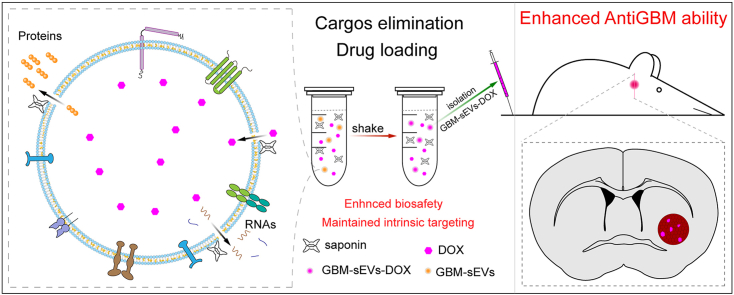
Abstract
Tumor derived small extracellular vesicles (TsEVs) display a great potential as efficient nanocarriers for chemotherapy because of their intrinsic targeting ability. However, the inherited risks of their original cargos (like loaded proteins or RNAs) from parent cancer cells in tumor progression severely hinder the practical application. In this study, a saponin-mediated cargo elimination strategy was established and practiced in glioblastoma (GBM) cell-derived small extracellular vesicles (GBM-sEVs). A high eliminating efficacy of the cargo molecules was confirmed by systematic analysis of the original proteins and RNAs in GBM-sEVs. In addition, the inherited functions of GBM-sEVs to promote GBM progression vanished after saponin treatment. Moreover, the results of cellular uptake analysis and in vivo imaging analysis demonstrated that saponin treatment preserved the homotypic targeting ability of GBM-sEVs. Thus, we developed an efficient nanocarrier with improved biosafety for GBM suppression. Furthermore, doxorubicin (DOX) transported by the saponin-treated GBM-sEVs (sa-GBM-sEVs) displayed an effective tumor suppression in both subcutaneous and orthotopic GBM models of mouse. Collectively, this study provides a feasible way to avoid the potential protumoral risks of TsEVs and can advance the clinical application of TsEVs in chemotherapy.
Keywords: Tumor derived small extracellular vesicles, Cargo elimination, Saponin, Glioblastoma, Drug delivery
Graphical abstract
Highlights
-
•
The cargo-eliminating strategy is developed to improve the biosafety of glioblastoma cell derived sEVs used as nanocarrier.
-
•
Saponin treatment can remove most of the cargo RNA and proteins from GBM-sEVs to diminish their protumoral functions.
-
•
The intrinsic targeting ability of GBM-sEVs is preserved after saponin treatment.
-
•
Cargo eliminated GBM-sEVs can be used as safe and highly efficient drug delivery vehicles for glioblastoma suppression.
1. Introduction
Efficient delivery of chemotherapeutic agents to tumor cells is critical to improve the outcome of tumor therapy. To this end, many kinds of artificial nano drug delivery systems (DDSs) have been developed over decades, such as liposomes, micelles, and nanogels [[1], [2], [3]]. However, their application is impeded by the limited biological barrier penetration ability, circulation instability, immunogenicity, and biosafety. Extracellular vesicles (EVs) are phospholipid membrane vesicles secreted by eukaryotes and act as intracellular messengers to tranrt bispoomolecules from their parent cells to recipient cells [4,5]. They can be mainly divided into large extracellular vesicles (lEVs, 200 nm ∼ 1000 nm, mainly composed of microvesicles) and small extracellular vesicles (sEVs, 30–150 nm, mainly composed of exosomes) according to size [6,7]. As natural nano DDSs, these vesicles exhibit advanced characteristics in circulation stability, biological barrier crossing and intrinsic targeting ability compared with artificial nanocarriers [[8], [9], [10]].
Recently, tumor cell-derived EVs (TEVs) have attracted great attention, because of the excellent outcome achieved in cancer therapy when TEVs are applied as nanocarriers of chemotherapeutic agents [11,12]. Drug resistance of stem cell like-cancer cells is reversed by chemotherapeutic agents transported by tumor cell-derived lEVs [13]. In addition, autologous lEVs from tumor cells loaded with methotrexate are demonstrated to be effective in relieving human lung cancer [14]. However, due to the large size of lEVs, certain conditions are needed for lEVs to penetrate into solid tumor tissues [15]. sEVs have achieved good therapeutic effect on solid tumors because they are easy to penetrate into solid tumors [16]. Thus, TsEVs may be a better candidate for drug delivery than lEVs in treatment of solid tumors. In addition to the excellent penetration ability, TsEVs usually have intrinsic targeting ability to their origin cells, leading to the improved distribution in specific tumor tissues and enhanced drug delivery efficacy [17]. In addition, TsEVs can elicit immunostimulatory function by the loaded tumor antigen repertoire [18]. Furthermore, the infinite proliferation of tumor cells is favorable for the production of sufficient autologous sEVs without immunogenicity, guaranteeing a sufficient dosage during therapy. Based on the above reasons, TsEVs are attractive candidate nanocarriers for chemotherapy.
Despite these advantages displayed by TsEVs, great concerns still exist in their application. Solid evidence has proved the functions of TsEVs in cancer progression and metastasis, which is closely related to the loaded proteins and RNAs in TsEVs [5,19,20]. For example, vesicular miR-1247–3p from high-metastatic hepatocellular carcinoma cells convert normal fibroblasts to cancer-associated fibroblasts by directly targeting beta-1,4-galactosyltransferase 3, activating β1-integrin–NF–κb signaling in fibroblasts [20]. Lymph node metastasis-associated transcript 2 in TsEVs stimulates human lymphatic endothelial cell tube formation in vitro and enhances tumor lymphangiogenesis in vivo in a VEGF-C-independent manner [21]. sEVs from liver cancer cells can build a favorable microenvironment for tumor progression by delivering PKM2 [22]. Thus, eliminating the risk of TsEVs in tumor progression is critical for their application in clinical cancer therapy.
Considering the protumoral ability of the content in TsEVs, we hypothesize that it would be helpful to reduce the protumoral risk of TsEVs by eliminating the original cargos of TsEVs. Glioblastoma (GBM) is the most common primary malignant tumor in the central nervous system [23]. The overall and long-term survival of glioblastoma is still dim [24]. Because of its strong invasive ability, it is difficult to achieve complete surgical resection of glioblastoma. Thus, postoperative synchronous radiotherapy and chemotherapy are very important to GBM patients. However, a large percentage of glioblastoma patients develop resistance to temozolomide due to elevated expression of the DNA damage repair proteins [25], and other traditional chemotherapy is impaired by the limited delivery of therapeutic agents through the blood-brain barrier (BBB) [26]. Recently, several studies show that sEVs have the capability to penetrate the BBB [[27], [28], [29]], indicating that sEVs are suitable nanocarriers for chemotherapeutic agents for treating glioblastoma. Based on the above statement, in this study, the cargo eliminating strategy was carried out in glioblastoma cell-derived sEVs (GBM-sEVs) to diminish their protumoral functions. Saponin is a naturally derived surfactant with a good safety profile and can generate holes on phospholipid membrane by binding cholesterol, thus leading to enhanced permeation [30,31]. The saponin induced permeation was identified as a simple and effective way to achieve the cargo elimination in this study. The saponin-mediated cargos eliminated GBM-sEVs (sa-GBM-sEVs) were further used as targeting nano DDSs for glioblastoma suppression. Our results showed that most of the inner proteins and RNAs in GBM-sEVs could be removed by saponin treatment while the innate targeting ability of GBM-sEVs was maintained. In addition, the sa-GBM-sEVs were unable to promote GBM progression and could achieve efficient GBM suppression by targeting delivery of doxorubicin with improved biosafety. Collectively, this study demonstrates a simple saponin-mediated cargo eliminating strategy to improve the biosafety of TsEVs, which is of great referential value in the practical application of TsEVs.
2. Experimental section
2.1. Cell culture
Human embryonic kidney cell 293, murine glioblastoma cell line GL261, and human GBM cell line U87 and U251 cells, and HLF-1 cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All these cell lines were cultured in high glucose DMEM medium (Corning) containing 10% fetal bovine serum (FBS, Gibco). HLF-1 cells were cultured in F12 medium (Corning) containing 10% fetal bovine serum (FBS, Gibco) at 37 °C with 5% CO2. All the cells used were tested negatively for mycoplasma infection.
2.2. Isolation of sEVs
EV-depleted FBS was obtained by ultracentrifugation according to the procedures reported in a previous report [32]. To prepare and isolate sEVs, cells were planted at 60% confluence in the T75 plates (Thermo). After the cells were cultured in EV-depleted media for 72 h, the conditioned media were collected and centrifuged at 400 g for 10 min, 2000 g for 15 min, and 15,000 g for 30 min at 4 °C. Finally, the media underwent ultracentrifugation at 100,000 g for 75 min at 4 °C (Optima XPN-100 Ultracentrifuge, with a SW32 Ti rotor, Beckman Coulter Life Sciences), and the precipitate was resuspended in sterile phosphate-buffered saline (PBS), followed by another ultracentrifugation (100,000 g, 75 min, 4 °C) to collect the sEVs. The obtained sEVs were stored at −80 °C for use.
2.3. Characterizations of sEVs
sEVs were characterized according to the guidelines of the International Society for Extracellular Vesicles [33].
Size Distribution and Particle Concentration. The particle diameter and concentration of U87-sEVs, U251-sEVs and GL261-sEVs were detected by a nanoflow cytometer (N30 Nanoflow Analyzer; nanoFCM Inc, Xiamen, China). For particle concentration assay, standard polystyrene nanoparticles (200 nm, concentration: 1.58 × 108/mL, NanoFCM Inc, Xiamen, China) were used for nanoparticle quantification. The particle concentration of sEVs was calculated according to the particle number ratio between sEVs samples and the concentration known standard nanoparticles. For size distribution assay, a standard curve was created using the standard silica nanoparticles (diameter: 68, 91, 113, 155 nm) in the nanoflow cytometer. The sEV samples were diluted and loaded, and the size distribution was fitted to the standard curve and obtained.
Transmission electron microscopy (TEM). The morphologies of U87-sEV, U251-sEV, HEK293-sEV and GL261-sEV were detected by TEM (Hitachi H-7650, Tokyo, Japan). Briefly, 10 μL sEV solution (2 × 1010 particles/mL) was added onto a Formvarcarbon-coated grid (300 meshes) and dried for 20 min. Then, the grid was washed with sterile PBS once and fixed with 1% (w/v) glutaraldehyde for 5 min. After that, the grid was washed with deionized (DI) water and stained with 2% (w/v) saturated aqueous uranyl oxalate for 5 min. Finally, the sEV containing grid was dried for 10 min at room temperature, and the microstructure of sEVs was imaged.
Cryogenic transmission electron microscopy (cryo-TEM): The morphologies of U87-sEV, U251-sEV, HEK293-sEV and GL261-sEV with or without saponin treatment were detected by cryo-TEM (Leica, Wetzlar, Germany). 5 μL sEV solution (2 × 1010 particles/mL) was added onto a Lacey carbon-coated grid. Then, the grid was incubated at 85% humidity for 10 s and immersed in liquid ethanol (−196 °C). Next, the grid was put on a cryo-transfer sample holder at −175 °C, and observed at different acceleration voltages (80, 100, 120,160 and 200 kV).
