Abstract
The current outbreak of coronavirus disease-2019 (COVID-19) caused by SARS-CoV-2 poses unparalleled challenges to global public health. SARS-CoV-2 is a Betacoronavirus, one of four genera belonging to the Coronaviridae subfamily Orthocoronavirinae. Coronaviridae, in turn, are members of the order Nidovirales, a group of enveloped, positive-stranded RNA viruses. Here we present a systematic phylogenetic and evolutionary study based on protein domain architecture, encompassing the entire proteomes of all Orthocoronavirinae, as well as other Nidovirales. This analysis has revealed that the genomic evolution of Nidovirales is associated with extensive gains and losses of protein domains. In Orthocoronavirinae, the sections of the genomes that show the largest divergence in protein domains are found in the proteins encoded in the amino-terminal end of the polyprotein (PP1ab), the spike protein (S), and many of the accessory proteins. The diversity among the accessory proteins is particularly striking, as each subgenus possesses a set of accessory proteins that is almost entirely specific to that subgenus. The only notable exception to this is ORF3b, which is present and orthologous over all Alphacoronaviruses. In contrast, the membrane protein (M), envelope small membrane protein (E), nucleoprotein (N), as well as proteins encoded in the central and carboxy-terminal end of PP1ab (such as the 3C-like protease, RNA-dependent RNA polymerase, and Helicase) show stable domain architectures across all Orthocoronavirinae. This comprehensive analysis of the Coronaviridae domain architecture has important implication for efforts to develop broadly cross-protective coronavirus vaccines.
Keywords: Nidovirales, Coronaviridae, Orthocoronavirinae, Evolution, Phylogenetics, Phylogenomics, Protein domains, Genome, Hidden Markov models
1. Introduction
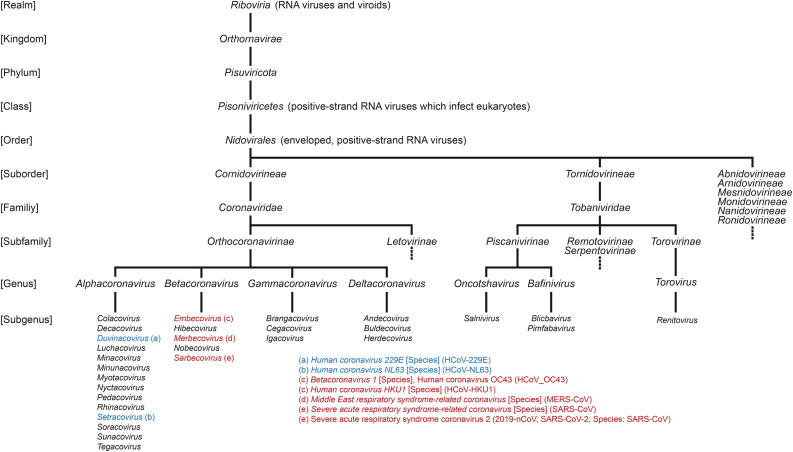
Coronaviridae is a family of enveloped, positive-strand RNA viruses that infect a wide variety of animals. The Coronaviridae family belongs to the suborder Cornidovirineae, which, together with Tornidovirineae belong to the order Nidovirales (enveloped, positive-strand RNA viruses) (Fig. 1 ). Recent phylogenetic studies based on RNA-directed RNA polymerases indicate that Nidovirales, together with Picornavirales, Caliciviridae, Astroviridae, and their relatives form a distinct supergroup of RNA viruses (Picornavirus supergroup) (Koonin et al., 2020; Wolf et al., 2018). Nidovirales can infect a wide range of animal hosts, including insects, mollusks, crustaceans, and vertebrates, suggesting horizontal virus transfer across metazoan species (Dolja and Koonin, 2020). Coronaviridae are divided into two subfamilies Letovirinae and Orthocoronavirinae, the latter of which are the main focus of this work. Orthocoronavirinae in turn are divided into four genera, Alpha-, Beta-, Gamma, and Deltacoronaviruses. Currently, there are seven Orthocoronavirinae species or sub-species, which have been found to infect humans, two members of the Alphacoronavirus genus: Human coronavirus 229E and Human coronavirus NL63, and five members of the Betacoronavirus genus: Human coronavirus OC43, Human coronavirus HKU1, Middle East respiratory syndrome-related coronavirus (MERS-CoV), Severe acute respiratory syndrome coronavirus (SARS-CoV), and Severe acute respiratory syndrome coronavirus 2 (2019-nCoV, SARS-CoV-2 (Andersen et al., 2020; Drosten et al., 2003; Fan et al., 2019; Fehr and Perlman, 2015).
Fig. 1.
Nidovirales taxonomy. This figure is based on the taxonomy established by the International Committee on Taxonomy of Viruses (ICTV) and currently used by the U.S. National Center for Biotechnology Information (NCBI) and the Universal Protein Resource (UniProt) databases. Viruses which infect humans are listed in blue (Alphacoronaviruses) and red (Betacoronaviruses). Their taxonomic level is indicated in square brackets. For some viruses, no taxonomic level has been established as of this writing. An example of this is Human coronavirus OC43.
