Abstract
The sequence of the blaARI-1 gene from imipenem-resistant Acinetobacter baumannii 6B92 has been determined. The structural gene encodes a 273-amino-acid protein which is most related to the OXA class D β-lactamases. The conserved S-T-F-K and K-T-G motifs were identified in the ARI-1 protein sequence, also named OXA-23, but significantly, a point mutation (Y→F) was identified in the Y-G-N conserved motif, also known to function in the active site.
Multiresistant Acinetobacter baumannii strains are now recognized as serious nosocomial pathogens (4, 5), and carbapenem-resistant strains are being reported increasingly (1, 3, 6, 11, 15, 18, 23). Imipenem-resistant A. baumannii 6B92 was isolated from a patient in Edinburgh, United Kingdom, in 1985 (15). Imipenem resistance was attributed to a novel serine β-lactamase, ARI-1 (15), and was subsequently demonstrated to be transferable to Acinetobacter junii (18). Imipenem resistance due to β-lactamases in A. baumannii has subsequently been reported worldwide, and two additional β-lactamases, ARI-2 (6) and an oxacillin-hydrolyzing enzyme (2, 11), defined only by their biochemical properties, have been described. In this paper we report the nucleotide and deduced amino acid sequences of ARI-1 carried by the R plasmid pUK1356.
Bacterial strains and plasmids.
The transconjugant A. junii BD413-2(pUK1356) was used as the source of ARI-1. Escherichia coli TG2 {supE hsdΔ5 thi Δ(lac-proAB) Δ(slr-recA) 306::Tn10(tetr) F′[traD36 proAB+ lacIq lacZΔM15]} (17) was used as a host for recombinant plasmids prepared in the vector pUC19 (24).
β-Lactamase purification and N-terminal amino acid sequencing.
Cell extracts of A. junii BD413-2(pUK1356) were loaded onto a Mono Q anion-exchange column (Pharmacia Co. Ltd., Uppsala, Sweden) equilibrated with 50 mM Tris-HCl (pH 8.2). Fractions were eluted with a gradient of 0 to 0.5 M NaCl in 50 mM Tris-HCl (pH 8.2). The ARI-1 β-lactamase, eluted in the unadsorbed fraction, was loaded onto a Superdex-75 gel filtration column (Pharmacia) equilibrated in 50 mM Tris-HCl (pH 7.5) containing 0.1 M NaCl. Fractions containing β-lactamase activity, detected by an assay with nitrocefin (Glaxo Group Research Ltd., Greenford, United Kingdom), were concentrated by centrifugation using VectaSpin microtube filters, with a molecular mass cutoff of 12 kDa (Whatman International Ltd., Maidstone, United Kingdom). Following native PAGE, ARI-1 was transferred to a Problot membrane (PE Applied Biosystems, Warrington, United Kingdom), stained with Coomassie blue, and cut out for N-terminal sequencing with a model 477A gas phase sequencer (PE Applied Biosystems). The 20 N-terminal amino acids were used to prepare the degenerate oligonucleotide probe ARI-N (Table 1).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide primera | Application | Sequenceb | Positions in nucleotide sequence shown in Fig. 1 |
|---|---|---|---|
| ARI-N | DNA hybridization | 5′-ACI GTI CAR CAY AAY YTI ATI AAY GA-3′ | 1026–1051 |
| P1 | PCR and sequencing | 5′-TGA ACA ATC TGA CTC GGG CT-3′ | 1072–1053 (complement) |
| P2 | PCR and sequencing | 5′-TGG AGA ACC AGA AAA CGG AT-3′ | 1240–1259 |
| Invrs2 | PCR and sequencing | 5′-TTT TGG AAA GAC TGG TTG GG-3′ | 1610–1629 |
| Invrs3 | PCR and sequencing | 5′-CTG CTG TCC AAT TTC AGC AT-3′ | 1451–1432 (complement) |
| Invrs1 | Sequencing | 5′-TTT GCA TGA GAT CAA GAC CG-3′ | 1404–1385 (complement) |
| P5 | PCR and sequencing | 5′-AAG CAT GAT GAG CGC AAA G-3′ | 785–803 |
| P6 | PCR and sequencing | 5′-AAA AGG CCC ATT TAT CTC AAA-3′ | 1850–1830 (complement) |
All oligonucleotides were purchased from Life Technologies Ltd.
I, inosine; R, any purine (A or G); Y, any pyrimidine (T or C).
Restriction enzymes and DNA cloning.
