierarchical clustering (n = 2 mice). Supporting proteomics data is provided in Supplementary Table 1: a, network annotation; b, protein expression ratios; c, temporal analysis; d, STRING network coordinates; e, network calculations. c, Mice were infected with ST and assessed for cfus in circulation and spleen (n = 6 mice). Limit of detection (LOD) = 100 cfu/mL blood; 50 cfu/gram spleen. d, Matched samples from the plasma proteome analysis were assessed for innate immune cytokine analysis via ELISA (d = days; n = 3 mice/time point). *P < 0.05; **P < 0.01; ***P < 0.001.
To this end, we identified 119 plasma proteins that exhibited a significant change in expression relative to uninfected animals, including specific time-dependent functional interactions that occur at early versus late stages of disease (Supplementary Figure 1; Supplementary Table 1a–e).20 A distinct pattern of coagulation factor protein abundance in the pre-septic state was identified including factor XI (FXI) and fibrinogen subunits (FGA, FGB, FGG), concomitant with a progressive reduction in abundance of protease inhibitors antithrombin-III (AT-III), HCF2, and alpha-1-antitrypsin (AAT) (Figure 1b). Further, the changes in protein abundance observed in ST murine sepsis generally have the same directionality (increased or decreased abundance) as reported in the literature for human sepsis (Supplementary Table 1b,c).21 Notably, this distinct pattern of coagulation factor abundance was predictive of coagulopathic activities that occurred during late-stage disease. Examples include increased fibrinogen as well as increased protein S, FII, FV, FVII, and FXI activities, coupled with diminished levels of AT-III, FX, and FXII activities during ST sepsis (Table 1).
Gram-negative infections elicited similar changes of coagulopathic activities during sepsis
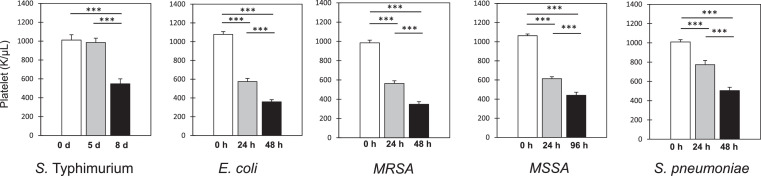
Significant overlap of ST coagulopathic activities was observed in Gram-negative E. coli sepsis, but not in Gram-positive sepsis models studied herein. Examples include increased fibrinogen as well as increased protein S, FII, FV, and FXI activities, while exhibiting diminished levels of AT-III activity (Table 1). In contrast, Gram-positive organisms, including methicillin-sensitive (MSSA) and methicillin-resistant (MRSA) S. aureus, elicited a general pattern of increased coagulation factor activation (e.g., FII, FV, FVII – XI), while S. pneumoniae elicited a specific pattern of decreased coagulation factor activation (e.g., FII, FV, FVII – FX). Additionally, contrary to the specific coagulation factor changes observed during Gram-negative sepsis, a shared moderate thrombocytopenia was elicited by all Gram-negative and Gram-positive organisms tested herein (Figure 2).
Figure 2.
Temporal changes to platelet abundance following infection with Gram-negative and Gram-positive organisms. Mice were infected with S. Typhimurium (gastric intubation; 107 cfu), E. coli (i.p.; 107 cfu), MSSA (i.v.; 5 × 108 cfu), MRSA (i.v.; 108 cfu) or S. pneumoniae (i.p.; 104 cfu) and blood was assessed for platelet abundance via Hemavet analysis. n = 16 mice/cohort. Data are presented as means ± SEM (***P < 0.001).
