Abstract
Radiomics is an emerging area in quantitative image analysis that aims to relate large-scale extracted imaging information to clinical and biological endpoints. The development of quantitative imaging methods along with machine learning has enabled the opportunity to move data science research towards translation for more personalized cancer treatments. Accumulating evidence has indeed demonstrated that non-invasive advanced imaging analytics, i.e., radiomics, can reveal key components of tumor phenotype for multiple three-dimensional lesions at multiple time points over and beyond the course of treatment. These developments in the use of CT, PET, US and MR imaging could augment patient stratification and prognostication buttressing emerging targeted therapeutic approaches. In recent years, deep learning architectures have demonstrated their tremendous potential for image segmentation, reconstruction, recognition, and classification. Many powerful open-source and commercial platforms are currently available to embark in new research areas of radiomics. Quantitative imaging research, however, is complex and key statistical principles should be followed to realize its full potential. The field of radiomics, in particular, require a renewed focus on optimal study design/reporting practices and standardization of image acquisition, feature calculation and rigorous statistical analysis for the field to move forward. In this article, the role of machine and deep learning as a major computational vehicle for advanced model building of radiomics-based signatures or classifiers, and diverse clinical applications, working principles, research opportunities and available computational platforms for radiomics will be reviewed with examples drawn primarily from oncology. We also address issues related to common applications in medical physics, such as standardization, feature extraction, model building, and validation.
Keywords: Quantitative image analysis, radiomics, machine learning, deep learning
I. Introduction
Radiomics is an emerging area in quantitative image analysis that aims to relate large-scale data mining of images to clinical and biological endpoints1. The fundamental idea is that medical images are much richer in information than what the human eye can discern. Quantitative imaging features, called also “radiomic features” can provide richer information about intensity, shape, size or volume, and texture of tumor phenotypes using different imaging modalities (e.g., MRI, CT, PET, ultrasound, etc.)2. Tumor biopsy-based assays provide limited tumor characterization as the extracted sample may not always represent the heterogeneity of the whole patient’s tumor, while radiomics can comprehensively assess the three-dimensional tumor landscape by means of extracting relevant imaging information3. It implies that, applying well-known machine learning methods to radiomic features extracted from medical images, it is possible to macroscopically decode the phenotype of many physio-pathological structures and, in theory, solve the inverse problem of inferring the genotype from the phenotype, providing valuable diagnostic, prognostic or predictive information4,5.
The term radiomics originated from other –omics sciences (e.g., genomics and proteomics) and conveys the clear intent to invoke personalized medicine based on medical images. It traces its roots to computer-aided detection/diagnosis (CAD) of medical images6,7. However, with recent advances and the diversity of medical imaging acquisition technologies and processing, radiomics is establishing itself as an indispensable image analysis and understanding tool with applications that transcend diagnosis into prognosis and prediction approaches for personalizing patients’ management and their treatment. One of the main differentiators from CAD consists of the link that radiomics has to establish between the current features of a physio-pathological structure at the time of investigation and its temporal evolution in order to personalize the therapeutical approach8. The recent availability of large databases of digital medical images and annotated information (e.g., evolution over time or response to treatment with a given prescription, clinical and survival information), the increase of computational power based on advanced hardware (e.g., GPU, cluster or cloud computing) as well as the tremendous mathematical and algorithmic development in areas like machine or deep learning have created favorable conditions to untap the potential of the enormous amount of imaging data wealth that is being generated.
Certainly, the complementarity of other information such as clinical or laboratory data as well as interaction measurements (e.g., radiogenomics9, relating imaging to genomics, or exposomics, that is the complementary information from the interaction of the patient with environmental variables) will play a key role to drive future success of radiomics, such as accuracy and reproducibility, to levels that are acceptable for routine clinical practice.
Radiomics has been applied to many diseases including cancer and neurodegenerative diseases to name a few. Although the examples drawn here are from the cancer field, the principles presented here are generally universal across the medical imaging domain. The number of publications issued in the last years has grown almost exponentially. Although there are many review articles already about radiomics, its definition, technical details, and applications in different areas of medicine, the view of radiomics as an image mining tool lends itself naturally to application of machine/deep learning algorithms as computational instruments for advanced model building of radiomics-based signatures9,10. This will be the main subject of this article, addressing issues related to common applications in medical physics, standardization, feature extraction, model building, and validation.
II. Overview of Research and Clinical Applications of cancer Radiomics
In this section the applications of radiomics to tumor detection and characterization and prediction of outcome will be reviewed. All the studies described are retrospective and mono-institutional, except where noted.
A. Radiomics in Diagnosis
a. Cancer detection and auto contouring
The radiomics approach of combining the extraction of radiomic features with machine learning, can be used either to detect/diagnose cancer or to automatically contour the tumor lesion. Methods for radiomics-driven automatic prostate tumor detection typically use a supervised method trained on a set of features calculated from multi-modality images11. For detection of prostate cancer, features were computed in a 3×3 pixels sliding window in multimodal MRI of prostate. The voxels were tagged as cancerous or non-cancerous using a support vector machine (SVM) classifier12. In Algohary et al.13, the prostate was segmented into areas according to the aggressiveness between malignant and normal regions in the training groups. A voxel-wise random forest model (RF) with a conditional random field spatial regulation was used to classify the voxels in multimodal MRI (T1, Contrast – Enhanced (CE) T1, T2 and FLAIR) of the brain of glioblastoma multiforme (GBM) patients into five classes: non-tumor region and four tumor subregions including necrosis, edema, non-enhancing area, and enhancing area14. area12. Convolutional neural networks have also been applied to segment organs at risk in head and neck cancer radiotherapy 15 and in lung16 and liver cancers17 compared to traditional methods.
b. Prediction of histopathology and tumor stage
Radiomics holds the potential to revolutionize the conventional tumor characterization and replace classic approaches based on macroscopic variables and can be used to distinguish between malignant and benign lesions3. Breast cancer lesions, automatically detected using connected component labelling and adaptive fuzzy region growing algorithm, were classified using radiomic features as benign mass or malignant tumor on digital mammography18, dynamic contrast enhanced (DCE) MRI, and ultrasound19. A radiomic model based on mean apparent diffusion coefficient (ADC), had better accuracy than radiologist assessment for characterization of prostate lesions as clinically significant cancer (Gleason grade group ≥ 2) during prospective MRI interpretation20 . A deep learning multiparametric MRI transfer learning method has also shown the ability to classify prostate cancer high grade/low or grade 21. Radiomic models based on CT images have been used to predict the histopathology (adenocarcinoma or squamous cell carcinoma)22,23 and PET tumor stage24 of lung cancer as well as micropapillary patterns in lung adercarcinomas25.
c. Microenvironment and intra-tumor partitioning
A radiomic signature combining features from CE-CT, and 18F-FDG PET was implemented for the presence of high level of hypoxia in head and neck cancer, defined in terms of maximum tumor-to-blood uptake ratio >1.4 in the 18F-FMISO PET26. Classification and clustering methods have been developed for tumor separation into subregions (habitat imaging), which contributes to the revelation of tumor heterogeneity, and potential selection of subregions to boost radiation dose27. A radiomics analysis focused on a characterization of GBM diversity, using various diversity indices to quantify habitat diversity of the tumor as well as to relate it to underlying molecular alterations and clinical outcomes28.
d. Tumor genotype
Significant associations between the radiomic features and gene-expression patterns were found in lung cancer patients3. A radiogenomic study demonstrated the associations of radiomic phenotypes with breast cancer genomic features as mitochondrial DNA (miRNA) expressions, protein expressions, gene somatic mutations, and transcriptional activities. In particular, tumor size and enhancement texture had associations with transcriptional activities of pathways and miRNA expressions29. Radiomic models were implemented for identification of Epithelial Growth Factor Receptor (EGFR) mutant status from CT through multiple logistic regression and pairwise selection 30 and to decode ALK (anaplastic lymphoma kinase), ROS1 (c-ros oncogene 1), or RET (rearranged during transfection) fusions in lung adenocarcinoma31.
Triple negative breast cancer (TNBC) is likely to be identified by considering heterogeneity of background parenchymal enhancement, characterized by quantitative texture features on DCE-MRI, adds value to such differentiation models as they are strongly associated with the TNBC subtype32. Furthermore, TNBC has been proven to be differentiated from fibroadenoma using ultrasound (US) radiomics. A radiomics score obtained by penalized logistic regression with a least absolute shrinkage and selection operator (LASSO) analysis showed significant difference between fibroadenoma and TNBC33. The extraction of radiomic features from MR of GBM was able to predict immunohistochemically identified protein expression patterns34.
Despite large evidence of association among radiomics and genomics, few preclinical studies have demonstrated causal relationship between tumor genotype and radiomic. In one study, HCT116 colorectal carcinoma cells were grown as xenografts in the flanks of NMRI-nu mice. Then overexpression of GADD34 gene was induced by administration of HCT116 doxycycline (dox), or placebo was given. The radiomic analysis demonstrated that that gene overexpression causes change in radiomic features, as many features differed significantly between the dox-treated and placebo groups4.
e. Clinical and macroscopic variables
Radiomic features, derived from T2-w and ADC MRI scan, correlate with clinical variables that are relevant for patient’s prognosis. These include prostate specific antigen (PSA) level35 in patients with prostate cancer, and Human Papilloma Virus (HPV) Status in head and neck squamous cell carcinoma36,37. Given the well-known behavior of HPV-positive head and neck cancer which is likely to respond at a lower dose of chemoradiation, this opens the way to a CT based patient stratification for a dose de-escalation.
B. Radiomics in therapy
Because radiomic features can describe histology22 and genetic footprint 29–31 of the tumor, which are correlated with the tumor aggressiveness, they can be used to build models to predict the outcome, in terms of local/distant control or survival, of cancer therapy performed with various therapeutic options (radio-, chemotherapy, targeted molecular therapy, immunotherapy, non – ionizing radiation) or a combination of them.
a. Local control, response, recurrence
Radiomics predicts response to neoadjuvant chemoradiation assessed at time of surgery for Non-Small Cell Lung Cancer (NSCLC) and locally advanced rectal cancer38. Local control in patients treated with stereotactic radiotherapy for lung cancer was described using a PET and CT signature developed by using supervised principal component analysis was developed using features from PET and CT39. A Radiomic model was developed using first-order statistics, GLCM, and geometrical measurements computed in T2-w and ADC 3T MRI by RF approach for biochemical recurrence of prostate cancer after radiotherapy35. A total of 126 radiomic features were extracted using GLCM, GLGCM, Gabor transform, and GLSZM from contrast-enhanced 3T MRI using T1-w, T2-w, and DWI sequences to predict the therapeutic response of nasopharyngeal carcinoma (NPC) to chemoradiotherapy40. Deep learning methods with radiomics are also proposed to predict outcomes after liver41 and lung cancers radiotherapy.
b. Distant metastases
Radiomic models to predict the development of distant metastases (DM) from NSCLC on patients treated with Stereotactic Body Radiotherapy (SBRT) patients for lung cancer were developed using features from CT42 or from PET -CT39. Vallières et al. used texture-based model for the early evaluation of lung metastasis risk in soft-tissue sarcomas43 from pre-treatment FDG-PET and MRI scans comprising T1-w and T2-w fat-suppressed sequences (T2FS). A radiomic signature was developed to predict DM after locally advanced adenocarcinoma44. Analysis of the peritumoral space can provide valuable information regarding the risk of distant failure, as more invasive tumors may have different morphologic patterns in the tumor periphery. An SVM classifier was trained to predict distant failure from radiomics analysis of the peritumoral space45.
