Abstract
The conventional approach for fabricating polydimethylsiloxane (PDMS) microfluidic devices is a lengthy and inconvenient procedure and may require a clean-room microfabrication facility often not readily available. Furthermore, living cells can't survive the oxygen-plasma and high-temperature-baking treatments required for covalent bonding to assemble multiple PDMS parts into a leak-free device, and it is difficult to disassemble the devices because of the irreversible covalent bonding. As a result, seeding/loading cells into and retrieving cells from the devices are challenging. Here, we discovered that decreasing the curing agent for crosslinking the PDMS prepolymer increases the noncovalent binding energy of the resultant PDMS surfaces without plasma or any other treatment. This enables convenient fabrication of leak-free microfluidic devices by noncovalent binding for various biomedical applications that require high pressure/flow rates and/or long-term cell culture, by simply hand-pressing the PDMS parts without plasma or any other treatment to bind/assemble. With this method, multiple types of cells can be conveniently loaded into specific areas of the PDMS parts before assembly and due to the reversible nature of the noncovalent bonding, the assembled device can be easily disassembled by hand peeling for retrieving cells. Combining with 3D printers that are widely available for making masters to eliminate the need of photolithography, this facile yet rigorous fabrication approach is much faster and more convenient for making PDMS microfluidic devices than the conventional oxygen plasma-baking-based irreversible covalent bonding method.
Keywords: Binding energy, 3D printing, Soft lithography, Microfluidics, Polydimethylsiloxane
Graphical abstract
A special formulation of PDMS with high noncovalent binding energy is discovered allowing for reversible assembly of PDMS microfluidic devices. Combining with 3D printing, this enables handy fabrication of leak-free microfluidic devices without clean-room facility and oxygen plasma. Cells can be conveniently loaded/seeded into the PDMS parts before their assembly into a final device and the device can be easily peeled open by hand to retrieve cells. This method my greatly facilitate the widespread utilization of PDMS microfluidic devices as a useful tool for various biomedical applications.
Highlights
-
•
The stability of noncovalent PDMS-PDMS binding is dependent on the binding energy instead of the binding strength.
-
•
The noncovalent binding of a special formulation of PDMS is sufficient for reversible assembly of leak-free microfluidic devices.
-
•
The noncovalent binding method enables loading multiple types of cells into PDMS parts before assembling into the final device.
-
•
The PDMS device can be easily dissembled due to the reversible nature of the noncovalent binding for retrieving cells.
1. Introduction
Microfluidic devices have been widely used for lab-on-a-chip cell culture, drug screening, and diagnostics, and the devices are often made of polydimethylsiloxane (PDMS) and fabricated with a procedure involving photolithography [[1], [2], [3], [4]]. PDMS is usually made by mixing its prepolymer and curing agent. When mixed, the prepolymer is crosslinked by the curing agent into a solid elastomer in ∼2 h of incubation at an elevated temperature (typically, ∼75 °C) [5]. The PDMS must be bound/bonded with various substrates like glass or other PDMS part(s) tightly for assembling into a microfluidic device with no leak [[6], [7], [8], [9], [10]]. This is commonly/conventionally done by treating PDMS surfaces with oxygen plasma or corona discharge to temporally activate the PDMS surfaces, immediately before device assembly [11]. Then, PDMS parts and/or substrates with activated surfaces are assembled into a device via irreversible covalent bonding between the surfaces, and they are usually baked at an elevated temperature for the covalent bonding to reach equilibrium before use [12]. However, these high-temperature and plasma/corona-discharge treatments can cause damage to living cells [13,14]. Therefore, living cells are usually loaded into the device via injection through the small inlet(s) or hole(s) made by punching after PDMS devices are assembled, which unfortunately makes it extremely hard to control the distribution of cells in the microfluidic channels/chambers of a microfluidic device [15]. Moreover, it is equally difficult to retrieve cells out of the PDMS devices assembled with the irreversible covalent bonding method for further culture or analysis.
Other less commonly used methods for assembling PDMS-based microfluidic devices include using binders/clips/screws, vacuum, and magnetism to physically bind and/or applying glue or other chemicals on the surfaces to enhance binding [[16], [17], [18], [19], [20], [21], [22], [23], [24]], as well as shortening the time for curing PDMS (i.e., partially cured PDMS with significant residual curing agent) [25]. However, all those alternative methods have their own issues. For example, physically binding PDMS parts with binders/clips for the assembly of microfluidic devices may cause device deformation, which could lead to leaking and make it difficult to image the samples in the device with a microscope. Processing the surface with chemicals such as glues [17] and poly-l-lysine [18] is also a lengthy procedure of soaking and drying, and the chemicals may be toxic to cells. Notably, the large amount of the residual curing agent in the partially cured PDMS has been reported to be toxic to cells [26,27]. In addition, the application of organic solvents such as benzophenone on the surface used for delay crosslinking is highly cytotoxic. Moreover, the methods of both using glue and partial curing still lead to irreversible covalent bonding and it is difficult to disassemble the glued or partially cured PDMS devices for retrieving cells. This may explain why none of the aforementioned studies reported loading cells into the PDMS microfluidic devices before their assembly. Therefore, a method is in need for assembling PDMS microfluidic devices that allows for convenient loading/seeding of living cells into the PDMS parts before their assembly and handy disassembly of the device to retrieve cells for further analysis, without cytotoxicity, device deformation/damage, blockage, and leak.
Herein, we report a method for facile fabrication of leak-free microfluidic devices without the need of plasma treatment, post-assembly baking at elevated temperature, any chemical including glue, or binders/clips/screws/vacuum/magnetism. This is enabled by our discovery that 1) the stability of the noncovalent binding between PDMS surfaces is dependent on their binding energy (i.e., the integration of binding strength over deformation) instead of the binding strength alone; and 2) if the PDMS is made by reducing the ratio of the curing agent to the PDMS prepolymer (compared to the commonly used ratio), the noncovalent binding energy is much increased to allow for facile yet stable binding of PDMS parts by simply hand-pressing them together. With this method, cells can be loaded into the PDMS parts before assembling them into a device. The binding is stable and tight enough to make PDMS-based microfluidic devices for both cell microencapsulation (requiring the capability of bearing high pressure) and long-term cell culture with no leaking. Furthermore, due to the reversible nature of the noncovalent binding, the assembled devices can be conveniently disassembled by simple hand-peeling, allowing for convenient retrieval of cells cultured in the devices. In addition, we utilized 3D printing to print molds for PDMS soft lithography, eliminating the need for expensive and often not readily available clean-room microfabrication facility for fabricating the molds. Collectively, our method is much more convenient and less expensive than the commonly used photolithography-plasma-baking-based approach for fabricating PDMS microfluidic devices and enables handy-yet-controlled loading and retrieval of cells in and out of the devices, which may be valuable to facilitate the wide-spread utility of PDMS microfluidic devices for various biomedical applications.
2. Materials and methods
2.1. Materials
Polydimethylsiloxane (PDMS, SYLGARD-184 Silicone Encapsulant) was purchased from Dow Corning (Midland, MI, USA). Fetal bovine serum (FBS) was purchased from Sigma-Aldrich (St. Louis, MO, USA). The DMEM, DMEM/F12, α-MEM basal medium, basal neural medium, nonessential amino acids (NEAA), 100x penicillin-streptomycin, and l-glutamine were purchased from Thermo Fisher (Gaithersburg, MD, USA). The Matrigel was purchased from Corning (NY, USA). All other materials were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless it is specifically noted otherwise.
2.2. 3D printing of resin masters
The resin masters were designed with the computer-assisted design software AutoCAD (Autodesk, Mill Valley, CA). Then, the designs were imported into the PreForm software of Formlabs (Somerville, MA, USA), to print the resin masters using a Formlabs Form 2 3D printer with the Formlabs RS-F2-GPCL standard clear resin.
2.3. Fabrication of microfluidic devices
The 3D printed resin masters were rinsed with ethanol, air-dried for 20 min, and placed in a 10 cm Petri dish for making the PDMS parts by soft lithography with the standard procedure reported in our previous work [14]. Different ratios of prepolymer to curing agent (10:1, 10:0.7, and 10:0.5) were used to make the PDMS parts. For the 10:1 and 10:0.7 PDMS parts, they were baked in an oven at 75 °C for 2 h. For the 10:0.5 PDMS parts, it was baked in an oven at 75 °C for 1 h. After baking, the PDMS parts were peeled off of the masters in a biosafety cabinet and trimmed. Then, inlets and outlets for injecting medium were punched with a 2 mm biopsy punch (Whatman, Maidstone, UK). Afterward, the PDMS parts were assembled into microfluidic devices within 13 h. When needed, oxygen plasma treatment of the 10:1 PDMS parts was done in a Harrick Plasma (Ithaca, NY, USA) PDC-32G plasma cleaner at 18 W and 27 Pa for 2 min. The 10:0.7 and 10:0.5 PDMS parts did not undergo oxygen plasma treatment. Either with or without oxygen plasma treatment, the PDMS parts were assembled in the same way by gentle hand pressing (i.e., causing no irreversible deformation) with particular attention to ensure no visible air bubble on the interface between the PDMS parts.
2.4. Fourier transform infrared spectroscopy (FTIR)
A total of 1 mL of the mixture of the PDMS prepolymer and curing agent at varying ratios of the prepolymer to the curing agent (10:1, 10:0.7, or/and 10:0.5) was pipetted onto a glass slide (75 × 50 mm). Any bubbles were removed by placing the slide under a bench vacuum for 15 min. After baking at 75 °C for a desired time given in the Results section for the specific studies, the PDMS forms a thin film of ∼250 μm in thickness. The PDMS samples were detached from the glass slides using a blade and cut into 1.5 cm × 1.5 cm sized samples for FTIR analysis using a Shimadzu (Tokyo, Japan) IR Prestige 21 spectrophotometer equipped with a KBr beam splitter, by following the manufacturer's instructions. Each PDMS sample was scanned 25 times at room temperature (RT). The range of wavenumber for data collection was 500–3600 cm−1. The data was plotted and further analyzed using the Microsoft (Seattle, WA, USA) Excel 2010 software.