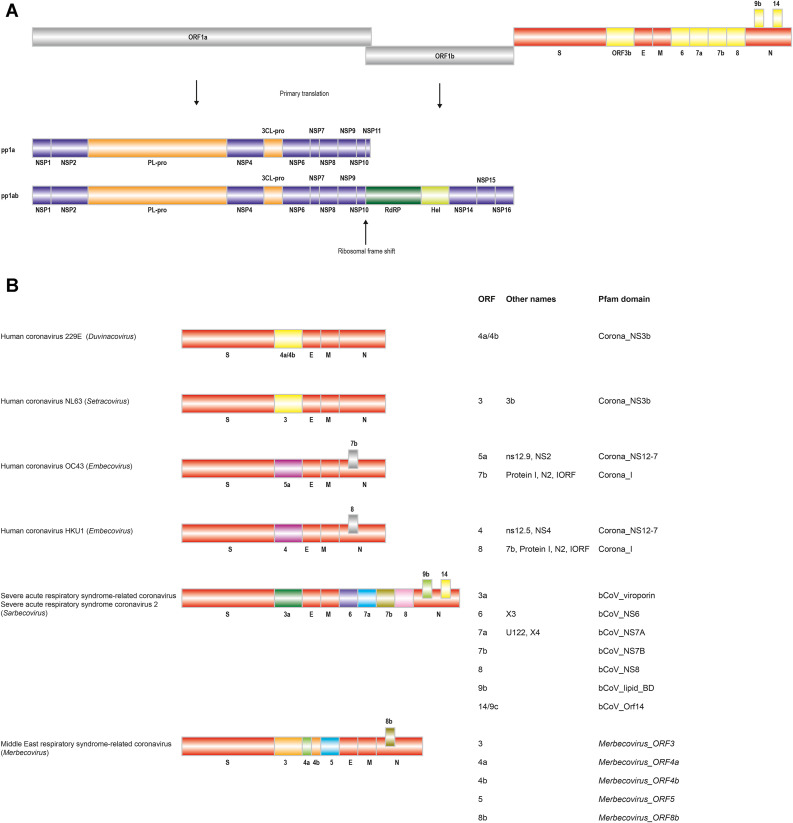
All Orthocoronavirinae viruses possess four shared structural proteins, the spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins. The genome is packed inside a helical capsid formed by the nucleoprotein N. This in turn is surrounded by an envelope containing the E and M proteins, which are involved in virus assembly, and the spike glycoprotein protein S, which mediates virus entry into host cells (McBride and Fielding, 2012). Orthocoronavirinae have relatively large viral genomes in comparison to other RNA viruses, with sizes ranging from 26 to 32 kilobases. The first two open reading frames, ORF1a and ORF1b, code for two overlapping large replicase-containing polyproteins, pp1a and pp1ab, with the larger pp1ab translated as a result of a -1 ribosomal frameshifting (Fig. 2 A). These large polyproteins are subsequently (self) cleaved into 15 or 16 mature proteins referred to as non-structural proteins (nsps). And while the PP1ab, S, E, M, and N proteins are found in all Coronaviridae family genomes, the individual protein domains show surprising diversity. In addition, depending on the specific strain, many coronaviruses contain additional ORFs coding for accessory proteins, many of which remain poorly characterized (Fig. 2B).
Fig. 2.
Coronaviridae genome organization. SARS-CoV-2 genome organization. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome is shown as an example of the Orthocoronavirinae genome organization. The abbreviations used are: pp: polyprotein, PL-pro: Papain-like protease, 3CL-pro: Cysteine protease, RdRP: RNA dependent RNA polymerase, Hel: Helicase, S: Spike protein, E: Envelope protein, M: Membrane protein, N: Nucleocapsid protein. Genome organization of human Orthocoronavirinae accessory proteins. The ORF-based names are shown, together with additional names and the corresponding Pfam domain. Note that the ORF-based names do not always match across taxonomic groups. For example, ORF5a in OC43 appears to be an ortholog of ORF4 in HKU1 given their conserved Pfam domain architecture. For two of the Merbecovirus accessory proteins for which no Pfam model exists, new hidden Markov models were developed (see Methods). These are labelled in italic fonts.
In this work, we performed a protein domain-centric evolutionary comparative genomics analysis of Coronaviridae genomes, revealing the complex domain architectures that have resulted from recombination and a complicated evolutionary history.
Homologs are genes that are related by shared ancestry. Orthologs were defined by Fitch in 1970 as homologous genes in different species that diverged by speciation. Genes that diverged by gene duplication, either in the same or different species, have been termed paralogs (Fitch, 1970, 2000). While the terms ortholog and paralog have no functional implications (Jensen, 2001), orthologs are often thought of as more functionally similar than paralogs at the same level of sequence divergence (Altenhoff et al., 2012; Eisen, 1998).
Protein domains are distinct functional and/or structural units of a protein. Domains tend to form stable compact three-dimensional structures that can often be independently folded. Many proteins are composed of multiple domains, with each domain having its own evolutionary history and biochemical function. Thus, the architecture of a protein is a product of the ordered arrangement of its constituent domains and their overall tertiary structure. During evolution, multiple domains can combine, creating a vast number of distinct domain combinations, even within the same species (Moore et al., 2008). Assembling multiple domains into a single protein creates an entity whose function can be more than the sum of its constituent parts. The generation of proteins with novel combinations of duplicated and then diverged domains is a major mechanism for rapid evolution of new functionality in genomes (Itoh et al., 2007; Peisajovich et al., 2010). This modular structure of proteins enables rapid emergence of a multitude of novel protein functions from an initially limited array of functional domains. Proteins can gain or lose domains via genome rearrangements; the domains themselves can be modified by small-scale mutations (Christian M. Zmasek and Godzik, 2012).
Here we use the Domain-architecture Aware Inference of Orthologs (DAIO) approach described in (Zmasek et al., 2019) to compare the arrangement of protein domains (and by extension, proteins) in polyproteins and ORFs from different Orthocoronavirinae sub-genera, updating and expanding our knowledge of Nidovirales genome evolution at the domain level, which, for example, has been review previously in (Gorbalenya et al., 2006). This approach places proteins into groups in which all members are not only orthologous to each other but also have the exact same domain architecture. This analysis resulted in the classification of Coronaviridae proteins into “Strict Ortholog Groups” (SOGs), in which all proteins are orthologous to each other (related by speciation events) and exhibit the same domain architecture. The SOG classification also enabled the development of an informative naming convention for each SOG that includes information about the protein's function (if known) and a suffix indicating the taxonomic group (such as Betacoronavirus) where a particular SOG is present. The SOG classification results are publicly available through the Virus Pathogen Resource (ViPR) (Pickett et al., 2012) at https://www.viprbrc.org.
2. Results and discussion
2.1. Nidovirales genome evolution: protein domain composition of extant and ancestral genomes
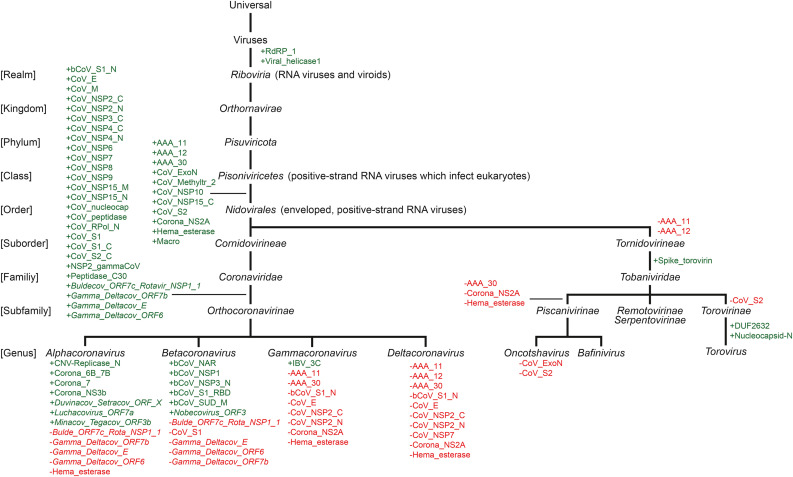
We analyzed complete sets of proteins for all publicly available Nidovirales genomes (for a total of roughly one million sequences, including ∼900,000 for SARS-CoV-2) for the presence of protein domains as defined by Pfam 34.0 (March 2021, 19179 entries) (El-Gebali et al., 2018). Within Orthocoronavirinae, the number of distinct protein domains varied from 9 in poorly studied viruses such as the White-eye coronavirus HKU16 (Deltacoronavirus) to 44 in SARS-CoV-2. Most Nidovirales not belonging to Orthocoronavirinae have even poorer coverage in Pfam. To understand the evolutionary history of the observed domain distribution in extant species, we reconstructed the domain content of ancestral genomes, specifically those lying at internal nodes corresponding to major branching points in the evolution of Nidovirales. Since independent evolution of the same domain more than once is highly unlikely, we used Dollo parsimony (https://doi.org/10.1093/sysbio/26.1.77), which, when applied to domain content, assumes that each domain can be gained only once and seeks to minimize domain losses, to reconstruct the Pfam domain repertoire of Nidovirales (Zmasek and Godzik, 2011) (see Fig. 3 and Supplementary Tables 1 and 2). It should be noted that these findings do not imply that every extant genome retained all domains gained on its path from its respective ancestor. In fact, similar to the situation in eukaryote evolution, domain losses are common (Zmasek and Godzik, 2011).
