Abstract
Computer-aided synthesis planning (CASP) aims to assist chemists in performing retrosynthetic analysis for which they utilize their experiments, intuition, and knowledge. Recent breakthroughs in machine learning (ML) techniques, including deep neural networks, have significantly improved data-driven synthetic route designs without human intervention. However, learning chemical knowledge by ML for practical synthesis planning has not yet been adequately achieved and remains a challenging problem. In this study, we developed a data-driven CASP application integrated with various portions of retrosynthesis knowledge called “ReTReK” that introduces the knowledge as adjustable parameters into the evaluation of promising search directions. The experimental results showed that ReTReK successfully searched synthetic routes based on the specified retrosynthesis knowledge, indicating that the synthetic routes searched with the knowledge were preferred to those without the knowledge. The concept of integrating retrosynthesis knowledge as adjustable parameters into a data-driven CASP application is expected to enhance the performance of both existing data-driven CASP applications and those under development.
Introduction
Since the 1960s, various computer-aided synthesis planning (CASP) applications have been developed to emulate chemists’ thinking and help organic synthesis chemists in their work.1−9 CASP applications have played an important role in the definable parts of synthesis (e.g., the characteristics of chemical structures and retrosynthetic tree size), whereas the indefinable parts of synthesis (e.g., chemists’ intuition) and opportunities to contribute to creativity in retrosynthetic analysis have been left to chemists.1 As an underlying chemists’ intuition, Corey formalized the concept of retrosynthesis (retrosynthesis knowledge) and major types of strategies (e.g., transform- and topology-based strategies). He stated that retrosynthetic analysis is most efficiently performed through the simultaneous use of as many different independent strategies as possible.10 For the selection of optimal strategies, the chemists’ knowledge of chemistry and their experiments are essential; the optimal strategies for a particular synthesis problem depend on the molecules, persons, and situations involved (e.g., lead optimization and large-scale synthesis of drug candidates).11
CASP approaches are generally classified into two types: knowledge-based8,12 and data-driven approaches.6,9 Knowledge-based approaches employ manually encoded (human-curated) transformations considering information, such as stereochemical and electronic effects.8 For instance, one excellent knowledge-based CASP application, Chematica8 (now rebranded as Synthia), provides a considerable discretion for chemists to perform retrosynthetic analysis based on their own ways of thinking using their own scoring functions (e.g., SMALLER, SELECTIVITY, and RINGS variables), and it is now used globally.8,13,14 However, knowledge-based approaches still require the great efforts of many experts, as the number of new reaction types discovered per year has been in the low few thousands.15
In contrast, data-driven CASP aims to automatically extract knowledge related to transformations from numerous reaction records to discover synthetic routes.16 Recent breakthroughs in deep learning (DL),17,18 along with the availability of reaction records19,20 and open-source codes,21−24 have improved the core techniques of data-driven CASP such as 1-step (retro)synthetic reaction prediction25−28 and multistep synthetic route searches.9,29−32 In the existing reaction prediction methods, various representations of molecules (e.g., fingerprints,25 Simplified Molecular Input Line Entry System (SMILES) strings,26,27,33 and graphs28,34) and their corresponding suitable DL techniques have been used, showing promising performance. Regarding search algorithms, the Monte Carlo tree search (MCTS),9,24,35,36 depth-first proof number search,31,37,38 and graph-based exploration methods30,32 have been used to obtain possible synthetic routes efficiently. Several outstanding data-driven CASP applications are being used practically in industries and laboratories,9,30,39 and these applications have led to a remarkable revival of interest in CASP research.40−42
However, in the case of actual chemical synthesis, most data-driven CASP applications are lacking in their ability to reflect or support flexible adaptation to individual chemists’ ways of thinking. The search algorithms used in such applications depend on naive scoring functions for evaluating whether one synthetic position found during a search is preferable to another.9,35 In addition, as for the 1-step retrosynthetic reaction prediction, large repositories of highly biased published reactions19,20 prevent the data-driven approaches from acquiring the chemical knowledge sufficiently because imbalanced data training is inherently difficult for AI.43,44 This implies that there are few opportunities to learn diverse strategies for retrosynthetic analysis. Moreover, the data-driven CASP approaches incorporating generally used various retrosynthesis knowledge have not been developed, and the effects of knowledge on search performance have yet to be investigated.