Restriction enzymes (Life Technologies Ltd., Paisley, United Kingdom) were used in accordance with the manufacturer's instructions. Plasmid pUK1356 was extracted, restricted, and cloned with the procedures described by Sambrook et al. (17). Recombinant clones were selected on nutrient agar (IDG Ltd., Bury, United Kingdom) containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 32.5 mg/liter), IPTG (isopropyl-β-d-thiogalactopyranoside; 7.8 mg/liter), and ampicillin (50 mg/liter; Sigma Aldrich Co. Ltd., Poole, United Kingdom).
Hybridization and identification of the 5′ end of the ARI-1 gene.
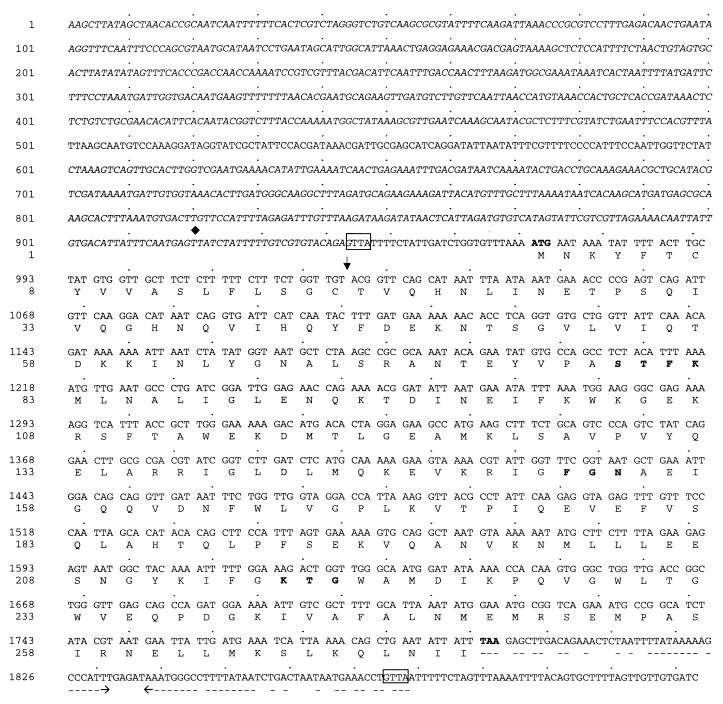
Enhanced chemiluminescence probe labelling, hybridization, and detection kits (Amersham Pharmacia Biotech UK Ltd., Amersham, United Kingdom) were used in accordance with the manufacturer's instructions. A partial library of pUK1356 recombinant clones was screened with an ARI-N probe 3′ end labelled with fluorescein-dUTP. Recombinant plasmid from a single positive clone was purified with a Qiagen (Crawley, United Kingdom) plasmid minikit and sequenced with an ABI PRISM 377 automated DNA sequencer (PE Applied Biosystems). The sequence (Fig. 1, nucleotides 1 to 1345) revealed 375 bp of the 5′ end of the blaARI-1 gene.
FIG. 1.
Nucleotide and deduced amino acid sequences of the ARI-1 gene and the ARI-1 protein from A. baumannii 6B92. The boldface ATG and TAA represent the initiation and termination codons, respectively. The β-lactamase active site S-T-F-K and the conserved motifs F-G-N and K-T-G are shown in boldface type. A proposed cleavage site generating a possible signal sequence is indicated with a vertical arrow. Nucleotide sequence in italics shows >90% homology with the phaBAc upstream region (19). ⧫ indicates the transcription start site identified by Schembri et al. (20). An imperfect inverted repeat sequence at the 3′ end of the gene is shown by a broken line, and possible cassette junctions are shown boxed.
PCR amplification and complete sequencing of the ARI-1 gene.
PCRs were carried out in 100-μl volumes containing 16 mM (NH4)2SO4, 67 mM Tris-HCl (pH 8.8), 0.1% Tween 20 buffer, 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (Amersham Pharmacia Biotech UK Ltd.), 0.1 to 0.5 μM each primer, and 1 U of BIOTAQ polymerase (Bioline UK Ltd., London, United Kingdom) or 1.25 U of Pfu DNA polymerase (Promega UK Ltd., Southampton, United Kingdom). Template DNA was boiled for 10 min before being added to the reaction mixture. DNA amplification was performed in an Omn-E thermal cycler (Hybaid Ltd., Teddington, United Kingdom) under the following cycle conditions: 94°C for 5 min, 60°C for 1 min, 72°C for 2 min (one cycle); 94°C for 15 s, 60°C for 1 min, 72°C for 2 min (30 cycles); and a final extension of 72°C for 5 min (one cycle). PCR products containing the 3′ end of the ARI-1 gene were obtained by inverse PCR of HincII or Sau3A fragments of pUK1356 by using the primer pairs P1-P2 and Invrs2-Invrs3 (Table 1). PCR products comprising the complete sequence of the blaARI-1 gene were generated by using the primer pair P5-P6 (Table 1). The ARI-1 gene sequence was determined from both strands with PCR products from three independent reactions, at least one of which was generated with Pfu DNA polymerase, being sequenced routinely.