The sEV markers CD63, TSG101, and Lamin A/C were detected by western blotting (WB), and the detailed information was shown in the section of western blotting (WB) analysis.
2.4. Western blotting analysis
Cells and isolated sEVs were lysed with radio-immunoprecipitation assay (RIPA, Beyotime Biotechnology, China) supplemented with phenylmethanesulfonyl fluoride (PMSF) (10 μL/mL of lysis buffer, Beyotime Biotechnology, China). Then, the protein concentration was measured by bicinchoninic acid assay (BCA, Beyotime Biotechnology, China). Briefly, the solution was mixed with 5 × SDS-PAGE loading buffer (Beyotime Biotechnology, China) and boiled at 95 °C for 10 min. Proteins were separated with SDS-PAGE and transferred to a 0.45 μm polyvinylidene difluoride (PVDF) membrane. After that, the membranes were blocked with 5% (w/v) non-fat milk for 90 min and incubated with primary antibody followed by secondary antibody. The primary antibodies used for immunoblotting: CD63 (1:1000, Abcam, ab134045), CD9 (1:1000, Abcam, ab92726), Alix (1:500, Santacruz, sc-53540), TSG101 (1:500, Santa Cruz Biotechnology, sc7964), LaminA/C (1:1000, Servicebio, GB11407), Actin (1:1000, Invitrogen, MA5-15739), ANXA6 (1:500, Santacruz, sc-271,859), ECM1 (1:1000, Abcam, ab126629), ITGB1 (1:500, Santacruz, sc-374,429). The second antibodies used for immunoblotting: anti-mouse IgG, HRP-linked antibody (1:2000, CST, 7076), anti-rabbit, IgG, HRP-linked antibody (1:2000, CST, 7074).
2.5. Cargo elimination
In this study, three methods (saponin, sonication, and freeze-thaw) were used to eliminate the cargos of GBM-sEVs. The isolated sEVs were resuspended in sterile PBS to a concentration of 1 × 1010 particles/mL for further cargo elimination.
For saponin treatment, 1 × 1010 particles/mL sEVs were incubated with different concentrations (0.1%, 0.2% and 0.5% w/v) of saponin in 1 mL PBS under room temperature for 30 min, 60 min, or 90 min. Then, the sEVs were diluted to 30 mL with sterile PBS and washed with sterile PBS twice.
For sonication treatment, 1 × 1010 particles/mL sEVs were sonicated (500 V, 2 kHz, 20% power, 4 s pulse/2 s pause) for 4 cycles by Qsonica Q500 (USA). Then, the sEVs suspensions were cooled on ice for 2 min and rotated for 15 min under room temperature at a speed of 30 rpm.
For freeze-thaw cycle treatment, 1 × 1010 particles/mL sEVs were rapidly frozen at −80 °C, and then thawed at room temperature for 30 min, rotating at a speed of 30 rpm. The freeze-thaw cycle was repeated for 4 cycles.
2.6. Detection of the total protein in sEVs
U87-sEVs (1 × 1010 particles) or U251-sEVs (1 × 1010 particles) samples or saponin treated samples (n = 3) were washed with PBS twice and obtained by ultracentrifugation. All sEVs were lysed by the same volume of RIPA (Beyotime Biotechnology, China) supplemented with PMSF (10 μL/mL of lysis buffer) and detected by Pierce BCA Protein Assay Kit (Thermo Scientific, Cat#23225) according to the instructions. The absorbance of the samples was detected by a plate reader (Bio-Rad, USA) at a wavelength of 562 nm. A standard curve was used to calculate the protein concentration of each sample.
2.7. Sliver staining
sEVs samples treated with or without different methods were washed with PBS twice and obtained by ultracentrifugation. All sEVs were lysed by the same volume of RIPA, and the solution was mixed with 5 × SDS-PAGE loading buffer (Beyotime Biotechnology, China) and boiled at 95 °C for 10 min 10 μL of the solution was added to the gel followed by electrophoresis. At the end of the electrophoresis, the gel was taken out, put into a fix solution (50 mL ethanol, 10 mL acetic acid and 40 mL Milli-Q grade pure water), and incubated overnight at room temperature at a shaking speed of 60–70 rpm. Then, the experiments were conducted according to the instructions of a silver stain kit (Beyotime Biotechnology, China). Finally, the bands of these gels were detected by ChemiDoc MP imaging system (Bio-Rad).
2.8. Detection of the total RNA in sEVs
U87-sEVs (1 × 1010 particles) or U251-sEVs (1 × 1010 particles) treated with or without different methods were washed with PBS twice and obtained by ultracentrifugation (n = 3). These sEVs were suspended in the same volume of sterile PBS, and 100 μL suspension was used to detect the total RNA in the sEVs according to the instruction of SYTO® RNA Select™ Green Fluorescent Cell Stain kit (Thermo Fisher Scientific, USA). As an RNA specific fluorescent dye, SYTO™ RNA Select Green dye can penetrate lipid membrane and emit fluorescence signal after binding to RNA molecules, leading to its ability in RNA quantification. The absorbance of the samples was detected by a plate reader (Bio-Rad, USA) at a wavelength of 530 nm.
2.9. Quantitative proteomic analysis
Quantitative proteomic analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific) that was coupled to Easy nLC (Proxeon Biosystems, now Thermo Fisher Scientific) for 60/120/240 min. The peptides were loaded onto a reverse phase trap column (Thermo Scientific Acclaim PepMap100, 100 μm × 2 cm, nanoViper C18) connected to the C18-reversed phase analytical column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3 μm resin) in buffer A (0.1% Formic acid) and separated with a linear gradient of buffer B (84% acetonitrile and 0.1% Formic acid) at a flow rate of 0.3 μL/min controlled by IntelliFlow technology. The mass spectrum data were acquired using a data-dependent top10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for HCD fragmentation. The Dynamic exclusion duration was 40.0 s. Survey scans were acquired at a resolution of 70,000 at m/z 200 and resolution for HCD spectra was set to 17,500 at m/z 200, and isolation width was 2 m/z. The raw data of each sample were combined and searched using the MaxQuant software (version 1.5.3.17) for identification and quantitation analysis. Related parameters and instructions are listed in supplemetaryTable 5.
2.10. In vitro sEVs internalization assay
sEVs (1 × 1010 particles/mL) were labeled with 10 μM DiO (Thermo Fisher Scientific, USA) at 37 °C for 20 min, filtered by a 0.22 μm membrane, washed with PBS twice, and finally isolated by ultracentrifugation (100,000 g, 75 min, 4 °C). Then, an incremental concentration of the DiO labeled sEVs were added into the culture medium of U87, U251, GL261, and HEK293 cells, and the cells were further cultured for another 12 h. Meanwhile, a parallel control group was set to eliminate the false positive induced by free DiO. In the parallel control group, 10 μM DiO in PBS was processed with the same procedures, and the tube bottom was rinsed with PBS. And the above PBS was added to the culture medium. Before fluorescence microscopy observation, the cells were fixed with 4% paraformaldehyde for 20 min, permeabilized in 0.1% Triton-X 100 for 10 min, and stained with 4′, 6-diamidino-2- phenylindole (DAPI, Beyotime Biotechnology, China). The stained cells were observed with a confocal fluorescence microscope (Leica Microsystems, Wetzlar, Germany). For flow cytometry analysis, at the end of culture, the cells were washed with PBS twice and suspended with PBS before they were detected by flow cytometry with a flow cytometer (Cytoflex, USA).
2.11. RNA isolation and real-time quantitative polymerase chain reaction (RT-qPCR)
RNAs in sEVs are mainly composed of miRNAs and mRNAs, and some of these RNAs are critical to tumor progression. Therefore, the proliferation-related mRNAs (TIMP1, IDH1) and miRNAs (miR-19b, miR-21, miR-1246) [[34], [35], [36], [37]], angiogenesis-related mRNA (VGER) [38], invasion-related mRNA (EGFR) and miRNAs (miR-20 and miR-320) [[39], [40], [41]], migration-related mRNAs (TGFB1, FGF2) [42], and drug resistance-related mRNAs (APNG, ABCC3 and BIRC5) were selected [43,44] for further evaluation of the RNA eliminating efficacies. Equal amount of sEVs (1 × 1010 particles/mL) were used in each group, and the total RNA was extracted from the sEVs using TRIzol Reagent (Invitrogen) according to the manufacturer's instructions, and the total RNAs were resuspended in the same volume of RNA-free water. cDNA was synthesized using PrimeScript™ RT Master Mix (TaKaRa, Japan) with the same volume of RNA suspension. Reverse transcription was performed on a proflex PCR system (life technology). Quantification of miRNAs and mRNAs expression was performed using a TB Green Premix Ex Taq kit (TaKaRa, Japan) on a 7900HT Fast Real-Time PCR instrument (life technology). The primers of miRNAs and mRNA are listed in supplementary Table 6. All of these reactions were run in triplicate. The data were calculated as 2-△CT expression.
2.12. CCK-8 assay
U87 cells were planted on 96 well plates at a density of 8000 cells/well, and U251 cells were planted on 96 well plates at a density of 10,000 cells/well for 24 h, and GL261 cells were planted on 96 well plates at a density of 6000 cells/well (n = 3). Then, sEVs administrations were taken. Briefly, different concentrations of U87-sEVs (5 × 108, 1 × 109, and 5 × 109 particles/mL) treated with or without saponin were incubated with U87 cells for 48 h. Meanwhile, 1 × 109 particles/mL U87-sEVs treated with or without saponin were incubated with U87 cells for different time points (24 h, 48 h and 72 h). Similar experiments were conducted to U251 cells and GL261 cells with sEVs from their own. At the end of incubation, cell viability was measured by a Cell counting kit-8 (CCK8; Dojindo) assay performed according to the manufacturer's protocol. The absorbance of each cell sample was detected at 450 nm by a plate reader (Bio-Rad, USA).
2.13. DOX loading
Doxorubicin (DOX) was chosen as a model drug because of its broad-spectrum anti-cancer ability and the limited BBB penetration ability. 1 × 1010 sEVs were incubated with 100 μM DOX (Selleck) in 0.2% saponin in PBS at room temperature for 4 h. Next, the solution was diluted and washed with PBS by ultrafiltration centrifuge tube (100 KD, Millipore, USA) for five times. Then, the DOX containing sEVs were suspended with sterile PBS. Finally, the fluorescence of sEVs-DOX was detected by a multifunctional enzyme label instrument (Varioskan LUX, Thermo). The detection wavelength was 480 nm (Ex.)/595 nm (Em.). We established a standard quantification curve of DOX in the same detection condition as:
| y = 0.6367x-0.0299 (R2 = | 0.984) |
where y refers to the DOX concentration, and x refers to the fluorescence signal value of DOX. The DOX loading efficiency was calculated by the formula:
Loading efficiency = The amount of DOX in sEVs/Total DOX × 100%.
