Abstract
Nanostructured devices and nanoparticles have fundamentally reshaped the development of precision healthcare in recent decades. Meanwhile, mass spectrometry (MS)-based proteomics has evolved from simple protein sequencing to a powerful approach that identifies disease patterns and signatures, reveals molecular mechanisms of pathological processes, and develops therapeutic or preventive drugs. Significantly, the two distinct disciplines have synergized and expanded our knowledge about human health and disease, as evidenced by a variety of nanotechnology-assisted sample processing strategies, facilitating in-depth proteome profiling and post-translational modifications (PTMs) characterization. This review summarizes recent advances in nanoparticle design for better enrichment of marker proteins and their PTMs from various bio-specimens and emerging nanotechnologies that are applied to MS-based proteomics for precision medicine discovery.
Keywords: Nanoparticle, nanotechnology, mass spectrometry, biomarkers, proteomics, post- translational modifications
Introduction
Precision medicine, also known as "personalized medicine", is an innovative approach to tailoring disease prevention and treatment based upon individual clinical characteristics, including genes, proteins, metabolites, environment, and lifestyle 1, 2. Multiple layers of information derived from the genome, transcriptome, and interactome are integrated to help better understand the complexity of human health and diseases that cannot be explained by any single approach 3. Proteome refers to the complete set of proteins expressed in cells, tissues, or organisms, and proteomics investigates the proteome composition and activities to obtain a system-level understanding of disease occurrence, cell metabolism, and other processes 4. Proteins are the main executors of cellular activities and are among the most critical macromolecules in precision medicine 5. Thus, developing methods to measure alterations of proteins in the context of their abundance, localization, interaction, and modifications could provide novel insights into the pathophysiology of cancer and other diseases and ultimately aid the development of precision medicine 6.
In the past decade, mass spectrometry (MS)-based proteomics has witnessed tremendous advances in accuracy, sensitivity, automation, and throughput and made remarkable contributions to novel biomarker discovery and our understanding of underlying molecular mechanisms of a variety of diseases 7, 8. More recently, proteomics has shifted its focus from the global characterization of protein expression to investigations of the spatial and temporal organization of the proteome, for instance, using targeted proteomics 9. However, challenges still exist as the proteome is dynamic and condition-dependent, leading to overwhelming complexity and heterogeneity of the abundance of proteins and their post-translational modifications (PTMs). The latter has been shown to mediate a wide variety of cellular processes, such as signal transduction and subcellular localization 10, significantly expanding the size of predicted human proteome (10-100 times) 11.
Efficient pretreatment methods are desired prior to MS analysis to reduce sample complexity and obtain deep proteome coverage. Nanostructured devices and nanoparticles (NPs) have unique characteristics, such as extremely small size, good biocompatibility, abundant active affinity sites, and large specific surface area, making them especially suitable for proteomic applications. For instance, magnetic nanocomposites have been used to purify proteins, significantly reducing the processing time. Mesoporous NPs, due to the adjustable pore size and large specific surface area, have been explored in size-based separation and membrane proteomics 12, 13. From the technical point of view of material science, several recent review articles have summarized the manufacturing of NPs and their applications in biomedical science 14. In this review, we intend to fill the gap from a proteomics perspective by focusing on the most pressing challenges currently facing MS-based proteomics (Figure 1). We illustrate how recent nanotechnological innovations, mainly the studies that were published within five years, have aided in addressing these challenges. We specifically detail significant advances in precision medicine development and provide our vision of the future for nanotechnology-enabled proteomics.
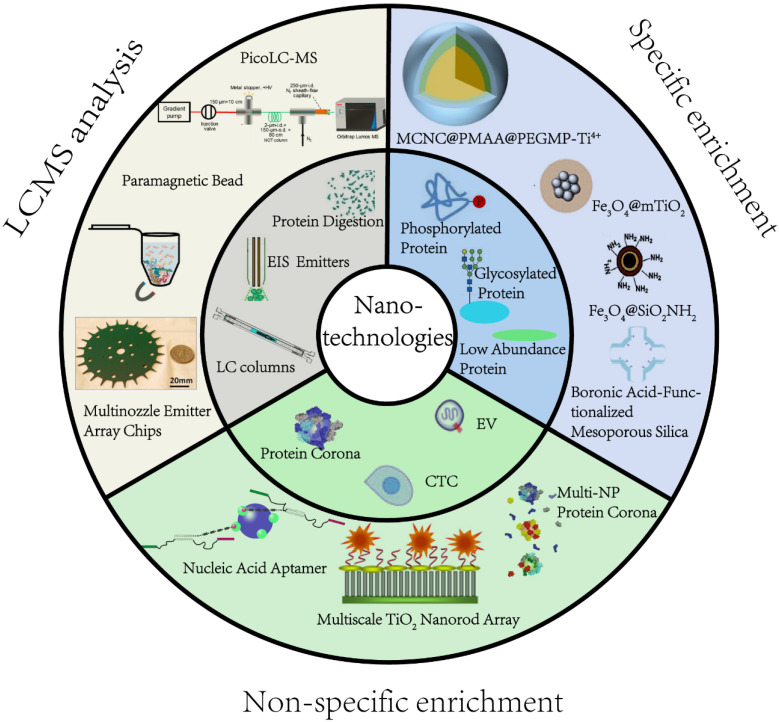
Figure 1.
An overview of application of nanomaterials in proteomics-driven precision medicine (PDPM).
2. Nanotechnology-enabled targeted and non-targeted proteome analysis
Proteins can be analyzed on MS with full-length and intact forms, called top-down proteomics, or peptides after proteolytic digestion, also known as bottom-up or shotgun proteomics 15. Either way, proteins of interest must be isolated from the complex background before introducing into MS. For instance, protein acetylation is known to play a key role in regulating chromatin accessibility and gene transcription (e.g., histones) as well as protein localization and enzymatic activity (e.g., non-histone proteins) 16. To analyze protein acetylation using proteomic approaches, antibody-based enrichment is usually performed to separate acetylated proteins or peptides from non-acetylated matrix 17. Hence, signal suppression can be minimized or avoided during MS acquisition. Such targeted enrichment processes can significantly enhance the chance of identifying important but low-abundance modified proteins. Some NPs (e.g., iron and titanium oxide) have unique physiochemical properties and can specifically bind proteins with certain PTMs, such as phosphorylation and glycosylation, facilitating the identification of these events. On the other hand, other NPs can non-selectively bind proteins when they encounter biospecimens 18. Such non-targeted enrichment offers an interesting alternative to global proteome profiling. In the following sections, we summarize recent developments in these areas.
2.1 Specific enrichment of protein PTMs
Protein modifications play essential roles in a wide range of physiological and pathological processes 19. Information on protein abundance together with PTM alterations has been shown to uncover disease markers 20. From a purely technical perspective, the major challenge to study post-translationally modified proteins by MS is their low abundance 21. Here, we focus on the two most widely studied PTMs, phosphorylation and glycosylation.
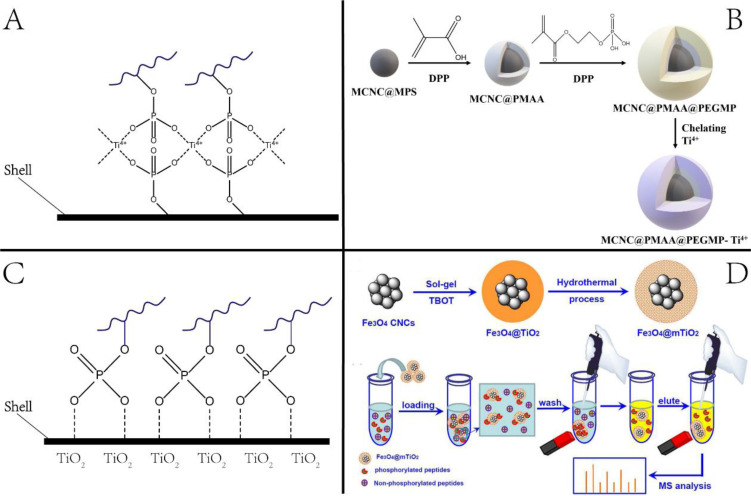
2.1.1 Phosphorylation is recognized as one of the most common PTMs. It regulates protein function and cell signaling and plays a central role in disease-causing aberrations of kinase signaling networks 22. However, proteomic analysis of phosphorylation remains challenging due to its low stoichiometry. Magnetic NPs, such as immobilized metal ion affinity chromatography (IMAC) and metal oxide affinity chromatography (MOAC), have been widely used for phosphoproteomic studies 23. For the IMAC approach, metal ions, such as Al3+, Ga3+, Fe3+, Ti4+, Zr4+, are functionalized onto NPs. Since the metal ions are typical Lewis acids under acidic conditions, they can interact with phosphate groups through bridged two-dentate chelation and then capture the phosphopeptides. The principle of enrichment and a representative example are illustrated in Figure 2. The enrichment condition is usually adjusted to pH 2-3 to neutralize the acidic residues for avoiding non-specific adsorption of non-phosphopeptides with multiple acidic residues onto the IMAC or MOAC.