2.5. Contact angle analysis
A droplet of water (5 μL) was pipetted onto the PDMS surface and photographed under RT, and photos of the droplet on the PDMS surface were taken using an iPhone 6 with a macro-lens attachment (APL 0.45WM, 140°/12.5x) (APEXEL, Shenzhen, Guangdong, China). A total of 4 water droplets were studied to measure the contact angle for each PDMS part (n = 4). The photos of the water droplet on the PDMS were analyzed and the contact angle was measured with the angle analysis tool in the NIH ImageJ software (v 1.52a).
2.6. Airburst test
The volumetric binding energy (or burst pressure) of the PDMS surfaces was measured by the airburst test based on a published protocol [26], as illustrated in Fig. S1. Briefly, two 10:1 (with or without plasma treatment as aforementioned), 10:0.7, or 10:0.5 PDMS parts (2.5 cm x 2.5 cm x 8 mm) were bound together by hand-pressing at different time points (0 h, 0.5 h, and 13 h) after peeled off the master either with or without plasma treatment), to make the final PDMS devices for the airburst test. A hole (diameter: 2 mm) was punched at the center of one of the PDMS slabs before assembly for connection with a tube (diameter: 2 mm) (Cole-Parmer, Vernon Hills, IL, USA). The tube was connected to a 5-mL syringe which was used to slowly compress the air to increase the air pressure in the tube. All the connections to the tubes were sealed with the 10:1 PDMS and cured at 37 °C for 24 h to prevent leaking. Two volume values on the syringe were recorded during the procedure: an initial volume (recorded as V1 = 5 mL) at the beginning of the test and a final volume (recorded as V2) at the time point when the two PDMS parts were observed to detach/burst apart at the bottom of the hole. The pressure (or volumetric binding energy) at the point of bursting the PDMS surface binding (n = 4) was calculated using the following equation:
| P2 = P1V1 / V2 | (1) |
where P2 is the burst pressure and P1 is the standard atmospheric pressure at ambient conditions (1 × 105 Pa). It is worth noting that although the volume of air in the syringe does change during the airburst test, the volume of air in the PDMS device does not change. To study the impact of the commonly used sterilization methods on the burst pressure, the PDMS slabs were sterilized by either rinsing with 75% ethanol for 5 min or autoclaving at 121 °C for 20 min before binding for the airburst test.
2.7. Mechanical testing
Mechanical testing of the binding between two PDMS surfaces was conducted via the single-lap joint test with an Instron mechanical testing system 33 R/44 65 (Norwood, MA, USA) [28]. Briefly, two PDMS slabs (diameter: 2.2 cm, and thickness: 0.5 cm) were bound together by simple hand-pressing, as shown in Fig. S2. The sample was then loaded into the system and pulled in opposite directions. The pulling force versus the displacement was recorded and the work required to detach the PDMS parts from each other was calculated by integrating the pulling force over the displacement from 0 displacement to the point of detachment of the two PDMS parts. A total of 3 bound PDMS parts were studied for each of the three (i.e., 10:1, 10:0.7, and 10:0.5) PDMS materials. The elastic moduli of the three PDMS materials were also determined by unidirectional tensile testing with respective PDMS slabs (4 cm x 1.2 cm x 0.2 cm) using the Instron mechanical testing system. A total of 3 PDMS parts were measured for each of the three types of PDMS materials.
2.8. Cell culture in microfluidic devices
The human bone marrow-derived stem cells (BM-MSCs) purchased from Lonza (Rockville, MD, USA), were cultured in the MSC medium consisting of α-MEM basal medium supplemented with 20% FBS, 1% NEAA, and 1% l-Glutamine [29,30]. BM-MSCs at passage 5 were used in this work. The MDA-MB-231 human triple-negative breast cancer cells purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA), were cultured in cancer cell culture medium made of DMEM supplemented with 10% FBS, 1% penicillin-streptomycin, and 1% l-glutamine. The human episomal induced pluripotent stem cells (eiPSCs) purchased from Wicell (Madison, WI, USA) [31], were cultured in the StemFlex medium (Thermo Fisher). Matrigel was used for coating the PDMS surface for 30 min at RT before loading eiPSCs and BM-MSCs into the chambers. The cells were enzymatically dissociated, suspended at a density of 5 × 104 cells mL−1 in 500 μL of medium, and pipetted into the chamber of the 10:0.5 PDMS part. The PDMS part with cells was put in an incubator (37 °C and 5% CO2) to culture overnight for cell seeding/attachment. Then, the medium was removed and another identical PDMS part punched with an inlet and an outlet was placed on top of the PDMS part with cells for assembling into the final device in a biosafety cabinet by simple hand-pressing to bind the two PDMS parts. Afterward, tubes were inserted into the inlet and outlet to flow medium into and out of the device, respectively. The cells in the PDMS devices (in a 10 mm Petri dish) were subsequently cultured in an incubator for 5 days. The PDMS parts used for cell culture in this work were sterilized by autoclaving at 121 °C for 20 min, after trimming and punching inlets/outlets [32].
2.9. Live/dead staining
Calcein AM and propidium iodide (PI) were used to stain live and dead (green and red) cells, respectively, according to the instructions of the manufacturer (Thermo Fisher). Briefly, 1 mL of DMEM containing the two dyes (1 μM for calcein AM and 1 μg mL−1 for PI) was flowed into the two chambers of the cell-laden PDMS devices and incubated for 5 min at 37 °C. The cells were imaged directly in the device with a Zeiss (Oberkochen, Germany) LSM 710 microscope.
2.10. Cell cycle analysis
The human eiPSCs were dissociated by 0.05% trypsin (Gibco) and fixed with 4% paraformaldehyde (PFA) at 4 °C overnight. The fixed cells were treated with 0.1% Triton X-100 (in saline) for 5 min and then the RNase from bovine pancreas (1 μg mL−1, Thermo Fisher) for 5 min at RT to remove any RNA. Afterward, the fixed cells were stained with PI (1 μg mL−1, Thermo Fisher) for 5 min at RT and rinsed with 1x PBS thrice. Lastly, the concentration of the stained cells was adjusted to 7 × 105 cells in 600 μL of 1x PBS per tube for cell cycle analysis with a BD (Franklin Lakes, NJ, USA) FACS Celesta flow cytometer. The data were analyzed using the BD Flowjo software (v10).
2.11. Cell migration and retrieval
The BM-MSCs and MDA-MB-231 cells were used for studying cell migration in the microfluidic devices. Briefly, the BM-MSCs and MDA-MB-231 cells in their suspensions were stained with 0.5 μM CMFDA (Thermo Fisher) and 1 μM Dil (Thermo Fisher) respectively, according to the manufacturer's instructions of the two fluorescence dyes of live cells. Afterward, the BM-MSCs (5 × 104 cells mL−1 in 500 μL of the BM-MSC medium) and MDA-MB-231 cells (5 × 104 cells mL−1 in 500 μL of the cancer cell medium) were loaded into chambers A and B of the PDMS parts, respectively, and incubated at 37 °C for 5-12 h to allow the cells to attach/seed on the respective chamber surfaces. Then, the medium was removed, identical PDMS parts (punched with inlet and outlet holes) without cells were assembled with the cell-laden PDMS parts by hand-pressing, and the cancer cell culture medium was flowed into the device. The assembled devices were put in an incubator (37 °C and 5% CO2) for 12 h. Then, cell migration was studied by taking fluorescence and difference interference contrast (DIC) images of cells in the two chambers and their connection channel using the Zeiss LSM 710 microscope. For retrieval of the cells from the microfluidic devices, the medium in the device was removed by withdrawing with a syringe and the two PDMS parts were separated by simple hand-peeling. Cancer cell medium (600 μL) was then pipetted into the PDMS part containing cells, and the MDA-MB-231 cells stained with Dil which migrated to the connecting channel between the two chambers were retrieved by pipetting with a 5-μL pipetting tip. The retrieved cells were transferred into a 6-well plate with the cancer cell culture medium for further culture to study their proliferation.
2.12. Scanning electron microscopy (SEM)
For SEM imaging, the 10:0.5 PDMS parts of 0.4 cm in thickness were cut into 1 cm × 1 cm sized samples. The PDMS samples were coated with gold using the Cressington-108 sputter coater at 15 mA for 3 min before imaging. The SEM images of the PDMS samples were acquired with a Hitachi (Tokyo, Japan) SU-70 FEG scanning electron microscope.
2.13. NPCs cultured on the surface of PDMS device with and without micro-pattern
The neural progenitor cells (NPCs) were derived from human eiPSCs using a previously reported protocol [33,34]. Briefly, the human eiPSCs in their monolayer were detached with Versene (Gibco, Gaithersburg, MD, US) at 37 °C for 2 min. The cells were collected and passed through a 70 μm cell strainer (Gibco). The resultant small eiPSC clumps were cultured in a medium consisting of mTeSR1 (STEMCELL Technologies, Vancouver, Canada) with 10 μM ROCK inhibitor (Y-27632) (Selleckchem, Houston, TX, USA) for 2 days for them to form spheroids in an ultra-low attachment dish (Corning). Then, the medium was changed to a neural differentiation medium consisting of DMEM/F12 and neural basal medium mixed at a ratio of 1:1, supplemented with 1x N2 (Gibco), 1x B27 (Gibco), 1% NEAA, and 1% l-glutamine for 10 days. Lastly, ∼20 spheroids in 500 μL of neural differentiation medium were transferred into the chambers of the 10:0.5 PDMS device with or without micro-patterns on the chamber surface for culturing. The chamber surface in the PDMS device was pre-coated with Matrigel at RT for 30 min before cell loading. The micro-patterns on the resin master surface formed as a result of the layer-by-layer printing process. To make PDMS devices without micro-patterns, the LANHU Electro-coated CW-1000 and CW-2500 sandpapers purchased from Amazon (Seattle, WA, USA) were used sequentially to sand down the resin master. Then, the resin master was rinsed with ethanol and air-dried for 20 min before use.