Treatment with matrix metalloprotease inhibitors prevented aberrant inflammatory and coagulopathic activities and increased the frequency of survival post ST infection
Host matrix-metalloproteinases (MMPs) play important roles in endothelial cell function, inflammation and coagulation.31, 32, 33 Thus, we questioned whether the administration of the MMP inhibitor, GM6001,26 could prevent aberrant inflammatory and coagulopathic activities post-ST infection. Daily intraperitoneal administration of GM6001 prevented the induction of host neuraminidase (Neu) activity and associated desialylation and accelerated clearance of host anti-inflammatory alkaline phosphatase (AP) activity involved in LPS detoxification (Figure 3a,b).20 The presence of MMP inhibitor also reversed coagulopathic changes as evidenced by blocking the induction of fibrinogen and FXI activity and preventing consumption of multiple coagulation factors (e.g., FII, FV, FIX, FX) (Table 2). Additionally, the presence of GM6001 reduced bacterial cfus in circulation and spleen (Figure 4a), and improved mouse survival post-ST infection (Figure 4b). Co-incubation of GM6001 with ST in vitro had no effect on bacterial viability or proliferation, which is consistent with GM6001 therapeutic effects acting through host molecular targets vs. direct action on the bacterium (Figure 4c).
Figure 3.
Effect of MMP inhibitor (GM6001) treatment on host alkaline phosphatase and neuraminidase activities during S. Typhimurium sepsis. Mice were infected via gastric intubation with S. Typhimurium (ST) (5 × 106 cfu). At 2 h post-infection, mice were treated in the presence and absence of GM6001, administered i.p. at a dose of 200 µg per mouse per day for 8 days; vehicle control mice were administered GM6001 diluent on the same treatment regimen. Mice were assessed for (a) serum AP activity (n = 10 mice per condition) and (b) plasma Neu activity (n = 6 mice per condition). Data are presented as means ± SEM (***P < 0.001).
Figure 4.
Effect of MMP inhibitor treatment on bacterial cfus and survival post-S. Typhimurium infection. Mice were infected via gastric intubation with S. Typhimurium (5 × 106 cfu). At 2 h post-infection, mice were treated in the presence and absence of MMP inhibitor, GM6001, administered i.p. at a dose of 200 µg per mouse per day for 8 days; vehicle control mice were administered GM6001 diluent on the same treatment regimen. Mice were assessed for (a) cfus in circulation and spleen (n = 8–10 mice/cohort) and (b) mouse survival (n = 20 mice/cohort). *P <0.05. c, ST was co-incubated in the presence/absence of GM6001 in vitro and assessed for viability.
Antibiotic treatment regimens initiated after specific changes arise in the plasma proteome were predictive of increased disease relapse and death after cessation of antibiotic treatment
Antibiotic treatment was initiated before and after blood proteome remodeling and presence of cfus in circulation. As expected, daily treatment with ciprofloxacin (Figure 5a) or ceftriaxone (Figure 5b) initiated at day 3, 4, 5, or 6 post ST-infection effectively cleared cfus from circulation in nearly all mice by day 8 (119/120 mice; Supplementary Table 2). Such treatment resulted in little-to-no overt disease symptoms through day 12 when treatment was ceased in all mouse cohorts. In comparison, 12/15 untreated mice showed high blood cfus at day 8 (>102–107 cfus/mL), with all (15/15) untreated mice succumbing to infection between days 8 and 11. Thus, as observed in the clinical setting,34,35 antibiotic treatment was effective in promoting bacterial clearance and survival even when initiated later in the disease process (day 5, 6), despite extensive blood proteome remodeling and cfus in circulation and/or spleen at the time of treatment (Figure 1a–c).
Figure 5.
Timing of initiation of antibiotic treatment and resultant disease relapse and death after cessation of antibiotics. Mice were infected via gastric intubation with S. Typhimurium (107 cfu) and treated twice daily with (a) ciprofloxacin (CIP; 30 mg/kg/day) or (b) ceftriaxone (CFX; 50 mg/kg/day), starting on either day 3, 4, 5, or 6 (green arrows) and continuing through day 12 post-infection, after which antibiotics were removed at day 13 (red arrow). Survival was monitored up to 25 d post-infection. n = 15 mice/cohort. *P <0.05; ***P <0.001.