Figure 2.
Nanotechnologies for enriching phosphopeptides. (A, B) Principle and example of phosphopeptides enrichment by Immobilized metal ion affinity chromatography (IMAC) method. (C, D) Principle and example of phosphopeptides enrichment by metal oxide affinity chromatography (MOAC) method. Adapted with permission from Ref. 27. Copyright 2012 American Chemical Society.]
A large class of IMAC materials has been developed by varying the nature of immobilized metal and ligands to afford abundant metal sites 24. We previously synthesized Ti4+-immobilized magnetic composite microspheres using a high-magnetic-response magnetic colloid nanocrystal cluster (MCNC) core, a poly(methyl acrylic acid) (PMAA) interim layer, and a Ti4+-immobilized poly(ethylene glycol methacrylate phosphate) (PEGMP) shell (Figure 2) 25. Due to the pure phosphate-Ti4+ interface and high Ti4+ loading amount, the resulting MCNC@PMAA@PEGMP-Ti4+ composite microspheres demonstrated rapid (< 5 min) and efficient (phosphopeptide/nonphosphopeptide = 1:500) separation of phosphopeptides 25. As for the MOAC material, the main strategy for its synthesis was in situ growth of metal oxides on magnetic nanoparticles/microspheres 26. Porous structured MOAC was designed to improve the enrichment efficiency, as it has a large specific surface area and can offer abundant reaction sites for anchoring phosphopeptides 27. Except for TiO2, other nanomaterials with titanium have been reported that can be used to enrich phosphopeptides, but their application is limited, and is not discussed herein.
Application of the enrichment strategy to biomedical research, especially for biomarker discovery, is one of the most promising areas of the proteomics field. Jiang and coworkers recently applied titanium dioxide (TiO2) beads to enrich phosphopeptides from 110 paired tumor and non-tumor tissues of early-stage hepatocellular carcinoma (HCC). Nearly 30,000 phosphorylation sites were identified in total, and hyper-phosphorylation of signaling pathways in HCC was revealed 28. Chen et al. used Ti4+-functionalized dendrimer to enrich phosphopeptides from human plasma-derived extracellular vesicles. Among the over 10,000 unique phosphopeptides, they reported 144 phosphoproteins significantly high in breast cancer patients 29. These findings suggested that nanomaterial-based enrichment strategies can identify additional disease biomarkers compared to conventional shotgun proteomics.
2.1.2 Glycosylation is another important and most structurally complicated protein modification 30. Depending on the linkage sites, there are two common types of protein glycosylation, N-glycosylation, where glycans are attached to asparagines via N-acetylglucosamine residues, and O-glycosylation, where the glycans are attached to serine or threonine through acyl linkages. The cancer glycoproteome varies at different levels, including alterations in the glycosylation sites and/or glycan chain composition, offering tremendous potential for targeted therapeutics against glycosylation 31. There is growing evidence that cancer cells have a unique glycosylation profile, and alterations in glycosylation are regarded as a hallmark of cancer 32. Moreover, studies have shown that protein glycosylation can significantly improve the clinical value of classical biomarkers 33.
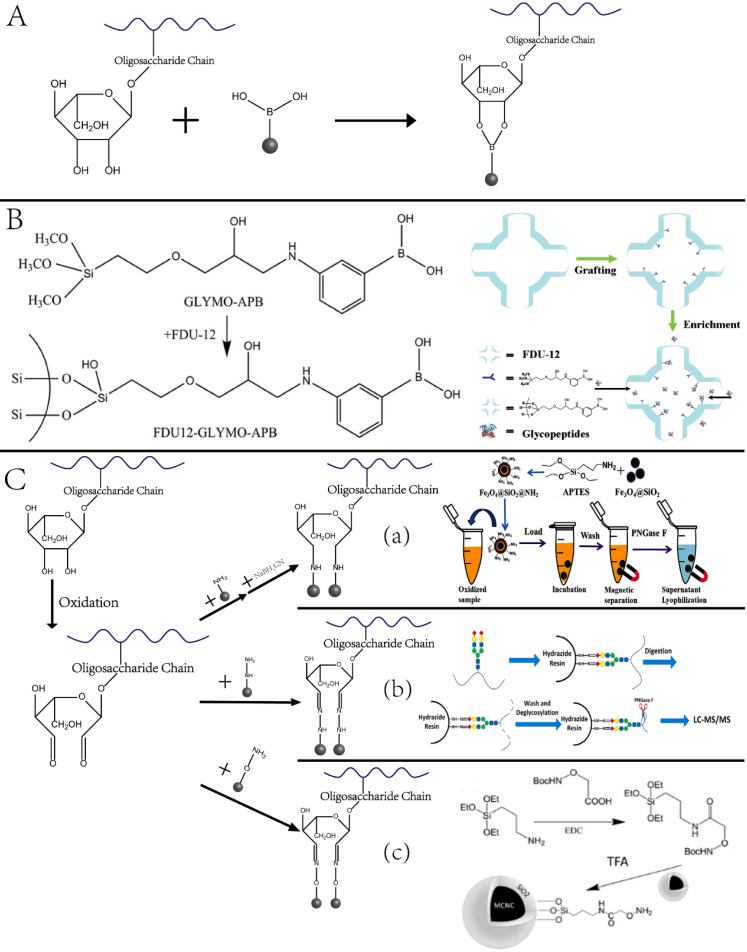
Glycosylation enrichment using nanomaterials is based on affinity-based and covalent binding-based enrichment. In affinity-based methods, lectin is usually immobilized onto the surface of NPs, and glycopeptides are enriched by hydrophilic interaction between the particles and sugar chains. In the method based on covalent interaction, glycopeptides are mainly enriched by the interaction between the boronic acid and the cis-diol-containing glycoproteins/glycopeptides 34 or the interaction between aldehydes produced by the oxidation of sugar chains and hydrazine, hydroxylamine, and other functional groups on the surface of solid materials 35-37. The principle and representative examples are displayed in Figure 3. As summarized in a recent review article, a variety of functional materials have been investigated, including magnetic and mesoporous materials, metal frame compounds, graphene, and dendrimers 38.
Figure 3.
Nanotechnologies for enriching glycoproteins. (A, B) Principle and example of glycoproteins enrichment by boric acid functionalized materials. Adapted with permission from Ref. 34. Copyright 2009 American Chemical Society.] (C) Aldhyde-based covalent bond formation assisted enrichment methods. a) Amine-functionalized nanoparticles. [Adapted with permission from Ref. 35. Copyright 2013 American Chemical Society]. b) Hydrazide Chemistry. [Adapted with permission from Ref. 36 Copyright 2014 American Chemical Society] c) Oxime Click Chemistry. [Adapted with permission from Ref.37. Copyright 2014 American Chemical Society]
Our group designed and synthesized new aminooxy-functionalized magnetic nanoparticles using oxime click reaction-based enrichment 37. The oxidized glycan chains on the glycopeptides could conjugate with aminooxy groups through oxime click reaction, retaining the glycopeptides on the magnetic NPs. The oxime click chemistry-based method renders excellent enrichment performance within one hour, and the magnetic core brings the advantage of the fast separation of glycopeptide with good reproducibility. Notably, because cis-diol-containing glycoproteins/glycopeptides can form stable and reversible covalent bonds with boronate by adjusting pH, functionalized nano-systems using boronic acid have been established to enrich glycoproteins. A typical example is the fabrication of dendrimeric boronic acid-functionalized magnetic nanoparticles for enriching glycoproteins from a complex sample 39. Due to the dendrimer-assisted multivalent synergistic binding, the boronate avidity material exhibited more than three orders of magnitude higher dissociation constants than the affinities of single boronic acid binding towards glycoproteins, leading to significantly efficient enrichment of glycoproteins 36.
