Abstract
miRNAs are small noncoding RNAs that may contribute to common diseases through epigenetic regulation of gene expression. Little is known regarding the role of miRNAs in type 2 diabetes (T2D). We performed miRNA sequencing and transcriptomic profiling of peripheral monocytes from the longitudinal Multi-Ethnic Study of Atherosclerosis (MESA) (N = 1,154). We examined associations between miRNAs and prevalent impaired fasting glucose and T2D and evaluated the T2D-associated miRNA effect on incident T2D. Of 774 detected miRNAs, 6 (miR-22-3p, miR-33a-5p, miR-181c-5p, miR-92b-3p, miR-222–3p, and miR-944) were associated with prevalent T2D. For five of the six miRNAs (all but miR-222-3p), our findings suggest a dose-response relationship with impaired fasting glucose and T2D. Two of the six miRNAs were associated with incident T2D (miR-92b-3p: hazard ratio [HR] 1.64, P = 1.30E-03; miR-222-3p: HR 1.97, P = 9.10E-03) in the highest versus lowest tertile of expression. Most of the T2D-associated miRNAs were also associated with HDL cholesterol concentrations. The genes targeted by these miRNAs belong to key nodes of a cholesterol metabolism transcriptomic network. Higher levels of miRNA expression expected to increase intracellular cholesterol accumulation in monocytes are linked to an increase in T2D risk.
Introduction
Epigenetic mechanisms such as miRNAs may contribute to the development of common diseases, including type 2 diabetes (T2D) (1–4). miRNAs, which are small (∼22 nucleotides) noncoding RNAs, have emerged as important regulators of gene expression in a cell-type–specific manner, acting predominantly to repress target gene expression at the posttranscriptional level by binding complementary mRNA (5). With increasing use of high-throughput sequencing to survey small RNA populations, >1,800 miRNAs have been identified to date in various human tissues (6). However, little human evidence is available regarding the role of miRNAs in T2D.
A significant barrier to understanding the role of epigenetic regulation in the pathogenesis of T2D is the limited accessibility of human-relevant tissues for metabolism (adipose, pancreatic islet cells, and skeletal muscle) in large population studies. Monocyte-derived macrophages are implicated in the development of T2D given their role as effector cells in the initiation of inflammation, including the obesity-related inflammation that contributes to T2D (7). Given shared characteristics, such as inflammatory states, between monocytes and monocyte-derived macrophages, analyses of peripheral monocytes may capture molecular changes in monocyte-derived macrophages that contribute to T2D. Alternatively, analyses of monocytes may identify systemic epigenetic and transcriptional changes relevant to T2D pathogenesis. Work by our group and others have demonstrated the power of studying monocytes to better understand the molecular mechanisms of biological aging, cardiovascular disease, and T2D (8–13). The objectives of the current work were to use miRNA sequencing of monocytes from the large, community-based Multi-Ethnic Study of Atherosclerosis (MESA) to 1) identify miRNA signatures of T2D, 2) evaluate their association with incident T2D, and 3) explore the potential mechanisms underlying these associations by investigating expression of the miRNA experimentally validated target genes that are differentially expressed in T2D.
Research Design and Methods
Study Design
MESA is a prospective, longitudinal cohort study designed to identify factors associated with the development of cardiovascular disease. MESA participants without known cardiovascular disease were recruited between 2000 and 2002 at six locations across the U.S. (14). The current analysis includes data from 1,154 participants obtained at MESA examination visits 5 (2010–2012) and 6 (2016–2018) from three study sites (Johns Hopkins University, University of Minnesota, and Columbia University). Included participants were 51% women, 22% African Americans, 32% Hispanic Americans, and 46% White Americans. Given that there are no other extant data sets of monocyte miRNAs to replicate our findings, we first evaluated cross-sectional associations of monocyte miRNAs with impaired fasting glucose (n = 522) and T2D (n = 264) at examination visit 5. To better understand the role of the T2D-associated miRNAs identified in the cross-sectional analysis, we excluded participants with prevalent T2D and evaluated the effect on risk of T2D from examination visits 5–6 (over 6 years of follow-up).
Clinical and laboratory data, including BMI, hemoglobin A1c (HbA1c), HDL cholesterol, and T2D status were obtained from MESA examination visits 5 and 6. Serum interleukin 6 (IL-6) was measured at examination visit 1 by ultrasensitive ELISA (Quantikine HS Human IL-6 Immunoassay; R&D Systems, Minneapolis, MN). Methods used to measure additional covariates are described in detail elsewhere (13). Presence of T2D was defined on the basis of self-reported T2D, fasting glucose ≥7.00 mmol/L, or the use of hypoglycemic medications. Impaired fasting glucose was defined by a fasting glucose of 5.55–6.99 mmol/L and not using hypoglycemic drugs (15). MESA was approved by the institutional review boards at each site, and participants provided written informed consent.
Blood Cell Count and Purification of CD14+ and CD4+ Cells
We purified RNA from peripheral monocytes as previously described (13). In brief, blood samples collected into EDTA were obtained for complete blood count with differential analysis. Sodium heparin–containing Vacutainer CPT cell separation tubes (Becton Dickinson, Rutherford, NJ) were used to isolate peripheral blood mononuclear cells within 2 h of blood draw. Monocytes were isolated with anti-CD14 monoclonal antibody–coated magnetic beads, using an autoMACS automated magnetic separation unit (Miltenyi Biotec, Bergisch Gladbach, Germany). For quality control (QC), flow cytometry analysis of 18 specimens was performed, including samples from all three MESA field centers, which were found to be consistently >90% pure.