2.14. Cytotoxicity assays
U87 and U251 cells were planted in 96 well plates at a density of 10,000 cells/well for 24 h respectively (n = 3). U87 cells were incubated with free DOX or U87-sEVs-DOX (DOX concentration of 0.2, 0.5, 1, 2, 4 and 10 μM) for 24 h, and U251 cells were incubated with free DOX or U251-sEVs-DOX (DOX concentration of 0.2, 0.5, 1, 2, 4 and 10 μM) for 24 h. At the end of incubation, cell viability was measured by Cell counting kit-8 (CCK8; Dojindo) assay according to the manufacturer's protocol. Finally, the absorbance at 450 nm of each sample was detected by a plate reader (Bio-Rad, USA).
2.15. Cell apoptosis assay
5 × 105 U87 and U251 cells were planted on a six well plate for 24 h (n = 3). U87-sEVs (concentration of 5 × 108 and 5 × 109 particles/mL) or sa-U87-sEVs (concentration of 5 × 108 and 5 × 109 particles/mL) were added to the culture medium of U87 cells and incubated with U87 cells for 24 h. Meanwhile, U251-sEVs (concentration of 5 × 108 and 5 × 109 particles/mL) or sa-U251-sEVs (concentration of 5 × 108 and 5 × 109 particles/mL) were added into the culture medium of U251 cells and incubated with U251 cells for 24 h. For DOX administration, U87 cells were incubated with free DOX or U87-sEVs-DOX (concentration of 0.2, 0.4 and 0.6 μM) for 24 h, and U251 cells were incubated with free DOX or U251-sEVs-DOX (concentration of 0.2, 0.4 and 0.6 μM) for 24 h. At the end of incubation, cells were washed with PBS and stained with Annexin V-PE/PI kit (Beyotime Biotechnology, China) according to the direction, then analyzed by a flow cytometer (Cytoflex, USA).
2.16. Wound healing assay
Cells were seeded into a 6-well plate to create a confluent monolayer (n = 3). The cell monolayer was then scraped in a straight line to create a “scratch” with a p200 pipette tip, and the images were acquired. After that, the U87 cells were incubated with U87-sEVs (concentration of 5 × 108 and 5 × 109 particles/mL) or sa-U87-sEVs (concentration of 5 × 108 and 5 × 109 particles/mL). Similarly, the U251 cells were incubated with U251-sEVs (concentration of 5 × 108 and 5 × 109 particles/mL) or sa-U251-sEVs (concentration of 5 × 108 and 5 × 109 particles/mL). The images were taken at 12 h and 24 h after sEVs treatment. The wound closure percentage was defined as the below formula:
Wound closure percentage = the migrated cell surface area/total surface area × 100%.
2.17. Transwell assay
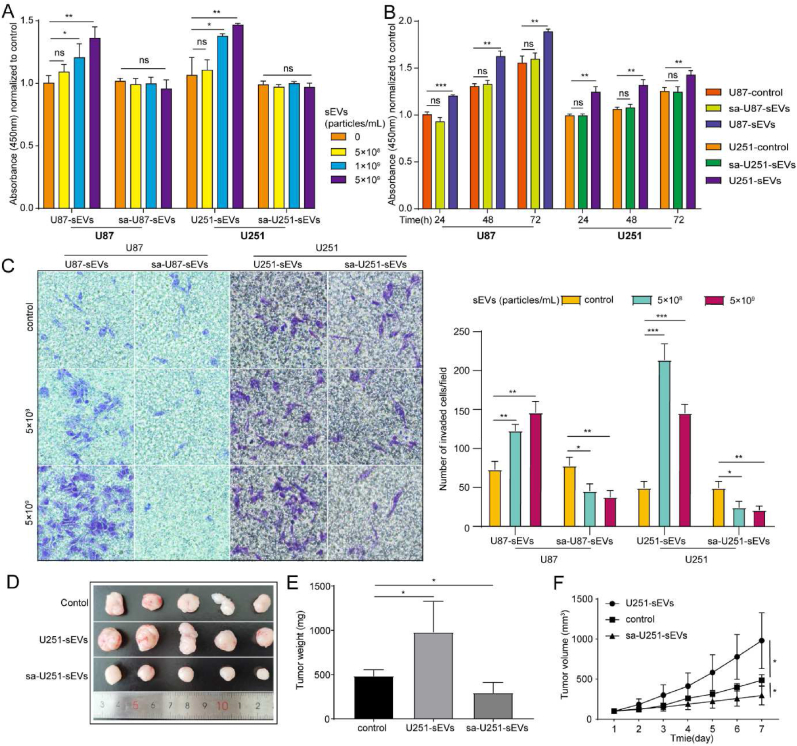
Transwell assay was used to evaluate the function of sEVs on the invasion of cancer cells. Briefly, Matrigel (Corning) was mixed with PBS (1:8, v/v), and 30 μL of the mixture was added to the upper chamber of the transwell plate (24-well plates, 8-μm pores; Corning). After that, the transwell plates were incubated in the incubator for 4 h. Next, 5 × 104 U87 cells together with U87-sEVs (5 × 108 and 5 × 109 particles/mL) or sa-U87-sEVs (5 × 108 and 5 × 109 particles/mL) were seeded in the upper chamber. 24 h later, the cells migrated to the bottom of the chamber were fixed and stained with 1% crystal violet solution before being observed with a microscope. Similar experiments were also conducted to U251 and GL261 cells with sEVs from their own. Three independent experiments were conducted.
2.18. In vivo assay
All animal experimental protocols were approved by the Animal Research Committee of Shanghai Sixth People's Hospital (SYXK (Shanghai, China) 2011–0128, January 1, 2011). Four weeks old female nude mice and C57 mice were purchased from SLAC Laboratory Animal Company (Shanghai, China).
Subcutaneous xenograft tumor model. U251 cells (1 × 107, 100 μL) were injected in the right flank of nude mice to construct subcutaneous xenograft tumor model. When the tumor volume reached about 100 mm3, sEVs administration was carried out. Briefly, the tumor bearing nude mice were randomly divided into three groups (n = 5). PBS (100 μL), U251-sEVs (1 × 1011 particles/mL, 100 μL), or sa-U251-sEVs (1 × 1011 particles/mL, 100 μL) were injected intravenously into mice daily for a week. These mice were sacrificed on the eighth day, and the tumors were collected. At the same time, DOX administration was carried out when the tumor volume of nude mice reached 100 mm3. Briefly, the tumor bearing nude mice were randomly divided into three groups (n = 5). PBS (100 μL), DOX (2 μg/kg, 100 μL) or U251-sEVs-DOX (2 μg/kg, 100 μL) were injected intravenously into mice daily for a week. These mice were sacrificed on the eighth day and the tumors were collected. Tumor volumes were measured by caliper and calculated by the formula:
| tumor volume = L × W2/2 |
Orthotopic brain tumor model of C57 mouse and nude mouse. U251 cells (2 × 105, 1 μL) were injected into the right striatum of nude mice by a stereotactic instrument (Neurostar, SD581). GL261 cells (1 × 105, 1 μL) were injected into the right striatum of C57 mice by a stereotactic instrument. Three weeks later, one mouse was randomly selected and sacrificed, and the brain of this mouse was cut into pieces to confirm the formation of tumor in the brain.
In vivo biodistribution of sEVs: The tumor bearing nude mice were randomly divided into three groups (n = 3). HEK293-sEVs, U251-sEVs and sa-U251-sEVs were labeled with DiR in a concentration of 10 μM at 37 °C for 30 min, filtered by 0.22 μm membrane, and washed with PBS. Then, 200 μL sEVs suspension (1 × 1011 particles/mL) were intravenously injected into orthotopic glioma-bearing model mice. Mice were imaged with a Bioluminescence imaging system (PerkinElmer, USA) after 12 h and 24 h. And mice were sacrificed 24 h after injection, the mean fluorescence intensity of main organs, including brain, liver, lung, and small intestine, were measured and the weights of these tissues were weighed. The fluorescence of each organ was calculated with the formula: mean fluorescence intensity/weight.
Administration. The tumor bearing C57 mice were randomly divided into three groups (n = 5). PBS (100 μL), DOX (2 μg/kg, 100 μL), or GL261-sEVs-DOX (2 μg/kg, 100 μL) were injected intravenously into each group for a week every day. These mice were sacrificed the next day after the treatment finished. Then, the extracted tumors were stained with HE for histological examination. The DOX distribution was visualized by confocal microscopy. The tumor volume was measured by caliper and calculated by the below formula [45]:
| Tumor volume= (long axis) × (short axis)2 × π/6 |
For survival analysis, orthotopic brain tumor bearing C57 mice were constructed as described above and randomly divided into three groups (n = 5). PBS (100 μL), DOX (2 μg/kg, 100 μL) or GL261-sEVs-DOX (2 μg/kg, 100 μL) were injected intravenously into mice daily for a week. Finally, the survival times of these mice were recorded.
2.19. Hematoxylin and eosin (HE) staining
The brains of orthotopic tumor bearing C57 mice, and the livers, kidneys and hearts of tumor bearing nude mice were rapidly isolated and fixed with 4% paraformaldehyde in PBS overnight at 4 °C and embedded in paraffin. The fixed brains were sectioned into slices with thicknesses of 20 μm. The samples were then stained with hematoxylin and eosin and imaged by DM6 microscopy (LEICA, Germany).
2.20. Statistical analysis
Statistical analysis was performed with GraphPad Prism software (version 8.0.1). All results were expressed as mean ± standard deviation. Statistical analysis was performed using Student's t-test for comparison between two groups. Differences were considered significant when *P < 0.05, **P < 0.01, ***P < 0.001, or ****P < 0.0001, ns indicates no statistical significance.
3. Results
3.1. Saponin-mediated elimination of the original cargos in GBM-sEVs
We isolated sEVs from the cultured medium of U87 and U251 by differential centrifugation. And the characterizations of these sEVs were shown in Figs. S1A–C which demonstrated the successful isolation of GBM-sEVs. To eliminate the original cargo of GBM-sEVs, saponin was selected as an effective tool to enhance the permeability of bilayer phospholipid membrane, inducing the leakage of the cargo molecules from the internal cavity of sEVs.