In this study, we developed a data-driven CASP application integrated with rule-based techniques called “Retrosynthesis planning application using retrosynthesis knowledge (ReTReK),” which introduces retrosynthesis knowledge into the evaluation of promising search directions to obtain promising synthetic routes considering the knowledge of synthetic chemists. ReTReK is based on a data-driven framework of retrosynthetic reaction prediction by deep learning and path search by MCTS. To explicitly introduce retrosynthesis knowledge into ReTReK, referring to previous works,4,7,8,13,14 we formulated four scores that aimed to explore the ideally shortest synthetic route or select a reaction that ideally provides only the desired product. A graph convolutional network (GCN) technique, which tolerates the biased reactions data set,28 was used to build the retrosynthetic reaction prediction model. The Reaxys reaction database19 was used to construct the ReTReK model. We evaluated the performance of ReTReK using drug-like molecules45−50 for demonstrations and molecules from the ChEMBL database51 for quantitative evaluations. We successfully demonstrated that synthetic routes designed using ReTReK with retrosynthesis knowledge were preferable to those designed without retrosynthesis knowledge. Furthermore, we quantitatively showed that retrosynthesis knowledge improved the performance when solving certain target molecules, and it successfully guided the search direction in MCTS. The ReTReK application is publicly available on GitHub at https://github.com/clinfo/ReTReK. The proposed concept of integrating retrosynthesis knowledge, in the form of adjustable parameters, into a data-driven CASP application is expected to enhance the performance of both existing data-driven CASP applications and those under development.
Results and Discussion
Construction of ReTReK
To implement a data-driven CASP application that can reflect retrosynthesis knowledge, ReTReK was constructed using MCTS, the GCN technique, and the four retrosynthesis knowledge scores introduced earlier (Figure 1). When designing a synthetic route for a target molecule, the following three factors are described as basic points:52 the construction of the required carbon skeleton considering regiochemistry and stereochemistry; ideally the shortest synthetic route to the molecule; reaction that ideally gives only the desired product in each step. Thus, we formulated the four scores: a convergent disconnection score (CDScore), an available substances score (ASScore), a ring disconnection score (RDScore), and a selective transformation score (STScore). The CDScore and ASScore are designed to favor convergent synthesis, which is an efficient strategy in multistep chemical synthesis. The RDScore is designed to reflect a ring construction strategy. This strategy is preferred if the target compound has complex ring structures because the construction of ring structures in a synthetic route tends to result in simple and easily available starting materials. The STScore is designed to reflect the number of possible products from a reaction because a synthetic reaction with few byproducts is preferred, considering the yield. Additionally, to handle stereochemistry and regiochemistry when applying a retrosynthetic reaction predicted by the GCN to a molecule, the Reactor in the ChemAxon API53 was used. The basic MCTS algorithm comprises four steps: selection, expansion, rollout, and update. For the selection step, a tree policy is used to select a promising retrosynthetic tree position. The policy of the ReTReK also considers the retrosynthesis knowledge scores. A GCN-based model (Figure 2a) was used for the 1-step retrosynthetic reaction prediction as a policy network in the expansion and rollout steps. Reaxys reaction records19 were used to train the model and to prepare the starting materials, and the compounds obtained from the ZINC database54 were used as the starting materials. By iterating through the four steps listed above, a retrosynthetic tree is expanded, thus attempting to identify a promising synthetic route. ReTReK without the retrosynthesis knowledge scores can be regarded as a basic data-driven CASP model such as the approach proposed by Segler et al.9 and AiZynthFinder24 although there are some minor differences in the policy networks and evaluation terms of the MCTS. Moreover, when comparing the performance of the CASP approaches, it should be noted that what is used for reaction templates and starting materials will affect the performances.
Figure 1.

Complete workflow of ReTReK. ReTReK combines a path-finding algorithm (MCTS) and GCN technique, and retrosynthesis knowledge is incorporated into the selection step of the MCTS procedure. The retrosynthesis knowledge is formalized using four scores: the CDScore, STScore, RDScore, and ASScore.
Figure 2.
(a) Model architecture of the GCN-based policy network. (b) Top-n accuracies of the model for n values ranging from 1 to 1000. Specifically, the top-1, top-50, top-100, top-300, and top-500 accuracies are 0.361, 0.906, 0.938, 0.968, and 0.976, respectively.
Top-n Accuracies of the GCN-Based Policy Network
To determine the effective size for the expansion step in the MCTS procedure, the top-n accuracies (for n up to 1000) of the GCN-based policy network are calculated as shown in Figure 2b. The 1-step retrosynthetic reaction prediction model aimed to prioritize 19 633 reaction templates for application to an input molecule. In this study, we used reaction templates considering a reaction center, first-degree neighbors, and protecting groups because a previous study proposed this type of reaction template to maintain chemical integrity.35 Accordingly, the top-1, top-50, top-100, top-300, and top-500 accuracies were found to be 0.361, 0.906, 0.938, 0.968, and 0.976, respectively. Beyond the top-500 accuracies, an increase in the prediction performance was not significant. Considering the results of a previous study28 on reaction templates of different sizes, the prediction performance is assumed to be equivalent to or better than that of the previous template-based 1-step retrosynthetic reaction prediction model.9 Based on the results for the top-n accuracies, we evaluated the effect of the MCTS expansion sizes and the retrosynthesis knowledge on the performance of solving for target molecules using the top 50, 100, 300, and 500 predicted templates.