c. Survival
Aerts et al.3 built a radiomic signature consisting of a combination of four features in a retrospective lung cancer cohort, which was predictive for survival in head and neck and NSCLC independent cohorts. One textural feature calculated from GLCM, SumMean46, was identified using the LASSO procedure as an independent predictor of overall survival that complements metabolic tumor volume (MTV) in decision tree47. A radiomic signature was built from PET-CT for survival after SBRT for lung cancer 39. Deep learning was also proposed to stratify NSCLC patients according to mortality risk using standard of care CT48.
d. Molecular targeted therapy
Many tumors commonly overexpress oncogenes such as the EGFR and respond to molecular targeted therapies such as EGFR tyrosine kinase inhibitor. From the change in features between the CT acquisitions before and three weeks after therapy it was possible to identify NSCLC patients responding to treatment with gefitinib49. A radiomic prediction model was designed to stratify patients according to progression-free and overall survival after therapy with antiangiogenic for GBM 50.
e. Immunotherapy
Cancer immunotherapy by immune checkpoint blockade is a promising treatment modality that is currently under strong development, and there is a great need for models to select patients responding to immunotherapy. In a retrospective multicohort study, an eight-feature radiomic signature predictive of the presence of CD8 T cells, which is related to the tumor-immune phenotype, was developed from CE-CT images, using elastic-net regularized regression method51. The signature was successfully validated on external cohorts for discrimination of immune phenotype, and for the prediction of survival and response to anti-PD-1 or PD-L1 immunotherapy.
f. Delta-radiomics
The longitudinal study of features and of their change during the treatment, with the goal of predicting response to therapy, is called delta-radiomics. Features calculated from pretreatment and weekly intra-treatment CT change significantly during radiation therapy (RT) for NSCLC52. Delta-radiomics could possibly be performed by the Cone Beam CT (CBCT) devices for image guidance of radiotherapy treatment, thus allowing large-scale study of tumor response to total dose, fractionation and fraction dose. It has been shown that reproducible features can be extracted from CBCT53 predictive for overall survival in NSCLC patients as much as features from CT54. Nevertheless, the studies on CBCT delta-radiomics are still limited to assessment of feasibility and reproducibility55.
g. Prediction of side effects
Radiomics-based models can help early identify the development of side effects such as radiation induced lung injury (RILI). The change from pre- to post-treatment (at 3, 6, and 9 months) CT features significantly correlates with lung-injury as scored by oncologist post-SBRT for lung cancer and was found to be correlated with dose and fractionation56.
A logistic regression–based classifier was constructed to combine information from multiple features to identify patients that will develop grade ≥2 radiation pneumonitis among those who received RT for esophageal cancer57. The addition of normal lung image features produced superior model performance with respect to traditional dosimetric and clinical predictors of radiation pneumonitis (RP), suggesting that pre-treatment CT radiomic features should be considered in the context of RP prediction. CT radiomic features were extracted from the total lung volume defined using the treatment planning scan for RP 58.
h. Differentiation of recurrence from benign changes
The differentiation of tumor recurrence from benign radiation-induced changes in follow-up images can be a major challenge for the clinician. A radiomic signature consisting of 5 image-appearance features from CT demonstrated high discriminative capability to differentiate recurrence of lung tumor from consolidation and opacities in SBRT patients59. Similarly, a combination of five radiomic features from CE-T1w and T2w MR were found to be capable of distinguishing necrosis from progression in follow up MR images in patients treated with Gamma Knife radiosurgery for brain metastases60.
i. Non ionizing radiation and other therapies
Radiomic features in MRI respond differently when Laser interstitial thermal therapy (LITT), a highly promising focal strategy for low-grade, organ-confined prostate cancer, is performed on cancer or healthy prostate tissue. A radiomic signature then could allow to assess if prostate cancer is successfully ablated61. A radiomic model was predictive of complete response after transcatheter arterial chemoembolisation combined with high-intensity focused ultrasound treatment in hepatocellular carcinoma62.
III. Radiomics Analysis with Machine and Deep Learning Methods
A. Preprocessing
Prior to radiomics analysis, preprocessing steps need to be applied to the images, which aim at reducing image noise, enhancing image quality, enabling the reproducible and comparable radiomic analysis. For some imaging modalities, such as PET, the images should be converted to a more meaningful representation (standardized uptake value, SUV). Image smoothing can be achieved by average or Gaussian filters63. Voxel size resampling is important for datasets that have variable voxel size64. Specifically, isotropic voxel size is required for some texture feature extraction. There are two main categories of interpolation algorithms: Polynomial and spline interpolation. Nearest neighbor is a zero-order polynomial method that assigns grey-level values of the nearest neighbor to the interpolated point. Bilinear or trilinear interpolation and bicubic or tricubic interpolation are often used for 2D in-plane interpolation or 3D cases. Cubic spline and convolution interpolation are third order polynomial method that interpolates smoother surface than linear method, while being slower in implementation. Linear interpolation is a rather commonly used algorithm, since it neither leads to the rough blocking artifacts images that are generated by nearest neighbors, nor will it cause out-of-range grey levels that might be produced by higher order interpolation65.
In the context of feature-based radiomics analysis, as discussed below, the computation of textures would require discretization of the grey levels (intensity values). There are two ways to do the discretization: fixed bin number N and fixed bin width B. For fixed bin number, we first decide a fixed number of N bins, and the grey levels will be discretized into these bins using the formula below:
| (1) |
where Xgl,k is the intensity of kth voxel.
For fixed bin width, starting at a minimum Xgl,min, a new bin will be assigned for every intensity interval of wb. Discretized grey levels are calculated as follow:
| (2) |
The fixed bin number method is better when the modality used is not well calibrated. It maintains the contrast and makes the images of different patients comparable, but loses the relationship between image intensity, while fixed bin size method keeps the direct relationship with the original scale. Some investigations about the effect of both methods have shown that fixed bin size method offered better repeatability and thus may be suitable for intra- and inter- patient studies, however, this remains a subject of ongoing research66,67. In CT-radiomics the image pixel intensity maps to the Hounsfield Units (HU) and thus is much more directly comparable and interpretable. MRI-related modalities are more challenging since the pixel intensities are not directly interpretable, rather need to normalized relative to some standard reference (e.g., contralateral brain, or normal appearing white matter in neuroimaging, psoas muscle in abdominal imaging, etc.).
B. Machine and Deep Learning Algorithms for Radiomics
Machine and deep learning algorithms provide powerful modeling tools to mine the huge amount of image data available, reveal underlying complex biological mechanisms, and make personalized precision cancer diagnosis and treatment planning possible. Hereafter, two main types - feature-engineered (conventional radiomics) and non-engineered (deep learning-based) radiomics modeling methods – will be briefly introduced. Generally speaking, machine learning methods can also be divided into supervised, unsupervised and semi-supervised for both feature-based and featureless methods. Each of these categories will be briefly discussed in the following sections. A workflow diagram illustrating the radiomics analysis process after image acquisition is shown in Fig. 1.
Fig. 1.
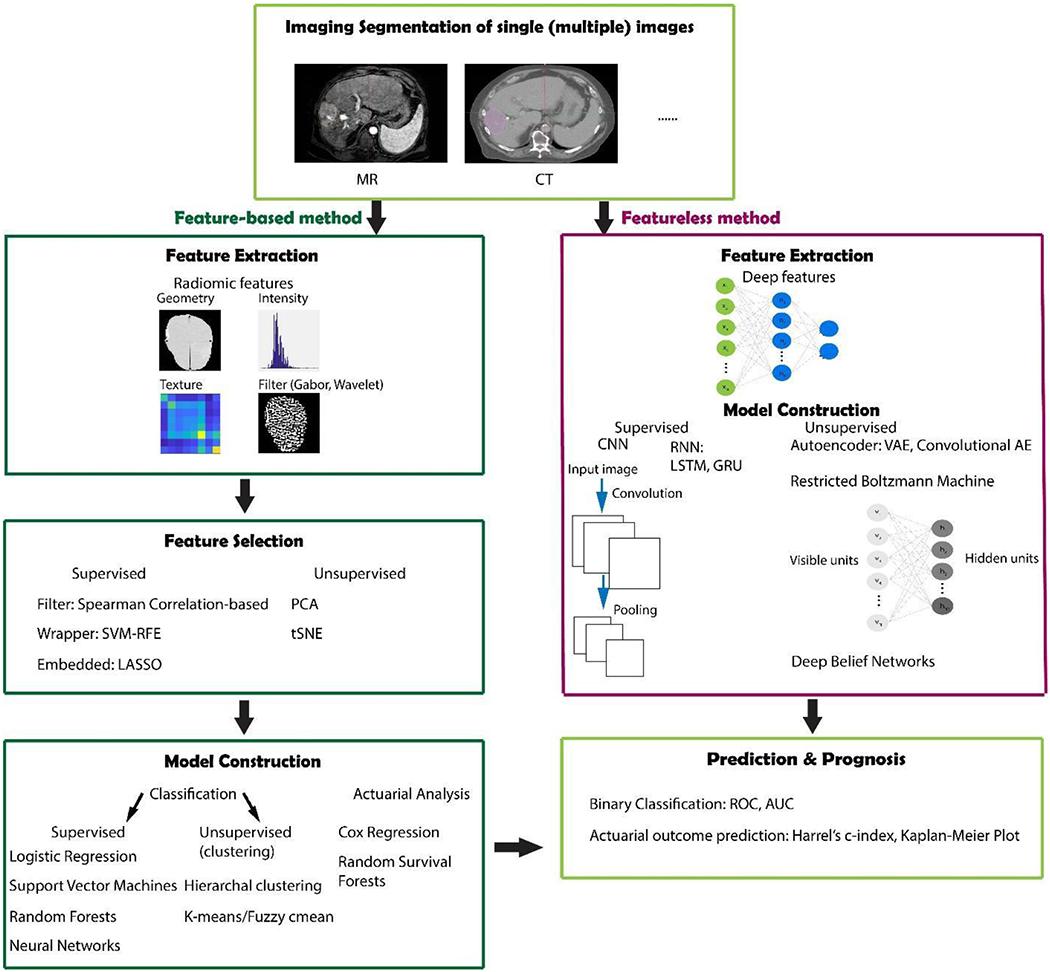
Workflow for radiomics analysis with feature-based (conventional machine learning) and featureless (deep learning) approaches.
a. Feature-engineered radiomics methods
Traditionally, the radiomic features being extracted are hand-crafted features that capture characteristic patterns in the imaging data, including shape-based, first-, second-, and higher order statistical determinants and model-based (e.g. fractal) features. Feature-based methods require a segmentation of the region of interest (ROI), either through a manual, semi-automated, or automatic methods. Shape-based features are external representations of a region, that characterize the shape, size and surface information of the ROIs68. Typical metrics include sphericity, and compactness3,43,69,70. First-order features (e.g. mean, median) describe the overall intensity and variation of the ROIs, while ignoring spatial relations8,24. Second-order (texture) features in contrast can provide inter-relationships among voxels. Textural features can be extracted from different matrices, e.g. grey-level co-occurrence matrix (GLCM), grey-level run-length matrix (GLRLM), etc35,46,71. Semantic features are another type of feature that can be extracted from medical images. These features describe qualitative features of the image typically used in the radiology workflow.