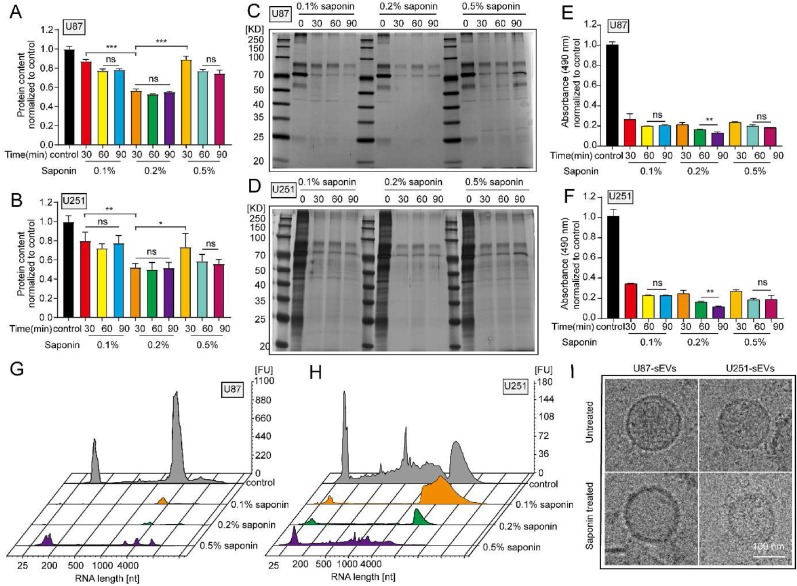
Proteins and RNAs are two predominant components of extracellular vesicles. Thus, these biomacromolecules were employed to indicate the eliminating efficacy of the cargos inside sEVs. After the isolated GBM-sEVs were incubated with increasing saponin concentrations from 0.1 wt% to 0.5 wt% for different durations (30 min, 60 min, and 90 min), bicinchoninic acid assay (BCA) was taken to detect the protein concentration. The results of BCA (Fig. 1A and B) showed that 0.2 wt% saponin treatment exhibited the highest protein eliminating efficacy in both U87-sEVs (47.8% ± 4.0% eliminating efficacy) and U251-sEVs (43.4% ± 1.8% eliminating efficacy). In addition, there was no significant difference in protein eliminating efficacies when the sEVs were treated for different durations, suggesting that 30 min treatment with 0.2 wt% saponin was sufficient to eliminate proteins in sEVs. Moreover, the results of silver staining (Fig. 1C and D) also showed an obvious decline of protein contents in all the saponin treating groups compared with that of untreated sEVs. Thereafter, several analyses were used to evaluate the RNA eliminating efficacy by saponin treatment. The results of RNA quantification by the SYTO™ RNA Select Green dye confirmed that over 70% RNAs in U87-sEVs and U251-sEVs were eliminated under all these saponin treatment conditions (Fig. 1E and F). In addition, both U87-sEVs (87.8% ± 1.2%) and U251-sEVs (88.6% ± 0.8%) treated with 0.2 wt% saponin for 90 min exhibited the highest eliminating efficiency. While, the RNA eliminating efficiency was not increased when the treating time prolonged to 120 min (Figs. S1D–E). Next, U87-sEVs and U251-sEVs were treated with saponin at different concentrations for 90 min, and the RNAs in sEVs were detected by the RNA enrichment analysis (Fig. 1G and H). Significant decrease of RNA was observed after saponin treatment, and 0.2 wt% saponin treatment obtained the highest efficiency. Because 30 min and 90 min saponin treatment achieved similar protein eliminating efficacy, we chose the condition of 90 min 0.2 wt% saponin treatment for the cargo elimination in the following experiments. In addition, some other methods could also increase membrane permeability for exogenous molecules to load into sEVs. Thus, we further evaluated the sEVs cargo eliminating efficacy of sonication and freeze-thaw methods. The results (Figs. S1F–H) showed that the highest protein and RNA removal efficiency was achieved by saponin and sonication, while the removal efficiency of freeze-thaw cycle is far lower than that of the other two methods.
Fig. 1.
Saponin-mediated elimination of original cargos in GBM-sEVs. (A and B) The total proteins in U87-sEVs and U251-sEVs detected by BCA assay. (C and D) The total proteins in U87-sEVs and U251-sEVs detected by sliver staining. (E and F) The total RNAs in U87-sEVs and U251-sEVs detected by SYTO RNA select dye. (G and H) RNA enrichment analysis depicted in fluorescence units (FU) per nucleotide (nt) of the RNA content of different groups. U87-sEVs and U251-sEVs were treated with saponin at different concentration for 90 min. (I) The shape of sEVs detected bycryo-EM. Scale bar: 100 nm *P < 0.05, **P < 0.01, ***P < 0.001, ns indicates no statistical significance.
In order to validate saponin treatment is a mild method to change the permeability of phospholipid bilayer membrane to achieve the cargo elimination in GBM-sEVs. The main physical properties of these GBM-sEVs after saponin treatment were investigated. There was no significant change in the size distribution, concentration, and zeta potential of U87-sEVs and U251-sEVs after saponin treatment (Figs. S1I–S1J). While, it was found that the buoyant densities of U87-sEVs and U251-sEVs were slightly decreased from 1.17 g/mL to 1.12 g/mL after saponin treatment (Fig. S1M), which should be caused by the elimination of cargo proteins and RNAs. Furthermore, the morphology and structure before and after saponin treatment of these sEVs did not show obvious difference, and the integrity of these membrane was preserved after saponin treatment (Fig. 1I). These results suggested that saponin treatment was compatible for the preservation of the main physical properties of sEVs.
3.2. Maintenance of the inherent targeting ability of GBM-sEVs after saponin treatment
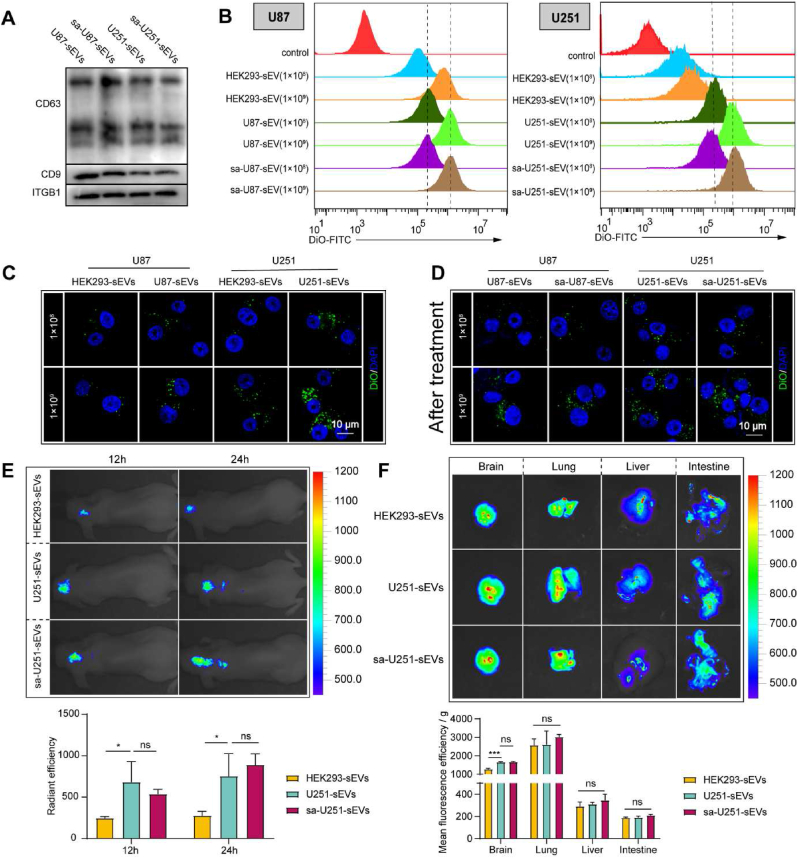
The cancer targeting ability is quite important to achieve high transportation of chemotherapeutic agents to tumor sites when using TsEVs as nano DDSs. Therefore, we evaluated whether saponin treatment could preserve the targeting ability of GBM-sEVs. Considering the critical role of surface proteins in rendering the targeting properties, the expressions of some surface proteins were detected after saponin treatment by WB analysis. The WB analysis results (Fig. 2A and Fig. S2A) showed that there was no significant difference in the expressions of CD9, CD63, and integrin β1 (ITGB1) between the control group and saponin treated group, suggesting a preservation of surface proteins after saponin treatment.
Fig. 2.
Evaluation of targeting and accumulating ability of GBM-sEVs after saponin treatment. (A) CD63, CD9 and ITGB1 detected by WB. (B) Representative histogram of DiO fluorescence in U87 and U251 cells. (C and D) Confocal fluorescence microscopic images of U87 and U251 cells. Scale bar: 10 μm. (E) In vivo fluorescence images of the orthotopic brain tumor model by an IVIS Spectrum/CT imaging system (upper). Statistical analysis of the DiR fluorescence in vivo (lower). (F) Fluorescence images of the main organs of the orthotopic brain tumor bearing mice were captured 24 h after DiR-labeled sEVs were intravenously injected (upper). Statistical analysis of the DiR fluorescence of the main organs (lower). *P < 0.05, ***P < 0.001, ns indicates no statistical significance.
According to the previous studies, tumor cells are prone to uptake autologous sEVs [46,47]. Therefore, cellular uptake assay by DiO labeled sEVs was applied to further evaluate the preservation of the homotypic recognition ability after saponin treatment. HEK293 cell-derived sEVs (HEK293-sEVs) were chosen as non-GBM cell-derived sEVs controls. To exclude the possibility of the false positive signal of DiO clusters, a parallel experiment was conducted, and the cellular uptake results in the parallel group (Fig. S2B) showed no fluorescent signal of DiO cluster inside cells. As shown in Fig. 2B and C, the cellular uptake and flow cytometry analysis results demonstrated that both U87 and U251 cells exhibited a higher uptake efficacy of their autologous sEVs (DiO labeled) than HEK293-sEVs, confirming the homotypic targeting ability of these sEVs to their parent cells. As shown in Fig. 2B and D, the cellular uptake ability of saponin treated sEVs was barely affected compared with that of the natural GBM-sEVs, indicating that the saponin treatment has no influence on the targeting ability of GBM-sEVs. The above results demonstrate that saponin treatment is favorable for the maintenance of the homotypic targeting ability of TsEVs. In addition, although sonication method showed high efficacy in the cargo elimination, both U87-sEVs and U251-sEVs lost their targeting ability after sonication treatment (Figs. S2C–D), implying a destroy of this method to the function of sEVs.
Several reports have demonstrated the BBB penetration ability of sEVs and their accumulation in tumor site by enhanced permeability and retention effect. Thus, the in vivo targeting ability of saponin treated GBM-sEVs was further evaluated using an orthotopic brain tumor model of nude mice. Fig. 2E showed the in vivo accumulation of HEK293-sEVs, U251-sEVs, or saponin treated U251-sEVs (sa-U251-sEVs) 12 h and 24 h after their intravenous injection. Obviously, the radiant efficacy of the interesting area (ROI) between U251-sEVs and sa-U251-sEVs group showed no significant difference, and the ROI value of the two groups was significantly higher than that of HEK293-sEVs group at both time points. In addition, the main organs (brain, lung, liver, and intestine) were extracted for further detection. As shown in Fig. 2F, the average ROI value in the brain was higher than that in the liver and intestine, but lower than that in the lung. And a significantly higher ROI value of U251-sEVs and sa-U251-sEVs treated groups than that of HEK293-sEVs treated group was also demonstrated. Collectively, the above results prove the homotypic targeting ability of GBM-sEVs to glioblastoma in vivo, and saponin treatment exerts no negative impact on this property.