Effects of Expansion Sizes and Retrosynthesis Knowledge on the Performance of Solving for Target Molecules
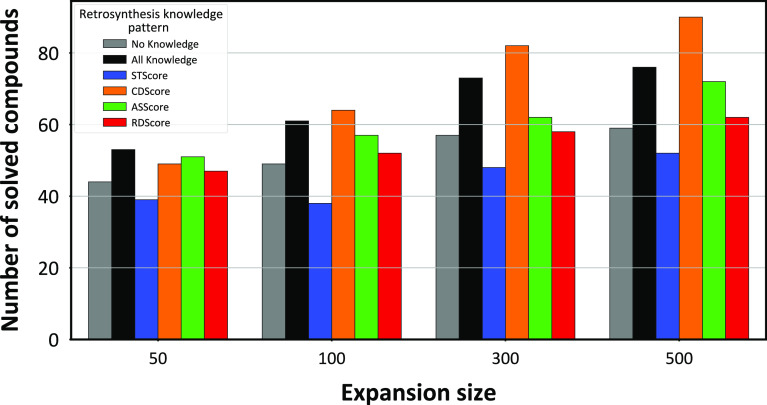
Figure 3 shows how the expansion sizes and retrosynthesis knowledge influenced the performance of solving for target molecules. The 161 molecules from the preprocessed ChEMBL data set were used as the target molecules. The searches were performed using different expansion sizes (50, 100, 300, and 500) and six retrosynthesis knowledge patterns: no retrosynthesis knowledge (no knowledge), the STScore, CDScore, ASScore, RDScore, and all the four retrosynthesis knowledge scores (all knowledge). In most of the cases, the solution performance was improved in proportion to the expansion size. However, the case of the STScore pattern and an expansion size of 100 resulted in a lower number of solved molecules than the case of the same pattern and an expansion size of 50. This result is attributed to a relative lack of MCTS iterations because of the increase in the expansion size. Regarding the retrosynthesis knowledge, all the knowledge patterns, except the STScore pattern, resulted in an increase in the number of solved molecules compared to the no-knowledge pattern. The CDScore pattern with an expansion size of 500 showed the best solution performance, yielding 90 solved molecules, whereas the no-knowledge pattern with the same expansion size resulted in 59 solved molecules. Although the STScore pattern resulted in fewer solved molecules than the no-knowledge pattern, this result was considered reasonable because the STScore focused on reactions with few byproducts. This often leads to strict conditions for retrosynthesis. To compare the ReTReK with the other data-driven CASP model, ASKCOS23 was applied to the 161 molecules, and the solving performance was shown in Figure S1. This result shows that the solving performances of ReTReK without retrosynthesis knowledge and ASKCOS were comparable, which suggests that the retrosynthesis knowledge score may improve other data-driven CASP approaches.
Figure 3.
Comparison of the numbers of solved molecules with different expansion sizes and retrosynthesis knowledge patterns. The gray, black, blue, orange, green, and red bars correspond to the no-knowledge, all-knowledge, STScore, CDScore, ASScore, and RDScore patterns, respectively.
Moreover, the search times necessary for solution are compared between the different expansion sizes and six retrosynthesis knowledge patterns as shown in Figure S2. The search time increases in proportion to the expansion size because an increase in the expansion size expands the search space for MCTS. The median search times for expansion sizes of 50, 100, 300, and 500 are 32, 45, 133, and 294 s, respectively. The STScore pattern requires shorter search times than the no-knowledge pattern although all the other knowledge patterns, except STScore, result in longer search times. These results suggest that synthetic routes can be more efficiently identified under the STScore pattern than the other patterns, although the STScore pattern results in lower solution performance.
Effects of Retrosynthesis Knowledge on the Search Directions in MCTS
Figure 4a shows how the six retrosynthesis knowledge patterns influence the characteristics of the searched synthetic routes, in terms of four route scores (rSTScore, rCDScore, rASScore, and rRDScore). Each route score was defined as the average corresponding retrosynthesis knowledge score in each step of the searched synthetic route. For ease of comparison, each route score for each of the five knowledge patterns was standardized with respect to the corresponding score for the no-knowledge pattern. The standardized mean values of the rSTScore for the STScore pattern, rCDScore for the CDScore pattern, rASScore for the ASScore pattern, and rRDScore for the RDScore pattern were 0.178, 0.555, 0.130, and 0.309, respectively. All the values were positively shifted compared to the values for the no-knowledge pattern, indicating that all the four retrosynthesis knowledge scores successfully guided the search directions in the MCTS according to the characteristics of each type of knowledge. The CDScore pattern caused MCTS to select more transformation-oriented searches compared to the STScore pattern. The mean values of the CDScore and STScore were 0.299 and 0.178, respectively. A convergent-disconnection-oriented search is assumed to have more chances of splitting the reactive centers into divided molecules because the CDScore attempts to minimize the sizes of each divided molecule simultaneously. Figure 4b–d shows the parts of the exemplary synthetic routes found by the ReTReK using all-knowledge and no-knowledge patterns. Each case shows that the ReTReK with retrosynthesis knowledge successfully chooses the preferable retrosynthetic reactions than the ReTReK without the knowledge, in terms of the STScore, CDScore, ASScore, and RDScore. Considering that the all-knowledge pattern shows a higher rSTScore than the CDScore and STScore patterns, these results suggest the existence of synergistic effects of retrosynthesis knowledge; however, this hypothesis needs further analysis.