Next, we assessed whether antibiotic treatment regimens initiated after specific changes arise in the plasma proteome were predictive of increased disease relapse and death after cessation of antibiotic treatment. Antibiotics were removed at day 13 in all treated cohorts. Ciprofloxacin treatment initiated on day 3 was significantly more effective at preventing disease relapse and death relative to regimens that initiated treatment later post-infection (day 3 vs. 4, 5, 6) (Figure 5a). Notably, day 3 precedes blood proteome remodeling, with nearly all mice exhibiting below-detectable levels of cfus in the spleen (5/6 mice) and in circulation (6/6 mice) when antibiotic treatment was initiated (Figure 1a–c). Nevertheless, ∼40% of mice which initiated treatment on day 3 succumbed to infection after removal of ciprofloxacin presumably due to low-level cfus seeding antibiotic-privileged tissues such as the gallbladder.36 A similar trend was observed following ceftriaxone treatment (day 3, 4 vs. 5, 6) (Figure 5b). These data indicate that antibiotic treatment regimens initiated after specific changes arise in the plasma proteome were predictive of increased disease relapse and death after termination of antibiotic therapy.
Discussion
An alternative to the current symptom-based approach used to diagnose sepsis is a precise assessment of blood proteomic changes during the onset and progression of sepsis to identify biomarkers at early stages of disease to minimize host-driven pathology. Herein, we identified a distinct pattern of coagulation factor protein abundance in the pre-septic state– prior to overt disease symptoms or bacteremia– that was predictive of the dysregulation of fibrinolytic and anti-coagulant activities and resultant consumptive coagulopathy during ST sepsis. Significant overlap of ST coagulopathic activities was observed in Gram-negative EC sepsis but not among Gram-positive sepsis models studied herein. Sepsis can thus be potentially stratified by different disease mechanisms elicited by discrete pathogens, dispelling the prevailing notion that sepsis manifests as a stereotypical process spanning inflammation and coagulopathy. MMP inhibitor therapeutic intervention prevented aberrant inflammatory and coagulopathic activities, reduced microbial cfus in blood and tissues, and improved survival post-ST infection. Antibiotic treatment regimens initiated after blood proteomic changes were predictive of an increase in disease relapse and death after cessation of antibiotics, and may reflect inadequate bacterial clearance associated with increased host-driven pathology. Such treatment failure provides a rationale as to why delays in antibiotic administration in human sepsis are associated with increased risk for death.
Marked changes to the blood proteome occurred prior to bacteremia, suggesting that tissue colonization and resultant pathogen-associated molecular patterns (PAMPs) and/or damage associated molecular patterns (DAMPs) trigger copious changes to the blood proteome at early stages post-ST infection.37 Such altered blood proteomics provides a platform to develop rapid and easy-to-perform tests to forecast sepsis for early intervention via biomarker panels that can be incorporated into existing blood tests prompted by patient presentation with general malaise and to stratify Gram-negative and Gram-positive infections for appropriate treatment. Clinical benefit is evident by the acquisition of risk profile data well-before overt disease symptoms that are necessary for SOFA and qSOFA scores used to evaluate ICU and non-ICU patients, respectively.38
Validation of this approach was established by the early-onset detection of the acute-phase immune response proteins, CRP and LBP, which are benchmark biomarkers for human sepsis;7,8,18 early-onset increased abundance of immune modulator proteins CD14 and MPO that stimulate TLR4 and neutrophil activation;29,30 and marked overlap of proteomic changes with recently discovered pathogenic-proteomic-signatures for prediction of SA patient mortality.22 Additionally, blood from sepsis patients has been used for (i) the development of DNA-based tests to identify microbes and their resistance, (ii) host-response RNA, (iii) novel biomarkers and rapid tests that detail risk profiles39; and (iv) the discovery that the source of infection contributes to the heterogeneity of the host response.40