RNA Extraction
RNA was isolated from samples using the AllPrep DNA/RNA Mini Kit (QIAGEN, Hilden, Germany). RNA QC metrics were obtained by optical density measurements (NanoDrop spectrophotometer) and evaluation of the integrity of 18s and 28s rRNA (Agilent 2100 Bioanalyzer using RNA 6000 Nano Chips; Agilent Technology, Santa Clara, CA) according to the manufacturer’s instructions. RNA with an RNA integrity score >9.0 was used for global expression microarrays. The median RNA integrity score for our 1,154 samples was 9.4.
Small RNA Sequencing Library Construction and HiSeq Sequencing
miRNA sequencing was performed at the Genomics Research Laboratory of the Fralin Life Sciences Institute at Virginia Tech. RNA adapters were ligated to the 5′-phosphate and the 3′-hydroxyl ends of small RNAs using TruSeq Small RNA Library Preparation Kit (RS-200-0012; Illumina) from 1 μg of miRNA-enriched total RNA. The ligated small RNAs were reverse transcribed and barcoded with PCR amplification for 11 cycles. Samples were run on an Agilent BioAnalyzer 2100 to confirm miRNA libraries. Equal volumes of 12 individually barcoded samples were pooled, and the 147-base pair (miRNAs) and 157-base pair (PIWI-interacting and some miRNAs) fractions were extracted using Pippin Prep (Sage Science, Beverly, MA). Pools were cleaned using Agencourt AMPure XP magnetic beads (A63880; Beckman Coulter) and quantitated using Quant-iT dsDNA high-sensitivity assay kit (Invitrogen), and sizes were validated on an Agilent BioAnalyzer 2100. Libraries were clustered onto flow cells using TruSeq SR Cluster Kit v3 (GD-401-3001; Illumina) and sequenced for 50 cycles using TruSeq SBS Kit v3-HS (FC-401-1002; Illumina) on an Illumina HiSeq 2500 system.
Global mRNA Expression Quantification
The Illumina HumanHT-12 v4 Expression BeadChip and BeadArray Reader were used to perform monocyte genome-wide expression analysis in a sample of MESA participants that included those with monocyte miRNA data, as previously described (8). These data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE56047).
QC and Preprocessing of Microarray Data
Data preprocessing and QC analyses were performed in R (https://www.r-project.org) using Bioconductor (https://www.bioconductor.org) packages, as previously described (8,13).
Statistical Analysis
We used logistic regression to determine the association of monocyte-derived miRNAs and T2D status at MESA examination visit 5. We adjusted for age, sex, race/ethnicity, study site, batch effects, and cell contamination. We calculated odds ratios and 95% CIs. We conducted additional sensitivity analyses that adjusted for the covariates smoking status, total weekly MET-minutes of intentional physical activity, lipid profile (HDL and LDL cholesterol), presence/absence of hypertension, BMI, serum IL-6, and statin use. We evaluated models stratified by age (<60 and ≥60 years), sex, and race/ethnicity. Given that MESA participants with normal fasting glucose may have impaired glucose tolerance, we repeated our analysis using HbA1c as an outcome, with pre-T2D defined as an HbA1c ≥5.7% (39 mmol/mol) and <6.5% (48 mmol/mol) and T2D as ≥6.5% (48 mmol/mol) or use of hypoglycemic medications. For miRNAs significantly associated with T2D at examination visit 5, we used linear regression to evaluate the association with HOMA of insulin resistance (HOMA-IR), adjusting for age, sex, race/ethnicity, study site, batch effects, and cell contamination. All statistical analyses were using R statistical software.
For the prospective analysis, we constructed Cox proportional hazards models to evaluate the association of miRNA expression and T2D from examination visits 5–6 (over 6 years of follow-up), adjusting for age, sex, race/ethnicity, study site, batch effects, and cell contamination and with or without adjustment for fasting glucose at visit 5. We calculated hazard ratios (HRs) and 95% CIs. MESA participants with both normal and impaired fasting glucose at visit 6 were included as nonevents. We repeated the prospective analysis using HbA1c as an outcome for the same reason and followed the same approach as described in the cross-sectional analysis. We also evaluated the association of the T2D-associated miRNAs with incident impaired fasting glucose, excluding MESA participants with impaired fasting glucose and T2D at visit 5.
We used linear regression to separately evaluate the association of monocyte-derived miRNAs with HDL cholesterol and BMI at examination visit 5, adjusting for age, sex, race/ethnicity, study site, batch effects, and cell contamination. To control for multiple testing, P values were adjusted using the false discovery rate (FDR) method. For miRNAs significantly associated with HDL cholesterol at visit 5, we conducted an additional sensitivity analysis adjusting for fasting glucose levels.
To investigate miRNAs as a potential molecular link between BMI and T2D status, we performed mediation analyses under an assumed causal model in which BMI leads to a change in miRNA expression, which at least partially mediates the effects of BMI on T2D status, controlling for biological and technical covariates. Mediation analyses were performed using robust structural equation modeling as implemented in the R package lavaan, as previously described (12,16).