Fig. 3.
Domain gains and losses during Nidovirales evolution. Gained Pfam domains are shown in green, whereas lost domains are shown in red, as inferred by Dollo parsimony. For members of suborder Tornidovirineae only select examples are shown (due to limited genome and Pfam HMM data availability). Data for subgenera is not shown. Detailed lists of gained and lost domain are available in Supplementary Tables 1 and 2.
The two most ancestral proteins are strongly associated with the realm of Ribovira (RNA viruses and viroids): RNA-dependent RNA polymerase (RdRP_1) and viral RNA helicase (Viral_helicase1). RNA-dependent RNA polymerase (RdRp) is an essential protein encoded in the genomes RNA viruses which catalyzes synthesis of the RNA strand complementary to a given RNA template (De et al., 1996). Viral RNA helicase is a member of the P-loop containing nucleoside triphosphate hydrolase superfamily and has multiple roles at different stages of viral RNA replication (Koonin et al., 1993).
11 domains are associated with the evolution of Nidovirales from Pisoniviricetes (positive-strand RNA viruses which infect eukaryotes). Four of these are domains are also found in eukaryotes and bacteria. These are the AAA domains AAA_11, AAA_12, and AAA_13 as well as the Macro domain. AAA_11, AAA_12, AAA_30 are members of the P-loop containing nucleoside triphosphate hydrolase superfamily (same as the viral RNA helicase mentioned above), which often perform chaperone-like functions that assist in the assembly, operation, or disassembly of protein complexes (Neuwald et al., 1999). In Orthocoronavirinae, these domains are part of the RNA helicase (Gomez de Cedro è et al., n.d.). In Orthocoronavirinae, the Macro domain is part of the papain-like peptidase protein (PL-pro), together with domains CoV_peptidase and NSP3_C (as well as other, genus-specific domains). The Hema_esterase domain appears both in some Nidovirales (Embecovirus and Torovirus) as well as in Herpesvirales (dsDNA viruses) and Influenza C and D viruses. Together with the Hema_HEF domain, this domain is part of the Haemagglutinin-esterase fusion glycoprotein found in Embecoviruses. It has been speculated that Haemagglutinin-esterases have been acquired from viral host lectins, although it is unclear whether this acquisition happened in a putative ancestral virus followed by speciation and gene loss or by multiple independent acquisitions (Chen and Li, 2013). The Corona_NS2A domain is found in various Riboviria, although from the genomes studied in this work it is present in only select Orthocoronavirinae genomes. The Corona_NS2A domain can be found in Rotaviruses (double-stranded RNA viruses in the family Reoviridae), where together with the Rotavirus_VP3 domain, they form a multifunctional enzyme, the VP3 protein, involved in mRNA capping (Zhang et al., 2013). In Berne Virus (Tornidovirineae, Torovirinae) Corona_NS2A is found encoded in the polyprotein (pp1ab). Interestingly, in Embecovirus and Luchacovirus it is encoded as an individual ORF. The remaining five domains are unique to Nidovirales and are found in (some) Orthocoronavirinae as well as in (some) Tobaniviridae. These are the proofreading exoribonuclease (CoV_ExoN), the 2′-O-methyltransferase (CoV_Methyltr_2), the RNA synthesis protein NSP10 (CoV_NSP10), the uridylate-specific endoribonuclease (CoV_NSP15_C), and the S2 subunit of the Spike protein (CoV_S2).
The main finding from this analysis is that, during Coronaviridae evolution, the largest number of domain gains (26) occurred on the branch leading from Nidovirales to Orthocoronavirinae. These domain gains include the small envelope protein E (with CoV_E domain), the matrix/glycoprotein M (CoV_M), nucleocapsid N (CoV_nucleocap), and three domains of the spike glycoprotein (bCoV_S1_N, CoV_S1_C, and CoV_S2_C). These gains also include numerous domains encoded within the polyproteins Pp1a and Pp1ab, namely the CoV_peptidase and CoV_NSP3_C domains, which are part of the papain-like peptidase (PL-pro), domain Peptidase_C30, which is the single domain of the 3C-like proteinase (3CL-pro), the N-terminal domain of the RNA-dependent RNA polymerase RdRP (CoV_RPol_N), as well as NSP2 (CoV_NSP2_C and CoV_NSP2_N domains), NSP4, NSP6, NSP7, NSP9, and two domains of NSP15 (CoV_NSP15_N and CoV_NSP15_M). A more detailed analysis of protein domain changes in the Pp1ab polyprotein and the spike glycoproteins in the Orthocoronavirinae family is provided below.
Besides the domains and proteins discussed above, the distribution of which can be best explained with (ancestral) gains and subsequent loss, there are also several domains present in Orthocoronavirinae most likely resulting from horizontal gene transfer or recombination (due to them being present in only a few Orthocoronavirinae genomes as well as being present in very distantly related species). Orthoreo_P10 (Orthoreovirus membrane fusion protein p10) is thought to be a multifunctional protein that plays a key role in virus-host interaction (Bodeló et al., 2002) and is currently only found in Rousettus bat coronavirus GCCDC1 (Nobecovirus), as well as in some Spinareovirinae genomes and in various Eukaryotes. PRK (Phosphoribulokinase/Uridine kinase family) is found in Cegacovirus species as well as in numerous bacteria and Eukaryotes. The Astro_capsid_p (Turkey astrovirus capsid protein) domain which has been described as part of capsid proteins from various astrovirus strains (Tang et al., 2005) is found in Cegacovirus species as well as Human, astrovirus-1 and select Eukaryotes.