2.14. Immunostaining
For immunostaining, the cell samples were fixed with 4% paraformaldehyde (PFA) at RT for 30 min after removing the top PDMS part of the device by hand peeling. The cells were rinsed twice with 1x PBS, and then incubated in a solution of 0.2% Triton X-100 and 5% normal goat serum (Invitrogen, Carlsbad, CA) in 1x PBS for 1 h at RT to block nonspecific binding. Afterward, the samples were incubated with primary antibodies of MUSASHI and β-TUBUBLIN (R&D Systems, Minneapolis, MN, USA, 1:500 dilution) at 4 °C. After overnight incubation, the cells were rinsed with 1x PBS for 3 times and subsequently incubated with a secondary antibody in 1x PBS for 1.5 h at RT. The secondary antibodies for MUSASHI and β-TUBUBLIN were goat anti-rabbit PE and goat anti-mouse FITC (Invitrogen), respectively. Lastly, the samples were rinsed with 1x PBS for 4 times, and the cell nuclei were stained with DAPI (0.5 mg mL−1 in 1x PBS, 5 min at RT) for imaging. The images were taken using the Zeiss LSM 710 microscope.
2.15. Cell encapsulation in core-shell microcapsules
The procedure for the generation of cell-laden core-shell microcapsules with microfluidic devices is based on a protocol published previously [35]. Briefly, the fluids in the core (from inlet I-1) and shell (from inlet I-2) microchannels were 1% high-viscosity sodium carboxymethyl cellulose and 2% sodium alginate (purified using a previously published protocol [36]), respectively. The mineral oil was emulsified with a 1.0 g mL−1 aqueous calcium chloride solution (5:1, v v−1) and 1% SPAN-80 using a Branson Digital Sonifier 550 (EMERSON, St. Louis, MO, USA) and the resultant emulsion was introduced into the oil channel (from inlet I-3). A 1% medium-viscosity sodium carboxymethyl cellulose solution was used as the extraction solution that was introduced into the device via inlet I-4. All fluids were injected into the microfluidic device using Harvard Apparatus 11 Elite syringe pumps (Holliston, MA, USA) at ambient conditions. MDA-MB-231 cells were used for cell encapsulation and culture in core-shell microcapsules in this study. Briefly, the cells were dissociated with 0.25% trypsin and rinsed with saline twice. Afterward, the cells were resuspended in the 1% high-viscosity sodium carboxymethyl cellulose and the cell density was adjusted to 1.5 × 107 cells mL−1. The flow rates of the core, shell, oil, and extraction fluids were 300 μL h−1, 1.4 mL h−1, 8 mL h−1, and 8 mL h−1, respectively. The oil emulsion flowed out of the microfluidic device via a tube connected to the outlet (O-1) while the microcapsules were collected from the tube connected to the other outlet (O-2) of the microfluidic device. The microcapsules were collected into a 50-mL tube, centrifuge at 47 g for 3 min, and then rinsed with saline twice. Lastly, the cell-laden microcapsules were cultured in the same way as aforementioned for culturing the MDA-MB-231 cells for 5 days. The encapsulated cells on day 0 and day 5 were washed in DMEM medium with 2% FBS twice and the cell viability was determined using the live/dead assay mentioned above.
2.16. Statistical analysis
Quantitative data were collected from at least three independent experiments and all data were presented as mean ± standard deviation. All data were processed using the Excel 2010 software and analyzed with the Student's t-test for comparisons between two groups. A p-value less than 0.05 was considered to be statistically significant and labeled with one asterisk. A p-value less than 0.01 was labeled with two asterisks.
3. Results
3.1. Stable PDMS-PDMS noncovalent binding and its dependence on the ratio of PDMS prepolymer to curing agent for making the PDMS elastomer
To investigate the effect of the curing agent on the binding property of the resultant crosslinked PDMS elastomer, different ratios of the PDMS prepolymer to curing agent (10:1 that has been commonly used as recommended by the manufacturer/Dow, 10:0.7, and 10:0.5) are examined. The three types of PDMS elastomers were studied with Fourier-transform infrared spectroscopy (FTIR) first. The two characteristic chemical bonds examined on the FTIR spectra are the terminal CH=CH2 (with peak at 3070 cm−1 due to stretching) and the Si–H (with peak at ~2100 cm−1 due to vibration) bonds in the prepolymer and curing agent, respectively (Fig. 1A) [[37], [38], [39]]. With the increase in the ratio of the PDMS prepolymer to curing agent from 10:1 to 10:0.7 and 10:0.5, the peak of the Si–H bond in the resultant PDMS elastomers after 2 h (a commonly used baking time) of baking decreases while the peak of the CH=CH2 bond increases (Fig. 1B). Reducing the amount of curing agent results in fewer residual Si–H bonds of the curing agents while increasing the residual/unreacted CH=CH2 terminal groups of the PDMS prepolymer. The FTIR spectra of the 10:0.5 ratio mixture baked for 30, 45, 60, and 90 min at 75 °C are shown in Fig. 1C. The peaks of the Si–H and CH=CH2 bonds do not change after 45 min, indicating the crosslinking is completed after 45 min for the 10:0.5 mixture baked at 75 °C. This is much shorter than the 2 h baking time commonly used for the 10:1 mixture, presumably because the baking time needed for the crosslinking reaction (Fig. 1A) to complete is reduced with the decreased curing agent in the 10:0.5 mixture. Therefore, a curing time of 1 h (with and additional period of 15 min for being safe) was used to ensure the crosslinking of all curing agents for making the 10:0.5 PDMS in this study, to reduce the overall time needed for fabricating PDMS devices with the 10:0.5 formulation. We still used 2 h for curing the 10:1 and 10:0.7 mixtures to ensure sufficient curing with negligible residual cytotoxic curing agent.
Fig. 1.
Characterization of PDMS surfaces made by using different ratios of prepolymer to curing agent and their noncovalent binding. (A) The chemical structures of PDMS prepolymer and curing agent together with the formation of the PDMS elastomer as a result of the curing reaction (i.e., crosslinking of the PDMS prepolymers by the curing agents). (B) FTIR spectra of PDMS made with different ratios (10:1, 10:0.7, and 10:0.5) of prepolymer to curing agent. The absorbance peak of the CH=CH2 (blue dashed line-boxed) stretching is the highest for the 10:0.5 PDMS that has the lowest absorbance peak of the Si–H vibration (red dashed line-boxed), which indicates the lowest curing agent and the highest unreacted prepolymer in the 10:0.5 PDMS. (C) FTIR spectra of the 10:0.5 PDMS elastomer after baking at 75 °C for 30, 45, 60, and 90 min. The absorbance peaks of the CH=CH2 stretching (blue dashed line-boxed) and Si–H vibration (red dashed line-boxed) do not change after 45 min of baking at 75 °C, which indicates crosslinking of the prepolymer by the curing reagent is complete in no more than 45 min of baking at 75 °C. (D) Contact angle of water droplets on the surfaces of 10:1 (without oxygen plasma treatment by default), 10:0.7, and 10:0.5 PDMS together with the 10:1 PDMS with oxygen plasma treatment (10:1 plasma). The contact angle on the PDMS surface decreases as the ratio of prepolymer to curing agent increases, and oxygen plasma treatment greatly reduces the contact angle. (E) Results from airburst test of two identical PDMS slabs bound together either immediately (0 h) or waiting for up to 13 h after peeling off the master, by simple hand-pressing without and with (for the 10:1 PDMS only) oxygen plasma treatment. The pressure (i.e., volumetric energy) required to burst the surface binding (i.e., burst pressure) increases with the increase of the ratio of prepolymer to curing agent in the absence of plasma treatment, and is independent of the waiting time. In contrast, the burst pressure of the oxygen plasma-treated surface of the 10:1 PDMS decreases quickly with the waiting time (in no more than 30 min), although plasma treatment could greatly increase the burst pressure for the immediately bound (via covalent bonding) PDMS (10:1) slabs. **, p < 0.01.
Since crosslinking of PDMS prepolymer increases the hydrophobicity of the PDMS surface [[40], [41], [42]], the reduced crosslinking of the prepolymer in the 10:0.7 and 10:0.5 formulations (which contains more unreacted terminal groups of CH=CH2 in the resultant elastomer) should increase the hydrophilicity of the 10:0.7 and 10:0.5 PDMS surfaces compared to the 10:0.1 PDMS surface. This is confirmed by quantifying the contact angle of water drops (5 μL) on the three different PDMS surfaces. As shown in Fig. 1D, the contact angles of the water droplets on the 10:0.7 and 10:0.5 PDMS surfaces are significantly smaller than that of the water droplets on the 10:1 PDMS surface. This indicates that the 10:0.7 and 10:0.5 PDMS surfaces are significantly more hydrophilic than the 10:1 surface. Nonetheless, they are not as hydrophilic as the 10:1 PDMS surface immediately after oxygen plasma treatment and still quite hydrophobic to prevent water-spreading/wetting on their surfaces (Fig. 1D).
The oxygen plasma-treated surface of the 10:1 PDMS with high hydrophilicity has been conventionally used for tightly binding two PDMS parts due to the formation of covalent bond on their interface [42,43]. This led us to study the noncovalent surface binding property of the three different PDMS elastomers, which was done with the air burst testing of two PDMS parts of the same type of PDMS bound together by simple hand-pressing (press-bound). As shown in Fig. 1E, the pressure (or volumetric work) needed to burst (i.e., detach at where the air pressure is applied, Fig. S1) the noncovalent binding between the press-bound 10:0.7 PDMS parts (222 kPa on average) or 10:0.5 PDMS parts (274 kPa on average) is significantly higher than that of the press-bound 10:1 PDMS parts (145 kPa on average), although it is not as high as that (503 kPa on average) for the 10:1 PDMS parts covalently bonded together immediately (i.e., 0 h) after the oxygen plasma treatment.