We also associated examination visit 5 monocyte mRNA expression with T2D status, following the same approach that we used for the cross-sectional miRNA analysis described above. We used miRTarBase (https://miRTarBase.cuhk.edu.cn) to identify experimentally validated genes that are regulated by the T2D-associated miRNAs (17). On the basis of findings from our previous work demonstrating that alterations of the cholesterol metabolism transcriptional network (CMTN) are associated with obesity-related T2D (12), we focused on genes involved in cholesterol metabolism.
Data and Resource Availability
Data used in this analysis can be obtained through the MESA data coordinating center (https://www.mesa-nhlbi.org) and Database of Genotype and Phenotype (accession no. phs000209, MESA cohort). The data sets generated and/or analyzed during this study are available from the corresponding author upon reasonable request.
Results
Clinical Data and Sample Characteristics
Descriptive statistics for MESA participants at examination visit 5 in this sample are shown in Table 1. Participants with impaired fasting glucose were more likely to be male, African American, Hispanic American, former smokers, overweight or obese, and hypertensive with lower HDL cholesterol and greater statin use compared with participants with normal fasting glucose. Participants with T2D were more likely to be African American, Hispanic American, overweight or obese, and hypertensive with lower LDL and HDL cholesterol and greater statin use compared with participants with normal fasting glucose. Age, current smoking status, and physical activity were not significantly different between participants with impaired fasting glucose or T2D compared with participants with normal fasting glucose.
Table 1.
MESA cohort characteristics by fasting glucose and T2D status
| Variable | Normal fasting glucose (n = 368) | Impaired fasting glucose (n = 522) | T2D (n = 264) |
Impaired vs. normal P | T2D vs. normal P |
|---|---|---|---|---|---|
| Age (years) | 69.0 (9.63) | 69.7 (9.60) | 70.4 (8.53) | 0.34 | 0.06 |
| Female | 55 | 49 | 49 | 0.04 | 0.12 |
| African American | 14 | 21 | 35 | 1.3E-03 | 7.3E-04 |
| Hispanic | 26 | 34 | 38 | 6.7E-14 | 4.5E-09 |
| Former smoker | 54 | 47 | 50 | 0.03 | 0.59 |
| Current smoker | 9 | 9 | 12 | 0.54 | 0.29 |
| BMI (kg/m2) | 28 (5) | 30 (5) | 32 (6) | 9.6E-09 | <2E-32 |
| Fasting glucose (mmol/L) | 4.94 (0.33) | 5.49 (0.56) | 7.44 (2.66) | <2E-32 | <2E-32 |
| Hypertension | 50 | 60 | 79 | 5.2E-03 | 1.5E-12 |
| LDL cholesterol (mmol/L) | 2.85 (0.80) | 2.80 (0.26) | 2.41 (0.88) | 0.31 | 3.1E-10 |
| HDL cholesterol (mmol/L) | 1.50 (0.47) | 1.40 (0.41) | 1.27 (0.31) | 2.7E-04 | 2.3E-13 |
| Statin use (%) | 27 | 38 | 55 | 5.0E-04 | 2.3E-12 |
| Physical activity (MET-min/week) | 3,059 (4,013) | 2,806 (3,524) | 2,818 (5,345) | 0.34 | 0.43 |
Data are % or mean (SD). P value is shown for impaired fasting glucose and T2D group differences compared with normal fasting glucose.
Cross-Sectional Analysis
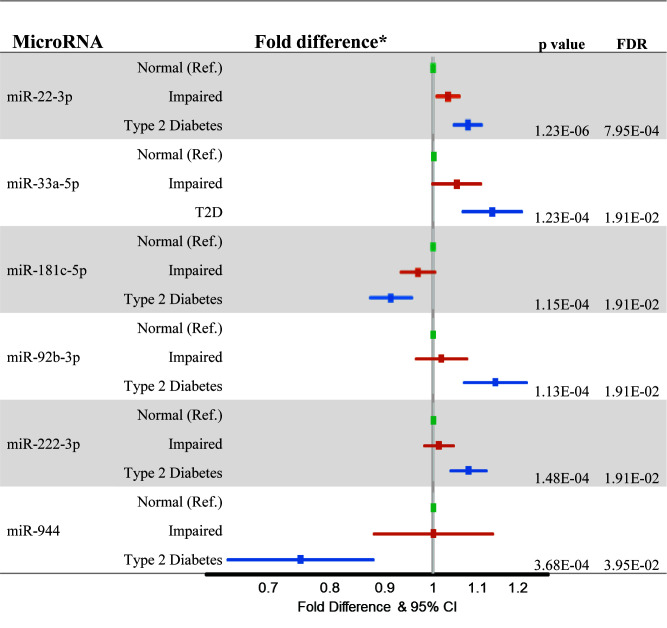
miRNA-enriched RNA sequencing analysis of monocytes detected 774 miRNAs, 6 of which were associated with a diagnosis of T2D at MESA examination visit 5 at an FDR threshold of 0.05 (Fig. 1). miR-22-3p was positively associated with HOMA-IR (P = 2.18E-06) and impaired fasting glucose status (P = 0.010). While miR-22-3p was the only miRNA associated with impaired fasting glucose status at an FDR threshold of 0.05, five of the six miRNAs (all but miR-222-3p) suggested a dose-response relationship with impaired fasting glucose status and T2D (Fig. 1). With the exception of miR-33a-5p, the remaining miRNAs were also significantly associated with HOMA-IR. miR-92b-3p and miR-222-3p were positively associated with HOMA-IR (P = 9.63E-08 and 6.63E-08, respectively), and miR-181c-5p and miR-944 were negatively associated with HOMA-IR (P = 2.15E-07 and 4.89E-03, respectively). Using HbA1c as an outcome produced the same results (data not shown). Inclusion of covariates (smoking status, total weekly MET-minutes of intentional physical activity, lipid profile [HDL and LDL cholesterol], presence/absence of hypertension, BMI, serum IL-6, and statin use) did not significantly affect the observed associations, and we observed consistent associations stratified by age, sex, and race (data not shown).