In this context, several recent glycoproteomic studies have reported novel glycoproteins that could serve as noninvasive biomarkers. From a cohort of 74 aggressive and 68 non-aggressive prostate cancer patients, a glycoproteomic study proposed a three-glycoprotein panel (ACPP, CLU, and PSA) that could distinguish between aggressive and non-aggressive prostate cancers with an AUC of 0.86 40. Pan et al. investigated the role of protein glycosylation in high-grade serous ovarian carcinoma (HGSC) by extracting glycoproteins from 119 TCGA HGSC tissues using two independent approaches, solid-phase extraction of glycoside-containing peptides (SPEG) and intact glycopeptides for investigating glycoside-specific glycans (IGPs). The study discovered that glycosylation with specific glycol-form features was associated with disease progression and severity, indicating that a deep understanding of the proteome glycosylation might provide important clues for precision medicine 41.
2.1.3 Other PTMs. Cysteine is prone to post-translational modifications because of its high reactivity, including affinity and redox sensitivity. Common PTMs on cysteine include redox-dependent nitrosylation (SNO), lipid-derived electrophilic 4-hydroxynonenal (HNEs), and isoprene lipid modifications. For example, a novel approach based on the fluorous solid-phase extraction of SNO-peptides using nanographite fluoride was developed to analyze the SNO-proteome. In this strategy, a fluorous tag, which could increase the ionization efficiency of SNO-peptides and facilitate the following enrichment, was introduced into the SNO modification sites. Subsequently, the fluorescence-tagged SNO-peptides were captured by nanographite fluoride specifically through fluorous-fluorous interactions. Taking advantage of the highly fluorinated level and the high surface area of nanographite fluoride, the enrichment approach was shown to have remarkable selectivity, good sensitivity, high post-enrichment recovery, and large enrichment capacity 42, 43.
2.2 Specific enrichment of low abundance proteins
Blood is one of the most common body fluids used for laboratory-based analyses. It contains a wide range of molecules, such as electrolytes, small molecules, drugs, and proteins, routinely tested in clinical settings. Although extremely challenging, MS-based proteomics is an unbiased way to identify novel blood biomarkers. The dynamic range of the blood proteome is more than ten orders of magnitude, and the top 10 proteins with the highest abundance account for nearly 90% of the total protein content 8. Therefore, without depletion, fractionation, and/or enrichment, only a few hundred proteins could be identified by LC-MS/MS 8, 44; hence, nanomaterials have found wide applications in blood proteomics. Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) is a classical method based on antibody-immobilized magnetic nanomaterials to capture specific protein biomarkers from blood 45. Recent developments of the SISCAPA include automation 46, cost-effectiveness 47, and quantitation 48, 49. The latter included the recently developed targeted proteomics approach, multiple reaction monitoring (MRM), and demonstrated a lower incidence of false-negative findings in the early detection of hepatocellular carcinoma 49.
Another important application of specific enrichment of blood proteins using NPs is the recently designed surface-functionalized superparamagnetic iron-oxide (magnetite, Fe3O4), which specifically enriched a well-established cardiac biomarker troponin I (cTnI). The study analyzed cTnl using the top-down approach and demonstrated an association between molecular fingerprints of diverse cTnI forms and pathophysiology 50.
2.3 Non-specific enrichment of proteins and cells
While specific enrichment of proteins of interest has shown great selectivity and broad application in biomedical research, emerging non-specific enrichment approaches with NPs offer interesting alternatives to conventional global and targeted proteomic analysis.
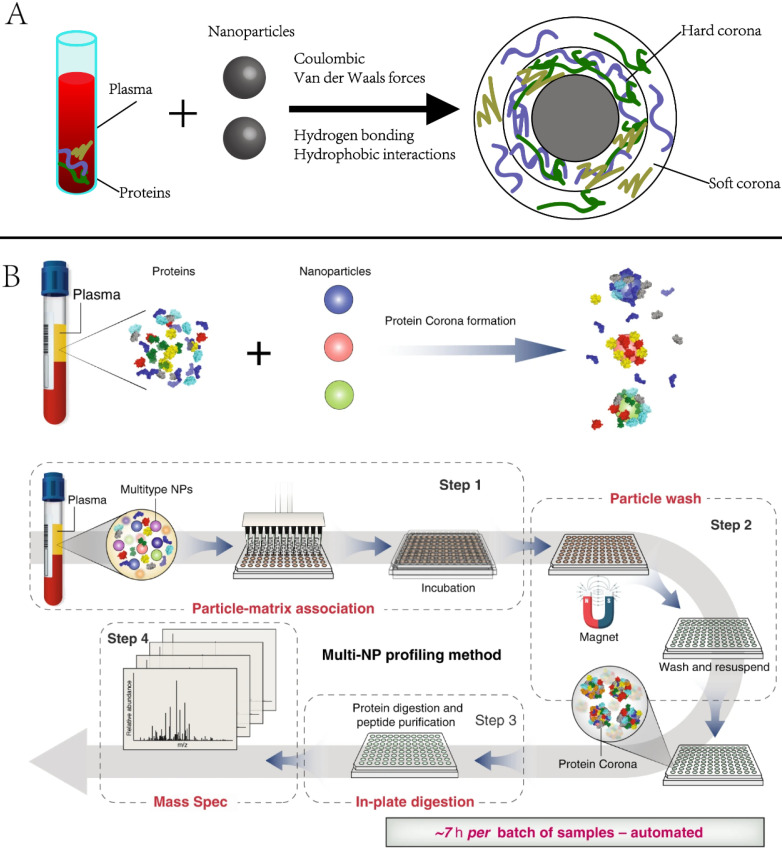
2.3.1 Protein corona. Protein corona is the group of proteins that are spontaneously and non-specifically adsorbed to NP surfaces when they encounter biospecimens or are introduced to the physiological environment 51, 52. It alters the physicochemical properties of NPs and eventually affects their circulation, bio-distribution, and toxicity in living systems. Numerous studies have investigated the physicochemical properties (e.g., size, shape, and coating) of NPs in the context of in vivo imaging, drug delivery, or targeted therapies 53-56. However, the protein corona has not been often used for biomarker discovery. Recently, Blume et al. reported comprehensive profiling of the protein corona pulldown by NPs, representing a subset of the plasma proteome. 53. As the protein corona varies with NP properties, combining multiple different types of engineered NPs is expected to capture distinct protein corona patterns, enabling deep profiling of plasma proteome (Figure 4). The investigators screened 43 types of magnetic nanoparticles and demonstrated that using a panel of 10 could achieve in-depth plasma proteome profiling across more than seven orders of magnitude, including the identification of 53 FDA-approved protein biomarkers in a single pooled plasma. They further applied the panel to profile the plasma proteome of early-stage non-small-cell lung cancer (NSCLC) patients and age- and gender-matched healthy controls. A multi-protein classifier was identified to distinguish NSCLC patients from healthy controls with a high average area under the curve (AUC) of 0.91
Figure 4.
Principle and example of using nanoparticle protein corona in protein biomarker discovery. Adapted with permission from Ref. 53 Copyright 2020 Spring Nature.
Although proteins are the major components of the NP corona, recent studies revealed that non-protein entities, such as saccharides and lipids, could also be detected in the corona 57. Therefore, multiomics investigation of the NP corona would enable a better understanding of its composition and biological identity, eventually enabling precise NP designs and the development of safe nanomedicines.