Amazingly, the pressure needed for bursting the noncovalent binding between the surfaces of the PDMS parts made from all the three different ratios does not change significantly even after keeping the parts at 37 °C for 13 h in a cell culture incubator and then binding them together by simple hand-pressing. In contrast, the pressure or volumetric work needed to burst the binding between the 10:1 PDMS surfaces with plasma treatment and a pre-binding incubation of 0.5 h (or 13 h) at 37 °C, reduces to values (154 kPa and 146 kPa on average for 0.5 and 13 h, respectively) close that between the 10:1 PDMS surfaces without plasma treatment (Fig. 1E). This is because the plasma-activated PDMS surface is not stable and returns to that before plasma treatment shortly [44,45]. This may explain why the 10:1 PDMS parts need to be assembled quickly after the plasma treatment, to prevent leaking.
It is worth noting that no significant change is observable for the burst pressure of the 10:0.5 PDMS surfaces (with no plasma treatment) after sterilization with either 75% ethanol or autoclave (i.e., heating at 121 °C), compared with the control surfaces without any sterilization treatment (Fig. S3). This shows the 10:0.5 PDMS is compatible with the commonly used sterilization treatments for fabricating PDMS devices for biomedical applications.
3.2. Mechanical testing of the noncovalent binding between the PDMS elastomers
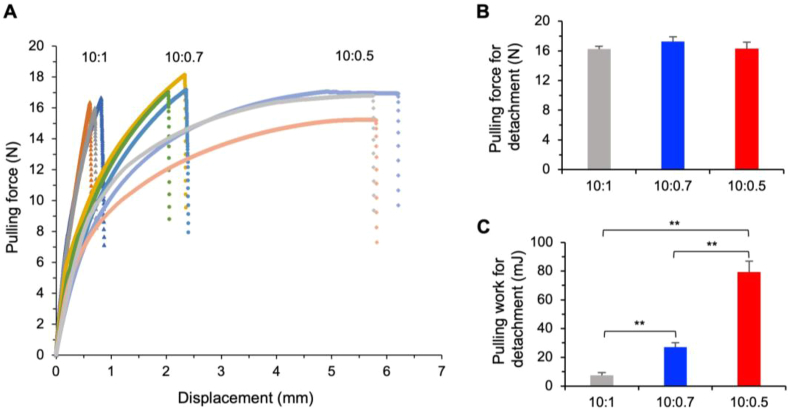
To further the understanding on how the 10:0.5 PDMS devices assembled by simple press-binding by hand without forming covalent bond on the interface, we conducted mechanical testing of the surface binding of the 10:0.7 and 10:0.5 PDMS elastomers as compared to the 10:1 PDMS. As illustrated in Fig. S2, this was done by binding two slabs of the same PDMS elastomer and then pulling the two slabs from the two opposite sides, to measure the pulling (tensile) force versus displacement (extension) till the two slabs detach from each other using the Instron mechanical testing system. The curves of pulling force versus displacement for the three different PDMS elastomers are shown in Fig. 2A. Surprisingly, the maximum force needed for pulling apart the two 10:1 PDMS slabs is not significantly different from that for the 10:0.7 and 10:0.5 PDMS slabs (Fig. 2B).
Fig. 2.
Mechanical testing of the noncovalent binding of PDMS-PDMS surfaces done by simple hand-pressing. (A) Pulling force versus displacement (extension) for the three different PDMS elastomers till the two surfaces detach. Three independent runs are shown. (B) The maximum pulling force required for detaching the two PDMS surfaces noncovalently bound together by simple hand pressing. (C) The pulling work required for detaching the two PDMS surfaces noncovalently bound together by simple hand pressing, showing the surface binding energy increases with the increase of the ratio of prepolymer to curing agent for making the PDMS elastomers. **, p < 0.01.
However, the 10:0.7 and 10:0.5 PDMS slabs are much more ductile than the 10:1 PDMS slabs (Fig. 2A). The high ductility or deformability of the 10:0.5 PDMS slabs may provide some cushioning effect to prevent them from detaching from each other, given the same stress/force applied on them. Therefore, we further calculated the pulling work needed to detach the bound PDMS slabs by integrating the pulling force over displacement. As shown in Fig. 2C, the 10:0.5 PDMS slabs require the highest pulling work for detachment compared to the 10:0.7 and 10:1 PDMS slabs. This trend of the increase in the binding energy with the decrease of the curing agents follows closely to the trend of the increase in the burst pressure with the reduction of the curing agent shown in Fig. 1E, for making the PDMS elastomer. This is not surprising because the burst pressure is actually the volumetric work or energy (J m−3 = Pa) needed to burst the binding between two PDMS slabs, according to the way by which it is measured (i.e., the volume of air in the hole of the top slab for the air burst test is constant (Fig. S1). Taken together, these data indicate that it is the binding energy (indicating toughness) rather than the binding force (indicating strength) between the PDMS surfaces determines the resistance to detachment.
3.3. Facile fabrication of PDMS microfluidic devices via noncovalent binding
The aforementioned high burst pressure (or volumetric work) and binding energy of the noncovalent binding of two 10:0.5 PDMS surfaces/parts and its independence of the waiting time for at least 13 h are of particular interest. This is because it may enable us to make microfluidic devices by simple press-binding of two or more PDMS parts without any surface treatment and post-assembly baking at an elevated temperature for biomedical applications involving living cells that can't survive the elevated temperature or oxygen plasma treatment. Therefore, we developed a method for fabricating PDMS microfluidic devices with a 5-step procedure (Fig. 3). It includes 3D printing of a resin master (1), casting the mixture of PDMS prepolymer and curing agent on the master and baking at 75 °C for 1 h (2), peeling off the PDMS parts from the master (3), trimming and punching inlet and outlet holes, autoclaving, and loading/seeding cells (4), and binding the PDMS parts together by hand-pressing (5) for further use (e.g., cell culture and analysis). The method for fabricating a PDMS microfluidic device is much more convenient and cost-effective than the 12-step procedure (Fig. S4) for the commonly used conventional photolithography-based approach. Importantly, the PDMS devices assembled with our reversible noncovalent binding method can be easily disassembled by hand-peeling for convenient retrieval of the desired cells in the device, as illustrated in Fig. 3. To confirm these, we chose two representative and common biomedical applications of microfluidics: cell culture and cell microencapsulation. The former is chosen to demonstrate the capability of loading, seeding, and culturing cells (for at least 13 h) before device assembly and conveniently retrieving the cells out of the devices after a long-term cell culture, while both applications require the ability to withstand high pressure and high flow rates without leaking, as detailed below.
Fig. 3.
A schematic illustration of the facile fabrication of PDMS microfluidic devices via noncovalent binding to allow for convenient cell loading/seeding and retrieval. A 3D printed master is created, followed by casting and baking with PDMS. Once the PDMS sets, the PDMS parts can be peeled off from the master. The PDMS parts can be trimmed and punched with holes for inlets and outlets, autoclaved, and loaded/seeded with cells. Then, the PDMS parts can be assembled into the final PDMS device by simple hand pressing without any other treatment for cell culture or microencapsulation with no leaking. The whole process of fabricating a PDMS device for cell culture/analysis takes 2–3 days, which is much shorter than the time (1–2 weeks) needed by the conventional photolithography-based fabrication of PDMS devices. Furthermore, this new method enables conveniently loading cells into the PDMS parts before assembling them into the final device because no plasma treatment or high-temperature baking is needed to bind the parts for assembly. Moreover, the assembled device can be reversibly disassembled by hand-peeling for convenient retrieval of cells out of the device for further culture and/or off-chip analysis.
3.4. Leak-free microfluidic devices assembled by noncovalent binding with simple press-binding for convenient loading of multiple types of cells into, long-term cell culture of the cells in, and handy retrieval of the cells out of the devices
The cell culture device made using the method given in Fig. 3 is shown in Fig. 4A. After peeling off the master, the PDMS parts were used either within 1.5 h or after waiting overnight (12 h). The two PDMS parts had the same chamber (depth: 2 mm, and diameter: 1 cm) and channel (depth: 2 mm, width: 2 mm, length: 1.2 cm) designs and were aligned and hand-pressed against each other for assembling into the final device. After being injected with medium using a syringe at 3 mL h−1, neither the 10:0.5 nor the 10:0.7 PDMS devices showed any leaking during static incubation at 37 °C in a cell culture incubator for 7 days (Table 1), whereas the medium leaked out of the devices made with the 10:1 PDMS during the first 24 h of incubation. The 10:0.5 PDMS devices (n = 4) were further tested and found to withstand a flow rate as high as 11 mL h−1 without leaking.
Fig. 4.
Convenient cell loading/seeding/retrieval and leak-free cell culture with the 10:0.5 PDMS microfluidic devices made by noncovalent binding. (A) A real image of the noncovalently assembled PDMS microfluidic device for cell culture containing two chambers infused with cell culture medium. Scale bar: 1 cm. (B) Morphology of human MDA-MB-231 cells, BM-MSCs, and eiPSCs grown in the chambers of the PDMS microfluidic device shown in (A). The cells are highly viable as shown by the live/dead (green/red) stain. Scale bar: 100 μm. (C) Representative peaks (left and middle panels) and quantitative data (right panel) from flow cytometry analyses (n = 3 independent runs), showing no statistically significant difference in the cell cycle distribution between the human eiPSCs grown in the device and Petri dish (control). (D) Cancer cell migration in the PDMS microfluidic device after 12 h of culture post device assembly. The human BM-MSCs and MDA-MB-231 cells were stained with CMFDA (green) and Dil (red) dyes, respectively. After 12 h of culture, the MDA-MB-231 cancer cells initially seeded in chamber B were observed to migrate into chamber A initially seeded with BM-MSCs only. Scale bar: 100 μm. (E) A cell retrieved from the PDMS microfluidic device after disassembling the device by simple hand-peeling was further cultured in Petri dish, showing proliferation of the cancer cell into a cell colony of ∼28 cells in ∼5.5 days. The MDA-MB-231 cells stained with Dil were retrieved from the connection channel between the two chambers in the device. Scale bar: 100 μm.