Hundreds or even thousands of radiomic features are not uncommon when we deal with outcome modeling. Feature selection and/or extraction thus is a crucial step that aims at obtaining the optimal feature subset or feature representation that correlates most with the endpoint and meanwhile correlates least between each other. After the feature subset is obtained, various machine learning algorithms can be applied based on them. 14 feature selection and 12 classification methods were evaluated in terms of their predictive performance on two independent lung cancer cohorts72. Sometimes, the feature selection and model construction can be implemented together, called the embedded method, such as least absolute shrinkage and selection operator (LASSO)73. In contrast, wrapper methods select the features based on the models’ performance for different subsets of features, for which we need to rebuild the model again after features are selected, for instance, recursive feature elimination support vector machines (SVM-RFE). Filter method also separates the feature selection and model construction processes, whose uniqueness of it is its independence of the classifier being used for the subsequent model building, such as Pearson correlation-based feature ranking. In any feature selection method, it is essential to ensure that there is no “double dipping” into the training data for both feature selection, hyperparameter optimization and model selection. Rather the methods of “nested cross validation” should be used in order to prevent overfitting or incorrect estimates of generalization. According to whether or not the labels (ground truths) are used, feature selection and extraction can be divided into supervised, unsupervised and semi-supervised ways. The three feature selection methods discussed above are mostly supervised. Examples of unsupervised methods are principle component analysis (PCA)74, clustering and t-Distributed Stochastic Neighbor Embedding (t-SNE)75. PCA uses an orthogonal linear transformation to convert the data into a new coordinate system so that large variances are projected to orthogonal coordinates. Clustering is another feature extraction algorithm which aims at finding relevant features and combining them by their cluster centroids based on some similarity measure, such as K-means and hierarchical clustering 76. Unsupervised consensus clustering identified robust imaging subtypes using dynamic CE-MRI data for patients with breast cancer77. tSNE is a dimension reduction method capable of retaining the local structure (pairwise similarity) of data, while revealing some important global structure.
In the medical field, two types of questions are mainly investigated, binary problems (classification), such as whether or not a disease has recurred, the patient is alive beyond certain time threshold, etc; and survival analysis, that is able to show if a risk factor or treatment affects time to event. For the classification problem logistic regression fits the coefficients of the variables to predict a logit transformation of the probability of the presence of the event. SVM, frequently used in Computed Aided Diagnosis (CAD)6 and radiomics32,59,76,78, learns an optimal hyperplane that separates the classes as wide as possible, while trying to balance with misclassified cases. SVM can also perform non-linear classification using the “kernel trick” -- different basis functions (e.g. radial basis function), mapping to higher dimensional feature space. The hyperplane maximizes the margin between the two classes in a non-linear feature space. SVM also tolerates some points on the wrong side of the boundary, thus improving model robustness and generalization79. RF is based on decision trees, a popular concept in machine learning especially in the field of medicine, because their representation of hypotheses as sequential “if-then” resembles human reasoning80. RF applies bootstrap aggregating to decision trees and improve the performance by lowering the high variance of the trees81. Risk assessment models (classification and survival) were constructed via RFs and imbalance adjustment strategies for locoregional recurrences and distant metastases in head and neck cancer82
Neural networks, though usually used in the featureless context, can also be used in conventional feature selection and modeling22,38,78. These algorithms are mainly for supervised learning, while in particular in the medical field, there are a lot of data without labeling, in these cases, semi-supervised learning can be applied to make use of the unlabeled data combined with the small amount of labeled data. The self-training is bootstrapped with additional labelled data obtained from its predictions83. The transductive SVM (TSVM) tries to keep the unlabeled data as far away from the margin as possible84. Graph-based methods construct a graph connecting similar observations and enable the class information being transported through the graph85.
For the survival analysis, Cox regression86, random survival forests87 and support vector survival88 methods are also available to investigate the presence of a set of variables that may affect survival time. Due to the length limit, we will not go into the details. Interested readers can refer to the references to read more about these algorithms.
b. Non feature-engineered radiomics methods
Though hand-crafted features introduced above provide prior knowledge, they also suffer from the tedious designing process and may not faithfully capture the underlying imaging information. Alternatively, with the development of deep learning technologies based on multi-layer neural networks, especially the convolutional neural networks (CNN), the extraction of machine learnt features is becoming widely applicable recently. In deep learning, the processes of data representation and prediction (e.g, classification or regression) are performed jointly89. In such a case, multi-stack neural layers of varying modules (e.g., convolution or pooling) with linear/non-linear activation functions perform the task of learning the representations of data with multiple levels of abstraction and subsequent fully connected layers are tasked with classification, for instance. A typical scenario to get such features is to use the data representation CNN layers as feature extractor. Each hidden layer module within the network transforms the representation at one level. For example, the first level may represent edges in an image oriented in a particular direction, the second may detect motifs in the observed edges, the third could recognize objects from ensembles of motifs89. Patch-/pixel-based machine learning (PML) methods use pixel/voxel values in images directly instead of features calculated from segmented objects as in other approaches 90,89. Thus PML removes the need for segmentation, one of the major sources of variability of radiomic features. Moreover, the data representation removes the feature selection portion eliminating associated statistical bias in the process. For the CNN network, either self-designed (from scratch) or existing structures, e.g. VGG91, Resnet92, can be used. Depending on the data size, we can choose to fix the parameters or fine tune the network using our data, also called transfer learning. Instead of using deep networks as feature extractors, we can use them directly for the whole modeling process. Similarly to the conventional machine learning methods, there are also supervised, unsupervised and semi-supervised methods. CNN are similar to regular neural networks, but the architecture is modified to fit to the specific input of large-scale images. Inspired by the Hubel and Wiesel’s work on the animal visual cortex93, local filters are used to slide over the input space in CNNs, which not only exploit the strong local correlation in natural images, but also reduce the number of weights significantly by sharing weights for each filter. Recurrent neural networks (RNN) can use their internal memory to process sequence inputs and take the previous output as inputs. There are two popular types of RNN – Long short-term memory (LSTM)94 and Gated recurrent units (GRU)95. They were invented to solve the problem of vanishing gradient for long sequences by internal gates that are able to learn which data in the sequence is important to keep or discard. Deep autoencoders (AE), which are unsupervised learning algorithms, have been applied to medical imaging for latent representative feature extraction. There are variations to the AEs, such as variational autoencoders that resemble the original AE and variational Bayesian methods to learn a probability distribution that represents the data96, convolutional autoencoders that preserve spatial locality97, etc. Another unsupervised method is the restricted Boltzmann machine (RBM), which is consists of visible and hidden layers98. The forward pass learns the probability of activations given the inputs, while the backward pass tries to estimate the probability of inputs given activations. Thus, the RBMs lead to the joint probability distribution of inputs and activations. Deep belief networks can be regarded as a stack of RBMs, where each RBM communicates with previous and subsequent layers. RBMs are quite similar with AEs, however, instead of using deterministic units, like RELU, RBMs use stochastic units with certain distribution. As mentioned above, labeled data is limited, especially in the medical field. Neural network based semi-supervised approaches combine unsupervised and supervised learning by training the supervised network with an additional loss component from the unsupervised generative models (e.g. AEs, RBMs)99.
Machine learning methods are highly effective with large number of samples; however, they suffer from overfitting pitfalls with limited training samples. For deep learning, data augmentation (e.g. by affine transformation of the images) during training is commonly implemented. Transfer learning is another way to reduce the difficulty in training. Using deep models trained on other dataset (natural images) and then fine-tune on the target dataset. The structures of the networks can also be modified to reduce overfitting, such as, by adding dropout and batch normalization layers. Dropout randomly deactivates a fraction of the units during training and can be viewed as a regularization technique that adds noise to the hidden units100. Batch normalization reduces the internal covariate shift by normalizing for each training mini-batch101.
Comparing with feature-based methods, deep learning methods are more flexible and can be used with some modifications in various tasks. In addition to classification, segmentation, registration, and lesion detection are widely explored by deep learning techniques. Fully CNN (FCN), trained end-to-end, merge features learnt from different stages in the encoders and then upsampling low resolution feature maps by deconvolutions102. Unet, built upon FCN, with the pooling layers being replaced by upsampling layers, resulted in a nearly symmetric U-shaped network103. Skipping structures combines the context information with the unsampled feature maps to achieve higher resolution. CNN, trained end-to-end from clinical images were directly used for binary classification of skin cancer and achieved performance on par with experts104. Chang et al proposed a multi-scale convolutional sparse coding method that provides an unsupervised solution for learning transferable base knowledge and fine-tuning it towards target tasks105.
C. Validation and Benchmarking of Radiomics Models
Once models are developed using the selected predictors, quantifying the predictive ability of the models (validation) is necessary. Based on the TRIPOD criteria106, there are 4 types of validation: 1a. Developing and validating on the same data, which gives apparent performance. This evaluation is usually optimistic estimation of the true performance. 1b. Developing the models using all the data, then using resampling techniques to evaluate the performance. 2a. Randomly split the data into 2 groups for development and validation separately. 2b. Split the data non-randomly (e.g. by location or time), which is stronger than 2a. 3 & 4. Develop the model using one data set and validate on separate data. It is ideal if there is a separate data set for external validation, however, in the frequent case that only a single data set is available, internal validation (1b) is required. Two popular resampling methods are bootstrapping and cross-validation. Feature selection, which is required before machine learning, should precede cross-validation, or it will lead to a selection bias due to the leak of information by the pre-filtering of the features107.
Radiomic classifiers output a score that indicates the likelihood of one event to happen, and a threshold, to generate positive or negative predictions according to the task at hand. For example, fewer false positives would be required if we are implementing a conservative experiment, thus larger threshold will be preferred. Classifiers are evaluated using either a numeric metric (e.g. accuracy), or the so-called confusion matrix, or a graphical representation of performance, such as a receiver operating characteristic curve (ROC), a two-dimensional graph with true positive rate being the Y axis, and false positive rate the X axis. It has the advantage that they show classifier performance without regard to threshold and class distribution, thus widely used in model evaluation. The area under an ROC curve (AUC) is more convenient when comparing, and is equivalent to the probability that the classifier will rank a randomly chosen positive instance higher than a randomly chosen negative instance108. For survival analysis, Harrell’s C index109 is commonly used to measure discrimination ability of the model, which is motivated by Kendall’s tau correlation. Harrell defines the overall C index as the proportion of all usable pairs in which the predicted risk probabilities and outcomes are concordant (Usable pairs are two cases that at least one of them is event)110.
Kaplan-Meier (KM) curves are used to estimate the survival function from lifetime data, and also used to compare different risk groups. The risk groups can be patients that are treated with certain plan and the control group, or they can be the outputs from a survival model (e.g. Cox model) that divides the patients into high and low risk groups. It is highly recommended to visualize confidence intervals of the curves. The log rank test gives a quantitative evaluation of the statistical significance of the difference for different curves, which is also widely provided for KM curves111.
IV. Implementation in medical physics practice
A. Software tools for radiomics
In most published research studies in radiomics, in-house developed methods are used. However, some research groups developed image analysis/radiomic software tools, both commercial or open source, available to the scientific community. The main goals of these tools are: 1) to speed up the development of competences based on more recent skills on radiomics; 2) to allow reproducibility and comparability of results from different research groups, and 3) to standardize both feature definitions and computation methods to guarantee the reliability of radiomic results 112,113.
Table 1 shows a list of the software, web platforms, and toolkits available free of charge for the extraction of radiomics features, along with some of their main functionalities and relevant information. Given the high pace of radiomic developments, the list is not exhaustive and does not intend to cover all possible solutions. Furthermore, considering recent and increased interest in the radiomic field, many other dedicated tools are under development. All the open source solutions shown in this overview have been implemented by research teams (MaZda114, LifeX 115, ePAD116, HeterogeneityCAD3, PyRadiomics/Radiomics 117, QuantImage 118, the Texture Analysis Toolbox43, QIFE119, IBEX 120, and MedomicsLab) and are capable of analyzing CT, MRI, and PET, some of them can process also other medical images, such as mammography, radiography, or ultrasound.