Figure 4.

Evaluation of the effects of retrosynthesis knowledge on the search directions of MCTS, in terms of the four route scores. Synthetic routes solved with an expansion size of 500 were used for this evaluation. (a) Each route score standardized based on the corresponding mean and standard deviation of the no-knowledge pattern. The gray, black, blue, orange, green, and red plots represent the standardized route scores for the no-knowledge, all-knowledge, STScore, CDScore, ASScore, and RDScore patterns, respectively. The rhombuses represent the mean values for each case, and the confidence intervals at the 95% confidence level are also shown. (b–d) Parts of the exemplary synthetic routes found by the ReTReK using the all-knowledge and no-knowledge patterns. The circled symbol “S” indicates that a molecule is in the starting materials’ list. All the synthetic routes are shown in Figure S3. (b) Example of parts of the routes showing the STScore effect. The ReTReK with retrosynthesis knowledge successfully chose the reaction with fewer reactive centers than the ReTReK without the knowledge. (c) Example of parts of the routes showing the CDScore and ASScore effects. The ReTReK with retrosynthesis knowledge more successfully guided the convergent synthetic route than the ReTReK without knowledge. (d) Example of parts of the route showing the RDScore effect. The ReTReK with retrosynthesis knowledge successfully chose ring-opening retrosynthetic reaction, whereas the ReTReK without the knowledge found no routes.
Considering the results of Figure 3 and Figure 4, the following strategy to adjust the weight parameters can be considered. First, the search without the retrosynthesis knowledge is recommended to know the baseline result. If the target is not solved with these parameters, applying the CDScore is the reasonable choice to solve the target because the CDScore showed the best solution performance (Figure 3). Or, if the target seems to require ring-opening, applying the RDScore is the prospective choice to solve (Figure 4d). Additionally, if better quality synthetic routes are desired, applying the STScore and/or ASScore could improve the routes based on their concepts. The score weights should be adjusted gradually with positive integers to combinatorial explosion.
Demonstrations of ReTReK for Drug-like Molecules
To demonstrate retrosynthesis planning using ReTReK, we applied ReTReK to six drug-like molecules in the all-knowledge and no-knowledge patterns, and the results are presented in this section and Figure S4. The detailed parameters used in these demonstrations were described in the Methods section. Figure 5 panels a and b illustrate an exemplary retrosynthetic route to a molecule 1 known as a hepatitis B virus capsid inhibitor,45 found by ReTReK with and without retrosynthesis knowledge. The exploration with retrosynthesis knowledge suggested a convergent route, successfully reflecting the specified knowledge scores. In this route, the target molecule 1 is disconnected into two main segments, iodophthalazinone 7 and pyridylboronic acid 12, which can be converted into 1 by the Suzuki coupling reaction. The key intermediates 7 and 12 could be retrosynthetically divided into three representative materials: hydroxyphthalazine 2, benzyl alcohol 3, and trihalogenated pyridine 8. Iodination of 2 and subsequent N-benzylation with p-iodobenzyl iodide 5, which can be obtained from 3, would provide 2-(iodobenzyl)phthalazin-1-one 6. A reaction of 6 with copper cyanide would provide the intermediate 7. The other intermediate 12 would be prepared from 8 by three-step sequences (i.e., chlorination, incorporation of a boronic acid moiety, and amination with aminoalcohol 11). In contrast, a straightforward route was presented through the exploration without retrosynthesis knowledge. Friedel–Crafts acylation of trihalopyridine 14 with 2-(chlorocarbonyl)benzoic acid (13) would provide 2-(pyridinecarbonyl)benzoic acid 15, which is further reacted with aminoalcohol 11 to afford tricyclic ketone 16. The construction of the phthalazine ring could be performed by the reaction of 16 with p-bromobenzylhydrazine (17) to yield the precursor 18. Finally, the introduction of a nitrile group into the benzyl group of 18 would provide the target molecule 1. Additional demonstrations for two other drug-like molecules, kwakhurin47 and α7 nicotinic acetylcholine receptor silent agonist,48 are shown in Figure 5c,d and Figure 5e,f, respectively. To confirm the difference in each step’s score between ReTReK with and without retrosynthesis knowledge, four retrosynthesis knowledge scores were added to each step in the synthetic routes (Figure S5).
Figure 5.

Comparison of the synthetic route for three target compounds (a,b) hepatitis B virus capsid inhibitor,45 (c,d) kwakhurin,47 and (e,f) α7 nicotinic acetylcholine receptor silent agonist48) found by ReTReK with retrosynthesis knowledge (a, c, and e) and the corresponding route found without retrosynthesis knowledge (b, d, and f).