Sequence data analysis, alignment, and phylogeny.
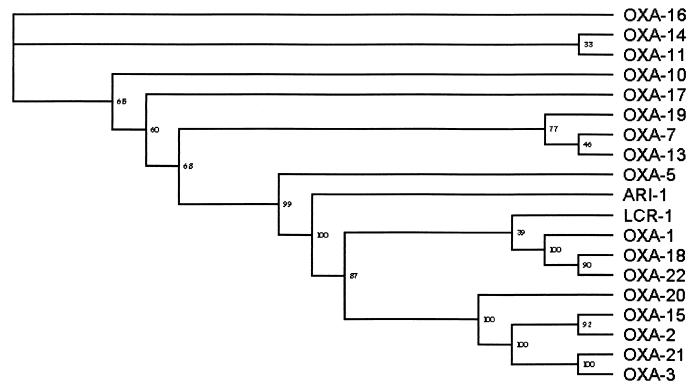
For computer analysis of sequence data, the software from the BCM Search Launcher was used (21). The blaARI-1 gene comprises an 822-bp open reading frame, between the initiation codon ATG (positions 972 to 974) and the stop codon TAA (positions 1791 to 1793), which translates into a protein of 273 amino acids (Fig. 1) showing sequence homology to Ambler class D (Bush group 2d) oxacillin-hydrolyzing enzymes. Some unique features and distinct biochemical properties were noted. Two highly conserved motifs, S-T-F-K and K-T-G (positions 79 to 82 and 216 to 218), believed to contribute to the function of the serine active site (7, 12), were identified (Fig. 1). A third motif, F-G-N at positions 152 to 154, which differs from the corresponding motif in all other OXA enzymes by the presence of phenylalanine instead of tyrosine. This unique substitution may have significant biochemical effects and is possibly a factor in the evolution of carbapenem resistance in this enzyme. A cladogram was constructed to relate ARI-1 to 18 other class D β-lactamases (Fig. 2). ARI-1 showed less than 40% identity with other OXA enzymes. Greatest identity was observed with OXA-5 and OXA-10 (36% identity), 35% identity was observed with OXA-7 and OXA-11, and 32% identity was observed with LCR-1 (8). ARI-1 is therefore a novel class D β-lactamase and has been assigned the alternative name OXA-23.
FIG. 2.
Cladogram relating ARI-1 to 18 other class D β-lactamases. Analysis was done with the CLUSTAL W Multiple Sequence Alignment Program (22), based on a progressive alignment using the minimum mutation matrix of Dayhoff for scoring (9). A gap penalty of 3 was applied, and 100 bootstrap replications gave node values.
Analysis of the genetic environment of blaARI-1.
The majority of oxa genes are contained within mobile cassettes (13, 14, 16) normally found inserted within integrons (10). The sequence immediately upstream of the blaARI-1 structural gene did not reveal cassette characteristics (i.e., consensus core sequence GTTRRRY). Nevertheless, a large region of 944 bases showing more than 90% homology with the phaBAc upstream region previously identified in the chromosome and plasmids of Acinetobacter strains isolated from sludge (19) was found. Schembri et al. (20) have shown a transcription start point within this region (position 920 in Fig. 1). This may indicate that the blaARI-1 gene has been specifically inserted downstream of an active promoter, enabling low-level constitutive expression of the gene in A. baumannii. Analysis of the region immediately downstream of blaARI-1 revealed an imperfect inverted repeat sequence with some of the characteristics of the 59-bp recombination sites found in cassette structures (16). Although characteristic of gene cassettes, it is possible that this sequence is functioning as a transcription terminator sequence since there was no evidence of an inverse core sequence (consensus sequence RYYYAAC). Furthermore, continuation of the published pha operon sequence could not be identified. Despite the apparent lack of evidence of an integron or cassette location for the blaARI-1 gene, two GTTA sequences (boxed in Fig. 1) were identified and may define the ends of a potential novel and unusual cassette structure.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the EMBL nucleotide database under accession no. AJ132105.
Acknowledgments
We thank Peter Rowell for assistance with the enzyme purification procedures.
This work was funded by the Scottish Office Home and Health Department (grant no. K/MRS/50/C2522).