Table I.
Open access software programs for radiomics analysis.
| Software/Toolbox | MaZda114 | lifeX115 | ePAD116 | QIFE119 | HeterogeneityCAD 3 | PyRadiomics/Radiomics117 | QuantImage 118 | Texture Analysis Toolbox43 | IBEX120 | MEDomicsLab |
|---|---|---|---|---|---|---|---|---|---|---|
| Research group | Institute of Electronics, Technical University of Lodz, Poland | IMIV, CEA, Inserm, CNRS, Univ. Paris-Sud, Université Paris Saclay | Rubin Lab, Stanford University | Sandy Napel, Stanford University | V.Narayan, J. Jagadeesan | Dana-Farber Cancer Institute, Brigham Women’s Hospital Harvard Medical School, Boston | University of Applied Science and Arts, Western Switzerland | M. Vallières | The University of Texas MD Anderson Cancer Center, Houston, Texas | MEDomics consortium |
| Image modalities | CT, MRI, PET | CT, MRI, PET, ultrasound | CT, MRI, radiography | CT, MRI, PET | CT, MRI, PET | CT, MRI, PET | CT, MRI, PET | CT, MRI, PET | CT, MRI, PET | |
| Segmentation | YES | YES | YES | NO | NO | NO | NO | NO | YES | NO |
| Segmentation methods | manual, automatic (threshold, flood-filling) | manual, automatic (threshold, snake) | Manual | / | / | / | / | / | manual, automatic (threshold) | / |
| Radiomic features: morphology | YES | YES | NO | YES | YES | YES | YES | YES | YES | YES |
| statistical 1° order | YES | YES | YES | YES | YES | YES | YES | YES | YES | YES |
| statistical 2° order | YES | YES | YES | YES | YES | YES | YES | YES | YES | YES |
| statistical 3° order | YES | YES | NO | YES | YES | YES | YES | YES | YES | YES |
| Filtering | YES | NO | NO | NO | NO | YES | NO | NO | YES | YES |
| Feature selection | YES | NO | NO | NO | NO | NO | NO | YES | NO | YES |
| Feature selection methods | Fisher score, classification error, corr. coeff, mutual informat., minimal classification error | / | / | / | / | / | / | Maximal information coefficient | / | False discovery avoidance, Elastic Net, minimum Redundancy Maximum Relevance |
| Stratification | NO | NO | NO | NO | NO | NO | NO | NO | NO | YES |
Four software programs (MaZda, LifeX, ePAD, IBEX) offer the possibility of manually or automatically segmenting medical images. Three toolkits (HeterogeneityCAD, PyRadiomics/Radiomics, QIFE) are designed exclusively for the extraction of features. They can be embedded in more complete solutions (e.g. 3D Slicer 121). Morphological, first, second and third order statistical features can be extracted by all software solutions, except for ePAD. Four of them (TexRAD, MaZda, PyRadiomics/Radiomics, IBEX) offer also the possibility of extracting features from filtered images. Of note, MEDomicsLab is an open-source software currently being developed by a consortium of research institutions, which will be available in the second half of 2019.
B. Commercial Programs for radiomics
Commercial software programs are also becoming increasingly available due to the interest of many medical device incumbents as well as newcomers such as commercial spin-off of research groups or de novo start-up companies. Such software programs can be divided into:
a. Research platforms
These platforms enable the discovery of new signatures by linking quantitative imaging biomarkers, clinical and –omics data to clinical endpoints. They are usually considered non-medical devices in that they do not affect the clinical routine, run usually on independent workstations, and are not used to drive clinical decisions. Their main differentiator from open access software consists of workflow optimization and efficiency improvements, enabling an automatic, end-to-end seamless processing pipeline. TexRad®, QIDS®, RadiomiX, iBiopsy® and EVIDENS offer research capabilities at a different level, ranging from simple features extraction to image filter application and machine learning modules. In the research mode, these software programs are usually open to process any 3D image, DICOM or not, up to 2D digital pathology images (histomics or pathomics).
b. Clinically validated software programs,
In order to use decision support systems (DSSs), based on an already discovered signatures and thoroughly validated on large independent datasets, also known as clinical grade DSS, in clinical practice, a regulatory clearance is usually needed, as they fall within the definition of medical devices in many regulatory systems, e.g., class I or II medical device as a function of their intended use (e.g. mere support to decision versus a computer aided diagnosis/prognosis). DSSs are usually limited to a specific modality, mostly CT, and to a specific disease in a specific body district: these constraints come primarily from the intended use definition to which these DSSs are subjected to be compliantly marketed.
Research tools or clinical grade DSSs can be embedded into more comprehensive platforms such as Picture Archive and Communication Systems (PACS), Hospital Information Systems (HIS), Oncology Information Systems (OIS) or Treatment Planning Systems (TPS), or being stand-alone. Usually, large medical device incumbents tend to embed DSSs into their research or clinical solutions, while newcomers often offer their solution as a standalone system.
It is not unusual that large medical device players embed open access or commercial software programs to provide their customers with the possibility of exploring or exploiting radiomic potential: examples are IntelliSPace Discovery (Philips, the Netherlands) which interfaces to Pyradiomics, Advantage Workstation (GE, Buc, France) which interfaces through a plugin to Quantib™ Brain or Syngo.via Frontier (Siemens, Erlangen, Germany) which interfaces to RadiomiX. It is also beneficial to mention the platform (www.envoyai.com) which offers the possibility of sharing applications and, once solutions reached the product maturity, to commercialize them.
V. Current challenges and recommendations
A. Interpretability issues
It is recognized that machine learning algorithms tend to generally trade interpretability for better prediction. Hence, clinicians are still reluctant to embrace these methods as part of their clinical practice, because they have long been perceived them as “black boxes”, meaning that it is difficult to determine how they arrive at their predictions. For example, it is difficult to understand deep neural networks due to the large number of interacting, non-linear parts 122,123. In order to improve interpretability of radiomics for the clinician, methods based on graph approaches can be utilized124, and in the context of deep learning better visualization tools are being developed such as maps highlighting regions of the tumor that impact the prediction of the deep learning classifier are also being proposed 123.
B. Repeatability and Reproducibility issues
In radiomics, repeatability is measured by extraction of features from repeated acquisition of images under identical or near-identical conditions and acquisition parameters125, whereas reproducibility or robustness, is assessed when measuring system or parameters differ. These can be assessed by use of digital or physical phantoms. Physical phantoms usually contain inserts of different with different density, shape or texture properties in order to produce a wide range of radiomics feature values. These phantoms allow to assess the reproducibility or robustness of the entire workflow, from image acquisition to extraction of radiomic features. Their major drawback is that they do not reflect the variability of human anatomy in the clinical scenario.
A phantom for radiomics was created for use with CT 113 or CBCT126 called Credence Cartridge Radiomics (CCR) Phantom. This consisted of 10 cartridges with different density and texture properties in order to produce a wide range of radiomics feature values: wood, rubber, cork, acrylic, and plaster. Phantoms for PET with heterogeneous lesions have been also proposed, e.g. with different 3D printed inserts reflecting different heterogeneities in FDG uptake127.
Digital phantoms are usually scans of patients acquired under controlled conditions. They are therefore realistic, but cannot be used for studying radiomic features’ sensitivity to the image acquisition and its parameters. A dataset consisting of 31 sets of repeated CT scans acquired approximately 15 minutes apart is now publicly accessible through The Reference Image Database to Evaluate Therapy Response (RIDER). This dataset allows “test-retest” analysis, a comparison of the results from images acquired within a short time on the same patient128.
C. Factors affecting stability
For CT, inter-scanner variability of image features produces differences in extracted features that are comparable to the variability in patient images acquired by the same scanner113. The choice of methods of reconstruction, such as filtered back projection or iterative algorithm, also affect radiomic feature129. Smoothing of the image and reducing the slice thicknesses can improve reproducibility of CT-extracted features128,130. In PET imaging, textural features are sensitive to different acquisition modes 131,132, reconstruction algorithms, and their user-defined parameters such as the number of iterations, the post-filtering level, input data noise, matrix size, and discretization bin size133,134.
Radiomic features extracted from MRI scans depend on the field of view, field strength, reconstruction algorithm and slice thickness. Results of the DCE MRI depend on the contrast agent dose, method of administration, and the pulse sequence used. The radiomic features extracted from DW-MRI depend on acquisition parameters and conditions as k-space trajectory, gradient strengths and b-values. The repeatability of MR-based radiomic features has been investigated135 using a ground truth digital phantom of brain glioma patients and an MRI simulator capable of generating images according to different acquisition (field strength, pulse sequence, arrangement of field coils) and reconstruction methods. It was found that some features are subject to small changes, compared with clinical effect size.
In presence of significant respiratory tumor motion as in the case of lung cancer, conventional PET images are influenced by motion as, because of their relatively long acquisition times, the counts measured are averaged over multiple breathing cycles. Respiratory-gated PET accounts for respiratory motion and textural features from gated PET have been found robust136.
Segmentation affects the radiomics workflow, regardless of the imaging technique, because many extracted features depend on the segmented region2,5. Semiautomatic segmentation algorithms may improve the stability of radiomic features137, and recently available fully automatic segmentation tools may be as accurate as manual segmentation by medical experts138.
The studies on the comparisons of the performance of many classifier and feature selection methods indicate that the choice of classification method is the most dominant source of models’ predictive performance variability 72. Fourteen feature selection algorithms were compared on a set of 464 lung cancer patients considering 440 radiomic variables 76. The feature selection method based on the Wilcoxon signed-rank (WLCX) test had the highest prognostic performance with high stability against data perturbation. Interestingly, WLCX is a simple univariate method based on ranks, which does not take into account the redundancy of selected features during feature ranking. In a comparison of performance of 24 feature selection methods for radiomic signature building for lung cancer histology it was shown that RELIEF with its variants were the best performing methods22.
D. Quality, Radiomics quality score
The workflow for radiomic studies involves several steps, from data acquisition, selection, and curation, to feature extraction, feature selection, and modelling. There is an important need that radiomics studies are properly designed and reported to ensure the field can continue to develop and produce clinically useful tools and techniques. A number of issues can arise providing misleading information, including imaging artifacts, poor study design, overfitting of data, and incomplete reporting of results8,139. Although imaging artifacts are inevitable in medical imaging, consistent imaging parameters may help reduce variability in radiomic features126. To minimize the potential of overfitting of radiomic models, 10 patients are needed for each feature in the final model140. Ideally, an independent external validation dataset is also used to confirm the prognostic ability of any radiomic model. The radiomics quality score (RQS) has recently been developed to assess all areas of a radiomic study and determine whether it is compliant with best practice procedures139, emulated from the TRIPOD initiative previously described.
E. Standardization and harmonization
Although research in the field of radiomics has drastically increased over the past several years, there still remains a lack of reproducibility and validation of current radiomic models. There are currently no guidelines and standard definitions for radiomic features and for constructing these features into clinical models. Current initiatives are underway to improve standardization and harmonization in radiomic studies.
As a part of radiomic signature validation, there are efforts to explore distributed feature sharing and model development across contributing institutions141. A key component in this exercise is the assessment and redressal of batch effects142 and confounding variables across contributing sites, so as to ameliorate systematic yet unmeasured sources of variation. Another key component is the use of methods to harmonize data as well as model parameters across study sites, with the intent of meaningful comparisons across clinical population143. Such efforts are necessary to enable the widespread and generalizable development of models that are transportable across institutions. In addition to the careful calibration and stability analysis of radiomic features within predictive models, there is also a need for ensuring model robustness through approaches like noise injection144. Adversarial training approaches from neural networks can have value in the modern deep learning modeling area by incorporating not only positive examples but negative ones too145. The workflow for computing features is complex and involves many steps, often leading to incomplete reporting of methodological information (e.g., texture matrix design choices and gray-level discretization methods). As a consequence, few radiomics studies in the current literature can be reproduced from start to end.