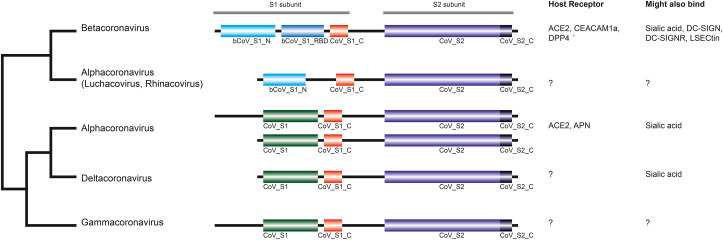
2.2. Evolution of spike glycoproteins
Coronaviridae spike proteins are multifunctional proteins that mediate viral entry into host cells. Composed of two subunits, S1 and S2, they first bind to a receptor on the host cell surface through their S1 subunit and then fuse viral and host membranes through their S2 subunit. In SARS-CoV-2 (but not in SARS-CoV1 or MERS-CoV) the two subunits S1 and S2 have been shown to be proteolytically cleaved by a Furin protease (Örd et al., 2020). The spike proteins of Coronaviridae are known to bind a broad range of cellular targets, including sialic acids, sugars, and proteins (Fig. 4 ). For example, Human coronavirus 229E (Alphacoronavirus, subgenus Duvinacovirus) binds aminopeptidase N, whereas Human coronavirus NL63 (Alphacoronavirus) and SARS-CoV (Betacoronavirus) bind to angiotensin converting enzyme 2 (ACE2) (Graham et al., 2013).
Fig. 4.
Phylogeny and domain organization of Coronaviridae spike glycoproteins. The phylogeny on the left side was calculated using a maximum likelihood approach applied to a MAFFT alignment of the CoV_S2 domains. Spike protein domain architecture for each genus is shown in the middle; for a description of the Pfam domains see Table 1. Host cell receptors and likely additional receptors are shown on the right side (Graham et al., 2013). The following abbreviations are used: ACE2, angiotensin converting enzyme 2; APN, aminopeptidase N; CEACAM1a, carcinoembryonic cell adhesion molecule 1a; DC-SIGN, dendritic cell-specific ICAM-grabbing non-integrin; DC-SIGNR, DC-SIGN-related protein; DPP4, dipeptidyl peptidase 4; LSECtin, liver and lymph node sinusoidal C-type lectin.
Sequence analysis shows that spike glycoproteins are composed of distinct combinations of six Pfam protein domains (Fig. 4 and Table 1 ). While the carboxy-terminal S2 subunit shows the same two-domain CoV_S2--CoV_S2_C arrangement for all Orthocoronavirinae genomes analyzed here, the amino-terminal S1 subunit differs significantly between Alpha-, Beta-. Gamma-, and Deltacoronaviruses (we use "--" to indicate connected domains in a protein). The S1 subunit of all Betacoronaviruses analyzed have a bCoV_S1_N--bCoV_S1_RBD--CoV_S1_C architecture, whereas Gamma-, and Deltacoronaviruses have a CoV_S1--CoV_S1_C arrangement (Gammacoronaviruses have a longer N-terminal extension). Surprisingly, the S1 subunits of Alphacoronaviruses differ between sub-genera. In Luchacovirus and Rhinacovirus, S1 has a bCoV_S1_N--CoV_S1_C arrangement and is thus similar to the arrangement found in Betacoronaviruses (but lack bCoV_S1_RBD), whereas the other sub-genera have the same architectures as in Gamma-, and Deltacoronaviruses, with differing lengths of the N-terminal extension. Interestingly, this split of Alphacoronaviruses is also found when analyzing the phylogenetic history of spike glycoproteins, both when performing phylogenetic inference on entire proteins (in which the phylogenetic signal is likely to be somewhat obscured by differences in domain architectures; data not show) as well as on CoV_S2 domains alone, as shown in Fig. 4. Interestingly, phylogenetic analysis of all other proteins does not show this split within Alphacoronaviruses. Therefore, this split is likely not the result of taxonomic misclassification, but rather some recombination event between some Alpha- and Betacoronavirus spike proteins and perhaps convergent evolution selecting for this architecture. This interesting difference in spike proteins within Alphacoronaviruses has been previously noted, for example for the Rhinolophus bat coronavirus HKU2 (Chinese horseshoe bat virus; Bat-CoV HKU2) from the Rhinacovirus subgenus (Lau et al., 2007).
Table 1.
Spike protein Pfam domains found in Orthocoronavirinae.
| Pfam Domain | Name | Function | Pfam Clan | Taxonomic Distribution |
|---|---|---|---|---|
| bCoV_S1_N | Betacoronavirus-like spike glycoprotein S1, N-terminal | Receptor binding | Concanavalin: carbohydrate binding domains and glycosyl hydrolase enzymes | Alphacoronavirus (Luchacovirus, Rhinacovirus) Betacoronavirus |
| bCoV_S1_RBD | Betacoronavirus spike glycoprotein S1, receptor binding | Receptor binding | Betacoronavirus | |
| CoV_S1_C | Coronavirus spike glycoprotein S1, C-terminal | Alphacoronavirus Betacoronavirus | ||
| Gammacoronavirus | ||||
| Deltacoronavirus | ||||
| CoV_S1 | Coronavirus spike glycoprotein S1 | Receptor binding | Alphacoronavirus Gammacoronavirus | |
| Deltacoronavirus | ||||
| CoV_S2 | Coronavirus spike glycoprotein S2 | Fusion | Fusion_gly: viral glycoproteins that mediate fusion with target membranes | Nidovirales, including: Alphacoronavirus Betacoronavirus |
| Gammacoronavirus | ||||
| Deltacoronavirus | ||||
| CoV_S2_C | Coronavirus spike glycoprotein S2, intravirion | Cysteine rich intravirion region, targets for palmitoylation | Alphacoronavirus Betacoronavirus | |