Table 1.
Leaking test under cell culture condition of various PDMS devices assembled by simple press-binding.
| PDMS | Waiting time before assembly [h] | Observation time post assembly [Days] | Leaking |
|---|---|---|---|
| 10:0.5 | 1.5 | 7 | No |
| 10:0.5 | 12 | 7 | No |
| 10:0.7 | 1.5 | 7 | No |
| 10:0.7 | 12 | 7 | No |
| 10:1 | 1.5 | 7 | Yesa) |
| 10:1 | 12 | 7 | Yesb) |
(observed within 24 h).
(observed within 24 h).
The data in Table 1 are consistent with Fig. 1E showing that waiting for 13 h before binding does not affect the pressure needed to burst the binding. This important feature of our press-binding approach with the 10:0.5 PDMS allows for device assembly after a long waiting (i.e., delay time), which should enable convenient loading/seeding of cells into the PDMS parts before assembling them into a device for long-term cell culture. To demonstrate this important feature, human eiPSCs, human BM-MSCs, and MDA-MB-231 human triple-negative breast cancer cells were cultured in the chambers A and B of the device made with two 10:0.5 PDMS parts (Fig. 4A). Before loading the cells, the surface of the open chambers A and B in the PDMS parts was soaked/coated with either 50% FBS in saline (for cancer cells) or 6 μg mL−1 Matrigel (for the two stem cells) for 30 min at room temperature (RT). The cells in 500 μL of their culture medium (5 × 104 cells mL−1) were then loaded into the open chambers of one of the two PDMS parts by simple pipetting and cultured overnight (12 h) in an incubator (at 37 °C and 5% CO2) for them to seed/attach on the bottom chamber surface. Afterward, the medium in the chamber was removed by gentle pipetting and the device was assembled by simply hand-pressing the two PDMS parts together. The assembled device was injected with fresh medium and cells in the device were cultured for 5 days. No leakage of medium was observed. The attached cells grew well in the chambers of the devices as indicated by the live and dead (green and red) stains with calcein AM and propidium iodide (PI), respectively, on day 5 (Fig. 4B). In addition, the cells maintained their respective typical morphologies: the eiPSC formed colonies, the BM-MSCs spread very thin with a spindle-like shape, and the MDA-MB-231 cancer cells attached and spread well although they appear rounder than the BM-MSCs. Moreover, cell cycle analysis shows the human eiPSCs grown in the chamber/device are similar to the control eiPSCs grown in Petri dish (Fig. 4C, left and middle), and there is no statistically significant difference between the two groups in terms of the distribution of cells in the different cell cycles (G1, S, and G2/M, Fig. 4C, right).
The capability of allowing for a delay/waiting time of at least 12-13 h between the creation of the PDMS parts and their assembly also enables convenient loading of two or more different types of cells inside the two different chambers for studying their interactions and chemotaxis-driven cell migration. For example, it has been shown that cancer cells respond to MSC-secreted chemokines and migrate toward MSCs [46,47]. To visualize this in vitro, we loaded the cell tracker dye CMFDA (with green fluorescence) stained BM-MSCs (5 × 104 cells mL−1 in the BM-MSC medium) and cell tracker dye Dil (with red fluorescence) stained MDA-MB-231 cells (5 × 104 cells mL−1 in the cancer cell medium) into the chamber A and B of one of the two PDMS parts, respectively. The samples could be retained in their respective chambers without mixing after an overnight (12 h) incubation of the parts in an incubator (Fig. S5). After the overnight incubation, the cells attached well to the chamber surfaces. Typical microscopic images of the two types cells attached separately in the two chambers with no MDA-MB-231 cells in chamber A and no BM-MSCs in chamber B, are shown in Fig. S6. The medium was then removed from the chambers and two PDMS parts were assembled into a device by simple hand-pressing. Of note, any residual moisture on the surface of the PDMS parts evaporates quickly (usually with ∼5 min) due to the air flow in the biosafety cabinet where the medium is removed and the PDMS parts are assembled into the final device, and it is important to ensure the surface of the PDMS parts is dry before assembling them. Fresh medium was injected and flowed from chamber A with BM-MSCs to chamber B with the cancer cells (Fig. 4A). After culturing the cells in the device overnight, we found that the MDA-MB-231 cells migrated into the connecting channel and chamber A initially seeded with only BM-MSCs (Fig. 4D). Furthermore, we can easily retrieve the migrated MDA-MB-231 cells from the connecting channel between chambers A and B by simply peeling apart the two PDMS parts (because of the reversible assembly via the noncovalent binding) with hand and pick up the cells by pipetting. The retrieved single cells can attach in a cell culture dish and proliferate from one cell into small colonies of 28 ± 3 cells in 5.5 days (Fig. 4E), with a doubling time of ∼1.1 days that is typical for the MDA-MB-231 cells [48].
Interestingly, the 3D printed resin master has a directional micropattern on its surface (Fig. S7) resulting from the layer-by-layer 3D-printing process, which can be imprinted on the PDMS surface created using the master (Fig. S8A, top). The micropattern consists of parallel fiber-like structure ranging over 1–10 μm in width. This micropattern could be conveniently removed from the surface of the 3Dprinted master by polishing it with sandpaper, so that the PDMS surface made with the sandpaper-polished master is smoother and more homogenous than that made using the master without any surface-polishing (Fig. S8A, bottom). We found that the micropattern could guide the directional growth of the neurites (i.e., the fiber-like neural processes including both axons and dendrites) from NPCs to extend in one direction, as shown by the images in the top row in Fig. S8B where MUSASHI is a neural-specific marker and is used together with β-TUBULIN for visualizing the NPCs (at the bottom of the images) and their neurites. In contrast, when the micropattern is removed by polishing the master with sandpaper for making the PDMS device, the neurites spread randomly on the smooth PDMS surface in all directions as shown by the images in the bottom row of Fig. S8B.
3.5. Leak-free microfluidic devices assembled by noncovalent binding of the 10:0.5 PDMS parts for cells microencapsulation
A sketch of the microchannel together with a real image of the microencapsulation device is shown in Fig. 5A. The microchannel design in the device is based on our previous studies on generating core-shell hydrogel microcapsules for cell and tissue microencapsulation, using non-planar (i.e., varying depths of the rectangular core, shell, oil, gelling, and extraction channels) microfluidic devices fabricated by the conventional photolithography-oxygen plasma-baking procedure (which requires multiple rounds of photoresist layering and alignment to fabricate the master, and plasma treatment to bind/assemble the device) [36,[49], [50], [51]]. In this study, the master was made by 3D printing in one step and the microencapsulation device was assembled by simple hand-pressing of two PDMS parts with the same channel design after alignment under a microscope without any treatment like oxygen plasma or post-assembly baking. It is worth noting that the microchannels in the PDMS device are circular in their cross-section (see the C⋯C cross-sectional view in Fig. 5A), for which the master can be easily made by 3D printing but very difficult (if not impossible) to fabricate using the conventional photolithography. The diameter of the channels is given in Fig. 5A and Table 2.
Fig. 5.
Leak-free 10:0.5 PDMS microfluidic devices fabricated with noncovalent binding for generating cell-laden hydrogel microcapsules. (A) A sketch together with a real picture showing the microchannel design and inlets and outlets in the PDMS microfluidic device for cell microencapsulation. The solid arrows indicate the directions of flows. The image of the C⋯C cross-sectional view shows the circular shape of the microchannel in the PDMS device. Scale bars: 1 cm for the device and 1 mm for the C⋯C cross-sectional view. (B) Image of MDA-MB-231 cells in the microcapsule on day 0 and day 5 post encapsulation with live/dead (green/red) staining. (C) Quantitative data of the MDA-MB-231 cell viability on day 0 (immediately after microencapsulation) and day 5 (after culture in the microcapsules). The PDMS microfluidic device assembled by simple hand-pressing is capable of sustaining the high pressure-driven fluid flows for generating microcapsules for hours without leaking. Scale bar: 200 μm.
Table 2.
The detailed information for generating the core-shell microcapsules with the PDMS microfluidic device. The table also shows that the PDMS microfluidic device assembled by simple hand-pressing is capable of sustaining the high pressure-driven fluid flows for generating microcapsules for hours without leaking.
| Channel | Diameter | Flow rate | Experiment time | Leaking (n = 7) |
|---|---|---|---|---|
| Core | 250 μm | 300 μL h−1 | 3 h | No |
| Shell | 350 μm | 1.4 mL h−1 | 3 h | No |
| Oil | 500 μm | 8 mL h−1 | 3 h | No |
| Extraction | 900 μm | 8 mL h−1 | 3 h | No |
To generate cell-laden core-shell microcapsules using the device, aqueous solutions of 1% high-viscosity carboxymethyl cellulose (containing MDA-MB-231 cells) and 2% sodium alginate in isotonic saline were used as the core and shell fluids, respectively. The core-shell microcapsules were formed with the flow-focusing mechanism [52], for which the flow of mineral oil emulsified with aqueous CaCl2 solution was used to pinch the aqueous core and shell flows into core-shell droplets at the flow-focusing junction (Fig. 5A) and to crosslink the sodium alginate in the shell of the droplets into stable core-shell microcapsules in the downstream gelling channel. The core fluid, shell fluid, and mineral oil were injected into the device via inlets I-1, I-2, I-3, respectively. An aqueous extraction flow of 1% medium-viscosity carboxymethyl cellulose in isotonic saline was injected into the device via inlet I-4 to extract the microcapsules from the mineral oil. This was achieved by utilizing the high surface-tension force between the hydrophilic alginate hydrogel microcapsules and hydrophobic oil to push the microcapsules from the oil flow into the aqueous extraction flow once they touch the oil-water interface between the two flows [36,53]. The mineral oil flows out the device at outlet O-1 and the extraction solution together with the extracted alginate hydrogel microcapsules flows out of the device at outlet O-2. The cell-laden microcapsules were collected in a 50-mL-tube, and centrifuged at 47 g for 3 min to remove the extraction solution, rinsed by saline, and resuspended in the fresh cell culture medium of the cancer cells. The MDA-MB-231 cells post encapsulation on days 0 and 5 were checked for their viability with live dead (green/red) staining (Fig. 5B). The cells are highly viable both immediately after microencapsulation on day 0 (97.5 ± 1.0%) and on day 5 (95.9 ± 1.2%) after culturing to form cell aggregates in the microcapsules (Fig. 5B–C). All the experimental parameters for generating the core-shell microcapsules are given in Table 2. The flow rate was as high as 8 mL h−1 (for oil) and the microfluidic devices (n = 7) were run for over 3 h without any leaking issue being observed for any of the fluids.