To accelerate the translation of radiomics methods to the clinical environment, the Image Biomarker Standardization Initiative (IBSI)65 has the goal to provide standard definitions and nomenclature for radiomic features, reporting guidelines, and to provide benchmark datasets and values to verify image processing and radiomic feature calculations. Figure 2 presents the standardized radiomics workflow defined by the IBSI. The IBSI aims at standardizing both the computation of features and the image processing steps required before feature extraction. For this purpose, a simple digital phantom was designed and used in Phase 1 of the IBSI to standardize the computation of 172 features from 11 categories. In Phase 2 of the IBSI, a set of CT images from a lung cancer patient was used to standardize radiomics image processing steps using 5 different combinations of parameters including volumetric approaches (2D vs 3D), image interpolation, re-segmentation and discretization methods. The initiative is now reaching completion and a consensus on image processing and computation of features has been reached over time.
Fig. 2.

Radiomics computation workflow as defined by the IBSI.
Overall, the use of standardized computation methods would greatly enhance the reproducibility of radiomics studies, and it may also lead to standardized software solutions available to the community. It would also be desirable that the code of existing software be updated to conform with standards established by the IBSI. Furthermore, it is essential to include in radiomics studies the comprehensive description of feature computation details as defined by the IBSI65 and Vallières et al146, as shown in Table 2. Ultimately, we envision the use of dedicated ontologies to improve the interoperability of radiomics analyses via consistent tagging of features, image processing parameters and filters. The Radiomics Ontology (www.bioportal.bioontology.org/ontologies/RO) could provide a standardized means of reporting radiomics data and methods, and would more concisely summarize the implementation details of a given radiomics workflow.
Table 2.
| GENERAL | |
| Image acquisition | Acquisition protocols and scanner parameters: equipment vendor, reconstruction algorithms and filters, field of view and acquisition matrix dimensions, MRI sequence parameters, PET acquisition time and injected dose, CT x-ray energy (kVp) and exposure (mAs), etc. |
| Volumetric analysis | Imaging volumes are analyzed as separate images (2D) or as fully-connected volumes (3D). |
| Workflow structure | Sequence of processing steps leading to the extraction of features. |
| Software | Software type and version of code used for the computation of features. |
|
| |
| IMAGE PRE-PROCESSING | |
| Conversion | How data were converted from input images: e.g, conversion of PET activity counts to SUV, calculation of ADC maps from raw DW-MRI signal, etc. |
| Processing | Image processing steps taken after image acquisition: e.g., noise filtering, intensity non-uniformity correction in MRI, partial-volume effect corrections, etc. |
|
| |
| ROI SEGMENTATION a;b | How regions of interests (ROIs) were delineated in the images: software and/or algorithms used, how many different persons and what expertise (specialty, experience), how a consensus was obtained if several persons carried out the segmentation, in automatic or semi-automatic mode, etc. |
|
| |
| INTERPOLATION | |
| Voxel dimensions | Original and interpolated voxel dimensions. |
| Image interpolation method | Method used to interpolate voxels values (e.g, linear, cubic, spline, etc.) as well as how original and interpolated grids were aligned. |
| Intensity rounding | Rounding procedures for non-integer interpolated gray levels (if applicable), e.g., rounding of Hounsfield units in CT imaging following interpolation. |
| ROI interpolation method | Method used to interpolate ROI masks. Definition of how original and interpolated grids were aligned. |
| ROI partial volume | Minimum partial volume fraction required to include an interpolated ROI mask voxel in the interpolated ROI (if applicable): e.g., a minimum partial volume fraction of 0.5 when using linear interpolation. |
|
| |
| ROI RE-SEGMENTATION | |
| Inclusion/exclusion criteria | Criteria for inclusion and/or exclusion of voxels from the ROI intensity mask (if applicable), e.g., the exclusion of voxels with Hounsfield units values outside a pre-defined range inside the ROI intensity mask in CT imaging. |
|
| |
| IMAGE DISCRETIZATION | |
| Discretization method | Method used for discretizing image intensities prior to feature extraction: e.g., fixed bin number, fixed bin width, histogram equalization, etc. |
| Discretization parameters | Parameters used for image discretization: the number of bins, the bin width and minimal value of discretization range, etc. |
|
| |
| FEATURE CALCULATION | |
| Features set | Description and formulas of all calculated features. |
| Features parameters | Settings used for the calculation of features: voxel connectivity, with or without merging by slice, with or without merging directional texture matrices, etc. |
|
| |
| CALIBRATION | |
| Image processing steps | Specifying which image processing steps match the benchmarks of the IBSI. |
| Features calculation | Specifying which feature calculations match the benchmarks of the IBSI. |
In order to reduce inter-observer variability, automatic and semi-automatic methods are favored.
In multimodal applications (e.g., PET/CT, PET/MRI, etc.) ROI definition may involve the propagation of contours between modalities via co-registration. In that case, the technical details of the registration should also be provided.
Finally, some guiding principles already exist to help radiomics scientists further implement the responsible research paradigm into their current practice. A concise set of principles for better scientific data management and stewardship, the “FAIR guiding principles”147, stating that all research objects should be findable, accessible, interoperable, and reusable. Implementation of the FAIR principles within the radiomics field could facilitate its faster clinical translation. First, all methodological details and clinical information must be clearly reported or described to facilitate reproducibility and comparison with other studies and meta-analyses. Second, models must be tested in sufficiently large patient datasets distinct from teaching (training and validation) sets to statistically demonstrate their efficacy over conventional models (e.g., existing biomarkers, tumor volume, cancer stage, etc.). To allow for optimal reproducibility potential and further independent testing, all data, final models and programming code related to a given study needs to be made available to the community. Table 3 provides guidelines that can help to evaluate the quality of radiomics studies146. More guidelines on reproducible prognostic modeling can also be found in the TRIPOD statement106.
Table 3.
| IMAGING | |
| Standardized imaging protocols | Imaging acquisition protocols are well described and ideally similar across patients. Alternatively, methodological steps are taken towards standardizing them. |
| Imaging quality assurance | Methodological steps are taken to only incorporate acquired images of sufficient quality. |
| Calibration | Computation of radiomics features and image processing steps match the benchmarks of the IBSI. |
|
| |
| EXPERIMENTAL SETUP | |
| Multi-institutional/external datasets | Model construction and/or performance evaluation is carried out using cohorts from different institutions, ideally from different parts of the world. |
| Registration of prospective study | Prospective studies provide the highest level of evidence supporting the clinical validity and usefulness of radiomics models. |
|
| |
| FEATURE SELECTION | |
| Feature robustness | The robustness of features against segmentation variations and varying imaging settings (e.g., noise fluctuations, inter-scanner differences, etc.) is evaluated. Unreliable features are discarded. |
| Feature complementarity | The inter-correlation of features is evaluated. Redundant features are discarded. |
|
| |
| MODEL ASSESSMENT | |
| False discovery corrections | Correction for multiple testing comparisons (e.g., Bonferroni or Benjamini- Hochberg) is applied in univariate analysis. |
| Estimation of model performance | The teaching dataset is separated into training and validation set(s) to estimate optimal model parameters. Example methods include bootstrapping, cross-validation, random sub-sampling, etc. |
| Independent testing | A testing set distinct from the teaching set is used to evaluate the performance of complete models (i.e., without retraining and without adaptation of cut-off values). The evaluation of the performance is unbiased and not used to optimize model parameters. |
| Performance results consistency | Model performance obtained in the training, validation and testing sets is reported. Consistency checks of performance measures across the different sets are performed. |
| Comparison to conventional metrics | Performance of radiomics-based models is compared against conventional metrics such as tumor volume and clinical variables (e.g., staging) in order to evaluate the added value of radiomics (e.g., by assessing the significance of AUC increase calculated with the DeLong test). |
| Multivariable analysis with non-radiomics variables | Multivariable analysis integrates variables other than radiomics features (e.g., clinical information, demographic data, panomics, etc.). |
|
| |
| CLINICAL IMPLICATIONS | |
| Biological correlate | Assessment of the relationship between macroscopic tumor phenotype(s) described with radiomics and the underlying microscopic tumor biology. |
| Potential clinical application | The study discusses the current and potential application(s) of proposed radiomics-based models in the clinical setting. |
|
| |
| MATERIAL AVAILABILITY | |
| Open data | Imaging data, tumor ROI and clinical information are made available. |
| Open code | All software code related to computation of features, statistical analysis and machine learning, and allowing to exactly reproduce results, is open source. This code package is ideally shared in the form of easy-to-run organized scripts pointing to other relevant pieces of code, along with useful sets of instructions. |
| Open models | Complete models are available, including model parameters and cut-off values. |
VI. Conclusions
The field of radiomics is constantly growing within the field of medical physics and is an exciting opportunity for the medical physics community to participate in novel research for the safe translation of quantitative imaging. Machine and deep learning-based models have the potential to provide clinicians with DSS to improve diagnosis, treatment selection, and response assessment in oncology. As the field expands, the need to associate radiomic features with other clinical and biological variables will become of increased importance. The field should also continue to strive for standardized data collection, evaluation criteria, and reporting guidelines in order to mature as a field. Data-sharing will be crucial to develop the large-scale datasets needed for proper validation of radiomic models and there will be a need for collaborations to validate models across multiple institutions. In order to move radiomic models into the clinical practice it is imperative to demonstrate improvements to the clinical workflow and decision making, through expert observer studies and eventually clinical trials. Future developments in the areas of machine and deep learning with their improved balance of interpretability and prediction will also continue to advance radiomic studies.