A distinct pattern of coagulation factor protein abundance in the pre-septic state was identified including FXI and fibrinogen subunits, concomitant with a progressive reduction in abundance of protease inhibitors AT-III, HCF2, and AAT. Moreover, the changes in protein abundance observed in ST murine sepsis generally have the same directionality (increased or decreased abundance) as reported in the literature for human sepsis. Additionally, coagulation factor abundance was predictive of coagulopathic activities that occurred during ST sepsis; e.g., increased fibrinogen as well as increased protein S, FII, FV, FVII, and FXI activities, coupled with diminished levels of AT-III, FX, and FXII activities. Significant overlap of ST coagulopathic activities was observed in Gram-negative E. coli sepsis. In contrast, Gram-positive organisms, including MSSA and MRSA elicited a general pattern of increased coagulation factor activation, while S. pneumoniae elicited a specific pattern of decreased coagulation factor activation. These data suggest that early-onset coagulation protein abundance is predictive of the dysregulation of fibrinolytic and anti-coagulant activities and resultant consumptive coagulopathy during Gram-negative sepsis. FXI plays a principal role in coagulopathy as its activation promotes thrombosis,41,42 is detrimental to the host in several experimental sepsis models,43,44 and has been explored as a target for inhibition in sepsis.45,46
Reduced platelet abundance is a key component of outcomes in Gram-positive sepsis as platelets contribute to both host thrombosis and microbial killing activity and thus the importance of platelet abundance and its regulation in the pathogenesis of sepsis can be protective or deleterious to the host depending upon the microbial source of infection.47, 48, 49 Contrary to the specific coagulopathic changes observed during Gram-negative sepsis, we show that a moderate thrombocytopenia is a shared response among all five Gram-negative and Gram-positive pathogens tested herein. A moderate thrombocytopenia has been previously linked to Gram-positive sepsis due to the desialylation of platelets by either the pathogen or the host in early sepsis followed by platelet clearance involving the hepatic Ashwell-Morell receptor (AMR).49, 50, 51
MMPs are key regulators of the inflammation/coagulation response and are potential targets for diagnostics and therapeutics.33 MMPs are increased > 8-fold in the circulation of sepsis patients and are correlated with increased mortality.31,52,53 Additionally, MMP activity is increased > 4-fold in circulation in a murine sepsis model31; and MMP inhibitors protect against vascular hyporeactivity in models of acute endotoxemia in rats.54,55 Herein, administration of GM6001 prevented increased bacterial cfus in circulation and spleen and improved mouse survival post-ST infection. Such therapeutic intervention is due, in part, to blocking the induction of host Neu activity, which facilitates clearance of desialylated anti-inflammatory AP enzymes by the hepatic AMR receptor,20 thereby restoring the host-protective mechanism of LPS de-toxification by circulating AP enzymes. Clinical significance is evidenced by human trials indicating that AP administration reduced anti-inflammatory effects in sepsis and ulcerative colitis.56, 57, 58 GM6001 administration also reversed coagulopathic changes as evidenced by blocking the induction of fibrinogen and FXI activity and preventing consumption of multiple coagulation factors (e.g., FII, FV, FIX, FX). Co-incubation of GM6001 with ST in vitro had no effect on bacterial viability or proliferation, suggesting the therapeutic effects act through host molecular targets. Our findings further support the hypothesis that investigation into the contribution of MMPs to sepsis pathogenesis may lead to new sepsis diagnostics and therapies.