2.3.2 Circulating tumor cells (CTCs). CTCs are shed into the circulation from primary or metastatic tumors. Documented evidence has suggested that detection of CTCs in peripheral blood offers a unique opportunity to identify prognostic markers for various cancers and better understand cancer metastasis and tumor heterogeneity 58, 59. The major challenge for CTC detection is their extreme paucity (1-10 cells per 10 ml peripheral blood) 59. Conventional protein-based technologies utilize antibodies to detect specific CTC markers. Immuno-magnetic beads (IMB) represent another promising alternative. Nano-sized beads have a higher specific surface area and thus higher cell separation efficiency than micron beads but may cause accumulation after high-speed centrifugation. Non-magnetic beads, such as TiO2 nanorods (160-300 nm diameter) on the F-doped SnO2 (FTO) substrate have also been examined 60. It is noteworthy that surface proteins on CTCs may change during the circulation, giving false-negative results. Furthermore, CTCs and normal cells have been shown to have different adhesion properties on rough surfaces. Therefore, a recent study attempted to fabricate glass surfaces with different roughness using ion etching, enabling enrichment of CTCs because of their higher adhesion to the surface than regular cells 61.
Recent studies also investigated the size-based separation of CTCs. The surface bombardment of polymer materials (e.g., polycarbonate film and polyp-xylene film) by micromachining can form nanometer to micron size holes. Since the size of CTCs is usually around 20 μm, they can be effectively separated by microfiltration, for instance, using 6-10 μm pore size membranes 62. However, uneven distribution of the pore size limits the capture efficiency of CTCs to only 50-60%. Yeh et al. managed to overcome the problem by fabricating a tandem flexible micro spring array (tFMSA) with micro mold. The resulting micro spring structure could separate CTCs from normal cells based on the size difference and deformability, thus increasing the CTC capture efficiency to 90%. Moreover, the study showed 86% of captured species were CTCs by measuring the marker of mesenchymal cells, vimentin, highlighting the advantage of microfilter separation 63. Other types of materials, such as graphene oxide (GO) and spiral microfluidics, have also been demonstrated to isolate CTCs from the blood 61, 62. In summary, NP-based enrichment of CTCs avoids potential losses of CTCs due to non-specific binding to antibody-coated beads and the resulting cytotoxicity. Thus, compared to traditional immunoaffinity-based and size-based filtration methods, NP-based CTC enrichment appears to have a broader application in biomedical research.
2.3.3 Extracellular vesicles. Nearly all cell types secrete lipid bilayer membrane-encapsulated vesicles ranging in diameter from 30 to 300 nm, referred to as extracellular vesicles (EVs) 64. Besides specific membrane lipids, EVs carry DNA and various RNAs, including miRNAs and proteins. EVs have diverse functions in immune tolerance 65, possess the ability to either promote or suppress cancer and promote the transfer of antimicrobial resistance proteins and genes 66, 67. Similar to the CTCs, EVs can be isolated by immunoaffinity-based methods using aptamers, short single-stranded nucleic acid sequences that function like antibodies but are economical, stable, and easy-to-modify, as demonstrated by recent EV separation studies 68, 69.
EVs can also be isolated by chelation. For instance, Sun et al. developed bifunctional magnetic beads (BiMBs) with a hydrophilic phosphate head for Ti4+ binding. The synergistic effect of Ti4+ with phosphate groups and 1,2-distearoyl-sn-glycero-3-phosphorylethanolamine enabled BiMBs to capture EVs from urine samples efficiently. Compared to conventional methods, BiMBs showed a much higher enrichment efficiency and lower sample consumption 70. Also, EVs could be separated free of antibodies based on their size by centrifugation. However, ultracentrifugation is time-consuming and requires expensive equipment. Studies that incorporated nano-membranes with different pore sizes during centrifugation 71 and utilized tangential flow filtration (TFF) technology solved the problem to some extent 72. Nevertheless, an inherent drawback of the size-based separation methods is that the isolated EVs may contain impurities of similar size. Therefore, the size-based separation techniques are often employed with other methods to achieve better separation results.
3. Nanotechnology and nanofabrication facilitate LC-MS/MS analysis
3.1 NP-assisted protein digestion
Digestion of biological samples is an important processing step upstream of the shotgun proteomic analysis for biomarker discovery and subsequent clinical development 73. Nanomaterials and/or microparticles are useful in facilitating proteomic sample digestion, especially for low-abundance proteins and those from protein-protein interaction complexes 74, 75. Several studies have investigated the on-bead digestion methodology in which the enriched proteins from cell lysate are digested directly on the magnetic 76, 77, chromatographic 78, or agarose beads 79. Hughes et al. developed a single-pot solid phase-enhanced sample-preparation technology, termed SP3, which utilizes interactions (e.g., hydrophilic, electrostatic repulsion) between proteins and the carboxyl or amine group of functionalized paramagnetic beads 74. Proteins can be non-selectively captured onto the beads, and after simple rinse and wash, are digested directly with minimal sample loss. Such one-pot, solid-phase-based methods tend to have fewer liquid handling steps and, therefore, are faster and more efficient than conventional gel-based and in-solution processing. This approach has identified novel interacting proteins and mechanisms to further our understanding of pathogenesis, such as in protein-misfolding disease and tumor regression 79, 80. Recently, this technique was implemented on a liquid handling robot for automated processing of samples in a 96-well format 81 and showed high sensitivity and good reproducibility in analyzing serum/plasma 82, FFPE tissues 83, and cell lines 84.
Immobilized enzyme reactor (IMER) has also been employed extensively to pre-treat samples for proteomic profiling 85. Generally, it uses a capillary column as a packing bed for NPs and immobilizes proteolytic enzymes via chemical coupling. Recently, covalent organic frameworks have been reported as a platform for enzyme immobilization 86. As the immobilization process reduces diffusion distance and increases the local enzyme concentration, the IMER significantly shortens the digestion time from hours to minutes and speeds up the entire proteomic analysis pipeline 85, 87. Fan et al. recently synthesized a thermo-responsive magnetic fluid (TMF) to expand the IMER application beyond classical tryptic digestion by immobilizing trypsin and the PNGase enzyme, enabling highly rapid and efficient deglycosylation 88.
Another interesting application of NPs for proteomics sample treatment is the proteome reactor. NPs with different chromatographic properties, for instance, reversed-phase, ion exchange, are immobilized or packed into pipette tips, which could be used directly for cell lysis, protein digestion, and peptide fractionation 89-91. Applications of such stop-and-go-extraction tips (StageTips) include analyses of global proteome 89, glycoproteome 91, interaction proteome 92, and phospho-proteome 93.
3.2 Electrospray ionization emitters
Liquid chromatography-MS (LC-MS) is central to any proteomic investigation 94, and electrospray (ESI) is the interface between the two systems, introducing biomolecules into the MS 95. A variety of ESI emitters of different shapes (e.g., tapered vs. non-tapered) and sizes (e.g., wide vs. narrow openings) have been developed, aimed at maximizing the transferring efficiency 96, 97. However, emitters are prone to clogging, especially for nanoESI. Another method, nanoflow LC that typically flows at 50-500 ml/min, has routinely been used for protein identification due to its high ionization efficiency and hence better sensitivity than ESI-MS 73. However, it lacks robustness and throughput for large-scale and complex proteome analysis 98. In contrast, microflow LC-MS with a flow rate between 1-50 μl/min usually shows robustness but at the cost of ionization efficiency and sensitivity 99. The microfabricated monolithic multi-nozzle emitter is a design that splits the microflow into multiple nanoflows, and, therefore, offers advantages of both microLC and nanospray in one LC-MS run, thereby boosting ionization stability and efficiency 100.
3.3 NanoLC columns
The separation efficiency of LC is one of the determining factors for the overall proteomic analysis. Although microcapillary columns have been mostly used, studies have shown that smaller inner diameters (i.d.) (from 75um to 30 um) could increase protein identification by 95% 101, 102. Microfabricated pillar array columns 102 and porous-layer open tubular columns 103 have been demonstrated to improve separation efficiency and increase proteome coverage for low-input samples. Recently, Xiang et al. reported that a 2-um i.d. narrow open tubular column running at picoflow (< 800 pl/min) could identify nearly 1,000 proteins from less than 100 pg tryptic digests 102. Also, Wang et al. reported a segmented microfluidic method to pack ultralong up to 5 m capillary columns 104. In contrast to the packed-bed capillary columns, a micro-pillar array column (μPAC) has recently been demonstrated to be more efficient than particle-packed columns, achieving up to one million theoretical plates at specific conditions 105. The μPAC leads to much lower back pressure, contributes to higher peak capacity, and identifies a greater number of peptides and proteins from complex samples.