REFERENCES
- 1.Avanzo M, Stancanello J, El Naqa I, “Beyond imaging: The promise of radiomics,” Phys. Med 38, 122–139 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ, “Radiomics: extracting more information from medical images using advanced feature analysis,” Eur. J. Cancer 48, 441–446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P, “Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach,” Nat. Commun 5, 4006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panth KM, Leijenaar RT, Carvalho S, Lieuwes NG, Yaromina A, Dubois L, Lambin P, “Is there a causal relationship between genetic changes and radiomics-based image features? An in vivo preclinical experiment with doxycycline inducible GADD34 tumor cells,” Radiother. Oncol 116, 462–466 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D, Goldgof DB, Hall LO, Lambin P, Balagurunathan Y, Gatenby RA, Gillies RJ, “Radiomics: the process and the challenges,” Magn. Reson. Imaging 30, 1234–1248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang J, Rangayyan RM, Xu J, El Naqa I, Yang Y, “Computer-aided detection and diagnosis of breast cancer with mammography: recent advances,” IEEE Trans. Inf. Technol. Biomed 13, 236–251 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Giger ML, Karssemeijer N, Schnabel JA, “Breast image analysis for risk assessment, detection, diagnosis, and treatment of cancer,” Annu. Rev. Biomed. Eng 15, 327–357 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Gillies RJ, Kinahan PE, Hricak H, “Radiomics: Images Are More than Pictures, They Are Data,” Radiology 278, 563–577 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, Tha KK, Xing L, Li R, “Radiomics and radiogenomics for precision radiotherapy,” J. Radiat. Res 59, i25–i31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Napel S, Mu W, Jardim-Perassi BV, Aerts HJWL, Gillies RJ, “Quantitative imaging of cancer in the postgenomic era: Radio(geno)mics, deep learning, and habitats,” Cancer 124, 4633–4649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoyanova R, Takhar M, Tschudi Y, Ford JC, SolÃrzano G, Erho N, Balagurunathan Y, Punnen S, Davicioni E, Gillies RJ, Pollack A, “Prostate cancer radiomics and the promise of radiogenomics,” Translational Cancer Research 5, 432–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khalvati F, Wong A, Haider MA, “Automated prostate cancer detection via comprehensive multi-parametric magnetic resonance imaging texture feature models,” BMC Med. Imaging 15, 27-015-0069-9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Algohary A, Viswanath S, Shiradkar R, Ghose S, Pahwa S, Moses D, Jambor I, Shnier R, Bohm M, Haynes AM, Brenner P, Delprado W, Thompson J, Pulbrock M, Purysko AS, Verma S, Ponsky L, Stricker P, Madabhushi A, “Radiomic features on MRI enable risk categorization of prostate cancer patients on active surveillance: Preliminary findings,” J. Magn. Reson. Imaging (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Bai H, Chen Y, Sun Q, Liu L, Zhou S, Wang G, Liang C, Li ZC, “A Fully-Automatic Multiparametric Radiomics Model: Towards Reproducible and Prognostic Imaging Signature for Prediction of Overall Survival in Glioblastoma Multiforme,” Sci. Rep 7, 14331-017-14753-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibragimov B and Xing L, “Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks,” Med. Phys 44, 547–557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lustberg T, van Soest J, Gooding M, Peressutti D, Aljabar P, van der Stoep J, van Elmpt W, Dekker A, “Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer,” Radiother. Oncol 126, 312–317 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Chlebus G, Schenk A, Moltz JH, van Ginneken B, Hahn HK, Meine H, “Automatic liver tumor segmentation in CT with fully convolutional neural networks and object-based postprocessing,” Sci. Rep 8, 15497-018-33860-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sapate SG, Mahajan A, Talbar SN, Sable N, Desai S, Thakur M, “Radiomics based detection and characterization of suspicious lesions on full field digital mammograms,” Comput. Methods Programs Biomed 163, 1–20 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Antropova N, Huynh BQ, Giger ML, “A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets,” Med. Phys 44, 5162–5171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonekamp D, Kohl S, Wiesenfarth M, Schelb P, Radtke JP, Gotz M, Kickingereder P, Yaqubi K, Hitthaler B, Gahlert N, Kuder TA, Deister F, Freitag M, Hohenfellner M, Hadaschik BA, Schlemmer HP, Maier-Hein KH, “Radiomic Machine Learning for Characterization of Prostate Lesions with MRI: Comparison to ADC Values,” Radiology 289, 128–137 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Yuan Y, Qin W, Buyyounouski M, Ibragimov B, Hancock S, Han B, Xing L, “Prostate cancer classification with multiparametric MRI transfer learning model,” Med. Phys 46, 756–765 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Wu W, Parmar C, Grossmann P, Quackenbush J, Lambin P, Bussink J, Mak R, Aerts HJ, “Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology,” Front. Oncol 6, 71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira Junior JR, Koenigkam-Santos M, Cipriano FEG, Fabro AT, Azevedo-Marques PM, “Radiomics-based features for pattern recognition of lung cancer histopathology and metastases,” Comput. Methods Programs Biomed 159, 23–30 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Ganeshan B, Abaleke S, Young RC, Chatwin CR, Miles KA, “Texture analysis of non-small cell lung cancer on unenhanced computed tomography: initial evidence for a relationship with tumour glucose metabolism and stage,” Cancer. Imaging 10, 137–143 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song SH, Park H, Lee G, Lee HY, Sohn I, Kim HS, Lee SH, Jeong JY, Kim J, Lee KS, Shim YM, “Imaging Phenotyping Using Radiomics to Predict Micropapillary Pattern within Lung Adenocarcinoma,” J. Thorac. Oncol 12, 624–632 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Crispin-Ortuzar M, Apte A, Grkovski M, Oh JH, Lee NY, Schoder H, Humm JL, Deasy JO, “Predicting hypoxia status using a combination of contrast-enhanced computed tomography and [(18)F]-Fluorodeoxyglucose positron emission tomography radiomics features,” Radiother. Oncol 127, 36–42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parra AN, Lu H, Li Q, Stoyanova R, Pollack A, Punnen S, Choi J, Abdalah M, Lopez C, Gage K, Park JY, Kosj Y, Pow-Sang JM, Gillies RJ, Balagurunathan Y, “Predicting clinically significant prostate cancer using DCE-MRI habitat descriptors,” Oncotarget 9, 37125–37136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J, Narang S, Martinez JJ, Rao G, Rao A, “Associating spatial diversity features of radiologically defined tumor habitats with epidermal growth factor receptor driver status and 12-month survival in glioblastoma: methods and preliminary investigation,” J. Med. Imaging (Bellingham) 2, 041006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Y, Li H, Guo W, Drukker K, Lan L, Giger ML, Ji Y, “Deciphering Genomic Underpinnings of Quantitative MRI-based Radiomic Phenotypes of Invasive Breast Carcinoma,” Sci. Rep 5, 17787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Kim J, Balagurunathan Y, Li Q, Garcia AL, Stringfield O, Ye Z, Gillies RJ, “Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas,” Clin.Lung Cancer, 17(5), 441–448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon HJ, Sohn I, Cho JH, Lee HY, Kim JH, Choi YL, Kim H, Lee G, Lee KS, Kim J, “Decoding Tumor Phenotypes for ALK, ROS1, and RET Fusions in Lung Adenocarcinoma Using a Radiomics Approach,” Medicine (Baltimore) 94, e1753 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Kato F, Oyama-Manabe N, Li R, Cui Y, Tha KK, Yamashita H, Kudo K, Shirato H, “Identifying Triple-Negative Breast Cancer Using Background Parenchymal Enhancement Heterogeneity on Dynamic Contrast-Enhanced MRI: A Pilot Radiomics Study,” PLoS One 10, e0143308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SE, Han K, Kwak JY, Lee E, Kim EK, “Radiomics of US texture features in differential diagnosis between triple-negative breast cancer and fibroadenoma,” Sci. Rep 8, 13546-018-31906-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, Aldape K, Cha S, Kuo MD, “Identification of noninvasive imaging surrogates for brain tumor gene-expression modules,” Proc. Natl. Acad. Sci. U. S. A 105, 5213–5218 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gnep K, Fargeas A, Gutierrez-Carvajal RE, Commandeur F, Mathieu R, Ospina JD, Rolland Y, Rohou T, Vincendeau S, Hatt M, Acosta O, de Crevoisier R, “Haralick textural features on T2 -weighted MRI are associated with biochemical recurrence following radiotherapy for peripheral zone prostate cancer,” J. Magn. Reson. Imaging , 45(1), 103–117 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Bogowicz M, Riesterer O, Ikenberg K, Stieb S, Moch H, Studer G, Guckenberger M, Tanadini-Lang S, “Computed Tomography Radiomics Predicts HPV Status and Local Tumor Control After Definitive Radiochemotherapy in Head and Neck Squamous Cell Carcinoma,” Int. J. Radiat. Oncol. Biol. Phys 99, 921–928 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Leijenaar RT, Bogowicz M, Jochems A, Hoebers FJ, Wesseling FW, Huang SH, Chan B, Waldron JN, O’Sullivan B, Rietveld D, Leemans CR, Brakenhoff RH, Riesterer O, Tanadini-Lang S, Guckenberger M, Ikenberg K, Lambin P, “Development and validation of a radiomic signature to predict HPV (p16) status from standard CT imaging: a multicenter study,” Br. J. Radiol 91, 20170498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nie K, Shi L, Chen Q, Hu X, Jabbour S, Yue N, Niu T, Sun X, “Rectal Cancer: Assessment of Neoadjuvant Chemo-Radiation Outcome Based on Radiomics of Multi-Parametric MRI,” Clin. Cancer Res 22(21), 5256–5263 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oikonomou A, Khalvati F, Tyrrell PN, Haider MA, Tarique U, Jimenez-Juan L, Tjong MC, Poon I, Eilaghi A, Ehrlich L, Cheung P, “Radiomics analysis at PET/CT contributes to prognosis of recurrence and survival in lung cancer treated with stereotactic body radiotherapy,” Sci. Rep 8, 4003-018-22357-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Mao Y, Li Z, Zhang D, Zhang Z, Hao S, Li B, “Use of texture analysis based on contrast-enhanced MRI to predict treatment response to chemoradiotherapy in nasopharyngeal carcinoma “ J. Magn. Reson. Imaging 44, 445–455 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Ibragimov B, Toesca D, Yuan Y, Koong A, Daniel C, Xing L, “Neural networks for deep radiotherapy dose analysis and prediction of liver SBRT outcomes,” IEEE J. Biomed. Health. Inform. IEEE J. Biomed. Health. Inform doi: 10.1109/JBHI.2019.2904078, (2019). [DOI] [PubMed] [Google Scholar]
- 42.Huynh E, Coroller TP, Narayan V, Agrawal V, Hou Y, Romano J, Franco I, Mak RH, Aerts HJ, “CT-based radiomic analysis of stereotactic body radiation therapy patients with lung cancer,” Radiother. Oncol 120(2), 258–66 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Vallieres M, Freeman CR, Skamene SR, El Naqa I, “A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities,” Phys. Med. Biol 60, 5471–5496 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RT, Hermann G, Lambin P, Haibe-Kains B, Mak RH, Aerts HJ, “CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma,” Radiother. Oncol 114, 345–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hao H, Zhou Z, Li S, Maquilan G, Folkert MR, Iyengar P, Westover KD, Albuquerque K, Liu F, Choy H, Timmerman R, Yang L, Wang J, “Shell feature: a new radiomics descriptor for predicting distant failure after radiotherapy in non-small cell lung cancer and cervix cancer,” Phys. Med. Biol 63, 095007-6560/aabb5e (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haralick RM, Shanmugam K, Dinstein I, “Textural Features for Image Classification “ IEEE Trans. Syst. Man Cybern 3, 610–621 (1973). [Google Scholar]