Time to antibiotic treatment is the principal tenet for improved patient outcome during emergency care for sepsis,34 and is consistent with the “Surviving Sepsis Campaign” recommendations for early and aggressive antibiotics for all patients with possible sepsis or septic shock.35 However, diagnostic uncertainty due to the current symptom-based approach used to diagnose sepsis runs the risk of damage to uninfected-but-suspected patients and virally-infected patients, who are subjected to antibiotic-related risks without any of the benefits.59 This study demonstrates that antibiotics are less effective in microbial clearance after the onset of altered blood proteomics as evidenced by increased disease relapse and death after termination of antibiotic therapy. Treatment failure may be due to altered bacterial / host-responses commensurate with increased host-driven pathology, providing insight into why delays in antibiotic administration in human sepsis are associated with increased risk for death. Delayed treatment may thus require prolonged duration for microbial clearance despite the prevailing notion of antibiotic de-escalation and shortened courses of antibiotics to improve drug stewardship.60
Data sharing statement
Proteomics data deposition. LUMOS-Orbitrap RAW datasets and the results output from the search engine MaxQuant have been deposited in the MassIVE proteomics repository at UCSD and given the identification code PXD021291 at: https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=1dc72a3bb34a43998f64505d209 29b61ba3. All data generated or analyzed during this study are included in this published article or in the supplement.
Contributors
Experiments were conducted by D.M.H., G.P., S.P.M., and W.H.Y. Data were analyzed by D.M.H., G.P., S.P.M., D.T.L., J.K.H., J.D.M., J.W.S. and M.J.M. The manuscript was prepared by D.M.H., S.P.M., and M.J.M. The study was planned and directed by D.M.H., G.P., S.P.M., J.W.S., and M.J.M. All authors had full access to all data in the study and had final responsibility for the decision to submit for publication.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
This research was funded by National Institutes of Health (NIH) HL131474 (D.T.L., J.D.M., J.W.S., and M.J.M) and U.S. Army Research Office via the Institute for Collaborative Biotechnologies cooperative agreement W911NF-19-2-0026 (M.J.M) and contract W911NF-19-D-0001-0013 (M.J.M). The funders had no role in the study design, execution or data interpretation.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103965.
Appendix. Supplementary materials
References
- 1.Singer M., Deutschman C., Seymour C., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudd K., Johnson S., Agesa K., et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet. 2020;395(10219):200–211. doi: 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwashyna T., Ely E., Smith D., Langa K. Long-term cognitive impairment and functional disability among survivors of severe sepsis. JAMA. 2010;304(16):1787–1794. doi: 10.1001/jama.2010.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prescott H., Angus D. Enhancing recovery from sepsis: a review. JAMA. 2018;319(1):62–75. doi: 10.1001/jama.2017.17687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung H.Y., Wickel J., Brunkhorst F., Geis C. Sepsis-associated encephalopathy: from delirium to dementia? J Clin Med. 2020;9(3):703. doi: 10.3390/jcm9030703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paoli C., Reynolds M., Sinha M., Gitlin M., Crouser E. Epidemiology and costs of sepsis in the United States—an analysis based on timing of diagnosis and severity level. Crit Care Med. 2018;46(12):1889–1987. doi: 10.1097/CCM.0000000000003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teggert A., Datta H., Ali Z. Biomarkers for point-of-care diagnosis of sepsis. Micromachines. 2020;11(3):286. doi: 10.3390/mi11030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pierrakos C., Velissaris D., Bisdorff M., Marshall J., Vincent J.L. Biomarkers of sepsis: time for a reappraisal. Crit Care. 2020;24(24):287. doi: 10.1186/s13054-020-02993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iba T., Levi M., Levy J. Sepsis-induced coagulopathy and disseminated intravascular coagulation. Semin Thromb Hemost. 2020;46(01):89–95. doi: 10.1055/s-0039-1694995. [DOI] [PubMed] [Google Scholar]