Furthermore, the effectiveness of retrosynthesis knowledge was confirmed in several cases of retrosynthetic analyses. In these cases, ReTReK with retrosynthesis knowledge succeeded in finding retrosynthetic routes to the target molecules, whereas no route was found using ReTReK without retrosynthesis knowledge (Figure 6a). Figure 6a shows the retrosynthetic route to a molecule 19 known as a Mycobacterium tuberculosis thymidylate kinase (MtbTMPK) inhibitor.46 In the suggested route, 19 is disconnected at the center of the molecule, giving imidazo[1,2-a]pyridine-3-carboxamide 25 and 1-(piperidin-4-yl)pyrimidine-2,4-dione 29. Compound 25 would be obtained from dibromide 24 by selective SNAr reaction with an organometallic reagent such as ethylmagnesium bromide, which is prepared from ethyl bromide. Dibromide 24 can be obtained from benzylamide 22 by stepwise SEAr bromination. Amide 22 would be provided from imidazopyridine-3-carboxylic acid (20) and N-Boc-benzylamine (21). Another intermediate 29 would be synthesized from 4-iodopiperidine (27) with pyrimidine-2,4-dione 28 by SN2 reaction. 27 would be easily prepared from N-Boc 26. From the viewpoint of the practical synthesis, deprotection of N-Boc group of the piperidine ring should be performed after N-alkylation of 28 with 26 to avoid oligomerization of 27 by self N-alkylation. Additional demonstrations for two other drug-like molecules, propolone49 and EGFR kinase inhibitor,50 are shown in Figure 6b and Figure S4, respectively. Further demonstrations for the molecules reported by Segler et al.9 are shown in Figure S6. For evaluating the performances of ReTReK and the other data-driven CASP approach on more advanced targets, ReTReK and ASKCOS were applied to 15 targets reported by previous research using Chematica.13,14,55 In terms of the solving performance, ReTReK found the retrosynthetic routes of 11 targets, while ASKCOS found those of 4 targets. As for the solved routes, the routes proposed by two data-driven applications had some skeptical steps that actually proceed, and the solutions were not close to the mature ones proposed by Chematica (Figure S7). According to the results, finding the sophisticated routes of advanced targets like Chematica did is still a challenging task for data-driven CASP applications.
Figure 6.

For two target compounds (a, MtbTMPK inhibitor46 and b, Propolone49) synthetic routes were found using the ReTReK with retrosynthesis knowledge, whereas no synthetic routes were found using the ReTReK without retrosynthesis knowledge.
These results clearly show that retrosynthesis knowledge effectively contributes to retrosynthetic analyses using ReTReK. However, the experimental validations56 with targets that chemists are interested in and blind assessments9 by trained chemists are not performed in this study; thus we plan to perform these validations to evaluate the performance of ReTReK on actual chemical synthesis in future work. Considering these evaluations and demonstrations, the ReTReK framework with integrated retrosynthesis knowledge has the potential to further improve the performance of data-driven CASP applications.
Conclusions
We developed ReTReK, a data-driven CASP application integrated with rule-based techniques, and it can flexibly reflect and apply retrosynthesis knowledge. Through the evaluation of ReTReK with and without retrosynthesis knowledge, we showed that the integration of such knowledge into data-driven CASP applications helps improve their performance and enhance the quality of the explored synthetic routes. We expect the concept of ReTReK to contribute to the further developments and improvements in data-driven CASP applications.
To allow for more realistic and preferable synthetic routes to be obtained in the future, we will address the further development of automatic reaction template extraction methods while maintaining the chemical integrity. In this study, orphan atoms (atoms appearing on only one side of the reaction arrow) were included in the reaction templates to automatically retain the protecting and leaving groups in the templates because these groups are often not recorded as reactants or products. Because such groups were manually defined in a previous study,22 this template definition (considering the orphan atoms) is expected to contribute to the further development of automatic template extraction methods. In addition, we may define the additional retrosynthesis knowledge scores to allow ReTReK to represent the chemists’ ways of thinking more extensively than in the current model. Furthermore, to facilitate the use of ReTReK, we will prepare a user-friendly interactive interface with functions such as range sliders for adjusting each retrosynthesis knowledge score and other tools for displaying the explored synthetic routes.
Methods
Data Sets
To create the ReTReK model, compounds obtained from the Reaxys reaction records19 and ZINC 15 database54 were used, and compounds were obtained from the ChEMBL 27 database51 and the literature45−50 to evaluate the performance of ReTReK.
Reaction Template Extraction
A set of approximately 50 million reaction records from Reaxys19 (1795–2019) was used to construct the 1-step retrosynthetic reaction prediction model. The model was designed to take a target or an intermediate molecule as input and was trained to predict a suitable reaction template for the input molecule. The purpose of a reaction template is to represent a generalized chemical reaction. In this study, a reaction template consists of a reactive center, orphan atoms, and their first-degree neighbors. An orphan atom is one that appears on only one side of the reaction arrow in ChemAxon.53 These atoms were identified using the Automapper in the ChemAxon API.