- 47.Ohri N, Duan F, Snyder BS, Wei B, Machtay M, Alavi A, Siegel BA, Johnson DW, Bradley JD, DeNittis A, Werner-Wasik M, El Naqa I, “Pretreatment 18F-FDG PET Textural Features in Locally Advanced Non-Small Cell Lung Cancer: Secondary Analysis of ACRIN 6668/RTOG 0235,” J. Nucl. Med 57, 842–848 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, Bussink J, Gillies RJ, Mak RH, Aerts HJWL, “Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study,” PLoS Med. 15, e1002711 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aerts HJ, “The Potential of Radiomic-Based Phenotyping in Precision Medicine: A Review,” JAMA Oncol, 2(12):1636–1642 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Kickingereder P, Gotz M, Muschelli J, Wick A, Neuberger U, Shinohara RT, Sill M, Nowosielski M, Schlemmer HP, Radbruch A, Wick W, Bendszus M, Maier-Hein KH, Bonekamp D, “Large-scale Radiomic Profiling of Recurrent Glioblastoma Identifies an Imaging Predictor for Stratifying Anti-Angiogenic Treatment Response,” Clin. Cancer Res 22(23), 5765–5771 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR, Verlingue L, Brandao D, Lancia A, Ammari S, Hollebecque A, Scoazec JY, Marabelle A, Massard C, Soria JC, Robert C, Paragios N, Deutsch E, Ferte C, “A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study,” Lancet Oncol. 19, 1180–1191 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Fave X, Zhang L, Yang J, Mackin D, Balter P, Gomez D, Followill D, Jones AK, Stingo F, Liao Z, Mohan R, Court L, “Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer,” Sci. Rep 7, 588-017-00665-z (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fave X, Mackin D, Yang J, Zhang J, Fried D, Balter P, Followill D, Gomez D, Jones AK, Stingo F, Fontenot J, Court L, “Can radiomics features be reproducibly measured from CBCT images for patients with non-small cell lung cancer?” Med. Phys 42, 6784–6797 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Timmeren JE, Leijenaar RT, van Elmpt W, Lambin P, “Interchangeability of a Radiomic Signature Between Conventional and Weekly Cone Beam Computed Tomography Allowing Response Prediction in Non-Small Cell Lung Cancer,” Int. J. Radiat. Oncol. Biol. Phys 96, S193 (2016). [Google Scholar]
- 55.van Timmeren JE, Leijenaar RTH, van Elmpt W, Reymen B, Lambin P, “Feature selection methodology for longitudinal cone-beam CT radiomics,” Acta Oncol. 56, 1537–1543 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Moran A, Daly ME, Yip SSF, Yamamoto T, “Radiomics-based Assessment of Radiation-induced Lung Injury After Stereotactic Body Radiotherapy,” Clin. Lung Cancer 18, e425–e431 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Cunliffe A, Armato SG 3rd, Castillo R, Pham N, Guerrero T, Al-Hallaq HA, “Lung texture in serial thoracic computed tomography scans: correlation of radiomics-based features with radiation therapy dose and radiation pneumonitis development,” Int. J. Radiat. Oncol. Biol. Phys 91, 1048–1056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krafft SP, Rao A, Stingo F, Briere TM, Court LE, Liao Z, Martel MK, “The utility of quantitative CT radiomics features for improved prediction of radiation pneumonitis,” Med. Phys 45, 5317–5324 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Mattonen SA, Palma DA, Johnson C, Louie AV, Landis M, Rodrigues G, Chan I, Etemad-Rezai R, Yeung TP, Senan S, Ward AD, “Detection of Local Cancer Recurrence After Stereotactic Ablative Radiation Therapy for Lung Cancer: Physician Performance Versus Radiomic Assessment,” Int. J. Radiat. Oncol. Biol. Phys 94, 1121–1128 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Zhang Z, Yang J, Ho A, Jiang W, Logan J, Wang X, Brown PD, McGovern SL, Guha-Thakurta N, Ferguson SD, Fave X, Zhang L, Mackin D, Court LE, Li J, “A predictive model for distinguishing radiation necrosis from tumour progression after gamma knife radiosurgery based on radiomic features from MR images,” Eur. Radiol 28, 2255–2263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Viswanath S, Toth R, Rusu M, Sperling D, Lepor H, Futterer J, Madabhushi A, “Identifying Quantitative In Vivo Multi-Parametric MRI Features For Treatment Related Changes after Laser Interstitial Thermal Therapy of Prostate Cancer,” Neurocomputing 144, 13–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu JY, Zhang HP, Tang ZY, Zhou J, He XJ, Liu YY, Liu XJ, Guo DJ, “Value of texture analysis based on enhanced MRI for predicting an early therapeutic response to transcatheter arterial chemoembolisation combined with high-intensity focused ultrasound treatment in hepatocellular carcinoma,” Clin. Radiol 73, 758.e9–758.e18 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Bagher-Ebadian H, Siddiqui F, Liu C, Movsas B, Chetty IJ, “On the impact of smoothing and noise on robustness of CT and CBCT radiomics features for patients with head and neck cancers,” Med. Phys 44, 1755–1770 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Shafiq-Ul-Hassan M, Zhang GG, Latifi K, Ullah G, Hunt DC, Balagurunathan Y, Abdalah MA, Schabath MB, Goldgof DG, Mackin D, Court LE, Gillies RJ, Moros EG, “Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels,” Med. Phys 44, 1050–1062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alex Z, Stefan L, Martin V, Steffen L, the,Image Biomarker Standardisation Initiative for, “Image biomarker standardisation initiative,” arXiv e-prints, ar:1612.07003 (2016). [Google Scholar]
- 66.van Velden FH, Kramer GM, Frings V, Nissen IA, Mulder ER, de Langen AJ, Hoekstra OS, Smit EF, Boellaard R, “Repeatability of Radiomic Features in Non-Small-Cell Lung Cancer [18F]FDG-PET/CT Studies: Impact of Reconstruction and Delineation,” Mol. Imaging Biol, 18(5):788–95 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leijenaar RT, Nalbantov G, Carvalho S, van Elmpt WJ, Troost EG, Boellaard R, Aerts HJ, Gillies RJ, Lambin P, “The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis,” Sci. Rep 5, 11075 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Julesz B, “Experiments in the visual perception of texture,” Sci. Am 232, 34–43 (1975). [DOI] [PubMed] [Google Scholar]
- 69.El Naqa I, Grigsby P, Apte A, Kidd E, Donnelly E, Khullar D, Chaudhari S, Yang D, Schmitt M, Laforest R, Thorstad W, Deasy JO, “Exploring feature-based approaches in PET images for predicting cancer treatment outcomes,” Pattern Recognit 42, 1162–1171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coroller TP, Agrawal V, Narayan V, Hou Y, Grossmann P, Lee SW, Mak RH, Aerts HJ, “Radiomic phenotype features predict pathological response in non-small cell lung cancer,” Radiother. Oncol, 119(3), 480–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tixier F, Hatt M, Valla C, Fleury V, Lamour C, Ezzouhri S, Ingrand P, Perdrisot R, Visvikis D, Le Rest CC, “Visual versus quantitative assessment of intratumor 18F-FDG PET uptake heterogeneity: Prognostic value in non-small cell lung cancer,” Journal of Nuclear Medicine 55, 1235–1241 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJ, “Machine Learning methods for Quantitative Radiomic Biomarkers,” Sci. Rep 5, 13087 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tibshirani R, “Regression Shrinkage and Selection via the Lasso,” Journal of the Royal Statistical Society.Series B (Methodological) 58, 267–288 (1996). [Google Scholar]
- 74.Wold S, Esbensen K, Geladi P, “Principal component analysis,” Chemometrics and Intelligent Laboratory Systems 2, 37–52 (1987). [Google Scholar]
- 75.van der Maaten Laurens and Hinton Geoffrey E., “Visualizing Data using t-SNE,” Journal of Machine Learning Research, 9, 2579–2605, (2008). [Google Scholar]
- 76.Parmar C, Leijenaar RTH, Grossmann P, Velazquez ER, Bussink J, Rietveld D, Rietbergen MM, Haibe-Kains B, Lambin P, Aerts HJWL, “Radiomic feature clusters and Prognostic Signatures specific for Lung and Head & Neck cancer,” Scientific Reports 5, 11044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu J, Cui Y, Sun X, Cao G, Li B, Ikeda DM, Kurian AW, Li R, “Unsupervised Clustering of Quantitative Image Phenotypes Reveals Breast Cancer Subtypes with Distinct Prognoses and Molecular Pathways,” Clin. Cancer Res 23, 3334–3342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ypsilantis PP, Siddique M, Sohn HM, Davies A, Cook G, Goh V, Montana G, “Predicting Response to Neoadjuvant Chemotherapy with PET Imaging Using Convolutional Neural Networks,” PLoS One 10, e0137036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen S, Zhou S, Yin FF, Marks LB, Das SK, “Investigation of the support vector machine algorithm to predict lung radiation-induced pneumonitis,” Med. Phys 34, 3808–3814 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.El Naqa I, Li R, Murphy MJ, eds.”Machine Learning in Radiation Oncology: Theory and Application”.Switzerland: Springer International Publishing;2015. 10.1007/978-3-319-18305-3, (2015) [DOI] [Google Scholar]
- 81.Yang D, Rao G, Martinez J, Veeraraghavan A, Rao A, “Evaluation of tumor-derived MRI-texture features for discrimination of molecular subtypes and prediction of 12-month survival status in glioblastoma “ Med. Phys 42, 6725–6735 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vallieres M, Kay-Rivest E, Perrin L, Liem X, Furstoss C, Aerts HJWL, Khaouam N, Nguyen-Tan P, Wang C, Sultanem K, Seuntjens J, El Naqa I, “Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer,” Scientific Reports 7, 10117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rosenberg Charles, Hebert Martial and Schneiderman Henry, “Semi-Supervised Self-Training of Object Detection Models,” in: WACV-MOTION ‘05 Proceedings of the Seventh IEEE Workshops on Application of Computer Vision (WACV/MOTION’05), Volume 1, 29–36 (2005). doi: 10.1109/ACVMOT.2005.107 [DOI] [Google Scholar]
- 84.Joachims Thorsten, “Transductive Inference for Text Classification Using Support Vector Machines,” in: ICML ‘99 Proceedings of the Sixteenth International Conference on Machine Learning, 200–209 (1999) doi: 10.1109/ACVMOT.2005.107. [DOI] [Google Scholar]
- 85.Blum Avrim, Lafferty John, Mugizi Robert Rwebangira and Rajashekar Reddy, “Semi-supervised Learning Using Randomized Mincuts” in ICML ‘04 Proceedings of the twenty-first international conference on Machine learning, 13 (2004) DOI: 10.1145/1015330.1015429. [DOI] [Google Scholar]
- 86.Cox DR, “Regression Models and Life-Tables,” Journal of the Royal Statistical Society.Series B (Methodological) 34, 187–220 (1972). [Google Scholar]
- 87.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS, “Random survival forests “ The Annals of Applied Statistics 2, 841–860 (2008). [Google Scholar]
- 88.Van Belle V, Pelckmans K, Van Huffel S, Suykens JAK, “Support vector methods for survival analysis: a comparison between ranking and regression approaches,” Artificial Intelligence in Medicine 53, 107–118 (2011). [DOI] [PubMed] [Google Scholar]
- 89.LeCun Y, Bengio Y, Hinton G, “Deep learning,” Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 90.Suzuki K, “Pixel-based machine learning in medical imaging,” Int. J. Biomed. Imaging 2012, 792079 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Simonyan K and Zisserman A, “Very Deep Convolutional Networks for Large-Scale Image Recognition,” arXiv:1409.1556 (2014). [Google Scholar]
- 92.He K, Zhang X, Ren S, Sun J, “Deep Residual Learning for Image Recognition,” presented at 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016. 770–778 (2016). [Google Scholar]
- 93.Hubel DH and Wiesel TN, “Receptive fields and functional architecture of monkey striate cortex,” J. Physiol. (Lond. ) 195, 215–243 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hochreiter S and Schmidhuber J, “Long Short-Term Memory,” Neural Comput. 9, 1735–1780 (1997). [DOI] [PubMed] [Google Scholar]
- 95.Cho K, van Merrienboer B, Gulcehre C, Bahdanau D, Bougares F, Schwenk H, Bengio Y, “Learning Phrase Representations using RNN Encoder-Decoder for Statistical Machine Translation,” in Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), 1724–1734 (2014) DOI: 10.3115/v1/D14-1179. [DOI] [Google Scholar]