4. Discussion
In this work, we discovered that it is the binding energy rather than the binding force between the surfaces of the PDMS elastomer that determines the PDMS-PDMS noncovalent binding stability, and the binding energy of the PDMS surface made with a 10:0.5 ratio of prepolymer to curing agent is much higher than that of the PDMS surface made with the commonly used 10:1 ratio of prepolymer to curing agent (Fig. 1, Fig. 2). This enables us to develop a simple 5-step method (Fig. 3) for facile yet rigorous fabrication of leak-free PDMS microfluidic devices reversibly assembled by simple hand-pressing, to achieve the long-term cell culture and microencapsulation applications without any leaking issue (Fig. 4, Fig. 5). It is worth noting that due to the reduced crosslinking, the elastic modulus of the 10:0.5 (0.9 ± 0.1 MPa) and 10:0.7 (1.7 ± 0.1 MPa) PDMS elastomers is significantly much lower than that (4.4 ± 0.2 MPa) of the 10:1 PDMS (Fig. S9). However, these decreased moduli may be beneficial for culturing most mammalian cells, because they are closer to the elastic modulus of mammalian tissues (i.e., the native home of mammalian cells) that are usually less than 100 kPa [54].
The contemporary methods for binding PDMS parts involve the use of oxygen plasma treatment and baking at an elevated temperature after binding (Fig. S4), which may kill living cells. The plasma-treated PDMS surfaces need to be bind quickly (in minutes) to ensure strong covalent bonding on the interface (Fig. 1E). Consequently, samples with cells are often loaded into the devices via tiny holes (i.e., inlets) after the PDMS parts are bound and assembled into the final devices, which unfortunately making controlling the distribution of the samples in the devices very difficult. Our data show the noncovalent binding between the 10:0.5 PDMS surfaces after simple hand-pressing is sufficient to prevent leaking for various biomedical applications, including culture of one or multiple types of cells requiring a long period of time (Fig. 4) and cell microencapsulation requiring high pressure and high flow rate (Fig. 5). Furthermore, unlike the bonding strength between the oxygen plasma-treated PDMS surfaces that decrease rapidly after plasma treatment, the noncovalent binding strength between the PDMS surfaces bound by simple hand-pressing does not change significantly in at least 12-13 h (Fig. 1E and Table 1). This finding enables conveniently loading and distributing different cells into the different chambers in the PDMS parts by simple pipetting, culturing the cells in the PDMS parts overnight at 37 °C in an incubator for them to attach/seed, and removing the medium by pipetting before assembling the PDMS parts into the final devices and infusing medium into the devices for further long-term cell culture (Fig. 4).
The independence of the reversible noncovalent PDMS-PDMS binding on waiting time (Fig. 1E) enables convenient loading and seeding cells in the PDMS parts before assembling them into a microfluidic device (Fig. 4). This is particularly important for constructing microphysiological systems to study the interactions between multiple organs. Conventionally, the multiple organs with different cells (e.g., hepatocytes, renal cells, and cardiomyocytes) are cultivated in different PDMS devices and connected to each other by tubes [55,56], probably because it is difficult to inject cells into the multiple chambers for different organs in one device made with the conventional 10:1 PDMS via the tiny holes after the oxygen plasma-assisted assembly of the final device for the microphysiological systems. However, leaking at the multiple connections between the tubes and multiple PDMS devices (i.e., organs) could be a headache, which could be addressed by our method of connecting chambers through channels in one PDMS device without the need for numerous inlets and connecting tubes (Fig. 4). Importantly, the PDMS parts in the assembled device can be separated by simple hand-peeling, which allows for convenient retrieval of the cells cultured in the device for further off-chip analysis (Fig. 4E).
Although photolithography is widely used to fabricate masters or molds for making PDMS-based microfluidic devices with high precision, it is a lengthy procedure with many steps (Fig. S4) [13,[57], [58], [59]]. This is even worse when fabricating devices with non-planar microfluidic channels (i.e., channels of varying depths), which requires multiple rounds of photoresist layering and alignment [15,49,[59], [60], [61], [62], [63], [64], [65]]. In contrast, the technique of 3D printing has rapidly developed over the past a few years into an affordable technology that has been utilized in teaching, manufacturing, medical device development, and personalized healthcare for easy fabrication of 3D objects [[66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78], [79]]. Hence, fabrication of masters by 3D printing is much faster than photolithography due to the elimination of procedures like photomask creation, photoresist layering, photomask aligning, and repeated developing and rinsing [80]. Moreover, it can be very challenging to create channels more than ∼400 μm using photolithography, and masters for non-planar channel designs are especially difficult and time-consuming to create using photolithography [14]. In contrast, there is no difficulty to create such large, non-planar, and/or non-polygonal channels with 3D printing (Fig. 4, Fig. 5A) [81,82]. Therefore, using the 3D printed resin master/molds to fabricate PDMS microfluidic devices is certainly desired for applications (including biomedical applications like cell culture and cell microencapsulation) where an ultrahigh precision of the microfluidic channels is not crucial.
Lastly, the 5-step procedure for creating a PDMS microfluidic device only takes at most 2–3 days to complete, as compared to the 12-step procedure (Fig. S4) for the commonly used conventional photolithography-based approach requiring at least 1–2 weeks to complete [14]. The latter involves time-consuming steps such as printing the photomask through a specialized company (usually takes 3–7 days depending on the specific companies and how busy they are at the time of request), fabricating the master in a clean-room microfabrication facility (step: 2–6, Fig. S4) which may need at least one day, and baking the device for 3 days after binding the PDMS parts. In addition, there are several common issues with photolithography including broken and partially cleaned wafers, uneven spin-coating, possible bubbles in photoresist layers, incorrect alignment, and possible under/over-development of the photoresist [55,83]. Any of these issues may require starting the process over with new materials, making the process even longer. In contrast, 3D printers are highly automated and efficient. Inputting the design for printing multiple devices only takes ∼20 min and there is no need to monitor the machine during the printing process. Photolithography is much more labor-intensive and usually requires at least 5 h of work in a microfabrication facility to fabricate one master. Therefore, our simple and cost-effective yet rigorous method for fabricating leak-free PDMS microfluidic devices allowing for convenient loading/seeding and retrieval of cells in and out of the devices, should facilitate the wide-spread utility of PDMS microfluidic devices for various biomedical applications.
5. Conclusions
In summary, our data show that increasing the ratio of the PDMS prepolymer to the curing agent from 10:1 to 10:0.5 greatly improves the noncovalent binding energy between the resultant PDMS parts, which enables reversible binding of the PDMS parts by simple hand-pressing without the need of any additional treatment to assemble the PDMS parts into the final leak-free PDMS devices. Furthermore, the noncovalent binding energy between 10:0.5 PDMS surfaces is independent of a waiting time of at least 12-13 h. This allows for convenient loading and culturing of living cells in the PDMS parts before assembling them by simple hand-pressing into the final cell-laden device for long-term cell culture, to study the growth of one type of cells and the interactions of multiple types of cells in one single device with no need for connecting tubes. In addition, we used 3D printing technology to fabricate the master for soft lithography of PDMS-based microfluidic devices, which is more convenient and cost-effective than conventional photolithography-based master fabrication. Therefore, the facile yet powerful approach reported in this work may be valuable to facilitate the widespread utilization of the 3D printing and microfluidic technologies for various biomedical applications like cell microencapsulation and cell culture including the complex microphysiological systems.
CRediT authorship contribution statement
Bin Jiang: Conceptualization, Methodology, Investigation, Writing, Writing – original draft, Formal analysis. Alisa White: Writing – original draft, Writing, Formal analysis. Wenquan Ou: Investigation. Sarah Van Belleghem: Methodology, Formal analysis. Samantha Stewart: Writing – original draft, Writing, Formal analysis. James G. Shamul: Writing – original draft, Writing, Formal analysis. Shaik O. Rahaman: Methodology. John P. Fisher: Resources, Methodology. Xiaoming He: Conceptualization, Methodology, Writing – original draft, Writing, Formal analysis, Supervision, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper, although provisional patent application was submitted for the new mtehods/materials reported in this work.