| Gammacoronavirus | ||||
| Deltacoronavirus |
2.3. Divergence of the polyprotein N-terminal domain/protein architecture
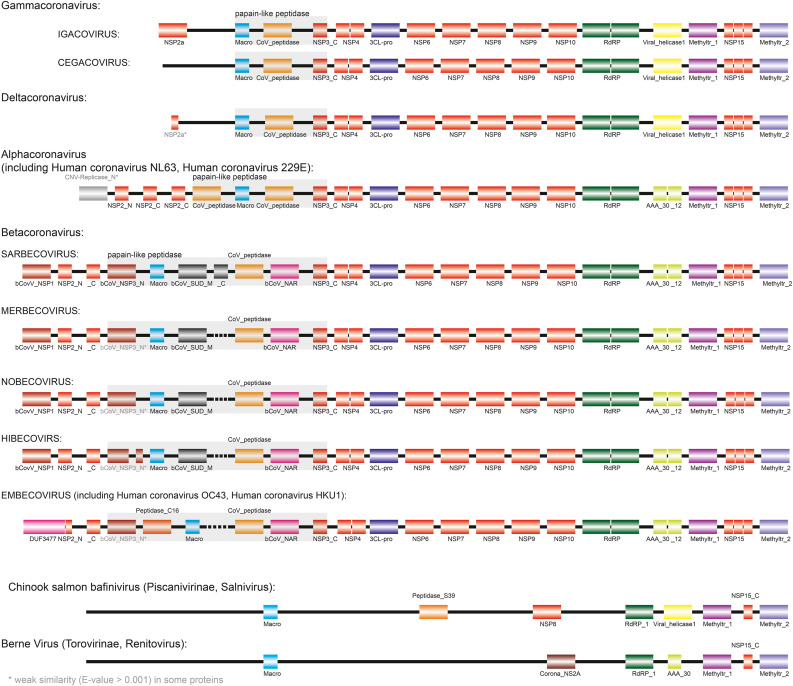
We used the DAIO approach to compare the arrangement of protein domains (and by extension, mature proteins) in polyproteins from different Orthocoronavirinae genera and sub-genera (Fig. 5 and Table 2 ). For comparison, we also included two example polyproteins from Tobaniviridae, which are currently not as well studied as Orthocoronavirinae and thus appear devoid of many proteins/domains. The polyproteins of all Alphacoronaviruses studied here exhibit an identical arrangement of domains/proteins, and the Gamma- and Deltacoronaviruses show a nearly identical arrangement. This is in sharp contrast to the Betacoronaviruses which show substantial variability between sub-genera, with Sarbecovirus and Embecovirus being the most divergent. The two main findings from this analysis are as follows. First, the central part of the polyprotein (NSP4--3CL-pro--NSP6-- … --NSP10--RdRP) is identical for all Orthocoronavirinae analyzed here. The same is true for the carboxy-terminal part (Viral_helicase1--Methyltr_1--NSP15--Methyltr_2) with the sole exception that for human (but not for all other animal hosts) Alpha- and Betacoronviruses an AAA_30--AAA_12 arrangement replaces Viral_helicase. However, this is very likely a sequence analysis-related artefact, since AAA_30, AAA_12, and Viral_helicase1 are related members of the P-loop containing nucleoside triphosphate hydrolase-superfamily (P-loop_NTPase Pfam clan) and match the same target sequences with similar E-values.
Fig. 5.
Arrangement of protein domains in polyproteins. Domains matching with a E-value of less than 0.001 are shown. Domains for which the E-values are larger than 0.001, or which are not present in all genomes of a given subgenus, are labelled in grey. In order to align corresponding domains, we introduced artificial insertions, shown with dashed lines. Domains making up the Papain-like peptidase are marked with a light grey box. Domains are not drawn to scale. For details of domains see Table 2.
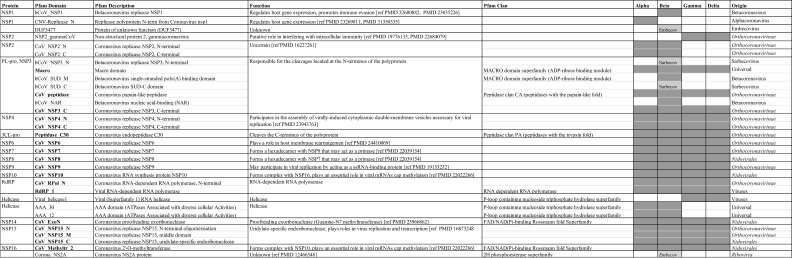
Table 2.
Pfam domains in polyproteins.
In contrast, the amino-terminal part of the polyprotein varies dramatically between genera and sub-genera of Orthocoronavirinae. Significantly, the papain-like peptidase protein in Gamma- and Deltacoronaviruses is smaller and simpler (three domains) than the form found in Alpha- and Betacoronaviruses (four to seven domains). It is particularly noteworthy that the papain-like peptidase in Embecoviruses has a different domain architecture, even when compared to other Betacoronaviruses, in that it has an additional domain belonging to the Peptidase C16 family (Peptidase_C16) and lacks the bCoV_SUD_M (single-stranded poly-A binding domain) domain. Also, Embecoviruses are unique in that they have domain of unknown function DUF3477 at the N-terminus. Sarbecoviruses are the only subgenus in which a bCoV_SUD_C domain is present in the papain-like peptidase. In addition, Alpha- and Betacoronaviruses have more mature peptides on the amino-terminal side of the papain-like peptidase protein.
2.4. Major differences between Orthocoronavirinae accessory proteins
We also used DAIO to compare the accessory proteins across Orthocoronavirinae subgenera and found that most accessory proteins are subgenus-specific, despite oftentimes having been given identical names such as “NS7” (non-structural protein 7) or “ORF7”. These names are based on the protein's placement in the genome and do not necessarily indicate homology or similarity in biological/molecular function. Therefore, these names cannot be used to compare or relate proteins between different subgenera, since for example, Hibecovirus ORF7 is not related to Cegacovirus ORF7 (also see Fig. 2B for additional examples).
Furthermore, most accessory proteins outside of the Betacoronavirus subgenera Embecovirus and Sarbecovirus do not have a corresponding Pfam entry [no profile Hidden Markov domain model (HMM)]. Accessory protein domains with an existing Pfam entry are listed in regular fonts in Table 3 (as are ORF1ab polyprotein, Spike glycoprotein, Membrane protein, Envelope small membrane protein, and Nucleoprotein). HMMs would be the ideal means to systematically classify accessory proteins since they represent a protein's molecular “signature” and are indifferent to the placement in a genome (Eddy, 2004). For this reason, HMMs were created for all accessory proteins that currently lack one. Domains defined by these new HMMs are in italic fonts in Table 3. Since these novel HMMs are representing sub-genus-specific domains, they do not appear in Fig. 3 and Supplementary Tables 1 and 2 as gains and losses.
Table 3.