Figure 1.
Cross-sectional associations of monocyte-derived miRNAs with impaired fasting glucose and T2D at examination visit 5. *Fold difference and 95% CI compared with MESA participants with normal fasting glucose. Raw and FDR-corrected P values are shown for the comparison of T2D with normal fasting glucose. Analyses were adjusted for age, sex, race/ethnicity, study site, and cell contamination (B cells, T cells, natural killer cells, neutrophils). Ref., reference.
Given previously published findings that inhibition of miR-33a/b raises plasma HDL and lowers VLDL triglycerides in mouse and nonhuman primate models (18), we evaluated whether the T2D-associated miRNAs were associated with plasma HDL cholesterol in MESA. We found that miR-33a-5p was inversely associated with plasma HDL cholesterol levels (P = 0.0005). Additionally, miR-222-3p, miR-92b-3p, and miR-22-3p were inversely associated with plasma HDL cholesterol levels (P = 1.04E-06, 1.31E-05, and 2.68E-05, respectively), and miR-181c-5p was positively associated with plasma HDL cholesterol levels (P = 0.006) (data not shown). These associations remained significant after adjusting for fasting glucose.
Given that obesity is a major risk factor for T2D, we evaluated whether the identified T2D-associated miRNAs mediated the cross-sectional association of BMI with T2D. We found that miR-222-3p, miR-22-3p, and miR-92b-3p were positively associated with BMI (P = 4.57E-04, 7.49E-07, and 4.69E-06, respectively) and miR-181c-5p was negatively associated with BMI (P = 2.17E-04) (data not shown). Mediation analyses showed that miR-222-3p explained 3.25% (95% CI 0.77, 6.43; P = 0.045) of the association of BMI with T2D status; there was a trend toward a significant mediation effect of miR-181c-5p (P = 0.057) and miR-944 (P = 0.097) with T2D, while this was not observed for miR-22-3p, miR-33a-5p, or miR-92b-3p.
Prospective Analysis
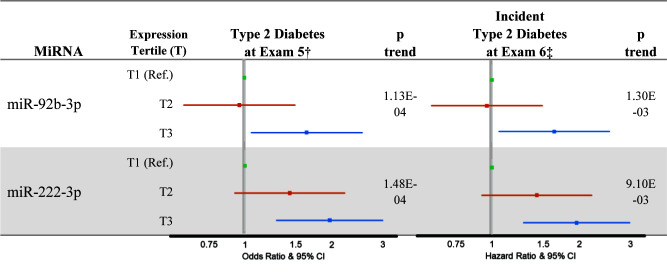
Of the 1,154 participants with data available at examination visit 5, 352 were lost to follow-up. In participants with data available at examination visits 5 and 6 (n = 802), excluding participants with T2D at visit 5 (n = 150), 65 participants developed incident T2D during 6 years of follow-up. Among the six T2D-associated miRNAs identified in the cross-sectional analysis, greater expression of miR-222-3p and miR-92b-3p was associated with incident T2D (miR-92b-3p: HR 1.64, P = 1.30E-03; miR-222-3p: HR 1.97, P = 9.10E-03) in the highest tertile of miRNA expression compared with the lowest tertile of miRNA expression (Fig. 2). Using HbA1c as an outcome produced the same results (data not shown). After adjusting for visit 5 fasting glucose levels, the association of miR-92-3p with T2D status remained unchanged (unadjusted β = 0.55, P = 0.015; adjusted β = 0.56, P = 0.066), while the association of miR-222-3p was attenuated (unadjusted β = 1.43, P = 0.009; adjusted β = 1.06, P = 0.054). Among participants with normal fasting glucose at visit 5 (n = 368), 92 developed impaired fasting glucose at visit 6. Neither miR-222-3p nor miR-92b-3p were associated with incident impaired fasting glucose (data not shown).
Figure 2.
Association of miR-92b-3p and miR-222-3p expression with T2D at examination visit 5 and incident T2D at examination visit 6. †For the cross-sectional analysis, MESA participants with impaired fasting glucose at visit 5 were excluded. ‡For the prospective analysis, MESA participants with a diagnosis of T2D at visit 5 were excluded. Analyses were adjusted for age, sex, race/ethnicity, study site, and cell contamination (B cells, T cells, natural killer cells, neutrophils). Ref., reference.