REFERENCES
- 1.Afzal-Shah M, Livermore D M. Worldwide emergence of carbapenem-resistant Acinetobacter spp. J Antimicrob Chemother. 1998;41:576–577. doi: 10.1093/jac/41.5.576. [DOI] [PubMed] [Google Scholar]
- 2.Afzal-Shah M, Villar H E, Livermore D M. Biochemical characteristics of a carbapenemase from an Acinetobacter baumannii isolate collected in Buenos Aires, Argentina. J Antimicrob Chemother. 1999;43:127–131. doi: 10.1093/jac/43.1.127. [DOI] [PubMed] [Google Scholar]
- 3.Amyes S G B. β-Lactam resistance and the use of inhibitor combinations. J Med Microbiol. 1997;46:728–731. [Google Scholar]
- 4.Bauernfeind A, Kljucar S, Jungwirth R. Overview of antibiotic resistance problems in Acinetobacter spp. J Med Microbiol. 1997;46:726–728. [Google Scholar]
- 5.Bergogne-Berezin E, Towner K J. Acinetobacter spp, as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown S, Bantar C, Young H-K, Amyes S G B. Limitation of Acinetobacter baumannii treatment by plasmid-mediated carbapenemase ARI-2. Lancet. 1998;351:186–187. doi: 10.1016/S0140-6736(05)78210-6. [DOI] [PubMed] [Google Scholar]
- 7.Bush K, Jacoby G A, Mederios A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couture F, Lachapelle J, Levesque R C. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol Microbiol. 1992;6:1693–1705. doi: 10.1111/j.1365-2958.1992.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 9.Dayhoff M O, Schwartz R M, Orcutt B C. A model of evolutionary change in proteins. In: Dayhoff M O, editor. Atlas of protein sequence and structure. 5, suppl. 5. Washington, D.C.: National Biomedical Research Foundation, Georgetown University Medical Center; 1978. pp. 345–352. [Google Scholar]
- 10.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 11.Hornstein M, Sautjeau-Rostoker C, Peduzzi J, Vessieres A, Hong L T H, Barthelemy M, Scavizzi M, Labia R. Oxacillin-hydrolyzing β-lactamase involved in resistance to imipenem in A. baumannii. FEMS Microbiol Lett. 1997;153:333–339. doi: 10.1111/j.1574-6968.1997.tb12593.x. [DOI] [PubMed] [Google Scholar]
- 12.Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mugnier P, Podglajen I, Goldstein F W, Collatz E. Carbapenems as inhibitors of OXA-13, a novel integron-encoded β-lactamase in Pseudomonas aeruginosa. Microbiology. 1998;144:1021–1031. doi: 10.1099/00221287-144-4-1021. [DOI] [PubMed] [Google Scholar]
- 14.Naas T, Sougakoff W, Casetta A, Nordmann P. Molecular characterization of OXA-20, a novel class D β-lactamase, and its integron from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:2074–2083. doi: 10.1128/aac.42.8.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paton R, Miles R S, Hood J, Amyes S G B. ARI-1: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 1993;2:81–88. doi: 10.1016/0924-8579(93)90045-7. [DOI] [PubMed] [Google Scholar]
- 16.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Scaife W, Young H-K, Paton R, Amyes S G B. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J Antimicrob Chemother. 1995;36:585–586. doi: 10.1093/jac/36.3.585. [DOI] [PubMed] [Google Scholar]
- 19.Schembri M A, Bayly R C, Davies J K. Cloning and analysis of the polyhydroxyalkanoic acid synthase gene from Acinetobacter sp.: evidence that the gene is both plasmid and chromosomally located. FEMS Microbiol Lett. 1994;118:145–152. doi: 10.1111/j.1574-6968.1994.tb06817.x. [DOI] [PubMed] [Google Scholar]
- 20.Schembri M A, Bayly R C, Davies J K. Phosphate concentration regulates transcription of the Acinetobacter polyhydroxyalkanoic acid biosynthetic genes. J Bacteriol. 1995;177:4501–4507. doi: 10.1128/jb.177.15.4501-4507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith R F, Wiese B A, Wojzynski M K, Davidson D B, Worley K C. BCM Search Launcher–an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J D, Higgens D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinbren M J, Johnson A P, Kaufmann M E, Livermore D M. Acinetobacter spp. isolates with reduced susceptibilities to carbapenems in a UK burns unit. J Antimicrob Chemother. 1998;41:574–576. doi: 10.1093/jac/41.5.574. [DOI] [PubMed] [Google Scholar]
- 24.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]