4. Conclusions and perspectives
The focus of this article was to summarize recent advances in nanotechnology-enabled proteome analysis and its applications to precision medicine development. Important aspects, such as the design and preparation of nanoparticles, the versatility of functionalization methods, and enrichment principles and efficiency, were discussed. Some of the proteomic applications by coupling various nano-techniques to mass spectrometry are summarized in Table 1. Due to space limitations, regrettably, other similar studies could not be included in the present review.
Table 1.
Summary of the nanotechnologies in PDPM discussed in this review.
| Nanotechnology | Target | Type of technology and material | Application | Main findings | Ref. | |
|---|---|---|---|---|---|---|
| Specific enrichment | Phosphorylated protein | Immobilized metal ion affinity chromatography (IMAC) | Ti4+ -immobilized magnetic composite microspheres | Milk and Human Serum | Efficient separation phosphopeptide/nonphosphopeptide = 1:500) | 25 |
| Metal oxide affinity chromatography (MOAC) | Titanium dioxide (TiO2) beads | Liver proteins | Nearly 30,000 phosphorylation sites and several hyper-phosphorylation of signaling pathways in HCC | 28 | ||
| Ti4+-functionalized dendrimer | Plasma Microvesicles and Exosomes | 144 phosphoproteins among the over 10,000 unique phosphopeptides are significantly higher in patients diagnosed with breast cancer compared with healthy controls | 29 | |||
| Glycosylated protein | Magnetic nanomaterials | Aminooxy-functionalized magnetic nanoparticle | Asialofetuin from fetal calf serum and myoglobin from horse heart | Highly efficient separation of N-glycoproteins with excellent sensitivity and be able to effectively analyze a small sample | 37 | |
| Boronic acid functionalized nano systems | Dendrimeric boronic acid-functionalized magnetic nanoparticles | Human saliva | Exhibited a strong avidity towards glycoproteins, which was 3-4 orders of magnitude higher than the conventional boronate affinity of a single binding. | 39 | ||
| SNO-proteome | Fluorous solid-phase extraction | nanographite fluoride | Human Umbilical Vein Endothelial Cell (HUVEC) | Better selectivity, lower limit of detection, and higher post enrichment recovery as well as large enrichment capacity. | 42, 43 | |
| Low abundance protein | Antibody-immobilized magnetic nanomaterial | Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) | Human plasma, human mammary epithelial cell line | Can enrich specific peptides from a mixture, the antibody-coupled beads can be reused consistently for up to 10 times. | 45-49 | |
| Magnetic nanomaterials | Surface-functionalized superparamagnetic iron-oxide (magnetite, Fe3O4) | Human plasma | Specifically enrich cardiac troponin I (cTnI), a well-established cardiac biomarker | 50 | ||
| Non-specific enrichment | Protein corona | Magnetic nanomaterials | 43 types of magnetic nanoparticles | Human plasma | 10 of them can achieve efficient plasma proteome profiling across more than seven orders of magnitude, including the identification of 53 FDA-approved protein biomarkers in a single pooled plasma. | 53 |
| Circulating tumor cells (CTCs) | Non-magnetic nanomaterials | TiO2 nanorods (160-300 nm diameter) on F-doped SnO2 (FTO) substrate | Artificial blood samples | Can effectively enhance the capture performance of target cancer cells even in a low cell density situation. | 60 | |
| Nanomaterials with different size | Tandem flexible micro spring array (tFMSA) | Human blood | Increasing the CTCs capture efficiency to 90% | 63 | ||
| Extracellular Vesicles | Magnetic nanomaterials | Bifunctional magnetic beads (BiMBs) | Urine | Higher enrichment efficiency and lower sample consumption | 70 | |
| Tangential flow filtration (TFF) | nano-membranes | Undiluted Serum | Operating pressures are orders-of-magnitude lower than membranes with conventional thicknesses (1-10 µm). Captured particles are associated with a surface, rather than trapped in a bulk-matrix. Higher efficiency of capture and release of particles. | 72 | ||
| LCMS analysis | Protein digestion | Magnetic, chromatographic or agarose beads | Functionalized paramagnetic beads | Drosophila embryo samples | Presented a novel protocol using paramagnetic beads, Single-Pot Solid-Phase-enhanced Sample Preparation (SP3). | 74 |
| Electrospray ionization emitters | Nanoflow LC | Microfabricated monolithic multinozzle emitter | Low-volume whole blood samples | Achieved a detection limit of less than 5 red blood cells | 100 | |
| LC columns | Microfabricated pillar array columns | A 2-um i.d. narrow open tubular column running at picoflow (< 800 pl/min) | Tryptic peptides | Provide nearly 1,000 protein identifications from only less than 100 pg tryptic digests | 102 | |
| Micro-pillar array column (μPAC) | Tryptic digest of a mixture of seven proteins with diverse mass and isoelectric point | High flow rates (600 nl/min) and higher protein identification rates. | 105 | |||
As discussed above, nanomaterials have distinct advantages over conventional assays, such as good biocompatibility, abundant active affinity sites, large specific surface area, and unique surface properties, providing a combination of important features for proteomic analysis. Currently, enrichment of low abundance proteins, protein modifications, and miniaturized devices for sample preparation are major areas that could benefit from the application of NPs. Although most enrichment processes are carried out in vitro, in the future, the development of in vivo enrichment technologies, linking the spatial location and functional information of proteins, and providing more accurate information for precision medicine development can be envisioned. With the emerging roles of protein modifications in human diseases 106-108, nanotechnologies with new types of materials, functionalization methods, and reagent generation will continue to play key roles in identifying and enriching modified proteins. Moreover, single-cell proteomics is emerging as a promising tool to uncover the heterogeneity between cells and identify new cell subtypes 109. However, single-cell proteomics faces extreme challenges for in-depth global analysis and investigation of their modifications. The unique surface properties and large specific surface area of nanomaterials to capture the proteome in single-cell samples with high efficiency offer advantages for developing sensitive, simple, and rapid detection methods necessary for proteomics-driven precision medicine.
Acknowledgments
This work is supported by National Key Research and Development Program of China (2020YFE0202200), National Science Foundation of China (Grants (21974026, 22174021 and 21775053), the innovative research team of high-level local university in Shanghai.