- 96.Kingma DP and Welling M, “Auto-Encoding Variational Bayes,” arXiv:1312.6114 (2013). [Google Scholar]
- 97.Li F, Qiao H, Zhang B, “Discriminatively boosted image clustering with fully convolutional auto-encoders,” Pattern Recognition 83, 161–173 (2018). [Google Scholar]
- 98.Hinton GE and Salakhutdinov RR, “Reducing the Dimensionality of Data with Neural Networks,” Science 313, 504–507 (2006). [DOI] [PubMed] [Google Scholar]
- 99.Kingma DP, Rezende DJ, Mohamed S, Welling M, “Semi-Supervised Learning with Deep Generative Models,” eprint arXiv:1406.5298 (2014). [Google Scholar]
- 100.E. HG, Nitish S, Alex K, Ilya S, R. SR, “Improving neural networks by preventing co-adaptation of feature detectors,” arXiv e-prints, ar:1207.0580 (2012). [Google Scholar]
- 101.Sergey I and Christian S, “Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift,” arXiv e-prints, ar:1502.03167 (2015). [Google Scholar]
- 102.Long J, Shelhamer E, Darrell T, “Fully convolutional networks for semantic segmentation,” presented at 2015 IEEE Conference on Computer Vision and Pattern Recognition. Published in EEE Transactions on Pattern Analysis and Machine Intelligence; 39(4): 640–651 doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 103.Ronneberger O, Fischer P, Brox T, “U-Net: Convolutional Networks for Biomedical Image Segmentation,” presented at Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2015. DOI: 10.1007/978-3-319-24574-4_28. [DOI] [Google Scholar]
- 104.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S, “Dermatologist-level classification of skin cancer with deep neural networks,” Nature 542, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chang H, Han J, Zhong C, Snijders AM, Mao JH, “Unsupervised Transfer Learning via Multi-Scale Convolutional Sparse Coding for Biomedical Applications,” IEEE Trans. Pattern Anal. Mach. Intell 40, 1182–1194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Collins GS, Reitsma JB, Altman DG, Moons KG, “Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement,” BMC Med. 13, 1-014-0241-z (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hastie T, Tibshirani R, Friedman JH, The Elements of Statistical Learning: Data Mining, Inference, and Prediction (Springer, New York, NY, 2009). [Google Scholar]
- 108.Ren J, Zhou J, Ding W, Zhong B, Zhou J, “Clinicopathological characteristics and imaging features of pulmonary adenocarcinoma with micropapillary pattern,” Zhonghua Zhong Liu Za Zhi 36, 282–286 (2014). [PubMed] [Google Scholar]
- 109.Harrell FE Jr, Lee KL, Mark DB, “Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors,” Stat. Med 15, 361–387 (1996). [DOI] [PubMed] [Google Scholar]
- 110.Pencina MJ and D’Agostino RB, “Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation,” Statist. Med 23, 2109–2123 (2004). [DOI] [PubMed] [Google Scholar]
- 111.Royston P and Altman DG, “External validation of a Cox prognostic model: principles and methods,” BMC Med. Res. Methodol 13, 33-2288-13-33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boellaard R, “Standards for PET image acquisition and quantitative data analysis,” J. Nucl. Med 50 Suppl 1, 11S–20S (2009). [DOI] [PubMed] [Google Scholar]
- 113.Mackin D, Fave X, Zhang L, Fried D, Yang J, Taylor B, Rodriguez-Rivera E, Dodge C, Jones AK, Court L, “Measuring Computed Tomography Scanner Variability of Radiomics Features,” Invest. Radiol 50, 757–765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Szczypinski PM, Strzelecki MFAU - Andrzej Materka, Materka AFAU - Artur Klepaczko, Klepaczko A, “MaZda--a software package for image texture analysis.” Computer methods and programs in biomedicine 94(1):66–76. DOI: 10.1016/j.cmpb.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 115.Nioche C, Orlhac F, Boughdad S, Reuze S, Goya-Outi J, Robert C, Pellot-Barakat C, Soussan M, Frouin F, Buvat I, “LIFEx: A Freeware for Radiomic Feature Calculation in Multimodality Imaging to Accelerate Advances in the Characterization of Tumor Heterogeneity,” Cancer Res. 78, 4786–4789 (2018). [DOI] [PubMed] [Google Scholar]
- 116.Rubin DL, Willrett D, O’Connor MJ, Hage C, Kurtz C, Moreira DA, “Automated tracking of quantitative assessments of tumor burden in clinical trials,” Transl. Oncol 7, 23–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL, “Computational Radiomics System to Decode the Radiographic Phenotype,” Cancer Res. 77, e104–e107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dicente Cid Y, Castelli J, Schaer R, Scher N, Pomoni A, Prior JO, Depeursinge A, “Chapter 12 - QuantImage: An Online Tool for High-Throughput 3D Radiomics Feature Extraction in PET-CT,” in Biomedical Texture Analysis, 349–377 (2017). DOI: 10.1016/B978-0-12-812133-7.00012-0. [DOI] [Google Scholar]
- 119.Echegaray S, Bakr S, Rubin DL, Napel S, “Quantitative Image Feature Engine (QIFE): an Open-Source, Modular Engine for 3D Quantitative Feature Extraction from Volumetric Medical Images,” J. Digit. Imaging 31, 403–414 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE, “IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics,” Med. Phys 42, 1341–1353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R, “3D Slicer as an image computing platform for the Quantitative Imaging Network,” Magn. Reson. Imaging 30, 1323–1341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yosinski J, Clune J, Nguyen A, Fuchs T, Lipson H, “Understanding Neural Networks Through Deep Visualization,” eprint arXiv:1506.06579 (2015). [Google Scholar]
- 123.Sankar V, Kumar D, Clausi DA, Taylor GW, Wong A, “SISC: End-to-end Interpretable Discovery Radiomics-Driven Lung Cancer Prediction via Stacked Interpretable Sequencing Cells,” eprint arXiv:1901.04641 (2019). [Google Scholar]
- 124.Luo Y, McShan D, Ray D, Matuszak M, Jolly S, Lawrence T, Kong F, Ten Haken R, El Naqa I, “Development of a Fully Cross-Validated Bayesian Network Approach for Local Control Prediction in Lung Cancer,” IEEE Transactions on Radiation and Plasma Medical Sciences 3, 232–241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hatt M, Tixier F, Pierce L, Kinahan PE, Le Rest CC, Visvikis D, “Characterization of PET/CT images using texture analysis: the past, the present… any future?” Eur. J. Nucl. Med. Mol. Imaging 44, 151–165 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fave X, Cook M, Frederick A, Zhang L, Yang J, Fried D, Stingo F, Court L, “Preliminary investigation into sources of uncertainty in quantitative imaging features,” Comput. Med. Imaging Graphics 44, 54–61 (2015). [DOI] [PubMed] [Google Scholar]
- 127.Pfaehler E, Beukinga RJ, de Jong JR, Slart RHJA, Slump CH, Dierckx RAJO, Boellaard R, “Repeatability of (18) F-FDG PET radiomic features: A phantom study to explore sensitivity to image reconstruction settings, noise, and delineation method,” Med. Phys 46, 665–678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leijenaar RT, Carvalho S, Velazquez ER, van Elmpt WJ, Parmar C, Hoekstra OS, Hoekstra CJ, Boellaard R, Dekker AL, Gillies RJ, Aerts HJ, Lambin P, “Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability,” Acta Oncol. 52, 1391–1397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim H, Park CM, Lee M, Park SJ, Song YS, Lee JH, Hwang EJ, Goo JM, “Impact of Reconstruction Algorithms on CT Radiomic Features of Pulmonary Tumors: Analysis of Intra- and Inter-Reader Variability and Inter-Reconstruction Algorithm Variability,” PLoS One 11, e0164924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhao B, James LP, Moskowitz CS, Guo P, Ginsberg MS, Lefkowitz RA, Qin Y, Riely GJ, Kris MG, Schwartz LH, “Evaluating Variability in Tumor Measurements from Same-day Repeat CT Scans of Patients with Non†“Small Cell Lung Cancer,” Radiology 252, 263–272 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Desseroit MC, Tixier F, Weber WA, Siegel BA, Cheze Le Rest C, Visvikis D, Hatt M, “Reliability of PET/CT shape and heterogeneity features in functional and morphological components of Non-Small Cell Lung Cancer tumors: a repeatability analysis in a prospective multi-center cohort,” J. Nucl. Med 58(3), 406–411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R, “Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters,” Acta Oncol. 49, 1012–1016 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lu L, Lv W, Jiang J, Ma J, Feng Q, Rahmim A, Chen W, “Robustness of Radiomic Features in [11C]Choline and [18F]FDG PET/CT Imaging of Nasopharyngeal Carcinoma: Impact of Segmentation and Discretization,” Mol. Imaging Biol 18(6), 935–945 (2016). [DOI] [PubMed] [Google Scholar]
- 134.Bailly C, Bodet-Milin C, Couespel S, Necib H, Kraeber-Bodéré F, Ansquer C, Carlier T, “Revisiting the robustness of PET-based textural features in the context of multi-centric trials,” PLoS ONE 11(7), e0159984 (2016) DOI: 10.1371/journal.pone.0159984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang F, Dogan N, Stoyanova R, Ford JC, “Evaluation of radiomic texture feature error due to MRI acquisition and reconstruction: A simulation study utilizing ground truth,” Phys. Med 50, 26–36 (2018). [DOI] [PubMed] [Google Scholar]
- 136.Grootjans W, Tixier F, van der Vos CS, Vriens D, Le Rest CC, Bussink J, Oyen WJ, de Geus-Oei LF, Visvikis D, Visser EP, “The impact of optimal respiratory gating and image noise on evaluation of intra-tumor heterogeneity in 18F-FDG positron emission tomography imaging of lung cancer,” J. Nucl. Med 57(11), 1692–1698 (2016). [DOI] [PubMed] [Google Scholar]
- 137.Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, Mitra S, Shankar BU, Kikinis R, Haibe-Kains B, Lambin P, Aerts HJ, “Robust Radiomics feature quantification using semiautomatic volumetric segmentation,” PLoS One 9, e102107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Echegaray S, Gevaert O, Shah R, Kamaya A, Louie J, Kothary N, Napel S, “Core samples for radiomics features that are insensitive to tumor segmentation: method and pilot study using CT images of hepatocellular carcinoma,” J. Med. Imaging (Bellingham) 2, 041011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S, “Radiomics: the bridge between medical imaging and personalized medicine,” Nat. Rev. Clin. Oncol 14, 749–762 (2017). [DOI] [PubMed] [Google Scholar]
- 140.Chalkidou A, O’Doherty MJ, Marsden PK, “False discovery rates in PET and CT studies with texture features: A systematic review,” PLoS ONE 10(5), e0124165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.AlBadawy EA, Saha A, Mazurowski MA, “Deep learning for segmentation of brain tumors: Impact of cross-institutional training and testing,” Med. Phys 45, 1150–1158 (2018). [DOI] [PubMed] [Google Scholar]
- 142.Johnson WE, Li C, Rabinovic A, “Adjusting batch effects in microarray expression data using empirical Bayes methods,” Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 143.Chang K, Balachandar N, Lam C, Yi D, Brown J, Beers A, Rosen B, Rubin DL, Kalpathy-Cramer J, “Distributed deep learning networks among institutions for medical imaging,” J. Am. Med. Inform. Assoc 25, 945–954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zur RM, Jiang Y, Pesce LL, Drukker K, “Noise injection for training artificial neural networks: a comparison with weight decay and early stopping,” Med. Phys 36, 4810–4818 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li S, Chen Y, Peng Y, Bai L, “Learning More Robust Features with Adversarial Training,” arXiv:1804.07757 (2018). [Google Scholar]
- 146.Vallieres M, Zwanenburg A, Badic B, Cheze Le Rest C, Visvikis D, Hatt M, “Responsible Radiomics Research for Faster Clinical Translation,” J. Nucl. Med 59, 189–193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJ, Groth P, Goble C, Grethe JS, Heringa J, ’t Hoen PA, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B, “The FAIR Guiding Principles for scientific data management and stewardship,” Sci. Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]