Accessory Proteins in Orthocoronavirinae. In addition to the Pp1ab polyprotein, Spike glycoprotein (S), Membrane protein (M), Envelope small membrane protein (E), and Nucleoprotein (N), this table lists all accessory proteins found in Orthocoronavirinae sub-genera. For each protein, the corresponding name of the Pfam HMM domain model is listed. Section A lists proteins/domains which are found in more than one subgenus (for example, bCoV_viroporin is found both in Hibecovirus and Sarbecovirus), whereas section B lists domains which are subgenus specific (for example, Decacovirus_ORF4a is specific to Decacovirus and bCoV_NS6 is specific to Sarbecovirus). This table does not list the detailed domain architectures for Spike proteins and polyproteins (Pp1a and Pp1ab) since these are presented in Fig. 4, Fig. 5, respectively. Normal fonts indicate domains that already exist in Pfam; italic fonts indicate domains in which a new HMM was created as part of this work. “v” means present with variable domain architectures (Pp1ab and S) and square brackets asterisk are used to indicate weak similarity.
2.5. The Nobecovirus sub-genus accessory proteins appear to be particularly prone to domain gain/loss
The Nobecovirus subgenus provides a more recent example of both domain gain and recurrent domain loss. In addition to the Orthoreo_P10 protein proposed to have been gained through recombination in the Rousettus bat coronavirus GCCDC1 species (Huang et al., 2016; Obameso et al., 2017; Paskey et al., 2020), various Nobecoviruses appear to have at least four other accessory proteins downstream of the nucleocapsid genome which we have designated as Nobecovirus_ORF7a, Nobecovirus_ORF7b, Nobecovirus_ORF7c, and Nobecovirus_ORF7d. A phylogenetic analysis of the RdRP domain of 27 Nobecovirus genomes revealed that domain loss was not necessarily associated with specific branches in the tree, indicating that domain loss is likely to have occurred repeatedly at discrete timepoints, and in multiple species. This has resulted genomes with varying combinations of accessory proteins even within the same species. Additionally, domain loss did not appear to be correlated with sampling timepoints, host species or the geographic location of isolation of viruses.
2.6. Classification of Orthocoronavirinae proteins into Strict Ortholog Groups
Using these newly developed HMMs, as well as the existing Pfam HMMs, we grouped Orthocoronavirinae proteins into Strict Ortholog Groups (SOGs), groups of orthologous proteins with the same domain architecture (Zmasek et al., 2019). Supplementary Table 3 provides on overview of the results from this analysis. The first column indicates the taxonomic distribution for each SOG (all uppercase descriptions are used for SOGs which are found in every member of a given taxonomic unit, whereas lowercase descriptions are used for SOGs which are present in some, but not all, members of a taxonomic unit). The domain architectures are also listed, using Pfam domain names for domains that have an entry in Pfam, and the names of our newly developed HMMs for domains that lack an entry in Pfam, as indicated by an asterisk in the fourth column (“--" is used to indicate domain connections in multidomain proteins). This table also lists the proposed names for each SOG [numbers in brackets are temporarily used to distinguish SOGs with same names but different domain architectures, currently a mixture of manually curated names and automatically inferred ones].
3. Conclusions
In this work we show that Orthocoronavirinae genomes evolved in what could be called three distinct 'modes'. (i) Certain sections of the genomes are stable and only differ by point mutations and small insertions and deletions. These sections include the central portion and the C-terminus of the ORF1ab polyprotein, encoding the 3C-like protease, NSP4, NSP6, NSP7, NSP8, NSP9, RNA-dependent RNA polymerase, Helicase (Hel), and NSP15 and the membrane protein M and nucleoprotein N, which are encoded by their own ORFs. These proteins are present and orthologous over all Coronaviridae genomes analyzed and thus help define this virus family.
-
(ii)
The spike proteins and the papain-like peptidases, in contrast, differ in their domain architectures between genera. Similarly, the N-terminus of the polyproteins differ in the proteins encoded between genera, and for Betacoronaviruses, even between sub-genera. The envelope small membrane protein E is orthologous across Alpha- and Betacoronaviruses, absent in Gammacoronaviruses, and encoded by a different, non-homologous gene in Deltacoronaviruses.
-
(iii)
The greatest variability is found in the accessory proteins. For these proteins, each sub-genus has its own unique set, with very little overlap between sub-genera. The only notable exception to this is NS3b which is present and orthologous over all Alphacoronaviruses.
In addition, we note the following:
The establishment of the Orthocoronavirinae family is associated with a large gain of domains. While this superficially appears as if these domains appeared at the same time, en bloc, the more likely explanation is that these domains were gained one domain at a time, but most viral species emerging from the branch leading from Nidovirales to Coronaviridae either went extinct and/or have not been discovered yet.
From a domain presence/absence perspective Alpha- and Betacoronaviruses are similar to each other, as are Gamma- and Deltacoronaviruses.
The only Coronaviridae which possess the Haemagglutinin-esterase fusion glycoprotein (composed of domains Hema_esterase and Hema_HEFG) are the Embecoviruses. Given that other viral species containing such proteins are phylogenetically distant (Torovirus, Herpesvirales, Influenza C and D viruses) it appears likely that this distribution pattern is the result of multiple, independent gene acquisitions from host species, instead of a acquisition by a putative ancestral virus followed by speciation and gene loss.
It is interesting that most of the differences in the polyproteins are towards the amino-terminal end, even though, when taking the need to keep coding sequences in-frame into account, mechanically a diverging carboxy-terminal end should be the favored “solution".
In the same context, is it noteworthy that proteins encoded at the amino-terminal end of the polyprotein appear to have functions related to modulating virus-host interaction and appear not as strictly essential as other proteins (such as the peptidases and RNA-dependent RNA polymerases). Examples are SARS-CoV-2 NSP1 which is believed to inhibits host translation (Thoms et al., 2020) and SARS-CoV-2 NSP2 which has been implicated in the modulation of host cell survival (Cornillez-Ty et al., 2009; Lei et al., 2018).
Finally, in the course of this work, we developed a consistent naming scheme for all Coronaviridae proteins as well as numerous novel hidden Markov models (HMMs) representing sub-genus specific accessory proteins. The resulting annotations of this efforts will be disseminated via the ViPR database (Pickett et al., 2012).
4. Materials and methods
We used a semi-automated software pipeline to analyze amino acid sequences for their protein domain-based architectures and to infer multiple sequence alignments and phylogenetic trees for the molecular sequences corresponding to these architectures, followed by gene duplication inference. This pipeline contains the following five major steps: (1) sequence retrieval; (2) domain architecture analysis, including the inference of the taxonomic distributions of domain architectures – each of which corresponds to one preliminary SOG - and manual naming of domain architectures/preliminary SOGs; (3) extraction of molecular sequences corresponding to domain architectures/preliminary SOGs; (4) multiple sequence alignment and phylogenetic inference; (5) gene duplication inference, to determine which preliminary SOGs contain sequences related by gene duplications and thus need to be divided into multiple, final SOGs. Links to all custom software programs developed for this work are available at https://sites.google.com/site/cmzmasek/home/software/forester/daio. The tools and methods used are described in more detail below.