Determination of T2D-Associated miRNA Target Genes and Their Differential Expression by T2D Status
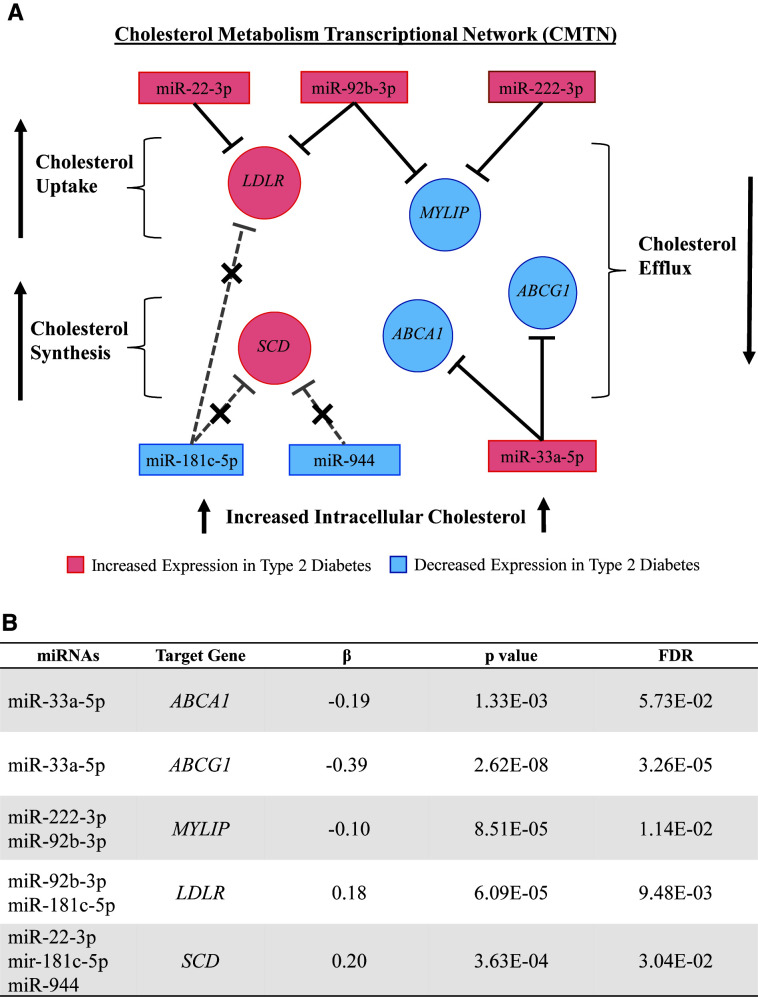
We used miRTarBase to identify experimentally validated genes that are regulated by the T2D-associated miRNAs (17). We identified mRNA targets of the T2D-associated miRNAs that constitute important nodes in cholesterol metabolism and would be expected to lead to increased intracellular cholesterol accumulation (12) (Fig. 3A). We identified five genes that are validated targets of the T2D-associated miRNAs. Four of the five T2D-associated miRNA target genes (ABCG1, MYLIP, LDLR, and SCD) were differentially expressed in T2D (FDR <0.05) (Fig. 3B).
Figure 3.
CMTN genes targeted by the T2D-associated miRNAs and their association with T2D. A: T2D miRNAs and their target CMTN genes, with a focus on genes related to cholesterol efflux, uptake, and synthesis. miRNAs and their target genes that were increased in T2D are colored pink, while miRNAs and their target genes that were decreased in T2D are colored blue. Greater miRNA expression is predicted to lead to decreased expression of its target genes, and lesser miRNA expression is predicted to lead to increased expression of its target genes. Therefore, the effects of miRNAs with decreased expression on their downstream gene targets are indicated with a short dashed line and an X. MYLIP encodes IDOL, which leads to degradation of the LDL receptor through its ubiquitin ligase activity. The activating effect of decreased MYLIP expression on the LDL receptor is indicated by the long dashed line and an X. B: Results of the differential gene expression analysis of the T2D-associated miRNA target genes by T2D status.
Discussion
In a large community-based cohort study, we found that greater monocyte expression of miR-33a-5p, miR-22-3p, miR-222-3p, or miR-92b-3p and lesser expression of miR-181c-5p or miR-944 were associated with prevalent T2D. With the exception of miR-222-3p, all the miRNAs showed a dose-response relationship with impaired fasting glucose and T2D. Additionally, all the miRNAs, with the exception of miR-33a-5p, were associated with HOMA-IR. Greater expression of miR-222-3p or miR-92b-3p was also associated with incident T2D over 6 years of follow-up. We found the same miRNA associations with prevalent and incident T2D using HbA1c as an outcome. Furthermore, we identified experimentally validated mRNAs targeted by these six miRNAs that were differentially expressed in T2D and constitute important nodes in a network of coexpressed cholesterol metabolism genes, referred to as the CMTN.
miRNA studies of T2D in humans, mainly using serum, plasma, or whole blood, are generally cross sectional and small scale. In the few large population studies published to date, investigators evaluated cross-sectional associations of miRNAs isolated from plasma or whole blood with measures of glycemia and insulin resistance (19,20). Given differences in sample type and study design, inconsistent results have been reported for miRNA associations with T2D outcomes in these studies, with only one miRNA (miR-122-5p) found to be associated with T2D outcomes in both studies. Additionally, none of the miRNAs identified in these studies overlapped with the monocyte miRNAs identified in our analysis, further enhancing the novelty of our findings.