- 10.Cinel I., Opal S. Molecular biology of inflammation and sepsis: a primer. Crit Care Med. 2009;37(1):291–304. doi: 10.1097/CCM.0b013e31819267fb. [DOI] [PubMed] [Google Scholar]
- 11.Rittirsch D., Flierl M., Ward P. Harmful molecular mechanisms in sepsis. Nat Rev Immunol. 2008;8(10):776–787. doi: 10.1038/nri2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhry H., Zhou J., Zhong Y., et al. Role of cytokines as a double-edged sword in sepsis. In Vivo. 2013;27(6):669–684. [PMC free article] [PubMed] [Google Scholar]
- 13.D'Elia R., Harrison K., Oyston P., Lukaszewski R., Clark C. Targeting the “cytokine storm” for therapeutic benefit. Clin Vaccine Immunol. 2013;20(3):319–327. doi: 10.1128/CVI.00636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulte W., Bernhagen J., Bucala R. Cytokines in sepsis: potent immunoregulators and potential therapeutic targets—an updated view. Mediat Inflamm. 2013;2013 doi: 10.1155/2013/165974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levi M., van der Poll T. Coagulation and sepsis. Thromb Res. 2017;149:38–44. doi: 10.1016/j.thromres.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Chang J. Disseminated intravascular coagulation: new identity as endotheliopathy-associated vascular microthrombotic disease based on in vivo hemostasis and endothelial molecular pathogenesis. Thromb J. 2020;18(1):25. doi: 10.1186/s12959-020-00231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seymour C., Liu V., Iwashyna T., et al. Assessment of clinical criteria for sepsis: for the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315(8):762–774. doi: 10.1001/jama.2016.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans T. Diagnosis and management of sepsis. Clin Med. 2018;18(2):146–149. doi: 10.7861/clinmedicine.18-2-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peri A., Stewart A., Hume A., Irwin A., Harris P. New microbiological techniques for the diagnosis of bacterial infections and sepsis in ICU including point of care. Curr Infect Dis Rep. 2021;23(8):1–11. doi: 10.1007/s11908-021-00755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang W., Heithoff D., Aziz P., et al. Accelerated aging and clearance of host anti-inflammatory enzymes by discrete pathogens fuels sepsis. Cell Host Microbe. 2018;24(4):500–513. doi: 10.1016/j.chom.2018.09.011. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pimienta G., Heithoff D., Rosa-Campos A., et al. Plasma proteome signature of sepsis: a functionally connected protein network. Proteomics. 2019;19(5) doi: 10.1002/pmic.201800389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wozniak J., Mills R., Olson J., et al. Mortality risk profiling of Staphylococcus aureus bacteremia by multi-omic serum analysis reveals early predictive and pathogenic signatures. Cell. 2020;182(5):1311–1327. doi: 10.1016/j.cell.2020.07.040. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y., Tan J., Sutton-Smith M., et al. Modeling human congenital disorder of glycosylation type IIa in the mouse: conservation of asparagine-linked glycan-dependent functions in mammalian physiology and insights into disease pathogenesis. Glycobiology. 2001;11(12):1051–1070. doi: 10.1093/glycob/11.12.1051. [DOI] [PubMed] [Google Scholar]
- 25.Grewal P. The Ashwell–Morell receptor. Methods Enzymol. 2010;479:223–241. doi: 10.1016/S0076-6879(10)79013-3. [DOI] [PubMed] [Google Scholar]
- 26.Galardy R., Cassabonne M., Giese C., et al. Low molecular weight inhibitors in corneal ulceration. Ann N Y Acad Sci. 1994;732(1):315–323. doi: 10.1111/j.1749-6632.1994.tb24746.x. [DOI] [PubMed] [Google Scholar]
- 27.RStudio Team. RStudio: integrated development environment for R. Boston, MA 2021. http://www.rstudio.com/. Accessed 21 January 2022.
- 28.Oksanen J, Blanchet F, Friendly M, et al. vegan: Community Ecology Package, R package version 2.5-7.2020. https://CRAN.R-project.org/package=vegan. Accessed 21 January 2022.