4.1. Sequence retrieval
Individual protein sequences were downloaded from the ViPR database (Pickett et al., 2012), while entire proteomes were downloaded from UniProtKB (Bateman et al., 2017).
4.2. Multiple sequence alignments
Multiple sequence alignments were calculated using MAFFT version 7.313 (with “localpair” and “maxiterate 1000” options) (Katoh and Standley, 2013). Prior to phylogenetic inference, multiple sequence alignment positions with more than 50% gaps were deleted.
4.3. Protein domain analysis
Protein domains were analyzed using hmmscan from HMMER v3.3.1 (Eddy, 2011) and the Pfam 33.1 (May 2020, 18259 entries) database (El-Gebali et al., 2018).
4.4. HMM construction
For ORFs lacking a defined Pfam domain, HMMs were constructed by first creating multiple sequence alignments of homologous sequences using MAFFT version 7.313 (with “localpair” and “maxiterate 1000” options) (Katoh and Standley, 2013). These multiple sequence alignments were then used as input for hmmbuild. Resulting HMMs where then tested against expected matching sequences, as well as against expected non-matching sequences.
4.5. Phylogenetic analyses
Phylogenetic trees were calculated for individual domain architectures (not full-length sequences). Distance-based minimal evolution trees were inferred by FastME 2.0 (Desper and Gascuel, 2006) (with balanced tree swapping and “GME” initial tree options) based on pairwise distances calculated by TREE-PUZZLE 5.2 (Schmidt and von Haeseler, 2007) using the WAG substitution model (Whelan and Goldman, 2001), a uniform model of rate heterogeneity, estimation of amino acid frequencies from the dataset, and approximate parameter estimation using a Neighbor-Joining tree. For maximum likelihood approaches, we employed RAxML version 8.2.9 (Stamatakis, 2006) (using 100 bootstrapped data sets and the WAG substitution model). Tree and domain composition diagrams were drawn using Archaeopteryx [https://sites.google.com/site/cmzmasek/home/software/forester]. Rooting was performed by the midpoint rooting method. Unless otherwise noted, Pfam domains are displayed with an E = 10−6 cutoff. Gene duplication inferences were performed using the SDI and RIO methods (Zmasek and Eddy, 2001, 2002). Automated genome wide domain composition analysis was performed using a specialized software tool, Surfacing version 2.002 (C M Zmasek and Godzik, 2012), a tool for the functional analysis of domainome/genome evolution [available at https://sites.google.com/site/cmzmasek/home/software/forester/surfacing]. All conclusions presented in this work are robust relative to the alignment methods, the alignment processing, the phylogeny reconstruction methods, and the parameters used. All sequence, alignment, and phylogeny files are available upon request.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Christian M. Zmasek: Conceptualization, Writing – original draft, Methodology, Software. Elliot J. Lefkowitz: Investigation, Writing – review & editing. Anna Niewiadomska: Investigation, Writing – review & editing. Richard H. Scheuermann: Investigation, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Acknowledgements
This work has been funded with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services - HHS75N93019C00076. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2022.03.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Domain gains during Nidovirales evolution.
Domain losses during Nidovirales evolution.
Strict Ortholog Groups (SOGs) in Orthocoronavirinae. Structural proteins are shown in bold. “--" is used to indicate domain connections in multidomain proteins in the second column.
References
- Altenhoff A.M., Studer R.A., Robinson-Rechavi M., Dessimoz C. Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020 doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Martin M.J., O'Donovan C., Magrane M., Alpi E., Antunes R., Bely B., Bingley M., Bonilla C., Britto R., Bursteinas B., Bye-AJee H., Cowley A., Da Silva A., De Giorgi M., Dogan T., Fazzini F., Castro L.G., Figueira L., Garmiri P., Georghiou G., Gonzalez D., Hatton-Ellis E., Li W., Liu W., Lopez R., Luo J., Lussi Y., MacDougall A., Nightingale A., Palka B., Pichler K., Poggioli D., Pundir S., Pureza L., Qi G., Rosanoff S., Saidi R., Sawford T., Shypitsyna A., Speretta E., Turner E., Tyagi N., Volynkin V., Wardell T., Warner K., Watkins X., Zaru R., Zellner H., Xenarios I., Bougueleret L., Bridge A., Poux S., Redaschi N., Aimo L., ArgoudPuy G., Auchincloss A., Axelsen K., Bansal P., Baratin D., Blatter M.C., Boeckmann B., Bolleman J., Boutet E., Breuza L., Casal-Casas C., De Castro E., Coudert E., Cuche B., Doche M., Dornevil D., Duvaud S., Estreicher A., Famiglietti L., Feuermann M., Gasteiger E., Gehant S., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Jungo F., Keller G., Lara V., Lemercier P., Lieberherr D., Lombardot T., Martin X., Masson P., Morgat A., Neto T., Nouspikel N., Paesano S., Pedruzzi I., Pilbout S., Pozzato M., Pruess M., Rivoire C., Roechert B., Schneider M., Sigrist C., Sonesson K., Staehli S., Stutz A., Sundaram S., Tognolli M., Verbregue L., Veuthey A.L., Wu C.H., Arighi C.N., Arminski L., Chen C., Chen Y., Garavelli J.S., Huang H., Laiho K., McGarvey P., Natale D.A., Ross K., Vinayaka C.R., Wang Q., Wang Y., Yeh L.S., Zhang J. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodeló G., Labrada L., Martínez-Costas J., Benavente J. 2002. Modification of Late Membrane Permeability in Avian Reovirus-Infected Cells VIROPORIN ACTIVITY of the S1-ENCODED NONSTRUCTURAL P10 PROTEIN*. [DOI] [PubMed] [Google Scholar]
- Chen L., Li F. Structural analysis of the evolutionary origins of Influenza virus Hemagglutinin and other viral lectins. J. Virol. 2013;87:4118–4120. doi: 10.1128/jvi.03476-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillez-Ty C.T., Liao L., Yates J.R., Iii, Kuhn P., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 2009;83:10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De P.M., Zanotto A., Gibbs M.J., Gould E.A., Holmes E.C. A reevaluation of the higher taxonomy of viruses based on RNA polymerases. J. Virol. 1996 doi: 10.1128/jvi.70.9.6083-6096.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desper R., Gascuel O. Current Protocols in Bioinformatics. 2006. Getting a tree fast: Neighbor Joining, FastME, and distance-based methods; pp. 6.3.1–6.3.28. [DOI] [PubMed] [Google Scholar]
- Dolja V.V., Koonin E.V. Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer. Virus Res. 2020;244:36–52. doi: 10.1016/j.virusres.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A.-M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.-C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.-D., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/nejmoa030747. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7 doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. See text for explanation; _computational BIOLOGY: 2004. P R I M E R what Is a Hidden Markov Model? Statistical Models Called Hidden Markov Models Are a Recurring Theme in Computational Biology. What Are Hidden Markov Models, and Why Are They So Useful for So Many Different Problems? 5 I E Start End Figure 1A Toy HMM for 5′ Splice Site Recognition. [Google Scholar]