References
- 1.Ashley EA. Towards precision medicine. Nat Rev Genet. 2016;17:507–22. doi: 10.1038/nrg.2016.86. [DOI] [PubMed] [Google Scholar]
- 2.Joyner MJ, Paneth N. Promises, promises, and precision medicine. J Clin Invest. 2019;129:946–8. doi: 10.1172/JCI126119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruiz C, Zitnik M, Leskovec J. Identification of disease treatment mechanisms through the multiscale interactome. Nat Commun. 2021;12:1796. doi: 10.1038/s41467-021-21770-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timp W, Timp G. Beyond mass spectrometry, the next step in proteomics. Sci Adv. 2020;6:eaax8978. doi: 10.1126/sciadv.aax8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roehrl MH, Roehrl VB, Wang JY. Proteome-based pathology: the next frontier in precision medicine. Expert Rev Precis Med Drug Dev. 2021;6:1–4. doi: 10.1080/23808993.2021.1854611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doll S, Gnad F, Mann M. The case for proteomics and phospho-proteomics in personalized cancer medicine. Proteomics Clin Appl. 2019;13:e1800113. doi: 10.1002/prca.201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacMullan MA, Dunn ZS, Graham N, Yang L, Wang P. Quantitative proteomics and metabolomics reveal biomarkers of disease as potential immunotherapy targets and indicators of therapeutic efficacy. Theranostics. 2019;9:7872–88. doi: 10.7150/thno.37373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geyer PE, Holdt LM, Teupser D, Mann M. Revisiting biomarker discovery by plasma proteomics. Mol Syst Biol. 2017;13:942. doi: 10.15252/msb.20156297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uzozie AC, Aebersold R. Advancing translational research and precision medicine with targeted proteomics. J Proteomics. 2018;189:1–10. doi: 10.1016/j.jprot.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 10.Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol Cell Proteomics. 2013;12:3444–52. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES. et al. How many human proteoforms are there? Nat Chem Biol. 2018;14:206–14. doi: 10.1038/nchembio.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, Jian R, Wang Y, Yang B, Zhao D, Shen C. et al. Mesoporous silica as sorbents and enzymatic nanoreactors for microbial membrane proteomics. ACS Appl Mater Interfaces. 2021;13:11571–8. doi: 10.1021/acsami.0c19528. [DOI] [PubMed] [Google Scholar]
- 13.Yao J, Sun N, Deng C. Recent advances in mesoporous materials for sample preparation in proteomics research. TrAC Trends in Anal Chem. 2018;99:88–100. [Google Scholar]
- 14.Sun N, Wu H, Shen X, Deng C. Nanomaterials in proteomics. Adv Funct Mater. 2019;29:1900253. [Google Scholar]
- 15.Chait BT. Mass spectrometry: bottom-up or top-down? Science. 2006;314:65–6. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 16.Spange S, Wagner T, Heinzel T, Krämer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–98. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 17.Korc M. Diabetes mellitus in the era of proteomics. Mol Cell Proteomics. 2003;2:399–404. doi: 10.1074/mcp.R300005-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Fredolini C, Pathak KV, Paris L, Chapple KM, Tsantilas KA, Rosenow M. et al. Shotgun proteomics coupled to nanoparticle-based biomarker enrichment reveals a novel panel of extracellular matrix proteins as candidate serum protein biomarkers for early-stage breast cancer detection. Breast Cancer Res. 2020;22:135. doi: 10.1186/s13058-020-01373-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karve TM, Cheema AK. Small Changes Huge Impact: The role of protein posttranslational modifications in cellular homeostasis and disease. J Amino Acids. 2011;2011:207691. doi: 10.4061/2011/207691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mnatsakanyan R, Shema G, Basik M, Batist G, Borchers CH, Sickmann A. et al. Detecting post-translational modification signatures as potential biomarkers in clinical mass spectrometry. Expert Rev Proteomics. 2018;15:515–35. doi: 10.1080/14789450.2018.1483340. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Zhang C, Jiang H, Yang P, Lu H. Fishing the PTM proteome with chemical approaches using functional solid phases. Chemical Society Reviews. 2015;44:8260–87. doi: 10.1039/c4cs00529e. [DOI] [PubMed] [Google Scholar]
- 22.Yang W, Freeman MR, Kyprianou N. Personalization of prostate cancer therapy through phosphoproteomics. Nat Rev Urol. 2018;15:483–97. doi: 10.1038/s41585-018-0014-0. [DOI] [PubMed] [Google Scholar]
- 23.Ochoa D, Jarnuczak AF, Viéitez C, Gehre M, Soucheray M, Mateus A. et al. The functional landscape of the human phosphoproteome. Nat Biotechnol. 2020;38:365–73. doi: 10.1038/s41587-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Y, Deng C. Recent advances in nanomaterials for sample pre-treatment in phosphoproteomics research. TrAC Trends in Anal Chem. 2019;120:115655. [Google Scholar]
- 25.Ma W, Zhang Y, Li L, Zhang Y, Yu M, Guo J. et al. Ti4+-immobilized magnetic composite microspheres for highly selective enrichment of phosphopeptides. Adv Funct Mater. 2013;23:107–15. [Google Scholar]
- 26.Luo B, Yu L, He J, Li Z, Lan F, Wu Y. Magnetic polymer nanomaterials for sample pretreatment in proteomics. Materials Advances. 2021;2:2200–15. [Google Scholar]
- 27.Ma WF, Zhang Y, Li LL, You LJ, Zhang P, Zhang YT. et al. Tailor-made magnetic Fe3O4@mTiO2 microspheres with a tunable mesoporous anatase shell for highly selective and effective enrichment of phosphopeptides. ACS Nano. 2012;6:3179–88. doi: 10.1021/nn3009646. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X. et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567:257–61. doi: 10.1038/s41586-019-0987-8. [DOI] [PubMed] [Google Scholar]
- 29.Chen I-H, Xue L, Hsu C-C, Paez JSP, Pan L, Andaluz H. et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci U S A. 2017;114:3175–80. doi: 10.1073/pnas.1618088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schjoldager KT, Narimatsu Y, Joshi HJ, Clausen H. Global view of human protein glycosylation pathways and functions. Nat Rev Mol Cell Biol. 2020;21:729–49. doi: 10.1038/s41580-020-00294-x. [DOI] [PubMed] [Google Scholar]
- 31.Fernandes E, Sores J, Cotton S, Peixoto A, Ferreira D, Freitas R. et al. Esophageal, gastric and colorectal cancers: Looking beyond classical serological biomarkers towards glycoproteomics-assisted precision oncology. Theranostics. 2020;10:4903–28. doi: 10.7150/thno.42480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in health and disease. Nat Rev Nephrol. 2019;15:346–66. doi: 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llop E, P EG, Duran A, Barrabés S, Massaguer A, José Ferri M. et al. Glycoprotein biomarkers for the detection of pancreatic ductal adenocarcinoma. World J Gastroenterol. 2018;24:2537–54. doi: 10.3748/wjg.v24.i24.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Wu Z, Zhang L, Lu H, Yang P, Webley PA. et al. Highly specific enrichment of glycopeptides using boronic acid-functionalized mesoporous silica. Anal Chem. 2009;81:503–8. doi: 10.1021/ac801912t. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Kuang M, Zhang L, Yang P, Lu H. An accessible protocol for solid-phase extraction of N-linked glycopeptides through reductive amination by amine-functionalized magnetic nanoparticles. Anal Chem. 2013;85:5535–41. doi: 10.1021/ac400733y. [DOI] [PubMed] [Google Scholar]
- 36.Zhu J, Sun Z, Cheng K, Chen R, Ye M, Xu B. et al. Comprehensive mapping of protein N-glycosylation in human liver by combining hydrophilic interaction chromatography and hydrazide chemistry. J Proteome Res. 2014;13:1713–21. doi: 10.1021/pr401200h. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Yu M, Zhang C, Ma W, Zhang Y, Wang C. et al. Highly selective and ultra fast solid-phase extraction of N-glycoproteome by oxime click chemistry using aminooxy-functionalized magnetic nanoparticles. Anal Chem. 2014;86:7920–4. doi: 10.1021/ac5018666. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Fang C, Bao H, Yuan W, Lu H. Discover the post-translational modification proteome using mass spectrometry. Chin J Chem. 2021;39:550–8. [Google Scholar]
- 39.Wang H, Bie Z, Lü C, Liu Z. Magnetic nanoparticles with dendrimer-assisted boronate avidity for the selective enrichment of trace glycoproteins. Chem Sci. 2013;4:4298–303. [Google Scholar]