- 29.Miller S., Ernst R., Bader M. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol. 2005;3(1):36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 30.Lau D., Mollnau H., Eiserich J., et al. Myeloperoxidase mediates neutrophil activation by association with CD11b/CD18 integrins. Proc Natl Acad Sci U S A. 2005;102(2):431–436. doi: 10.1073/pnas.0405193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tressel S., Kaneider N., Kasuda S., et al. A matrix metalloprotease-PAR1 system regulates vascular integrity, systemic inflammation and death in sepsis. EMBO Mol Med. 2011;3(7):370–384. doi: 10.1002/emmm.201100145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai X., Bai G., Tang L., Liu L., Li Y., Jiang W. Changes in MMP‑2, MMP‑9, inflammation, blood coagulation and intestinal mucosal permeability in patients with active ulcerative colitis. Exp Ther Med. 2020;20(1):269–274. doi: 10.3892/etm.2020.8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchimido R., Schmidt E., Shapiro N. The glycocalyx: a novel diagnostic and therapeutic target in sepsis. Crit Care. 2019;23(1):16. doi: 10.1186/s13054-018-2292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seymour C., Gesten F., Prescott H., et al. Time to treatment and mortality during mandated emergency care for sepsis. N Engl J Med. 2017;376(23):2235–2244. doi: 10.1056/NEJMoa1703058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhodes A., Evans L., Alhazzani W., et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med. 2017;43(3):304–377. doi: 10.1007/s00134-017-4683-6. [DOI] [PubMed] [Google Scholar]
- 36.Gunn J., Marshall J., Baker S., Dongol S., Charles R., Ryan E. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 2014;22(11):648–655. doi: 10.1016/j.tim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang D., Kang R., Coyne C., Zeh H., Lotze M. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol Rev. 2012;249(1):158–175. doi: 10.1111/j.1600-065X.2012.01146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koch C., Edinger F., Fischer T., et al. Comparison of qSOFA score, SOFA score, and SIRS criteria for the prediction of infection and mortality among surgical intermediate and intensive care patients. World J Emerg Surg. 2020;15(1):1–10. doi: 10.1186/s13017-020-00343-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Poll T. Molecular diagnosis and risk stratification of sepsis (MARS) 2013. https://www.clinicaltrials.gov/ct2/show/NCT01905033. Accessed 18 January 2022.
- 40.Peters-Sengers H., Butler J., Uhel F., et al. Source-specific host response and outcomes in critically ill patients with sepsis: a prospective cohort study. Intensive Care Med. 2022 doi: 10.1007/s00134-021-06574-0. https://pubmed.ncbi.nlm.nih.gov/34902047/ Accessed 18 January 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meijers J., Tekelenburg W., Bouma B., Bertina R., Rosendaal F. High levels of coagulation factor XI as a risk factor for venous thrombosis. N Engl J Med. 2000;342(10):696–701. doi: 10.1056/NEJM200003093421004. [DOI] [PubMed] [Google Scholar]
- 42.Wang X., Cheng Q., Xu L., et al. Effects of factor IX or factor XI deficiency on ferric chloride-induced carotid artery occlusion in mice. J Thromb Haemost. 2005;3(4):695–702. doi: 10.1111/j.1538-7836.2005.01236.x. [DOI] [PubMed] [Google Scholar]
- 43.Silasi R., Keshari R., Lupu C., et al. Inhibition of contact-mediated activation of factor XI protects baboons against S aureus–induced organ damage and death. Blood Adv. 2019;3(4):658–669. doi: 10.1182/bloodadvances.2018029983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tucker E.I., Verbout N.G., Leung P.Y., et al. Inhibition of factor XI activation attenuates inflammation and coagulopathy while improving the survival of mouse polymicrobial sepsis. Blood. 2012;119(20):4762–4768. doi: 10.1182/blood-2011-10-386185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mair G.E., Olson S.R., Shatzel J.J. The safety of novel drugs targeting factor XI and XII in human clinical trials: a systematic review. Blood. 2019 doi: 10.1182/blood-2019-130823. [DOI] [Google Scholar]