- Eisen J.a. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A., Sonnhammer E.L.L., Hirsh L., Paladin L., Piovesan D., Tosatto S.C.E., Finn R.D. The Pfam protein families database in 2019. Nucleic Acids Res. 2018;47:427–432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Zhao K., Shi Z.L., Zhou P. Bat coronaviruses in China. Viruses. 2019 doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: Methods and Protocols. Springer; New York: 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W.M. Homology. Trends Genet. 2000;16:227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- Fitch W.M. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. doi: 10.2307/2412448. [DOI] [PubMed] [Google Scholar]
- Gomez de Cedro è, M., Ehsani, N., Mikkola, M.L., Antonio Garc, J., Ka ëa ëria ëinen, L., n.d. RNA Helicase Activity of Semliki Forest Virus Replicase Protein NSP2. [DOI] [PubMed]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Liu W.J., Xu W., Jin T., Zhao Y., Song J., Shi Y., Ji W., Jia H., Zhou Y., Wen H., Zhao H., Liu H., Li H., Wang Q., Wu Y., Wang L., Liu D., Liu G., Yu H., Holmes E.C., Lu L., Gao G.F. A bat-derived putative cross-family recombinant coronavirus with a reovirus gene. PLoS Pathog. 2016;12:1–25. doi: 10.1371/journal.ppat.1005883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Nacher J.C., Kuma K., Goto S., Kanehisa M. Evolutionary history and functional implications of protein domains and their combinations in eukaryotes. Genome Biol. 2007;8:R121. doi: 10.1186/gb-2007-8-6-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.A. Orthologs and paralogs - we need to get it right. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-interactions1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V., Krupovic M., Varsani A., Wolf Y.I., Yutin N., Zerbini F.M., Kuhn J.H. Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 2020;84:1–33. doi: 10.1128/mmbr.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V., Morris T.J. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Wang M., Lam C.S.F., Xu H., Guo R., Chan K., hung, Zheng B., jian Yuen, yung K. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., Fielding B.C. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Viruses. 2012;4:2902–2923. doi: 10.3390/V4112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A.D., Björklund Å.K., Ekman D., Bornberg-Bauer E., Elofsson A. Arrangements in the modular evolution of proteins. Trends Biochem. Sci. 2008;33:444–451. doi: 10.1016/j.tibs.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Neuwald A.F., Aravind L., Spouge J.L., Koonin E.V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Obameso J.O., Li H., Jia H., Han M., Zhu S., Huang C., Zhao Yuhui, Zhao M., Bai Y., Yuan F., Zhao H., Peng X., Xu W., Tan W., Zhao Yingze, Yuen K.Y., Liu W.J., Lu L., Gao G.F. The persistent prevalence and evolution of cross-family recombinant coronavirus GCCDC1 among a bat population: a two-year follow-up. Sci. China Life Sci. 2017;60:1357–1363. doi: 10.1007/s11427-017-9263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Örd M., Faustova I., Loog M. 2020. The Sequence at Spike S1/S2 Site Enables Cleavage by Furin and Phospho-Regulation in SARS-CoV2 but Not in SARS-CoV1 or MERS-CoV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paskey A.C., Ng J.H.J., Rice G.K., Chia W.N., Philipson C.W., Foo R.J.H., Cer R.Z., Long K.A., Lueder M.R., Lim X.F., Frey K.G., Hamilton T., Anderson D.E., Laing E.D., Mendenhall I.H., Smith G.J., Wang L.F., Bishop-Lilly K.A. Detection of recombinant rousettus bat coronavirus GCCDC1 in lesser dawn bats (eonycteris spelaea) in Singapore. Viruses. 2020;12 doi: 10.3390/v12050539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisajovich S.G., Garbarino J.E., Wei P., Lim W. a. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science. 2010;328:368–372. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z., Zhou L., Larson C.N., Dietrich J., Klem E.B., Scheuermann R.H. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:593–598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H.A., von Haeseler A. Current Protocols in Bioinformatics. 2007. Maximum-likelihood analysis using TREE-PUZZLE; pp. 6.6.1–6.6.23. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tang Y., Murgia M.V., Saif Y.M. Molecular characterization of the capsid gene of two serotypes of Turkey astroviruses. Avian Dis. 2005;49:514–519. doi: 10.1637/7353-030305R.1. [DOI] [PubMed] [Google Scholar]
- Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Stuerzel C.M., Froehlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020:8665. doi: 10.1101/2020.05.18.102467. 2020, 05.18.102467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wolf Y.I., Kazlauskas D., Iranzo J., Lucía-Sanz A., Kuhn J.H., Krupovic M., Dolja V.V., Koonin E.V. Origins and evolution of the global RNA virome. mBio. 2018;9:1–31. doi: 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Jha B.K., Ogden K.M., Dong B., Zhao L., Elliott R., Patton J.T., Silverman R.H., Weiss S.R. Homologous 2?,5?-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc. Natl. Acad. Sci. U.S.A. 2013;110:13114–13119. doi: 10.1073/pnas.1306917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek C.M., Eddy S.R. RIO: analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinf. 2002;3:14. doi: 10.1186/1471-2105-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek C.M., Eddy S.R. A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics. 2001;17:821–828. doi: 10.1093/bioinformatics/17.9.821. [DOI] [PubMed] [Google Scholar]
- Zmasek C.M., Godzik A. This déjà vu feeling—analysis of multidomain protein evolution in eukaryotic genomes. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek C.M., Godzik A. This déjà vu feeling-analysis of multidomain protein evolution in eukaryotic genomes. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek C.M., Godzik A. Strong functional patterns in the evolution of eukaryotic genomes revealed by the reconstruction of ancestral protein domain repertoires. Genome Biol. 2011;12:R4. doi: 10.1186/gb-2011-12-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek C.M., Knipe D.M., Pellett P.E., Scheuermann R.H. Classification of human herpesviridae proteins using domain-architecture aware inference of orthologs (DAIO) Virology. 2019;529 doi: 10.1016/j.virol.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Domain gains during Nidovirales evolution.
Domain losses during Nidovirales evolution.
Strict Ortholog Groups (SOGs) in Orthocoronavirinae. Structural proteins are shown in bold. “--" is used to indicate domain connections in multidomain proteins in the second column.