Of the six T2D-associated miRNAs, miR-33 is the most widely studied in animal and in vitro studies. The metabolic syndrome is a significant risk factor for T2D (21), and overexpression of miR-33a adversely affects gene pathways influencing several primary risk factors for the metabolic syndrome, namely impaired insulin secretion, impaired insulin signaling, and low HDL (22–24). Conversely, inhibition of miR-33a/b in mice and nonhuman primates restores insulin secretion and raises plasma HDL (18,23,25). Consistent with this, in a small human study, serum miR-33a-5p was higher in 25 individuals with insulin resistance compared with 55 euglycemic control subjects (26). These studies also established a link between intracellular cholesterol concentrations and plasma HDL, with overexpression of miR-33a increasing intracellular cholesterol and decreasing plasma HDL (22) and inhibition decreasing intracellular cholesterol and raising HDL (25). There are limited experimental data regarding the role of the other five miRNAs in T2D pathogenesis. However, we did find that four of the five miRNAs (miR-222-3p, miR-92b-3p, miR-22-3p, and miR-181c-5p) are associated with important genes in the CMTN as well as with plasma HDL concentrations. Although a causal role of lesser plasma HDL on T2D risk has not been established (27), evidence from our group and others strongly support the role of intracellular cholesterol in T2D risk and pathogenesis.
An abundance of experimental data demonstrate that altered cellular cholesterol metabolism in multiple cell types (e.g., macrophages, pancreatic islets, adipocytes) leads to pathological changes that precede the development of T2D (28–35). In MESA, we previously reported that alterations in the CMTN are a cardinal feature of obesity and are significantly associated with T2D (12). The CMTN includes genes important for the regulation of cholesterol synthesis, uptake, and efflux and significantly associated with plasma LDL and HDL concentrations (12). Importantly, we showed that the directional changes in CMTN observed in MESA predicted increased intracellular cholesterol accumulation and mediated the association between obesity and T2D.
In the current study, we evaluated the association of the T2D-associated miRNAs with their experimentally validated target genes that were differentially expressed in T2D. We found that monocyte expression of miR-33a associates with ABCG1 and ABCA1, two key cholesterol efflux genes. This finding is consistent with in vitro and animal studies demonstrating that miR-33a regulates cellular cholesterol metabolism through targeting ABCG1 and ABCA1 in concert with its cotranscribed host gene, SREBP2 (23,25), and LXRα. Loss of function of Abca1 and Abcg1 in mouse models leads to increased islet cholesterol, reduced insulin secretion, decreased glucose tolerance, and increased fasting glucose (35,36). In humans, loss-of-function mutations in ABCA1 associates with impaired insulin secretion and increased T2D risk (37–39).
Our analyses of monocyte transcriptomic data show that the other five T2D-associated miRNAs (miR-222-3p, miR-92b-3p, miR-22–3p, miR-181c-5p, and miR-944) are also associated with key CMTN genes, particularly those that regulate cholesterol uptake and synthesis (Fig. 3). MYLIP and LDLR are key cholesterol metabolism genes that encode the inducible degrader of the LDL receptor (IDOL) and the LDL receptor, respectively, and are important for regulating cholesterol uptake (40). MYLIP is a direct target of miR-92b-3p and miR-222-3p and was downregulated in MESA participants with T2D (Fig. 3). Greater miR-92b-3p and miR-222-3p expression were also associated with an increased risk of T2D, suggesting that greater LDL receptor–mediated intracellular cholesterol accumulation may be one of the earliest epigenetic changes to contribute to T2D pathogenesis. Meanwhile, LDLR, a direct target of miR-92b-3p and miR-181c-5p, was upregulated in MESA participants with T2D likely because of decreased miR-181c-5p expression and the effect of miR-92b-3p on MYLIP expression, resulting in an overall increase in intracellular LDL cholesterol. SCD encodes stearoyl-CoA desaturase (12), which regulates cholesterol synthesis and is a direct target of miR-22-3p, miR-181c-5p, and miR-944. In MESA, SCD expression was upregulated, likely because of the effects of decreased miR-181c-5p and miR-944 outcompeting the effect of reduced miR-22-3p. Our findings provide evidence that miRNA-mediated epigenetic dysregulation of the CMTN, which is expected to increase intracellular cholesterol, is associated with plasma HDL cholesterol and prevalent and incident T2D in a longitudinal population cohort.
Our work has several strengths, including the use of a large, longitudinal, multiethnic cohort (22% African Americans and 32% Hispanic Americans) recruited from across the U.S., which enhances the generalizability of our findings. Our work is further strengthened by the use of purified monocytes for recovery of miRNAs and the inclusion of experimentally validated miRNA-targeted mRNAs. Additionally, few studies have addressed the role of epigenetic changes in T2D.