The reaction template extraction procedure comprises four steps. Figure S8 shows the workflow of the reaction template extraction.
In the first step, the reaction records were standardized by removing explicit hydrogen, aromatizing, and retaining the largest fragments.
In the second step, the reaction records were filtered based on three conditions: (1) the reaction was required to consist of a single step, (2) it must have a product and up to three reactants, and (3) the number of heavy atoms in the product was limited to 50 or fewer. Thereafter, the number of remaining reaction records was 22 337 137.
In the third step, the reaction templates were extracted from the reaction records, and sets consisting of a product and the corresponding reaction template were retained if the reaction template occurred at least 50 times. To prevent the occurrence of two or more fragments, a reaction template was retained only when all atoms on the product side of the template were connected.
In the final step, the sets consisting of a product and the corresponding reaction template were filtered on condition that the reaction template could be reversibly applied to the product and derived reactants. On the basis of this requirement, 7 589 744 product-template sets remained, and the number of unique reaction templates was 19 633. Referring to a previous study,57 a time-splitting strategy was employed to evaluate the neural network model performance. The sets published before 2017 were used for the training, and those published in 2017 and later were used for testing.
Preparation of Molecules for ReTReK Evaluations and Demonstrations
The molecules used for the ReTReK evaluations were obtained from ChEMBL 2751 and preprocessed via the following procedures. First, the molecules whose The United States Adopted Names’ (USAN) years ranged from 2017 to 2019 and for which chemical structure records were available were selected, resulting in a total of 219 compounds. Thereafter, the compounds were preprocessed by removing the explicit hydrogen, aromatizing, retaining the largest fragments, removing compounds with more than 50 atoms, and removing duplicates. The remaining 161 compounds were used for the evaluations (ChEMBL data set). For further evaluation of the ReTReK, six drug-like compounds45−50 were used for synthetic route search demonstrations.
Starting Materials
A set of compounds obtained from the ZINC database and Reaxys reaction records were used as the starting materials. A subset of 100 023 building blocks from major suppliers (Sigma-Aldrich, Alfa Aesar, and Acros) was obtained from the ZINC database. From the Reaxys reaction records, 649,130 compounds recorded as reactants with at least five occurrences before 2017 were used. All the compounds were stored in the canonical SMILES format calculated using RDKit.21
MCTS for Retrosynthesis
MCTS has been implemented in various CASP studies based on the achievements of Segler et al.9 MCTS is a search algorithm for exploring optimal solutions and comprises four steps: selection, expansion, rollout, and update.58 Following Segler’s implementation,9 a state consists of a set of molecules and is solved (the optimal solution) if all the molecules in the state are starting materials. In this study, retrosynthesis knowledge scores were incorporated into the evaluation term used in the selection step. The same policy network was used for both the expansion and rollout steps, similar to a previous study.35
Retrosynthesis Knowledge Used in ReTReK
We define four scores representing four types of retrosynthesis knowledge, namely, the CDScore, ASScore, RDScore, and STScore, inspired by previous studies.4,7,8
Convergent Disconnection Score
The CDScore is designed to favor convergent synthesis, which is known to be an efficient strategy in multistep chemical synthesis. The CDScore is calculated by evaluating how equally a product is divided among the reactants of a reaction {R1 + R2 + ... + Rn → P}, where Ri is a reactant and P denotes the product.
| 1 |
| 2 |
Here, a(P) and a(Ri) represent the number of atoms in the product and reactant, respectively, and MAE is the mean absolute error.
Available Substances Score
The ASScore, which serves a similar purpose as the CDScore, is defined to reflect the number of available substances generated in a reaction step and is calculated as
| 3 |
Here, b(S) and b(R) represent the numbers of available substances (starting materials) and reactants, respectively.
Ring Disconnection Score
A ring construction strategy is preferred if the target compound has complex ring structures because the construction of ring structures in a synthetic route tends to result in simple and easily available starting materials. The RDScore is calculated by checking whether the ring construction occurs in a reaction step as follows:
 |
4 |
Here, d(P) and d(Ri) represent the number of rings in the product and reactant, respectively.
Selective Transformation Score
A synthetic reaction with few byproducts is preferred, considering the yield. To reflect the number of possible products from a reaction, the STScore is calculated by focusing on the number of reactive centers in the reactants as follows:
| 5 |
Here, e(∑i=1nRi) represents the applicable number of patterns of products enumerated using the reactants and a certain reaction template.
Policy Network
In this study, a policy network is a template-based retrosynthetic reaction prediction model, and the same model is used in both the expansion and rollout steps. We employed a GCN model (a promising model for retrosynthesis found in a previous study28) as the retrosynthetic reaction prediction model. The model was trained using the data set prepared as described in the reaction template extraction section and comprised three graph convolutional layers with Leaky ReLU activation and a dropout ratio of 0.3, a graph-dense layer with Leaky ReLU activation, graph-gather layer with hyperbolic tangent activation, and dense layer with softmax activation. To confirm the effectiveness of the expansion sizes on the MCTS performance, the top-n accuracies were calculated for n values in the range from 1 to 1000. To implement this model, a graph-based deep learning framework kGCN59 was used.