3.3. Cargo eliminating evaluation of U251-sEVs by quantitative proteomic and RT-qPCR analysis
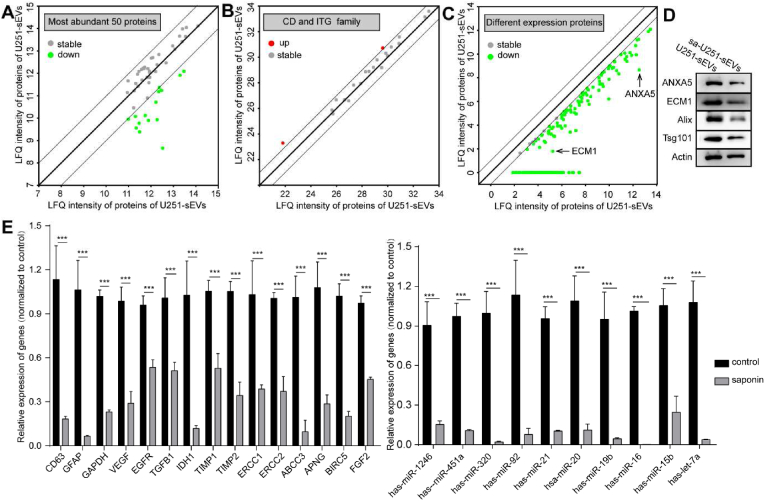
The cargo proteins and RNA play a critical role in endowing the TsEVs with protumor functions. Thus, we further studied the cargo eliminations of some representative proteins and specific RNA sequences of saponin treated U251-sEVs by quantitative proteomic analysis and qPCR in detail. Proteins in U251-sEVs and sa-U251-sEVs detected by quantitative proteomic analysis were compared based on the label-free quantification (LFQ) intensity. The scatter plot showed the LFQ intensity of proteins in U251-sEVs over sa-U251-sEVs (Fig. S3A, threshold: fold change >2, or fold change <0.5). 1369 proteins were detected both in U251-sEVs and sa-U251-sEVs group, 1089 and 1108 proteins were quantified in U251-sEVs group and sa-U251-sEVs group, respectively (Fig. S3, Supplementary Table 1), 280 and 261 proteins were identified without LFQ intensity in U251-sEVs group and sa-U251-sEVs group, respectively (Supplementary Table 7). The most abundant 50 proteins were selected and analyzed (Fig. 3A), which occupied more than half of the total amount of proteins. Because DDX11L8 and EFF1A1P5 cannot be recognized by Funrich software, these two proteins were deleted. These 48 proteins in U251-sEVs and sa-U251-sEVs can be mainly categorized into exosome, cytoplasm, lysosome, nucleus, plasma membrane, centrosome, and cytosol (Supplementary Table 2). Many clusters of differentiation (CD) proteins and integrins that expressed on sEVs surface are important for the cellular internalization of sEVs. Thus, proteins from CD and integrin family were classified, and the results showed that the expressions of these proteins were nearly unchanged during saponin treatment (Fig. 3B). In addition, 211 proteins were identified as differently expressed proteins between U251-sEVs and sa-U251-sEVs (Fig. 3C, Supplementary Table 3) (fold change <0.5). And it was found that these proteins could be mainly grouped into cytoplasm and exosomes (Supplementary Table 4). Finally, WB analyses of some representative proteins were performed to further validate the results of quantitative proteomic analysis. As shown in Fig. 2A, the expressions of CD9, CD63, and ITGB1 belonging to the membrane proteins were nearly unchanged after saponin treatment. While, the expressions of ECM1, ANXA5, Tsg101, and Alix that locate in the cavity of sEVs decreased significantly (Fig. 3D). These results clearly confirmed the effectiveness of saponin treatment in eliminating the inner cargo proteins of sEVs rather than membrane proteins. Furthermore, the saponin treatment slightly affected the expression of β-actin.
Fig. 3.
Quantitative proteomic analysis and qPCR analysis of U251-sEVs with and without saponin treatment. (A) Comparison of the most abundant 50 proteins between U251-sEVs and sa-U251-sEVs. (B) Comparison of the CD family and integrin family between U251-sEVs and sa-U251-sEVs. (C) The down regulated proteins in U251-sEVs after saponin detected by quantitative proteomic analysis. (D) The protein expression of ECM1, ANXA5, Alix, Tsg101 and Actin in U251-sEVs and sa-U251-sEVs detected by WB. (E) The mRNA expression of CD63, GFAP, GAPDH, VEGF, EGFR, TGFB1, IDH1, TIMP1, TIMP2, ERCC1, ERCC2 ABCC3, APNG, BIRC5, FGF2 were detected by qPCR, and the expression of let-7a, miR-15b, miR-16, miR-19b, miR-20, mir-21, miR-92, miR-320, miR-451a and miR-1246 were detected by qPCR. ***P < 0.001.
mRNA and miRNA occupy the majority of the total RNAs in sEVs. Thus, some specific mRNAs and miRNAs were selected for RNA eliminating efficiency analysis. As shown in Fig. 3E, compared with the control group, all of the selected mRNAs were decreased by 60% after saponin treatment, and the eliminating efficacies for these mRNA were in the range from 46.4% to 93.4%. Furthermore, the saponin treatment showed a more effective elimination in miRNAs, and the eliminating efficacies were in the range of 75.4%–99.7%. All these results proved the effective elimination of protumoral RNA molecules in sEVs after saponin treatment.
3.4. Eliminating the protumoral functions of GBM-sEVs by saponin treatment
The protumoral ability of TsEVs is mainly given by their original cargos. Thus, we assumed that these functions of GBM-sEVs would be diminished after saponin mediated cargo elimination. Therefore, the biological functions of saponin treated GBM-sEVs in tumor progression were evaluated. The results of cell proliferation assay showed that U87-sEVs promoted the proliferation of U87 cells, but this ability disappeared in sa-U87-sEVs independent of time and dosage, and it was also confirmed by the results in U251 cells (Fig. 4A and B). Next, the results of wound healing assays showed that U87-sEVs can promote U87 migration in a dosage-independent manner, but this phenomenon was not observed in sa-U87-sEVs group. And similar results were also obtained in U251 cells (Fig. S4A). The results of transwell migration assay showed that both U87-sEVs and U251-sEVs significantly promoted the invasion of U87 or U251, respectively. While sa-U87-sEVs and sa-U251-sEVs inhibited tumor cell invasion in a dosage-independent manner (Fig. 4C). The above results demonstrated that sa-GBM-sEVs showed no potential ability in the promotion of tumor progression.
Fig. 4.
Eliminating the original function of GBM-sEVs in tumor progression by saponin treatment. (A) Proliferation of U87 cells after 48 h incubation with U87-sEVs or sa-U87-sEVs at different concentration (left), proliferation of U251 cells after 48 h incubation with U87-sEVs or sa-U87-sEVs at different concentration (right). (B) Proliferation of U87 cells after 48 h incubation with U87-sEVs or sa-U87-sEVs for different time points (left), proliferation of U251 cells after 48 h incubation with U87-sEVs or sa-U87-sEVs for different time points (left) (C) Micrographs of the Transwell assay (left). The cell invasive ability of U87 and U251 cells after being treated with U87-sEVs, sa-U87-sEVs and U251-sEVs, sa-U251-sEVs, respectively, at different concentrations for different time points. Statistical analysis of the invasion of U87 and U251 cells after treatment (right). (D) Images of subcutaneous xenograft tumor-bearing mice treated with PBS, U251-sEVs or sa-U251-sEVs. (E) Weight of the subcutaneous xenograft tumor. (F) The volume of the subcutaneous xenograft tumor. *P < 0.05, **P < 0.01, ***P < 0.001, ns indicates no statistical significance.
To further test the improved biosafety of sa-GBM-sEVs in vivo, a subcutaneous xenograft tumor model of nude mice was established by injection of U251 cells. And U251-sEVs or sa-U251-sEVs were administrated daily by intravenous injection for a week. The in vivo results show that sa-U251-sEVs significantly inhibited the growth of solid tumor in mice, while U251-sEVs obviously promoted tumor progression (Fig. 4D–F). We hypothesized that the inhibition of sa-U251-sEVs to tumor growth might result from their disturbance to the regular communication mediated by EVs among tumor cells. Thus, a competing cellular internalization assay was carried out, and the results shown in Fig. S4B confirmed this hypothesis since the cellular uptake of natural U87-sEVs or U251-sEVs was significantly suppressed by the presence of sa-U87-sEVs or sa-U251-sEVs. Collectively, both these in vitro and in vivo results demonstrate that the cargo eliminated GBM-sEVs display no function in the promotion of GBM progression, thus improving the biosafety in application.
Additionally, the results in Fig. 2F showed a strong accumulation of sa-U251-sEVs in the lung tissue of the tumor bearing mice, thus the effect of sa-U251-sEVs on lung was also evaluated in vivo and in vitro. Immunohistochemistry (IHC) staining results show that there is no significant difference in the expression of Ki-67 between the control group and sa-U251-sEVs group (Fig. S4C). Next, a study shows that sEVs are mainly internalized by lung fibroblast cells [48]. The lung fibroblast cells HLF-1 was chosen to evaluate whether sa-U251-sEVs could affect their proliferation. And the CCK-8 results showed that the proliferation of HLF-1 cells was not increased by the incubation with sa-U251-sEVs (Fig. S4D). These results implied the safety profile of sa-U251-sEVs in other organs.
3.5. Cargo eliminated GBM-sEVs as efficient chemotherapeutic agent nanocarriers for glioblastoma suppression
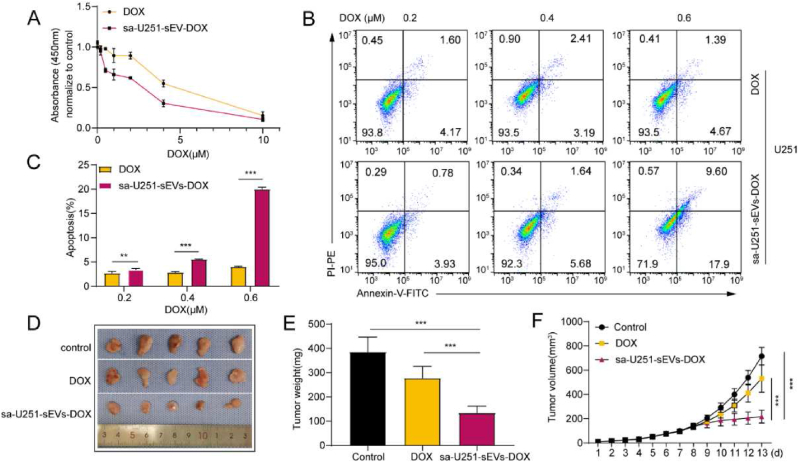
Next, we explored the potential of the cargo eliminated GBM-sEVs as nanocarriers for glioblastoma therapy. The saponin treatment can not only eliminate the cargo molecules from sEVs but also load external molecules into sEVs. Therefore, 0.2% saponin solution with DOX (100 μM) was used to achieve cargo elimination and drug loading synchronously in U251-sEVs. The drug loading content of U251 was about 1.16 μg/1010 particles (∼2 nmol/1010 particles). Then, the antitumor ability of sa-U251-sEVs-DOX was evaluated. As shown in Fig. 5A, sa-U251-sEVs-DOX exhibited a higher suppression rate than the free DOX nearly at all tested concentrations. Furthermore, sa-U251-sEVs-DOX was more effective in apoptosis induction of U251 cells than free DOX (Fig. 5B and C). The same tendency was also demonstrated in U87 cells (Figs. S5A–C). We speculated that the enhanced suppression in GBM cell growth might be caused by the more efficient cellular uptake of DOX mediated by sEVs. To prove this hypothesis, the antitumor efficacy of sa-U251-sEVs-DOX was evaluated by a subcutaneous xenograft tumor model of nude mice. As shown in Fig. 5D–F, both DOX and sa-U251-sEVs-DOX could significantly inhibit the growth of solid tumor in nude mice after continuous intravenous injection for 7 days. Further analysis of tumor weight and volume showed a significantly higher efficacy in tumor suppression by sa-U251-sEVs-DOX treatment (average weight: 134.54 ± 27.15 mg; average volume: 217.43 ± 47.60 mm3) than by free DOX treatment (average weight: 277.66 ± 48.43 mm3, p = 0.0004; average volume: 531.62,100.7476, p = 0.0005). Because the distribution of DOX can be influenced by sEVs, HE staining was used to detect the toxicity of DOX to the main organs of mice and the results are shown in Fig. S5D. It can be seen that neither DOX nor sa-U251-sEVs-DOX exhibited obvious toxicity to the main organs. Taken together, these results indicate that sa-U251-sEVs-DOX possess enhanced antitumor effect without increasing the toxicity to the main organs, which further ensures the biosafety of sa-U251-sEVs-DOX.