Limitations of our work include the lack of another data set with monocyte miRNAs to replicate our findings. We addressed this limitation by first performing a cross-sectional analysis of monocyte miRNA associations with T2D at MESA examination visit 5. After excluding participants with prevalent T2D cases at visit 5, we evaluated the association of monocyte miRNAs with incident T2D. There were a low number of cases of incident T2D (n = 65) over the follow-up period (after excluding prevalent cases at visit 5), which may have decreased our power to detect more significant associations in the prospective analysis. We found that the association of miR-222-3p with incident T2D was attenuated after adjusting for fasting glucose levels. Without experimental data, we cannot rule out that this finding was due to either mediation through fasting glucose levels or confounding by fasting glucose levels. Finally, while our findings may reflect a role of dysregulated epigenetic signaling in T2D, they may also be due to confounding (e.g., other chronic inflammatory diseases) and/or compensatory epigenetic changes in response to T2D.
In conclusion, we report for the first time that six monocyte-derived miRNAs are associated with T2D. We found that greater monocyte expression of miR-222-3p and miR-92b-3p are associated with incident T2D. Taken together with in vivo experimental studies supporting that cholesterol accumulation may contribute to diabetes pathogenesis (35,36), our findings suggest a hypothesis that epigenetic reprogramming of key metabolic genes/pathways results in alterations of the CMTN, ultimately leading to increased intracellular cholesterol accumulation and subsequent insulin resistance. Our findings further support the use of monocytes as an easily accessible cell type for identifying systemic epigenetic and transcriptional changes relevant to T2D pathogenesis. If validated in future studies, these miRNAs and their downstream-associated molecular pathways may serve as targets for prevention and treatment of T2D.
Article Information
Acknowledgments. MESA and the MESA SNP Health Association Resource (SHARe) project is conducted and supported by the National Heart, Lung, and Blood Institute in collaboration with MESA investigators.
Funding. Support for MESA is provided by National Heart, Lung, and Blood Institute grants 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1-TR-001881, and DK063491. The MESA Epigenomics and Transcriptomics Studies were funded by National Institutes of Health grants 1R01HL101250, 1RF1AG054474, R01HL126477, R01DK101921, and R01HL135009. This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases grant R01DK103531 to J.D. and Y.L. and National Heart, Lung, and Blood Institute grant R01HL119962 to J.S.P.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. D.C.P. researched data and wrote, reviewed, and edited the manuscript. M.W. contributed to the discussion and wrote, reviewed, and edited the manuscript. K.L. performed the statistical analyses. L.H. researched data. A.T.N. contributed to the discussion and reviewed and edited the manuscript. J.D. obtained funding, contributed to the discussion, and reviewed the manuscript. A.B., S.S., G.L.B., D.R.J., and W.P. reviewed the manuscript. D.C. and I.H. researched data and reviewed the manuscript. J.S.P. obtained funding and reviewed the manuscript. Y.L. conceptualized the study, obtained funding, developed the methodology, researched data, and wrote, reviewed, and edited the manuscript. Y.L. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1. De Rosa S, Curcio A, Indolfi C. Emerging role of microRNAs in cardiovascular diseases. Circ J 2014;78:567–575 [DOI] [PubMed] [Google Scholar]
- 2. Li R, Chung ACK, Yu X, Lan HY. MicroRNAs in diabetic kidney disease. Int J Endocrinol 2014;2014:593956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wendt A, Esguerra JL, Eliasson L. Islet microRNAs in health and type-2 diabetes. Curr Opin Pharmacol 2018;43:46–52 [DOI] [PubMed] [Google Scholar]
- 4. Esguerra JLS, Nagao M, Ofori JK, Wendt A, Eliasson L. MicroRNAs in islet hormone secretion. Diabetes Obes Metab 2018;20(Suppl. 2):11–19 [DOI] [PubMed] [Google Scholar]
- 5. Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 2007;8:93–103 [DOI] [PubMed] [Google Scholar]
- 6. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42(D1):D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lackey DE, Olefsky JM. Regulation of metabolism by the innate immune system. Nat Rev Endocrinol 2016;12:15–28 [DOI] [PubMed] [Google Scholar]
- 8. Liu Y, Ding J, Reynolds LM, et al. Methylomics of gene expression in human monocytes. Hum Mol Genet 2013;22:5065–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reynolds LM, Taylor JR, Ding J, et al. Age-related variations in the methylome associated with gene expression in human monocytes and T cells. Nat Commun 2014;5:5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reynolds LM, Wan M, Ding J, et al. DNA methylation of the aryl hydrocarbon receptor repressor associations with cigarette smoking and subclinical atherosclerosis. Circ Cardiovasc Genet 2015;8:707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reynolds LM, Ding J, Taylor JR, et al. Transcriptomic profiles of aging in purified human immune cells. BMC Genomics 2015;16:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ding J, Reynolds LM, Zeller T, et al. Alterations of a cellular cholesterol metabolism network are a molecular feature of obesity-related type 2 diabetes and cardiovascular disease. Diabetes 2015;64:3464–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Y, Reynolds LM, Ding J, et al. Blood monocyte transcriptome and epigenome analyses reveal loci associated with human atherosclerosis. Nat Commun 2017;8:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bild DE, Bluemke DA, Burke GL, et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol 2002;156:871–881 [DOI] [PubMed] [Google Scholar]