- 40.Dong M, Lih TM, Chen SY, Cho KC, Eguez RV, Höti N. et al. Urinary glycoproteins associated with aggressive prostate cancer. Theranostics. 2020;10:11892–907. doi: 10.7150/thno.47066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan J, Hu Y, Sun S, Chen L, Schnaubelt M, Clark D. et al. Glycoproteomics-based signatures for tumor subtyping and clinical outcome prediction of high-grade serous ovarian cancer. Nat Commun. 2020;11:6139. doi: 10.1038/s41467-020-19976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C, Tao T, Yuan W, Zhang L, Zhang X, Yao J. et al. Fluorous solid-phase extraction technique based on nanographite fluoride. Anal Chem. 2017;89:4566–72. doi: 10.1021/acs.analchem.6b05071. [DOI] [PubMed] [Google Scholar]
- 43.Zhang C, Xu Y, Wang G, Fang C, Bao H, Zhang Y. et al. FluoroTRAQ: quantitative analysis of protein S-nitrosylation through fluorous solid-phase extraction combining with iTRAQ by mass spectrometry. Anal Chem. 2020;92:15317–22. doi: 10.1021/acs.analchem.0c01706. [DOI] [PubMed] [Google Scholar]
- 44.Bhawal R, Oberg AL, Zhang S, Kohli M. Challenges and opportunities in clinical applications of blood-based proteomics in cancer. Cancers (Basel) 2020. 12. [DOI] [PMC free article] [PubMed]
- 45.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using stable isotope standards and capture by anti-peptide antibodies (SISCAPA) J Proteome Res. 2004;3:235–44. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 46.Neubert H, Shuford CM, Olah TV, Garofolo F, Schultz GA, Jones BR. et al. Protein biomarker quantification by immunoaffinity liquid chromatography-tandem mass spectrometry: current state and future vision. Clin Chem. 2020;66:282–301. doi: 10.1093/clinchem/hvz022. [DOI] [PubMed] [Google Scholar]
- 47.Zhao L, Whiteaker JR, Voytovich UJ, Ivey RG, Paulovich AG. Antibody-coupled magnetic beads can be reused in immuno-MRM assays to reduce cost and extend antibody supply. J Proteome Res. 2015;14:4425–31. doi: 10.1021/acs.jproteome.5b00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X, Naughton SX, Han Z, He M, Zheng YG, Terry AV. et al. Mass spectrometric quantitation of tubulin acetylation from pepsin-digested rat brain tissue using a novel stable-isotope standard and capture by anti-peptide antibody (SISCAPA) method. Anal Chem. 2018;90:2155–63. doi: 10.1021/acs.analchem.7b04484. [DOI] [PubMed] [Google Scholar]
- 49.Kim H, Sohn A, Yeo I, Yu SJ, Yoon JH, Kim Y. Clinical assay for AFP-L3 by using multiple reaction monitoring-mass spectrometry for diagnosing hepatocellular carcinoma. Clin Chem. 2018;64:1230–8. doi: 10.1373/clinchem.2018.289702. [DOI] [PubMed] [Google Scholar]
- 50.Tiambeng TN, Roberts DS, Brown KA, Zhu Y, Chen B, Wu Z. et al. Nanoproteomics enables proteoform-resolved analysis of low-abundance proteins in human serum. Nat Commun. 2020;11:3903. doi: 10.1038/s41467-020-17643-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monopoli MP, Aberg C, Salvati A, Dawson KA. Biomolecular coronas provide the biological identity of nanosized materials. Nat Nano. 2012;7:779–86. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 52.Docter D, Westmeier D, Markiewicz M, Stolte S, Knauer SK, Stauber RH. The nanoparticle biomolecule corona: lessons learned - challenge accepted? Chem Soc Rev. 2015. [DOI] [PubMed]
- 53.Blume JE, Manning WC, Troiano G, Hornburg D, Figa M, Hesterberg L. et al. Rapid, deep and precise profiling of the plasma proteome with multi-nanoparticle protein corona. Nat Commun. 2020;11:3662. doi: 10.1038/s41467-020-17033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corbo C, Molinaro R, Tabatabaei M, Farokhzad OC, Mahmoudi M. Personalized protein corona on nanoparticles and its clinical implications. Biomater Sci. 2017;5:378–87. doi: 10.1039/c6bm00921b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tenzer S, Docter D, Rosfa S, Wlodarski A, Kuharev J, Rekik A. et al. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS Nano. 2011;5:7155–67. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- 56.Wheeler KE, Chetwynd AJ, Fahy KM, Hong BS, Tochihuitl JA, Foster LA. et al. Environmental dimensions of the protein corona. Nat Nanotechnol. 2021;16:617–29. doi: 10.1038/s41565-021-00924-1. [DOI] [PubMed] [Google Scholar]
- 57.Phue WH, Bahadi M, Dynes JJ, Wang J, Kuppili VSC, Ismail A. et al. Protein-biomolecule interactions play a major role in shaping corona proteome: studies on milk interacted dietary particles. Nanoscale. 2021;13:13353–67. doi: 10.1039/d1nr03712a. [DOI] [PubMed] [Google Scholar]
- 58.Zhang L, Riethdorf S, Wu G, Wang T, Yang K, Peng G. et al. Meta-analysis of the prognostic value of circulating tumor cells in breast cancer. Clin Cancer Res. 2012;18:5701–10. doi: 10.1158/1078-0432.CCR-12-1587. [DOI] [PubMed] [Google Scholar]
- 59.Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14:623–31. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 60.Sun N, Li X, Wang Z, Zhang R, Wang J, Wang K. et al. A multiscale TiO2 nanorod array for ultrasensitive capture of circulating tumor cells. ACS Appl Mater Interfaces. 2016;8:12638–43. doi: 10.1021/acsami.6b02178. [DOI] [PubMed] [Google Scholar]
- 61.Chen W, Weng S, Zhang F, Allen S, Li X, Bao L. et al. Nanoroughened surfaces for efficient capture of circulating tumor cells without using capture antibodies. ACS Nano. 2013;7:566–75. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song Y, Tian T, Shi Y, Liu W, Zou Y, Khajvand T. et al. Enrichment and single-cell analysis of circulating tumor cells. Chem Sci. 2017;8:1736–51. doi: 10.1039/c6sc04671a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bankó P, Lee SY, Nagygyörgy V, Zrínyi M, Chae CH, Cho DH. et al. Technologies for circulating tumor cell separation from whole blood. J Hematol Oncol. 2019;12:48. doi: 10.1186/s13045-019-0735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fleshner M, Crane CR. Exosomes, DAMPs and miRNA: features of stress physiology and immune homeostasis. Trends Immunol. 2017;38:768–76. doi: 10.1016/j.it.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karlsson M, Lundin S, Dahlgren U, Kahu H, Pettersson I, Telemo E. "Tolerosomes" are produced by intestinal epithelial cells. Eur J Immunol. 2001;31:2892–900. doi: 10.1002/1521-4141(2001010)31:10<2892::aid-immu2892>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 66.Lee J, Lee E-Y, Kim S-H, Kim D-K, Park K-S, Kim KP. et al. Staphylococcus aureus extracellular vesicles carry biologically active β-lactamase. Antimicrob Agents Chemother. 2013;57:2589–95. doi: 10.1128/AAC.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rumbo C, Fernández-Moreira E, Merino M, Poza M, Mendez JA, Soares NC. et al. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55:3084–90. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tran PHL, Xiang D, Nguyen TNG, Tran TTD, Chen Q, Yin W. et al. Aptamer-guided extracellular vesicle theranostics in oncology. Theranostics. 2020;10:3849–66. doi: 10.7150/thno.39706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang T, Chen C, Larcher LM, Barrero RA, Veedu RN. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol Adv. 2019;37:28–50. doi: 10.1016/j.biotechadv.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 70.Sun J, Han S, Ma L, Zhang H, Zhan Z, Aguilar HA. et al. Synergistically bifunctional paramagnetic separation enables efficient isolation of urine extracellular vesicles and downstream phosphoproteomic analysis. ACS Appl Mater Interfaces. 2021;13:3622–30. doi: 10.1021/acsami.0c19400. [DOI] [PubMed] [Google Scholar]
- 71.Zhang M, Jin K, Gao L, Zhang Z, Li F, Zhou F. et al. Methods and technologies for exosome isolation and characterization. Small Methods. 2018;2:1800021. [Google Scholar]