Fig. 5.
Therapeutic effect of tumor-derived sEVs as DOX carriers for GBM suppression. (A) U251 viability after 24 h incubation with free DOX or sa-U251-sEVs-DOX. (B) Flow cytometry analysis of the apoptosis of U251 cells after treatment with free DOX or sa-U251-sEV-DOX for 24 h. Cell apoptosis was detected by Annexin V/PI as a maker. (C) Statistical analysis of the apoptosis rate. (D) Images of the extracted subcutaneous xenograft tumor treated with PBS, free DOX, or sa-U251-sEVs-DOX 7 d after treatments. The DOX or sa-U251-sEVs-DOX was administrated on the 7th day after U251 injection. (E) Tumor weight analysis. (F) Tumor volume analysis. **P < 0.01, ***P < 0.001.
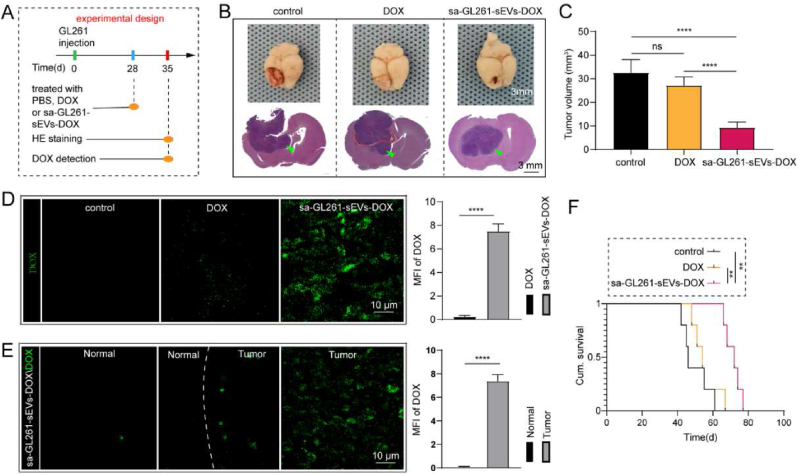
Because of the influence of the immunity system to tumor therapy and the obstruction of BBB to brain tumor therapy, orthotopic brain tumor model was further used to evaluate the ability of saponin treated GBM-derived sEVs as an efficient nanocarrier for GBM established in immunocompetent C56BL/6 mice. The experiment was conducted as the procedures illustrated in Fig. 6A. Correspondingly, GL261 derived sEVs (GL261-sEVs, Figs. S6A–C) were used as agent carriers to transport DOX to tumor sites. GL261-sEVs also presented a promotion function in GBM progression, which was also diminished by saponin mediated cargo elimination (Figs. S6D–E). Furthermore, the cellular uptake and further flow cytometry analysis show an intrinsic targeting ability of GL261-sEVs to their parent cells (Figs. S6F–G). These results confirmed the homotypic targeting ability of GL261-sEVs and the improved biosafety of GL261-sEVs by saponin treatment. Thus, saponin treated GL261-sEVs loaded with DOX (sa-GL261-sEVs-DOX) were used for the therapy of GBM in model mice. After continuous injections of sa-GL261-sEVs-DOX (2 μg/kg) or free DOX (2 μg/kg) for a week, the brain tissues were extracted for fluorescence and histological analysis. The extracted brain tissues were sectioned for histological analysis, and the representative images of the sections with the largest tumor area in each sample were shown in Fig. 6B. The tumor volume analysis also demonstrated a significantly smaller tumor volume in sa-GL261-sEVs-DOX treated group than that in the other two groups (Fig. 6C). It was explained by the higher DOX in the tumor region. As shown in Fig. 6D, a stronger fluorescence of DOX was observed in the tumor region of sa-GL261-sEVs -DOX group than that of the DOX group, while there was no fluorescence in the control group (treated with PBS). Moreover, the sa-GL261-sEVs-DOX treated group also presented an obvious aggregation in the tumor region rather than in normal brain tissue (Fig. 6E). These results confirmed the high DOX transportation efficacy of sa-GBM-sEVs to the orthotopic GBM region. Finally, the survival rate of orthotopic brain tumor-bearing mice was statistically analyzed by the Kaplan-Meier method. The results showed that sa-GL261-sEVs-DOX treated mice survived the longest average time among all groups (Fig. 6F). All these results confirm the saponin treated GBM-sEVs can be used as highly efficient nanocarriers to achieve GBM suppression.
Fig. 6.
Cargo eliminated GBM-sEVs as efficient drug vehicles in immunocompetent orthotopic brain tumor model. (A) Experimental design of the orthotopic tumor model. (B) Images of the orthotopic brain tumor model collected after treatment (upper), the tumors were pointed with green arrows. The tumor was detected by HE staining (lower). (C) Statistical analysis of the tumor volumes. (D) The fluorescence images of DOX in the brain of three groups (control group, DOX group and GL261-sEVs-DOX group) detected by confocal microscopy (upper). The statistical analysis was listed on the right. (E) The fluorescence images of DOX in normal brain region and tumor region of GL261-sEVs-DOX group, and the statistical analysis was listed on the right. (F) Kaplan-Meier analysis of the survival time of orthotopic brain tumor mice. **P < 0.01, ****P < 0.0001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Tumor cell-derived sEVs (TsEVs) show great advantages as effective nano DDSs in chemotherapy. However, their intrinsic protumoral functions lead to great concerns in their biosafety and severely hinder their clinical practice. To solve this issue, in the present study, a saponin-mediated cargo elimination strategy is developed and practiced successfully to GBM cell-derived sEVs to diminish their potential risks in tumor progression when they are used as effective DDSs for GBM suppression.
The GBM cell (U87, U251, and GL261) derived sEVs have been chosen to test the cargo eliminating strategy to improve the biosafety of TsEVs. GBM is a devastating and fetal cancer in which drug resistance can occur with high probability [23]. Due to the existence of BBB, there is an insistent demand for highly effective nano DDSs to transport broad anti-cancer drugs to the brain [49]. Endogenously derived sEVs have sparked great interest of medicine industry because of their superior performances in biocompatibility, delivery efficacy, and biological barrier penetration to synthetic nano DDSs. Up to now, sEVs from many cell types (eg., dendritic cells and embryonic cells) have been successfully used to achieve guest agent delivery to the brain [50,51]. Despite the BBB penetration ability of these sEVs, extra decorating steps of targeting moieties are also necessary to enhance their lesion accumulation in the brain, which complicates their producing processes. Recently, the homotypic targeting ability of tumor cell-derived EVs has been revealed in many studies [14,46,52], indicating the inherent targeting ability of TEVs as drug delivery vehicles for specific cancer therapy. And the underlying mechanism involves specific surface proteins (like integrin) and phospholipids [[53], [54], [55], [56]]. In this study, besides the BBB penetration ability, we have proved the homotypic targeting ability of GBM-sEVs in the orthotopic GBM model of mouse, suggesting the great potential application of GBM-sEVs as nano drug carriers for GBM therapy.
Over the past decades, numerous studies have constantly revealed TEVs' functions in angiogenesis, immunosuppression, tumor microenvironment development, and cancer metastasis, which undoubtably indicates a critical role of tumor cell secreted EVs in tumor progression [[57], [58], [59]]. Despite the intrinsic targeting ability, TEVs’ application can bring huge risks to further promote tumor progression rather than suppression. As the data presented in this study, the tumor promotion effects of GBM-sEVs are apparently displayed both in in vitro and in vivo evaluation, attesting the great biosafety issue when using them as nano DDSs directly. However, up to now, nearly all the related studies have usually neglected the potential risks of TEVs and used them directly. Thus, considering that the major mechanism of TEVs in tumor progression is usually achieved by cargo molecule transportation, the cargo eliminating strategy is put forward and practiced to GBM-sEVs for the first time in this study.
In this study, the saponin mediated permeation was successfully identified as an effective and large-scale processing method to eliminate the inner cargo proteins and RNAs from GBM-sEVs. Saponin is a naturally derived surfactant and can change the plasma membrane permeability by binding and removing cholesterol from the phospholipid membrane [30,60]. Meanwhile, it has been widely applied in loading guest molecules to EVs which are used as nano DDSs [61]. However, within our knowledge, whether this method can lead to the leakage of the original cargo molecules from EVs has never been noticed. According to the results of previous report, saponin can make holes in size of 40–50 Å on plasma membrane [31], and this value is in the same size range of most proteins and larger than mRNA and miRNA size. Thus, the leakage of the cargo molecules is inevitable, and we have creatively used saponin to eliminate cargo molecules in GBM-sEVs. Based on the discoveries of this study, three main advantages have been proved in eliminating the cargo molecules of GBM-sEVs. Firstly, saponin treatment exhibits a high efficacy in eliminating most of the inner cargo proteins and RNA molecules from GBM-sEVs. Many of the inner cargo proteins and RNAs in GBM-sEVs are tightly associated with tumor progression. For example, ECM1 is an oncoprotein and essential for the formation of invadopodia that can promote the invasion of cancer cells [62]. Overexpressed ANXA5 is positively correlated with the expression of CRKI/II and RAC1 and can promote the clinical progression and lymphatic metastasis [63]; miR-21 is a commonly recognized carcinogenic miRNA sequence. The WB and RT-qPCR results in Fig. 3 indicate a high efficacy of saponin treatment in eliminating all these tumor related cargos, thus laying the foundation of the elimination of the protumeral functions of GBM-sEVs. Secondly, saponin treatment can preserve the homotypic recognizing ability of GBM-sEVs, which is very critical for achieving the highly effective drug transportation of GBM-sEVs to GBM site in brain. Meanwhile, as a comparison, despite the high eliminating efficacy of sonication treatment in the inner cargo of GBM-sEVs, it severely affected the homotypic recognizing ability of GBM-sEVs. Surface protein ligand and receptors play an important role in the cell recognition of sEVs. The interacting targets of saponin is cholesterol in the plasma membrane rather than surface proteins. However, the sonication exhibits no target selecting ability, and several studies have proved the impact of high-power sonication on protein structure. Thirdly, the method can effectively eliminate the cargo molecules in all the three cell lines, suggesting the universality of this method to sEVs independent of parent cell types.