Selection
Starting from the root node, a tree policy was recursively applied to select the subsequent action, indicating that the simulation descended through the search tree gradually until an unvisited node with a nonterminal state was reached. The tree policy was based on the upper confidence bound (UCB) score, and retrosynthesis knowledge was incorporated into the policy as follows:
| 6 |
where wi represent the weights with values of w1 = 5.0, w2 = 0.5, w3 = 2.0, and w4 = 2.0, and n denotes the number of retrosynthesis knowledge scores used in a search (e.g., n is four if all four types of retrosynthesis knowledge are used).
| 7 |
Here, Q denotes an action value calculated in the update step; N and N–1 are the visit counts of the child and parent nodes, respectively; c denotes a constant value that is set to 10; P denotes the softmax probability obtained from the policy network; and K represents the mean of the retrosynthesis knowledge scores.
Expansion
Child nodes, the states of which are selected by the policy network, are added to the node selected by the tree policy. On the basis of the top-n accuracies in the policy network, in independent trials, the top 50, 100, 300, and 500 reaction templates were selected, and they were filtered on condition that the reaction templates could be successfully applied to an unsolved molecule in the state of the selected node.
Rollout
A simulation is implemented using the policy network if the state of a node is not proven or terminal.9 During the simulation, the following steps are recursively implemented for a maximum of five times: an unresolved molecule, which is not included in the starting materials, of the state is randomly sampled, the top 10 reaction templates of the molecule are obtained by the policy network, and a randomly sampled reaction template is applied to the molecule. At the end of each step, it is checked whether the state is proven or not.
A reward function, r, returns one of the three values as reward z, depending on the simulation result. Before the simulation is started, the reward is 10 if the state is proven and −1 if the state is terminated. After the simulation, the reward is equal to the ratio of the number of resolved molecules in the state to the total number of molecules.
Update
The reward obtained from the rollout step is backpropagated through the selected nodes to update their action values Q. On the basis of a previous study,9 the value of Q is defined as
| 8 |
| 9 |
where Lmax denotes the maximal branch length and is set to 10, L denotes the current branch length, and ∑i=1nPi denotes the sum of the softmax probabilities of the reaction templates in the selected nodes.
Evaluating the Effects of Expansion Sizes and Retrosynthesis Knowledge on MCTS Solution Performance
To investigate the effect of expansion sizes on the MCTS’ performance in solving for target molecules, both the number of solved molecules in the ChEMBL data set and times required to solve the molecules were compared for different expansion sizes and six retrosynthesis knowledge patterns. The expansion sizes were 50, 100, 300, and 500 and were determined by the policy network’s top-n accuracies. The six knowledge patterns were as follows: no retrosynthesis knowledge (no knowledge), the CDScore, ASScore, RDScore, STScore, and all the four retrosynthesis knowledge scores (all knowledge). In these experiments, the maximum number of iterations was set to 500 and the score weights for the CDScore, ASScore, RDScore, and STScore were fixed to 5.0, 2.0, 0.5, and 2.0, respectively.
Evaluating the Effects of Retrosynthesis Knowledge on the Search Directions in MCTS
To quantify the effects of the retrosynthesis knowledge on the search directions in MCTS, we defined a route score as the average value of the corresponding retrosynthesis knowledge scores in each step of a solved synthetic route. We calculated four types of route scores (rCDScore, rASScore, rRDScore, and rSTScore) for the solved synthetic routes under the corresponding retrosynthesis knowledge patterns. For comparisons, each route score for the five retrosynthesis knowledge patterns was standardized based on the corresponding mean and standard deviation for the no-knowledge pattern. In these experiments, the synthetic routes solved under the condition of an expansion size of 500 were used. The maximum number of iterations was set to 500, and the score weights for the CDScore, ASScore, RDScore, and STScore were fixed to 5.0, 2.0, 0.5, and 2.0, respectively.
Data and Software Availability
The ReTReK application is publicly available on GitHub at https://github.com/clinfo/ReTReK under the MIT License. The application is distributed in the model based on US Patent data set (10.6084/m9.figshare.5104873.v1) because Reaxys is a commercial database, which cannot be provided to the public. All the compounds used for the evaluations are available on https://github.com/clinfo/ReTReK/tree/master/data/evaluation_compounds. The README file in the GitHub repository provides information about how to setup and use the application.
Acknowledgments
The authors thank Dr. Nobuo Cho and Ms. Noriko Saito for their valuable discussions and comments from the perspective of expert synthetic organic chemists. This study was supported by the New Energy and Industrial Technology Development Organization (NEDO), MEXT, as part of the “Program for Promoting Researches on the Supercomputer Fugak” (MD-driven Precision Medicine).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jcim.1c01074.