- 46.Gailani D., Gruber A. Factor XI as a therapeutic target. Arterioscler Thromb Vasc Biol. 2016;36(7):1316–1322. doi: 10.1161/ATVBAHA.116.306925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deppermann C., Kratofil R., Peiseler M., et al. Macrophage galactose lectin is critical for Kupffer cells to clear aged platelets. J Exp Med. 2020;217(4) doi: 10.1084/jem.20190723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uchiyama S., Sun J., Fukahori K., et al. Dual actions of group B Streptococcus capsular sialic acid provide resistance to platelet-mediated antimicrobial killing. Proc Natl Acad Sci U S A. 2019;116(15):7465–7470. doi: 10.1073/pnas.1815572116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun J., Uchiyama S., Olson J., et al. Repurposed drugs block toxin-driven platelet clearance by the hepatic Ashwell-Morell receptor to clear Staphylococcus aureus bacteremia. Sci Transl Med. 2021;13(586):eabd6737. doi: 10.1126/scitranslmed.abd6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grewal P., Aziz P., Uchiyama S., et al. Inducing host protection in pneumococcal sepsis by preactivation of the Ashwell-Morell receptor. Proc Natl Acad Sci U S A. 2013;110(50):20218–20223. doi: 10.1073/pnas.1313905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grewal P., Uchiyama S., Ditto D., et al. The Ashwell receptor mitigates the lethal coagulopathy of sepsis. Nat Med. 2008;14(6):648–655. doi: 10.1038/nm1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lauhio A., Hästbacka J., Pettilä V., et al. Serum MMP-8,-9 and TIMP-1 in sepsis: high serum levels of MMP-8 and TIMP-1 are associated with fatal outcome in a multicentre, prospective cohort study. Hypothetical impact of tetracyclines. Pharmacol Res. 2011;64(6):590–594. doi: 10.1016/j.phrs.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 53.Lorente L., Martín M., Labarta L., et al. Matrix metalloproteinase-9,-10, and tissue inhibitor of matrix metalloproteinases-1 blood levels as biomarkers of severity and mortality in sepsis. Crit Care. 2009;13(5):R158. doi: 10.1186/cc8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cena J., Lalu M., Cho W., et al. Inhibition of matrix metalloproteinase activity in vivo protects against vascular hyporeactivity in endotoxemia. Am J Phys Heart Circ Phys. 2010;298(1):H45–H51. doi: 10.1152/ajpheart.00273.2009. [DOI] [PubMed] [Google Scholar]
- 55.Cena J., Lalu M., Rosenfelt C., Schulz R. Endothelial dependence of matrix metalloproteinase-mediated vascular hyporeactivity caused by lipopolysaccharide. Eur J Pharm. 2008;582(1-3):116–122. doi: 10.1016/j.ejphar.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 56.Pickkers P., Heemskerk S., Schouten J., et al. Alkaline phosphatase for treatment of sepsis-induced acute kidney injury: a prospective randomized double-blind placebo-controlled trial. Crit Care. 2012;16(1):R14. doi: 10.1186/cc11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parlato M., Charbit-Henrion F., Pan J., et al. Human ALPI deficiency causes inflammatory bowel disease and highlights a key mechanism of gut homeostasis. EMBO Mol Med. 2018;10(4):e8483. doi: 10.15252/emmm.201708483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lukas M., Drastich P., Konecny M., et al. Exogenous alkaline phosphatase for the treatment of patients with moderate to severe ulcerative colitis. Inflamm Bowel Dis. 2010;16(7):1180–1186. doi: 10.1002/ibd.21161. [DOI] [PubMed] [Google Scholar]
- 59.Klompas M., Calandra T., Singer M. Antibiotics for sepsis—finding the equilibrium. JAMA. 2018;320(14):1433–1434. doi: 10.1001/jama.2018.12179. [DOI] [PubMed] [Google Scholar]
- 60.Martínez M., Plata-Menchaca E., Ruiz-Rodríguez J., Ferrer R. An approach to antibiotic treatment in patients with sepsis. J Thorac Dis. 2020;12(3):1007–1021. doi: 10.21037/jtd.2020.01.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.