Acknowledgments
This work was partially supported by grants from the US National Science Foundation (CBET-1831019) and National Institutes of Health (NIH R01EB023632 and R01AI123661).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.02.031.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhang R.J., Larsen N.B. Stereolithographic hydrogel printing of 3D culture chips with biofunctionalized complex 3D perfusion networks. Lab Chip. 2017;17(24):4273–4282. doi: 10.1039/c7lc00926g. [DOI] [PubMed] [Google Scholar]
- 2.Sollier E., Murray C., Maoddi P., Di Carlo D. Rapid prototyping polymers for microfluidic devices and high pressure injections. Lab Chip. 2011;11(22):3752–3765. doi: 10.1039/c1lc20514e. [DOI] [PubMed] [Google Scholar]
- 3.Duffy D.C., McDonald J.C., Schueller O.J., Whitesides G.M. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane) Anal. Chem. 1998;70(23):4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 4.Tang Q., Li X., Lai C., Li L., Wu H., Wang Y., Shi X. Fabrication of a hydroxyapatite-PDMS microfluidic chip for bone-related cell culture and drug screening. Bioact Mater. 2021;6(1):169–178. doi: 10.1016/j.bioactmat.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berean K., Ou J.Z., Nour M., Latham K., McSweeney C., Paull D., Halim A., Kentish S., Doherty C.M., Hill A.J., Kalantar-zadeh K. The effect of crosslinking temperature on the permeability of PDMS membranes: evidence of extraordinary CO2 and CH4 gas permeation. Separ. Purif. Technol. 2014;122:96–104. [Google Scholar]
- 6.Unger M.A., Chou H.P., Thorsen T., Scherer A., Quake S.R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288(5463):113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 7.Tonin M., Descharmes N., Houdre R. Hybrid PDMS/glass microfluidics for high resolution imaging and application to sub-wavelength particle trapping. Lab Chip. 2016;16(3):465–470. doi: 10.1039/c5lc01536g. [DOI] [PubMed] [Google Scholar]
- 8.Tan H.Y., Loke W.K., Nguyen N.T. A reliable method for bonding polydimethylsiloxane (PDMS) to polymethylmethacrylate (PMMA) and its application in micropumps. Sensor. Actuator. B Chem. 2010;151(1):133–139. [Google Scholar]
- 9.Lee K.S., Ram R.J. Plastic-PDMS bonding for high pressure hydrolytically stable active microfluidics. Lab Chip. 2009;9(11):1618–1624. doi: 10.1039/b820924c. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya S., Datta A., Berg J.M., Gangopadhyay S. Studies on surface wettability of poly(dimethyl) siloxane (PDMS) and glass under oxygen-plasma treatment and correlation with bond strength. J Microelectromech S. 2005;14(3):590–597. [Google Scholar]
- 11.Rezai P., Selvaganapathy P.R., Rwohl G. Plasma enhanced bonding of polydimethylsiloxane with parylene and its optimization. J. Micromech. Microeng. 2011;21(6) [Google Scholar]
- 12.Palchesko R.N., Zhang L., Sun Y., Feinberg A.W. Development of polydimethylsiloxane substrates with tunable elastic modulus to study cell mechanobiology in muscle and nerve. PLoS One. 2012;7(12) doi: 10.1371/journal.pone.0051499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He X. Microfluidic encapsulation of ovarian follicles for 3D culture. Ann. Biomed. Eng. 2017;45(7):1676–1684. doi: 10.1007/s10439-017-1823-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agarwal P., Wang H., Sun M., Xu J., Zhao S., Liu Z., Gooch K.J., Zhao Y., Lu X., He X. Microfluidics enabled bottom-up engineering of 3D vascularized tumor for drug discovery. ACS Nano. 2017;11(7):6691–6702. doi: 10.1021/acsnano.7b00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skelley A.M., Voldman J. An active bubble trap and debubbler for microfluidic systems. Lab Chip. 2008;8(10):1733–1737. doi: 10.1039/b807037g. [DOI] [PubMed] [Google Scholar]
- 16.Zhao G., Zhang Z.G., Zhang Y.T., Chen Z.R., Niu D., Cao Y.X., He X.M. A microfluidic perfusion approach for on-chip characterization of the transport properties of human oocytes. Lab Chip. 2017;17(7):1297–1305. doi: 10.1039/c6lc01532h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin C., Sofla A.Y.N. Proceedings of the Asme International Mechanical Engineering Congress and Exposition. vol. 10. Imece; 2012. A method for bonding pdms without using plasma; pp. 557–560. [Google Scholar]
- 18.Harris J., Lee H., Vahidi B., Tu C., Cribbs D., Cotman C., Jeon N.L. Non-plasma bonding of PDMS for inexpensive fabrication of microfluidic devices. JoVE. 2007;9:410. doi: 10.3791/410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chueh B.H., Huh D., Kyrtsos C.R., Houssin T., Futai N., Takayama S. Leakage-free bonding of porous membranes into layered microfluidic array systems. Anal. Chem. 2007;79(9):3504–3508. doi: 10.1021/ac062118p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H., Huang B., Zare R.N. Construction of microfluidic chips using polydimethylsiloxane for adhesive bonding. Lab Chip. 2005;5(12):1393–1398. doi: 10.1039/b510494g. [DOI] [PubMed] [Google Scholar]
- 21.Gilmore S.F., Nanduri H., Parikh A.N. Programmed bending reveals dynamic mechanochemical coupling in supported lipid bilayers. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0028517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konda A., Taylor J.M., Stoller M.A., Morin S.A. Reconfigurable microfluidic systems with reversible seals compatible with 2D and 3D surfaces of arbitrary chemical composition. Lab Chip. 2015;15(9):2009–2017. doi: 10.1039/c5lc00026b. [DOI] [PubMed] [Google Scholar]
- 23.Rafat M., Raad D.R., Rowat A.C., Auguste D.T. Fabrication of reversibly adhesive fluidic devices using magnetism. Lab Chip. 2009;9(20):3016–3019. doi: 10.1039/b907957b. [DOI] [PubMed] [Google Scholar]
- 24.Anwar K., Han T., Kim S.M. Reversible sealing techniques for microdevice applications. Sensor. Actuator. B Chem. 2011;153(2):301–311. [Google Scholar]
- 25.Go J.S., Shoji S. A disposable, dead volume-free and leak-free in-plane PDMS microvalve. Sensor Actuat a-Phys. 2004;114(2–3):438–444. [Google Scholar]
- 26.Eddings M.A., Johnson M.A., Gale B.K. Determining the optimal PDMS-PDMS bonding technique for microfluidic devices. J. Micromech. Microeng. 2008;18(6) [Google Scholar]
- 27.Regehr K.J., Domenech M., Koepsel J.T., Carver K.C., Ellison-Zelski S.J., Murphy W.L., Schuler L.A., Alarid E.T., Beebe D.J. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip. 2009;9(15):2132–2139. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sina Ebnesajjad C.E. Adhes. Technol. Handb. second ed. William Andrew Publishing; Norwich, NY: 2009. Surface treatment of materials for adhesive bonding; pp. 273–287. [Google Scholar]
- 29.Yan L., Jiang B., Li E., Wang X., Ling Q., Zheng D., Park J.W., Chen X., Cheung E., Du X., Li Y., Cheng G., He E., Xu R.H. Scalable generation of mesenchymal stem cells from human embryonic stem cells in 3D. Int. J. Biol. Sci. 2018;14(10):1196–1210. doi: 10.7150/ijbs.25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan L., Jiang B., Niu Y., Wang H., Li E., Yan Y., Sun H., Duan Y., Chang S., Chen G., Ji W., Xu R.H., Si W. Intrathecal delivery of human ESC-derived mesenchymal stem cell spheres promotes recovery of a primate multiple sclerosis model. Cell Death Dis. 2018;5:28. doi: 10.1038/s41420-018-0091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin, Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mata A., Fleischman A.J., Roy S. Characterization of polydimethylsiloxane (PDMS) properties for biomedical micro/nanosystems. Biomed. Microdevices. 2005;7(4):281–293. doi: 10.1007/s10544-005-6070-2. [DOI] [PubMed] [Google Scholar]
- 33.Morizane A., Doi D., Kikuchi T., Nishimura K., Takahashi J. Small-molecule inhibitors of bone morphogenic protein and activin/nodal signals promote highly efficient neural induction from human pluripotent stem cells. J. Neurosci. Res. 2011;89(2):117–126. doi: 10.1002/jnr.22547. [DOI] [PubMed] [Google Scholar]
- 34.Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27(3):275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal P., Choi J.K., Huang H., Zhao S., Dumbleton J., Li J., He X. A biomimetic core-shell platform for miniaturized 3D cell and tissue engineering. Part. Part. Syst. Char. 2015;32(8):809–816. doi: 10.1002/ppsc.201500025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi J.K., Agarwal P., Huang H., Zhao S., He X. The crucial role of mechanical heterogeneity in regulating follicle development and ovulation with engineered ovarian microtissue. Biomaterials. 2014;35(19):5122–5128. doi: 10.1016/j.biomaterials.2014.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bokobza L. Spectroscopic techniques for the characterization of polymer nanocomposites: a review. Polymers. 2018;10(1) doi: 10.3390/polym10010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karlsson A., Singh S.K., Albertsson A.C. Controlled destruction of residual crosslinker in a silicone elastomer for drug delivery. J. Appl. Polym. Sci. 2002;84(12):2254–2264. [Google Scholar]
- 39.Carvalho Esteves d., Brokken-Zijp A.C., Huinink J.H.P., Van M.-P.W., de G. Influence of cross-linker concentration on the cross-linking of PDMS and the network structures formed. Polym. Bull. 2009;50(16):12. [Google Scholar]
- 40.McAuliffe C. Solubility in water of normal c9 and c10, alkane hydrocarbons. Science. 1969;163(3866):478–479. doi: 10.1126/science.163.3866.478. [DOI] [PubMed] [Google Scholar]
- 41.Mcauliffe C. Solubility in water of paraffin cycloparaffin olefin acetylene cycloolefin and aromatic hydrocarbons. J Phys Chem-Us. 1966;70(4):1267–1275. [Google Scholar]
- 42.Stafie N., Stamatialis D.F., Wessling M. Effect of PDMS cross-linking degree on the permeation performance of PAN/PDMS composite nanofiltration membranes. Separ. Purif. Technol. 2005;45(3):220–231. [Google Scholar]
- 43.Haubert K., Drier T., Beebe D. PDMS bonding by means of a portable, low-cost corona system. Lab Chip. 2006;6(12):1548–1549. doi: 10.1039/b610567j. [DOI] [PubMed] [Google Scholar]