- 15. Nettleton JA, Lutsey PL, Wang Y, Lima JA, Michos ED, Jacobs DR Jr. Diet soda intake and risk of incident metabolic syndrome and type 2 diabetes in the Multi-Ethnic Study of Atherosclerosis (MESA). Diabetes Care 2009;32:688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rosseel Y. lavaan: an R package for structural equation modeling. J Stat Softw 2012;48:1–36 [Google Scholar]
- 17. Huang H-Y, Lin Y-C-D, Li J, et al. miRTarBase 2020: updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res 2020;48:gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rayner KJ, Esau CC, Hussain FN, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 2011;478:404–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shah R, Murthy V, Pacold M, et al. Extracellular RNAs are associated with insulin resistance and metabolic phenotypes. Diabetes Care 2017;40:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mononen N, Lyytikäinen L-P, Seppälä I, et al. Whole blood microRNA levels associate with glycemic status and correlate with target mRNAs in pathways important to type 2 diabetes. Sci Rep 2019;9:8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ford ES, Li C, Sattar N. Metabolic syndrome and incident diabetes: current state of the evidence. Diabetes Care 2008;31:1898–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wijesekara N, Zhang LH, Kang MH, et al. miR-33a modulates ABCA1 expression, cholesterol accumulation, and insulin secretion in pancreatic islets. Diabetes 2012;61:653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rayner KJ, Suárez Y, Dávalos A, et al. miR-33 contributes to the regulation of cholesterol homeostasis. Science 2010;328:1570–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dávalos A, Goedeke L, Smibert P, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A 2011;108:9232–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Najafi-Shoushtari SH, Kristo F, Li Y, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010;328:1566–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Corona-Meraz F-I, Vázquez-Del Mercado M, Ortega FJ, Ruiz-Quezada S-L, Guzmán-Ornelas M-O, Navarro-Hernández R-E. Ageing influences the relationship of circulating miR-33a and miR- 33b levels with insulin resistance and adiposity. Diab Vasc Dis Res 2019;16:244–253 [DOI] [PubMed] [Google Scholar]
- 27. Haase CL, Tybjærg-Hansen A, Nordestgaard BG, Frikke-Schmidt R. HDL cholesterol and risk of type 2 diabetes: a Mendelian randomization study. Diabetes 2015;64:3328–3333 [DOI] [PubMed] [Google Scholar]
- 28. Bie J, Zhao B, Song J, Ghosh S. Improved insulin sensitivity in high fat- and high cholesterol-fed Ldlr-/- mice with macrophage-specific transgenic expression of cholesteryl ester hydrolase: role of macrophage inflammation and infiltration into adipose tissue. J Biol Chem 2010;285:13630–13637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bie J, Zhao B, Marqueen KE, Wang J, Szomju B, Ghosh S. Macrophage-specific transgenic expression of cholesteryl ester hydrolase attenuates hepatic lipid accumulation and also improves glucose tolerance in ob/ob mice. Am J Physiol Endocrinol Metab 2012;302:E1283–E1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Le Lay S, Ferré P, Dugail I. Adipocyte cholesterol balance in obesity. Biochem Soc Trans 2004;32:103–106 [DOI] [PubMed] [Google Scholar]
- 31. Yu B-L, Zhao S-P, Hu J-R. Cholesterol imbalance in adipocytes: a possible mechanism of adipocytes dysfunction in obesity. Obes Rev 2010;11:560–567 [DOI] [PubMed] [Google Scholar]
- 32. de Haan W, Bhattacharjee A, Ruddle P, Kang MH, Hayden MR. ABCA1 in adipocytes regulates adipose tissue lipid content, glucose tolerance, and insulin sensitivity. J Lipid Res 2014;55:516–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fryirs M, Barter PJ, Rye K-A. Cholesterol metabolism and pancreatic β-cell function. Curr Opin Lipidol 2009;20:159–164 [DOI] [PubMed] [Google Scholar]
- 34. Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature 2005;438:612–621 [DOI] [PubMed] [Google Scholar]
- 35. Brunham LR, Kruit JK, Pape TD, et al. β-Cell ABCA1 influences insulin secretion, glucose homeostasis and response to thiazolidinedione treatment. Nat Med 2007;13:340–347 [DOI] [PubMed] [Google Scholar]
- 36. Kruit JK, Wijesekara N, Westwell-Roper C, et al. Loss of both ABCA1 and ABCG1 results in increased disturbances in islet sterol homeostasis, inflammation, and impaired β-cell function. Diabetes 2012;61:659–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Villarreal-Molina MT, Flores-Dorantes MT, Arellano-Campos O, et al.; Metabolic Study Group . Association of the ATP-binding cassette transporter A1 R230C variant with early-onset type 2 diabetes in a Mexican population. Diabetes 2008;57:509–513 [DOI] [PubMed] [Google Scholar]
- 38. Acuña-Alonzo V, Flores-Dorantes T, Kruit JK, et al. A functional ABCA1 gene variant is associated with low HDL-cholesterol levels and shows evidence of positive selection in Native Americans. Hum Mol Genet 2010;19:2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Besseling J, Kastelein JJP, Defesche JC, Hutten BA, Hovingh GK. Association between familial hypercholesterolemia and prevalence of type 2 diabetes mellitus. JAMA 2015;313:1029–1036 [DOI] [PubMed] [Google Scholar]
- 40. Wang B, Tontonoz P. Liver X receptors in lipid signalling and membrane homeostasis. Nat Rev Endocrinol 2018;14:452–463 [DOI] [PMC free article] [PubMed] [Google Scholar]