According to the results of proteomics, WB, and qPCR analysis, saponin treatment can effectively eliminate the cargo proteins and RNA sequences with protumeral functions, resulting in the improvement in the biosafety of GBM-sEVs as nano DDS. The protumoral functions of the cargo eliminated GBM-sEVs (sa-GBM-sEV) are carefully evaluated both in in vivo and in vitro evaluation. And our results have clearly showed the dramatically diminished functions of sa-GBM-sEVs in promoting GBM progression, indicating the improved biosafety of these nanocarriers for drug delivery. Obviously, these results have doubtlessly verified the feasibility of saponin mediated cargo elimination strategy. To our surprise, although sa-U251-sEVs could not significantly inhibit the in vitro proliferation of U251 cells (Fig. 4), they showed an inhibition to GBM progression in the subcutaneous tumor model of mouse. Tumor growth is not only related to the proliferation of tumor cells but many other factors are also involved, like the invasion of tumor cells and angiogenesis, and sa-U251-sEVs showed significant effect on inhibiting the migration and invasion of U251 cells. Our further competing cellular uptake assay implied that the continuous injection of sa-U251-sEVs might interference the routine communication among GBM cells and led to tumor growth inhibition. Due to the persevered homotypic targeting ability and improved biosafety of sa-GBM-sEVs, they show great potential to be used as targeted nano DDSs for chemotherapeutic agent delivery to brain. Furthermore, during saponin mediated cargo elimination, exogenous molecules can be easily loaded, indicating a convenient and feasible process in practically engineering these nanocarriers for therapy. And DOX loaded sa-GBM-sEVs proved the high GBM suppression efficacy both in subcutaneous and orthotopic GBM model, confirming their feasibility as nanocarriers for GBM therapy.
5. Conclusion
In conclusion, a saponin-mediated cargo elimination strategy was successfully practiced in GBM-sEVs to remove their inherent auxo-actions in GBM progression to realize an improved biosafety when using GBM-sEVs as efficient nanocarriers for GBM suppression. The saponin treatment can effectively remove the original cargos (like proteins and RNAs) from GBM-sEVs, resulting in the significant elimination of their functions in promoting tumor progression. Furthermore, the inherent targeting ability of GBM-sEVs to GBM was preserved during saponin treatment. Moreover, the high effectiveness of the saponin treated GBM-sEVs in the transportation of DOX for GBM suppression was also confirmed in both subcutaneous xenograft and orthotopic brain tumor model of mouse. All in all, this study provides a simple and feasible way to improve the biosafety of TEVs, which presents a great referential value to achieve their clinical application as nano DDSs for chemotherapy.
CRediT authorship contribution statement
Yuhang Guo: wrote the manuscript. Guowen Hu: wrote the manuscript. HaiYan Li: made modification to the manuscript. Ji Yuan: wrote the manuscript. Juntao Zhang: wrote the manuscript. Yu Chen: wrote the manuscript. Yunlong Yang: wrote the manuscript, designed the experiment and provide funds. Yang Wang: designed the experiment and provide funds. Zhifeng Deng: designed the experiment and provide funds.
Declaration of competing interest
There is on conflict of interest.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 82071371 and 82072550).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.02.013.
Contributor Information
Yunlong Yang, Email: yunlong0424@outlook.com.
Yang Wang, Email: wangyang63@sjtu.edu.cn.
Zhifeng Deng, Email: zfdeng@sjtu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Chen Z.L.F., Chen Y., Liu J., Wang X., Chen A.T., Deng G., Zhang H., Liu J., Hong Z., Zhou J. Targeted delivery of CRISPR/Cas9-Mediated cancer gene therapy via liposome-templated hydrogel nanoparticles. Adv. Funct. Mater. 2017;27(46):1703036. doi: 10.1002/adfm.201703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin J.D.P.M., Wang C., Khan T.T., Martin M.R., Voutouri C., Toh K., Papageorgis P., Mpekris F., Polydorou C., Ishii G., Takahashi S., Gotohda N., Suzuki T., Wilhelm M.E., Melo V.A., Quader S., Norimatsu J., Lanning R.M., Kojima M., et al. Dexamethasone increases cisplatin-loaded nanocarrier delivery and efficacy in metastatic breast cancer by normalizing the tumor microenvironment. ACS Nano. 2019;13(6):6396–6408. doi: 10.1021/acsnano.8b07865. [DOI] [PubMed] [Google Scholar]
- 3.Ding F.M.Q., Ma Y., Pan G., Guo Y., Tong G., Choi C.H.J., Zhu X., Zhang C. A crosslinked nucleic acid nanogel for effective siRNA delivery and antitumor therapy. Angew Chem. Int. Ed. Engl. 2018;57(12):3064–3068. doi: 10.1002/anie.201711242. [DOI] [PubMed] [Google Scholar]
- 4.Nieland L.M.L., Broekman M.L.D., Breakefield X.O., Abels E.R. Extracellular vesicle-mediated bilateral communication between glioblastoma and astrocytes. Trends Neurosci. 2021;44(3):215–226. doi: 10.1016/j.tins.2020.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker A.T.B., Weiss J.M., Kim H.S., Peinado H., Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30(6):836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Niel G.D.A.G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 7.Mathieu M.M.-J.L., Lavieu G., Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019;21(1):9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 8.Erkan E.P.S.O. Extracellular vesicles as novel delivery tools for cancer treatment. Curr. Cancer Drug Targets. 2016;16(1):34–42. doi: 10.2174/1568009615666150923115439. [DOI] [PubMed] [Google Scholar]
- 9.Vader P.M.E., Pasterkamp G., Schiffelers R.M. Extracellular vesicles for drug delivery. Adv. Drug Deliv. Rev. 2016;15(Pt A):148–156. doi: 10.1016/j.addr.2016.02.006. 106. [DOI] [PubMed] [Google Scholar]
- 10.Elsharkasy O.M.N.J., Hagey D.W., de Jong O.G., Schiffelers R.M., Andaloussi S.E., Vader P. Extracellular vesicles as drug delivery systems: why and how? Adv. Drug Deliv. Rev. 2020;159:332–343. doi: 10.1016/j.addr.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Möller A.L.R. The evolving translational potential of small extracellular vesicles in cancer. Nat. Rev. Cancer. 2020;20(12):697–709. doi: 10.1038/s41568-020-00299-w. [DOI] [PubMed] [Google Scholar]
- 12.Bose RJC U.K.S., Zeng Y., Afjei R., Robinson E., Lau K., Bermudez A., Habte F., Pitteri S.J., Sinclair R., Willmann J.K., Massoud T.F., Gambhir S.S., Paulmurugan R. Tumor cell-derived extracellular vesicle-coated nanocarriers: an efficient theranostic platform for the cancer-specific delivery of anti-miR-21 and imaging agents. ACS Nano. 2018;12(11):10817–10832. doi: 10.1021/acsnano.8b02587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma J.Z.Y., Tang K., Zhang H., Yin X., Li Y., Xu P., Sun Y., Ma R., Ji T., Chen J., Zhang S., Zhang T., Luo S., Jin Y., Luo X., Li C., Gong H., Long Z., Lu J., Hu Z., Cao X., Wang N., Yang X., Huang B. Reversing drug resistance of soft tumor-repopulating cells by tumor cell-derived chemotherapeutic microparticles. Cell Res. 2016;26(6):713–727. doi: 10.1038/cr.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo M.W.F., Hu G., Chen L., Xu J., Xu P., Wang X., Li Y., Liu S., Zhang S., Huang Q., Fan J., Lv Z., Zhou M., Duan L., Liao T., Yang G., Tang K., Liu B., Liao X., Tao X., Jin Y. Autologous tumor cell-derived microparticle-based targeted chemotherapy in lung cancer patients with malignant pleural effusion. Sci. Transl. Med. 2019;11(474) doi: 10.1126/scitranslmed.aat5690. [DOI] [PubMed] [Google Scholar]
- 15.Liang Q.B.N., Yong T., Tang K., Shi X., Wei Z., Jia H., Zhang X., Zhao H., Huang W., Gan L., Huang B., Yang X. The softness of tumour-cell-derived microparticles regulates their drug-delivery efficienc. Nat Biomed Eng. 2019;3(9):729–740. doi: 10.1038/s41551-019-0405-4. [DOI] [PubMed] [Google Scholar]
- 16.Yong T.Z.X., Bie N., Zhang H., Zhang X., Li F., Hakeem A., Hu J., Gan L., Santos H.A., Yang X. Tumor exosome-based nanoparticles are efficient drug carriers for chemotherapy. Nat. Commun. 2019;10(1):3838. doi: 10.1038/s41467-019-11718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiao L.H.S., Huang K., Su T., Li Z., Vandergriff A., Cores J., Dinh P.U., Allen T., Shen D., Liang H., Li Y., Cheng K. Tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs. Theranostics. 2020;10(8):3474–3487. doi: 10.7150/thno.39434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuo B.Q.H., Lu Z., Chen L., Sun B., Yang R., Zhang Y., Liu Z., Gao X., You A., Wu L., Jing R., Zhou Q., Yin H. Alarmin-painted exosomes elicit persistent antitumor immunity in large established tumors in mice. Nat. Commun. 2020;11(1):1790. doi: 10.1038/s41467-020-15569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodrigues G.H.A., Kenific C.M., Matei I.R., Steiner L., Freitas D., Kim H.S., Oxley P.R., Scandariato I., Casanova-Salas I., Dai J., Badwe C.R., Gril B., Tešić Mark M., Dill B.D., Molina H., Zhang H., Benito-Martin A., Bojmar L., Ararso Y., et al. Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol. 2019;21(11):1403–1412. doi: 10.1038/s41556-019-0404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang T.L.H., Lv G., Li T., Wang C., Han Q., Yu L., Su B., Guo L., Huang S., Cao D., Tang L., Tang S., Wu M., Yang W., Wang H. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018;9(1):191. doi: 10.1038/s41467-017-02583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C.L.Y., He W., Zhao Y., Kong Y., Liu H., Zhong G., Li Y., Li J., Huang J., Chen R., Lin T. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J. Clin. Invest. 2020;130(1):404–421. doi: 10.1172/JCI130892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou P.P.L.L., Chen H.Z., Chen Q.T., Bian X.L., Wu S.F., Zhou J.X., Zhao W.X., Liu J.M., Wang X.M., Zhang Z.Y., Yao L.M., Chen Q., Zhou D., Wu Q. Ectosomal PKM2 promotes HCC by inducing macrophage differentiation and remodeling the tumor microenvironment. Mol. Cell. 2021;78(6):1192–1206. doi: 10.1016/j.molcel.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Tan A.C.A.D., López G.Y., Malinzak M., Friedman H.S., Khasraw M. Management of glioblastoma: state of the art and future directions. Ca - Cancer J. Clin. 2020;70(4):299–312. doi: 10.3322/caac.21613. [DOI] [PubMed] [Google Scholar]