- 44.Tan S.H., Nguyen N.T., Chua Y.C., Kang T.G. Oxygen plasma treatment for reducing hydrophobicity of a sealed polydimethylsiloxane microchannel. Biomicrofluidics. 2010;4(3):32204. doi: 10.1063/1.3466882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morra M., Occhiello E., Garbassi F., Maestri M., Bianchi R., Zonta A. The characterization of plasma-modified polydimethylsiloxane interfaces with media of different surface energy. Clin. Mater. 1990;5(2–4):147–156. doi: 10.1016/0267-6605(90)90014-m. [DOI] [PubMed] [Google Scholar]
- 46.Melzer C., Yang Y.Y., Hass R. Interaction of MSC with tumor cells. Cell Commun. Signal. 2016;14 doi: 10.1186/s12964-016-0143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartosh T.J., Ullah M., Zeitouni S., Beaver J., Prockop D.J. Cancer cells enter dormancy after cannibalizing mesenchymal stem/stromal cells (MSCs) Proc. Natl. Acad. Sci. U.S.A. 2016;113(42):E6447–E6456. doi: 10.1073/pnas.1612290113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sweeney K.J., Swarbrick A., Sutherland R.L., Musgrove E.A. Lack of relationship between CDK activity and G1 cyclin expression in breast cancer cells. Oncogene. 1998;16(22):2865–2878. doi: 10.1038/sj.onc.1201814. [DOI] [PubMed] [Google Scholar]
- 49.Huang H., Yu Y., Hu Y., He X., Berk Usta O., Yarmush M.L. Generation and manipulation of hydrogel microcapsules by droplet-based microfluidics for mammalian cell culture. Lab Chip. 2017;17(11):1913–1932. doi: 10.1039/c7lc00262a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H., Agarwal P., Xiao Y., Peng H., Zhao S., Liu X., Zhou S., Li J., Liu Z., He X. A nano-in-micro system for enhanced stem cell therapy of ischemic diseases. ACS Cent. Sci. 2017;3(8):875–885. doi: 10.1021/acscentsci.7b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agarwal P., Choi J.K., Huang H., Zhao S., Dumbleton J., Li J., He X. A biomimetic core-shell platform for miniaturized 3D cell and tissue engineering. Part. Part. Syst. Char. 2015;32(8):809–816. doi: 10.1002/ppsc.201500025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang H., He X. Fluid displacement during droplet formation at microfluidic flow-focusing junctions. Lab Chip. 2015;15(21):4197–4205. doi: 10.1039/c5lc00730e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang H., He X. Interfacial tension based on-chip extraction of microparticles confined in microfluidic Stokes flows. Appl. Phys. Lett. 2014;105:143704. doi: 10.1063/1.4898040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Discher D.E., Mooney D.J., Zandstra P.W. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324(5935):1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sontheimer-Phelps A., Hassell B.A., Ingber D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer. 2019;19(2):65–81. doi: 10.1038/s41568-018-0104-6. [DOI] [PubMed] [Google Scholar]
- 56.Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32(8):760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 57.Keskin D., Mokabbar T., Pei Y.T., van Rijn P. The relationship between bulk silicone and benzophenone-initiated hydrogel coating properties. Polymers. 2018;10(5) doi: 10.3390/polym10050534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuncova-Kallio J., Kallio P.J. PDMS and its suitability for analytical microfluidic devices. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006;1:2486–2489. doi: 10.1109/IEMBS.2006.260465. [DOI] [PubMed] [Google Scholar]
- 59.Isiksacan Z., Guler M.T., Aydogdu B., Bilican I., Elbuken C. Rapid fabrication of microfluidic PDMS devices from reusable PDMS molds using laser ablation. J. Micromech. Microeng. 2016;26(3) [Google Scholar]
- 60.Sun M., Durkin P., Toth T.L., He X. Label-free on-chip selective extraction of cell aggregate-laden microcapsules from oil into aqueous solution with optical sensor and dielectrophoresis. ACS Sens. 2018;3(2):410–417. doi: 10.1021/acssensors.7b00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adrega T., Lacour S.P. Stretchable gold conductors embedded in PDMS and patterned by photolithography: fabrication and electromechanical characterization. J. Micromech. Microeng. 2010;20(5) [Google Scholar]
- 62.Chen W.Q., Lam R.H.W., Fu J.P. Photolithographic surface micromachining of polydimethylsiloxane (PDMS) Lab Chip. 2012;12(2):391–395. doi: 10.1039/c1lc20721k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li C.W., Cheung C.N., Yang J., Tzang C.H., Yang M.S. PDMS-based microfluidic device with multi-height structures fabricated by single-step photolithography using printed circuit board as masters. Analyst. 2003;128(9):1137–1142. doi: 10.1039/b304354a. [DOI] [PubMed] [Google Scholar]
- 64.Halldorsson S., Lucumi E., Gomez-Sjoberg R., Fleming R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015;63:218–231. doi: 10.1016/j.bios.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 65.Malecha K., Gancarz I., Golonka L.J. A PDMS/LTCC bonding technique for microfluidic application. J. Micromech. Microeng. 2009;19(10) [Google Scholar]
- 66.Kamei K., Mashimo Y., Koyama Y., Fockenberg C., Nakashima M., Nakajima M., Li J., Chen Y. 3D printing of soft lithography mold for rapid production of polydimethylsiloxane-based microfluidic devices for cell stimulation with concentration gradients. Biomed. Microdevices. 2015;17(2):36. doi: 10.1007/s10544-015-9928-y. [DOI] [PubMed] [Google Scholar]
- 67.Huang Y., Leu M.C., Mazumder J., Donmez A. Additive manufacturing: current state, future potential, gaps and needs, and recommendations. J. Manuf. Sci. Eng. 2015;137(1) [Google Scholar]
- 68.Bhattacharjee N., Urrios A., Kang S., Folch A. The upcoming 3D-printing revolution in microfluidics. Lab Chip. 2016;16(10):1720–1742. doi: 10.1039/c6lc00163g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Costantini M., Jaroszewicz J., Kozon L., Szlazak K., Swieszkowski W., Garstecki P., Stubenrauch C., Barbetta A., Guzowski J. 3D-Printing of functionally graded porous materials using on-demand reconfigurable microfluidics. Angew. Chem. Int. Ed. 2019;58(23):7620–7625. doi: 10.1002/anie.201900530. [DOI] [PubMed] [Google Scholar]
- 70.Serex L., Bertsch A., Renaud P. Microfluidics: a new layer of control for extrusion-based 3D printing. Micromachines. 2018;9(2) doi: 10.3390/mi9020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shankles P.G., Millet L.J., Aufrecht J.A., Retterer S.T. Accessing microfluidics through feature-based design software for 3D printing. PLoS One. 2018;13(3) doi: 10.1371/journal.pone.0192752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Munshi A.S., Chen C., Townsend A.D., Martin R.S. Use of 3D printing and modular microfluidics to integrate cell culture, injections and electrochemical analysis. Anal. Methods. 2018;10(27):3364–3374. doi: 10.1039/C8AY00829A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou L., Gao Q., Fu J., Chen Q., Zhu J., Sun Y., He Y. Multimaterial 3D printing of highly stretchable silicone elastomer. ACS Appl. Mater. Interfaces. 2019;11:23573–23583. doi: 10.1021/acsami.9b04873. [DOI] [PubMed] [Google Scholar]
- 74.Shao H., Ke X., Liu A., Sun M., He Y., Yang X., Fu J., Liu Y., Zhang L., Yang G., Xu S., Gou Z. Bone regeneration in 3D printing bioactive ceramic scaffolds with improved tissue/material interface pore architecture in thin-wall bone defect. Biofabrication. 2017;9(2) doi: 10.1088/1758-5090/aa663c. [DOI] [PubMed] [Google Scholar]
- 75.Bertana V., De Pasquale G., Ferrero S., Scaltrito L., Catania F., Nicosia C., Marasso S.L., Cocuzza M., Perrucci F. 3D printing with the commercial UV-curable standard blend resin: optimized process parameters towards the fabrication of tiny functional parts. Polymers. 2019;11(2) doi: 10.3390/polym11020292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chung Y.J., Park J.M., Kim T.H., Ahn J.S., Cha H.S., Lee J.H. 3D printing of resin material for denture artificial teeth: chipping and indirect tensile fracture resistance. Materials. 2018;11(10) doi: 10.3390/ma11101798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Borrello J., Nasser P., Iatridis J., Costa K.D. 3D printing a mechanically-tunable acrylate resin on a commercial DLP-SLA printer. Addit Manuf. 2018;23:374–380. doi: 10.1016/j.addma.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han H., Cho S. Fabrication of conducting polyacrylate resin solution with polyaniline nanofiber and graphene for conductive 3D printing application. Polymers. 2018;10(9) doi: 10.3390/polym10091003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Derakhshanfar S., Mbeleck R., Xu K., Zhang X., Zhong W., Xing M. 3D bioprinting for biomedical devices and tissue engineering: a review of recent trends and advances. Bioact Mater. 2018;3(2):144–156. doi: 10.1016/j.bioactmat.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jina O. 3D Printing for Rapid Prototyping of Microfluidic Chips Microfluidics is a fast-growing field showing great potential for a wide range of applications. R. Mag. 2017;59(1):22–23. [Google Scholar]
- 81.Shallan A.I., Smejkal P., Corban M., Guijt R.M., Breadmore M.C. Cost-effective three-dimensional printing of visibly transparent microchips within minutes. Anal. Chem. 2014;86(6):3124–3130. doi: 10.1021/ac4041857. [DOI] [PubMed] [Google Scholar]
- 82.Dahlberg T., Stangner T., Zhang H.Q., Wiklund K., Lundberg P., Edman L., Andersson M. 3D printed water-soluble scaffolds for rapid production of PDMS micro-fluidic flow chambers. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-21638-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi J.K., Agarvval P., Huang H.S., Zhao S.T., He X.M. The crucial role of mechanical heterogeneity in regulating follicle development and ovulation with engineered ovarian microtissue. Biomaterials. 2014;35(19):5122–5128. doi: 10.1016/j.biomaterials.2014.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.