- 72.Dehghani M, Lucas K, Flax J, McGrath J, Gaborski T. Tangential flow microfluidics for the capture and release of nanoparticles and extracellular vesicles on conventional and ultrathin membranes. Adv Mater Technol. 2019;4:1900539. doi: 10.1002/admt.201900539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, Fonslow BR, Shan B, Baek M-C, Yates JR. Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013;113:2343–94. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hughes CS, Foehr S, Garfield DA, Furlong EE, Steinmetz LM, Krijgsveld J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol. 2014;10:757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turriziani B, Garcia-Munoz A, Pilkington R, Raso C, Kolch W, von Kriegsheim A. On-beads digestion in conjunction with data-dependent mass spectrometry: a shortcut to quantitative and dynamic interaction proteomics. Biology (Basel) 2014;3:320–32. doi: 10.3390/biology3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hughes CS, Moggridge S, Müller T, Sorensen PH, Morin GB, Krijgsveld J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat Protoc. 2019;14:68–85. doi: 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 77.Batth TS, Tollenaere MX, Rüther P, Gonzalez-Franquesa A, Prabhakar BS, Bekker-Jensen S. et al. Protein aggregation capture on microparticles enables multipurpose proteomics sample preparation. Mol Cell Proteomics. 2019;18:1027–35. doi: 10.1074/mcp.TIR118.001270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou H, Wang F, Wang Y, Ning Z, Hou W, Wright TG. et al. Improved recovery and identification of membrane proteins from rat hepatic cells using a centrifugal proteomic reactor. Mol Cell Proteomics. 2011;10:O111.008425. doi: 10.1074/mcp.O111.008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Serrels A, Lund T, Serrels B, Byron A, McPherson Rhoanne C, von Kriegsheim A. et al. Nuclear FAK controls chemokine transcription, tregs, and evasion of anti-tumor immunity. Cell. 2015;163:160–73. doi: 10.1016/j.cell.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mali GR, Yeyati PL, Mizuno S, Dodd DO, Tennant PA, Keighren MA. et al. ZMYND10 functions in a chaperone relay during axonemal dynein assembly. eLife. 2018;7:e34389. doi: 10.7554/eLife.34389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Müller T, Kalxdorf M, Longuespée R, Kazdal DN, Stenzinger A, Krijgsveld J. Automated sample preparation with SP3 for low-input clinical proteomics. Mol Syst Biol. 2020;16:e9111. doi: 10.15252/msb.20199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Messner CB, Demichev V, Wendisch D, Michalick L, White M, Freiwald A. et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020;11:11–24.e4. doi: 10.1016/j.cels.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Friedrich C, Schallenberg S, Kirchner M, Ziehm M, Niquet S, Haji M. et al. Comprehensive micro-scaled proteome and phosphoproteome characterization of archived retrospective cancer repositories. Nat Commun. 2021;12:3576. doi: 10.1038/s41467-021-23855-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruprecht B, Di Bernardo J, Wang Z, Mo X, Ursu O, Christopher M. et al. A mass spectrometry-based proteome map of drug action in lung cancer cell lines. Nat Chem Biol. 2020;16:1111–9. doi: 10.1038/s41589-020-0572-3. [DOI] [PubMed] [Google Scholar]
- 85.Wouters B, Currivan SA, Abdulhussain N, Hankemeier T, Schoenmakers PJ. Immobilized-enzyme reactors integrated into analytical platforms: Recent advances and challenges. TrAC Trends in Anal Chem. 2021;144:116419. [Google Scholar]
- 86.Zhong C, Ma W, He Y, Ouyang D, Li G, Yang Y. et al. Controllable synthesis of hollow microtubular covalent organic frameworks as an enzyme-immobilized platform for enhancing catalytic activity. ACS Appl Mater Interfaces. 2021;13:52417–24. doi: 10.1021/acsami.1c16386. [DOI] [PubMed] [Google Scholar]
- 87.Olsen C, Skottvoll FS, Brandtzaeg OK, Schnaars C, Rongved P, Lundanes E, Investigating monoliths (Vinyl Azlactone-co-Ethylene Dimethacrylate) as a support for enzymes and drugs, for proteomics and drug-target studies. Front Chem. 2019. 7. [DOI] [PMC free article] [PubMed]
- 88.Fan Z, Liu T, Zheng F, Qin W, Qian X. An ultrafast N-glycoproteome analysis method using thermoresponsive magnetic fluid-immobilized enzymes. Front Chem. 2021. 9. [DOI] [PMC free article] [PubMed]
- 89.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–24. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 90.Chen W, Wang S, Adhikari S, Deng Z, Wang L, Chen L. et al. Simple and integrated spintip-based technology applied for deep proteome profiling. Anal Chem. 2016;88:4864–71. doi: 10.1021/acs.analchem.6b00631. [DOI] [PubMed] [Google Scholar]
- 91.Chen Y-J, Yen T-C, Lin Y-H, Chen Y-L, Khoo K-H, Chen Y-J. ZIC-cHILIC-based stageTip for simultaneous glycopeptide enrichment and fractionation toward large-scale N-sialoglycoproteomics. Anal Chem. 2021;93:15931–40. doi: 10.1021/acs.analchem.1c03224. [DOI] [PubMed] [Google Scholar]
- 92.Mao Y, Chen P, Ke M, Chen X, Ji S, Chen W. et al. Fully integrated and multiplexed sample preparation technology for sensitive interactome profiling. Anal Chem. 2021;93:3026–34. doi: 10.1021/acs.analchem.0c05076. [DOI] [PubMed] [Google Scholar]
- 93.Chen W, Chen L, Tian R. An integrated strategy for highly sensitive phosphoproteome analysis from low micrograms of protein samples. Analyst. 2018;143:3693–701. doi: 10.1039/c8an00792f. [DOI] [PubMed] [Google Scholar]
- 94.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–55. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 95.Wilm M. Principles of electrospray ionization. Mol Cell Proteomics. 2011;10:M111.009407. doi: 10.1074/mcp.M111.009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu X, Oleschuk RD, Cann NM. Characterization of microstructured fibre emitters: in pursuit of improved nano electrospray ionization performance. Analyst. 2012;137:4150–61. doi: 10.1039/c2an35249d. [DOI] [PubMed] [Google Scholar]
- 97.Gibson GTT, Mugo SM, Oleschuk RD. Nanoelectrospray emitters: trends and perspective. Mass Spectrom Rev. 2009;28:918–36. doi: 10.1002/mas.20248. [DOI] [PubMed] [Google Scholar]
- 98.Bian Y, Zheng R, Bayer FP, Wong C, Chang YC, Meng C. et al. Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC-MS/MS. Nat Commun. 2020;11:157. doi: 10.1038/s41467-019-13973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bian Y, The M, Giansanti P, Mergner J, Zheng R, Wilhelm M. et al. Identification of 7 000-9 000 proteins from cell lines and tissues by single-shot microflow LC-MS/MS. Anal Chem. 2021;93:8687–92. doi: 10.1021/acs.analchem.1c00738. [DOI] [PubMed] [Google Scholar]
- 100.Mao P, Gomez-Sjoberg R, Wang D. Multinozzle emitter array chips for small-volume proteomics. Anal Chem. 2013;85:816–9. doi: 10.1021/ac3032965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu Y, Zhao R, Piehowski PD, Moore RJ, Lim S, Orphan VJ. et al. Subnanogram proteomics: impact of LC column selection, MS instrumentation and data analysis strategy on proteome coverage for trace samples. Int J Mass Spectrom. 2017;427:4–10. doi: 10.1016/j.ijms.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xiang P, Zhu Y, Yang Y, Zhao Z, Williams SM, Moore RJ. et al. Picoflow liquid chromatography-mass spectrometry for ultrasensitive bottom-up proteomics using 2-μm-i.d. open tubular columns. Anal Chem. 2020;92:4711–5. doi: 10.1021/acs.analchem.9b05639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li S, Plouffe BD, Belov AM, Ray S, Wang X, Murthy SK. et al. An integrated platform for isolation, processing, and mass spectrometry-based proteomic profiling of rare cells in whole blood. Mol Cell Proteomics. 2015;14:1672–83. doi: 10.1074/mcp.M114.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang X, Zhu J, Yang C, Qin F, Zhang B. Segmented microfluidics-based packing technology for chromatographic columns. Anal Chem. 2021;93:8450–8. doi: 10.1021/acs.analchem.1c00545. [DOI] [PubMed] [Google Scholar]
- 105.Tóth G, Panić-Janković T, Mitulović G. Pillar array columns for peptide separations in nanoscale reversed-phase chromatography. J Chromatogr A. 2019;1603:426–32. doi: 10.1016/j.chroma.2019.06.067. [DOI] [PubMed] [Google Scholar]
- 106.Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y. et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574:575–80. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Delaney K, Tan M, Zhu Z, Gao J, Dai L, Kim S. et al. Histone lysine methacrylation is a dynamic post-translational modification regulated by HAT1 and SIRT2. Cell Discov. 2021;7:122. doi: 10.1038/s41421-021-00344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang H, Zhang D, Weng Y, Delaney K, Tang Z, Yan C. et al. The regulatory enzymes and protein substrates for the lysine β-hydroxybutyrylation pathway. Sci Adv. 2021;7:eabe2771. doi: 10.1126/sciadv.abe2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Slavov N. Driving single cell proteomics forward with innovation. J Proteome Res. 2021;20:4915–8. doi: 10.1021/acs.jproteome.1c00639. [DOI] [PMC free article] [PubMed] [Google Scholar]