- 24.Ostrom Qt G.H., Truitt G., Boscia A., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro Oncol. 2018;1(suppl_4):iv1–iv86. doi: 10.1093/neuonc/noy131. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi G.Z., Huang G., Guo M., Zhang X., Wang H., Deng S., et al. Acquired temozolomide resistance in MGMT-deficient glioblastoma cells is associated with regulation of DNA repair by DHC2. Brain. 2019;142(8):2352–2366. doi: 10.1093/brain/awz202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aldape K., Brindle K.M., Chesler L., Chopra R., Gajjar A., Gilbert M.R., et al. Challenges to curing primary brain tumours. Nat. Rev. Clin. Oncol. 2019;16(8):509–520. doi: 10.1038/s41571-019-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morad G.C.C., Hagedorn E.J., Perlin J.R., Zon L.I., Mustafaoglu N., Park T.E., Ingber D.E., Daisy C.C., Moses M.A. Tumor-derived extracellular vesicles breach the intact blood-brain barrier via transcytosis. ACS Nano. 2019;13(12):13853–13865. doi: 10.1021/acsnano.9b04397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian T., Zhang H.X., He C.P., Fan S., Zhu Y.L., Qi C., et al. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials. 2018;150:137–149. doi: 10.1016/j.biomaterials.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Ha D., Yang N., Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm. Sin. B. 2016;6(4):287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moghimipour E., Handali S. Saponin: properties, methods of evaluation and applications. Annual Research & Review in Biology. 2015;5(3):207–220. [Google Scholar]
- 31.Seeman P., Cheng D., Iles G.H. Structure of membrane holes in osmotic and saponin hemolysis. J. Cell Biol. 1973;56:519–527. doi: 10.1083/jcb.56.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian Y.G.M., Hu Y., Liu H., Zhang W., Zhang M., Hu X., Aubert D., Zhu S., Wu L., Yan X. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles. 2019;9(1):1697028. doi: 10.1080/20013078.2019.1697028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Théry C.W.K., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., Ayre D.C., Bach J.M., Bachurski D., Baharvand H., Balaj L., Baldacchino S., Bauer N.N., Baxter A.A., Bebawy M., Beckham C., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedmann-Morvinski D.N.R., Xia Y., Myskiw C., Soda Y., Verma I.M. Targeting NF-κB in glioblastoma: a therapeutic approach. Sci. Adv. 2016;2(1) doi: 10.1126/sciadv.1501292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tönjes M.B.S., Park Y.J., Wang W., Schlotter M., Lindroth A.M., Pleier S.V., Bai A.H.C., Karra D., Piro R.M., Felsberg J., Addington A., Lemke D., Weibrecht I., Hovestadt V., Rolli C.G., Campos B., Turcan S., Sturm D., Witt H., Chan T.A., Herold-Mende C., Kemkemer R., König R., Schmidt K., Hull W.E., Pfister S.M., Jugold M., Hutson S.M., Plass C., Okun J.G., Reifenberger G., Lichter P., Radlwimmer B. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat. Med. 2013;19(7):901–908. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W.H.Y., Zhang A., Yang W., Wei W., Wang G., Jia Z. miR-19a/b promote EMT and proliferation in glioma cells via SEPT7-AKT-NF-κB pathway. Mol Ther Oncolytics. 2021;20:290–305. doi: 10.1016/j.omto.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaur A.B.H.S., Colburn N.H., Israel M.A. Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro Oncol. 2011;13(6):580–590. doi: 10.1093/neuonc/nor033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Tacchio M.M.J., Weissenberger J., Sommer K., Bähr O., Steinbach J.P., Senft C., Seifert V., Glas M., Herrlinger U., Krex D., Meinhardt M., Weyerbrock A., Timmer M., Goldbrunner R., Deckert M., Scheel A.H., Büttner R., Grauer O.M., Schittenhelm J., Tabatabai G., Harter P.N., Günther S., Devraj K., Plate K.H., Reiss Y. Tumor vessel normalization, immunostimulatory reprogramming, and improved survival in glioblastoma with combined inhibition of PD-1, angiopoietin-2, and VEGF. Cancer Immunol Res. 2019;7(12):1910–1927. doi: 10.1158/2326-6066.CIR-18-0865. [DOI] [PubMed] [Google Scholar]
- 39.Fang R.C.X., Zhang S., Shi H., Ye Y., Shi H., Zou Z., Li P., Guo Q., Ma L., He C., Huang S. EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nat. Commun. 2021;12(1):177. doi: 10.1038/s41467-020-20379-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z.W.B., Shi Y., Xu C., Xiao H.L., Ma L.N., Xu S.L., Yang L., Wang Q.L., Dang W.Q., Cui W., Yu S.C., Ping Y.F., Cui Y.H., Kung H.F., Qian C., Zhang X., Bian X.W. Oncogenic miR-20a and miR-106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP-2. Oncogene. 2015;34(11):1407–1419. doi: 10.1038/onc.2014.75. [DOI] [PubMed] [Google Scholar]
- 41.Li W.W.S., Shan B., Cheng X., He H., Qin J., Tang Y., Zhao H., Tian M., Zhang X., Jin G. CircHECTD1 regulates cell proliferation and migration by the miR-320-5p/slc2a1 Axis in glioblastoma multiform. Front. Oncol. 2021;11:666391. doi: 10.3389/fonc.2021.666391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brockmann M.A.U.U., Grüner K., Fillbrandt R., Westphal M., Lamszus K. Glioblastoma and cerebral microvascular endothelial cell migration in response to tumor-associated growth factors. Neurosurgery. 2003;52(6):1391–1399. doi: 10.1227/01.neu.0000064806.87785.ab. [DOI] [PubMed] [Google Scholar]
- 43.Shao H.C.J., Lee K., Balaj L., Min C., Carter B.S., Hochberg F.H., Breakefield X.O., Lee H., Weissleder R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skog J.W.T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Jr., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamoun W.S.L.C., Farrar C.T., Duyverman A.M., Lahdenranta J., Lacorre D.A., Batchelor T.T., di Tomaso E., Duda D.G., Munn L.L., Fukumura D., Sorensen A.G., Jain R.K. Edema control by cediranib, a vascular endothelial growth factor receptor-targeted kinase inhibitor, prolongs survival despite persistent brain tumor growth in mice. J. Clin. Oncol. 2009;27(15):2542–2552. doi: 10.1200/JCO.2008.19.9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villa A., Garofalo M., Crescenti D., Rizzi N., Brunialti E., Vingiani A., et al. Transplantation of autologous extracellular vesicles for cancer-specific targeting. Theranostics. 2021;11(5):2034–2047. doi: 10.7150/thno.51344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu L., Faruqu F.N., Liam-Or R., Abu Abed O., Li D., Venner K., et al. Design of experiment (DoE)-driven in vitro and in vivo uptake studies of exosomes for pancreatic cancer delivery enabled by copper-free click chemistry-based labelling. J. Extracell. Vesicles. 2020;9(1):1779458. doi: 10.1080/20013078.2020.1779458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang T.L.H., Lv G., Li T., Wang C., Han Q., Yu L., Su B., Guo L., Huang S., Cao D., Tang L., Tang S., Wu M., Yang W., Wang H. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018;15(1):191. doi: 10.1038/s41467-017-02583-0. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wa B. Characteristics of compounds that cross the blood-brain barrier. BMC Neurol. 2009;12(9):S3. doi: 10.1186/1471-2377-9-S1-S3. Suppl 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 51.Zhu Q., Ling X., Yang Y., Zhang J., Li Q., Niu X., et al. Embryonic stem cells-derived exosomes endowed with targeting properties as chemotherapeutics delivery vehicles for glioblastoma therapy. Adv. Sci. 2019;6(6):1801899. doi: 10.1002/advs.201801899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jc Bose R., Uday Kumar S., Zeng Y., Afjei R., Robinson E., Lau K., et al. Tumor cell-derived extracellular vesicle-coated nanocarriers: an efficient theranostic platform for the cancer-specific delivery of anti-miR-21 and imaging agents. ACS Nano. 2018;12(11):10817–10832. doi: 10.1021/acsnano.8b02587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamerkar S.L.V., Sugimoto H., Yang S., Ruivo C.F., Melo S.A., Lee J.J., Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;22(7659):498–503. doi: 10.1038/nature22341. 546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smyth T.J., Redzic J.S., Graner M.W., Anchordoquy T.J. Examination of the specificity of tumor cell derived exosomes with tumor cells in vitro. Biochim. Biophys. Acta. 2014;1838(11):2954–2965. doi: 10.1016/j.bbamem.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiao L., Hu S., Huang K., Su T., Li Z., Vandergriff A., et al. Tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs. Theranostics. 2020;10(8):3474–3487. doi: 10.7150/thno.39434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Pasquale D., Marino A., Tapeinos C., Pucci C., Rocchiccioli S., Michelucci E., et al. Homotypic targeting and drug delivery in glioblastoma cells through cell membrane-coated boron nitride nanotubes. Mater. Des. 2020;192:108742. doi: 10.1016/j.matdes.2020.108742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morad G., Moses M.A. Brainwashed by extracellular vesicles: the role of extracellular vesicles in primary and metastatic brain tumour microenvironment. J. Extracell. Vesicles. 2019;8(1):1627164. doi: 10.1080/20013078.2019.1627164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marar C., Starich B., Wirtz D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021;22(5):560–570. doi: 10.1038/s41590-021-00899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahmadi M., Rezaie J. Tumor cells derived-exosomes as angiogenenic agents: possible therapeutic implications. J. Transl. Med. 2020;18(1):249. doi: 10.1186/s12967-020-02426-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lorent J.L.L., Domenech Ò., Quetin-Leclercq J., Brasseur R., Mingeot-Leclercq M.P. Domain formation and permeabilization induced by the saponin α-hederin and its aglycone hederagenin in a cholesterol-containing bilayer. Langmuir. 2014;30(16):4556–4569. doi: 10.1021/la4049902. [DOI] [PubMed] [Google Scholar]
- 61.Vader P., Mol E.A., Pasterkamp G., Schiffelers R.M. Extracellular vesicles for drug delivery. Adv. Drug Deliv. Rev. 2016;106:148–156. doi: 10.1016/j.addr.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 62.Wu Q.C.D., Luo Q., Yang Q., Zhao C., Zhang D., Zeng Y., Huang L., Zhang Z., Qi Z. Extracellular matrix protein 1 recruits moesin to facilitate invadopodia formation and breast cancer metastasis. Cancer Lett. 2018;437:44–55. doi: 10.1016/j.canlet.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 63.Sun X.L.S., Wang J., Wei B., Guo C., Chen C., Sun M.Z. Annexin A5 regulates hepatocarcinoma malignancy via CRKI/II-DOCK180-RAC1 integrin and MEK-ERK pathways. Cell Death Dis. 2018;9(6):637. doi: 10.1038/s41419-018-0685-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.