Comparison of the numbers of solved molecules with ReTReK and ASKCOS; synthetic routes found by ReTReK and ASCKOS; comparison of the times necessary to solve compounds for different expansion sizes and retrosynthesis knowledge patterns; comparison of the synthetic route with each step’s retrosynthesis knowledge score; and workflow of reaction template extraction (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Corey E. J.; Wipke W. T. Computer-assisted design of complex organic syntheses. Science 1969, 166, 178–192. 10.1126/science.166.3902.178. [DOI] [PubMed] [Google Scholar]
- Corey E. J.; Wipke W. T.; Cramer R. D.; Howe W. J. Computer-assisted synthetic analysis. Facile man-machine communication of chemical structure by interactive computer graphics. J. Am. Chem. Soc. 1972, 94, 421–430. 10.1021/ja00757a020. [DOI] [Google Scholar]
- Wipke W.; Ouchi G. I.; Krishnan S. Simulation and evaluation of chemical synthesis—SECS: An application of artificial intelligence techniques. Artif. Intell. 1978, 11, 173–193. 10.1016/0004-3702(78)90016-4. [DOI] [Google Scholar]
- Hendrickson J. B.; Toczko A. G. SYNGEN program for synthesis design: basic computing techniques. J. Chem. Inf. Model. 1989, 29, 137–145. 10.1021/ci00063a001. [DOI] [Google Scholar]
- Ihlenfeldt W.-D.; Gasteiger J. Computer-Assisted Planning of Organic Syntheses: The Second Generation of Programs. Angew. Chem., Int. Ed. Engl. 1996, 34, 2613–2633. 10.1002/anie.199526131. [DOI] [Google Scholar]
- Satoh K.; Funatsu K. A Novel Approach to Retrosynthetic Analysis Using Knowledge Bases Derived from Reaction Databases. J. Chem. Inf. Comput. Sci. 1999, 39, 316–325. 10.1021/ci980147y. [DOI] [Google Scholar]
- Law J.; Zsoldos Z.; Simon A.; Reid D.; Liu Y.; Khew S. Y.; Johnson A. P.; Major S.; Wade R. A.; Ando H. Y. Route Designer: A Retrosynthetic Analysis Tool Utilizing Automated Retrosynthetic Rule Generation. J. Chem. Inf. Model. 2009, 49, 593–602. 10.1021/ci800228y. [DOI] [PubMed] [Google Scholar]
- Szymkuć S.; Gajewska E. P.; Klucznik T.; Molga K.; Dittwald P.; Startek M.; Bajczyk M.; Grzybowski B. A. Computer-Assisted Synthetic Planning: The End of the Beginning. Angew. Chem., Int. Ed. 2016, 55, 5904–5937. 10.1002/anie.201506101. [DOI] [PubMed] [Google Scholar]
- Segler M. H.; Preuss M.; Waller M. P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature 2018, 555, 604–610. 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- Corey E. J. The Logic of Chemical Synthesis: Multistep Synthesis of Complex Carbogenic Molecules (Nobel Lecture). Angew. Chem., Int. Ed. Engl. 1991, 30, 455–465. 10.1002/anie.199104553. [DOI] [Google Scholar]
- Satyanarayanajois S. D.; Hill R. A. Medicinal chemistry for 2020. Future Med. Chem. 2011, 3, 1765–1786. 10.4155/fmc.11.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey E.; Long A.; Rubenstein S. Computer-assisted analysis in organic synthesis. Science 1985, 228, 408–418. 10.1126/science.3838594. [DOI] [PubMed] [Google Scholar]
- Klucznik T.; Mikulak-Klucznik B.; McCormack M. P.; Lima H.; Szymkuć S.; Bhowmick M.; Molga K.; Zhou Y.; Rickershauser L.; Gajewska E. P.; Toutchkine A.; Dittwald P.; Startek M. P.; Kirkovits G. J.; Roszak R.; Adamski A.; Sieredzińska B.; Mrksich M.; Trice S. L.; Grzybowski B. A. Efficient Syntheses of Diverse, Medicinally Relevant Targets Planned by Computer and Executed in the Laboratory. Chem. 2018, 4, 522–532. 10.1016/j.chempr.2018.02.002. [DOI] [Google Scholar]
- Mikulak-Klucznik B.; Gołębiowska P.; Bayly A. A.; Popik O.; Klucznik T.; Szymkuć S.; Gajewska E. P.; Dittwald P.; Staszewska-Krajewska O.; Beker W.; Badowski T.; Scheidt K. A.; Molga K.; Mlynarski J.; Mrksich M.; Grzybowski B. A. Computational planning of the synthesis of complex natural products. Nature 2020, 588, 83–88. 10.1038/s41586-020-2855-y. [DOI] [PubMed] [Google Scholar]
- Szymkuć S.; Badowski T.; Grzybowski B. A. Is Organic Chemistry Really Growing Exponentially?. Angew. Chem. 2021, 133, 26430–26436. 10.1002/ange.202111540. [DOI] [PubMed] [Google Scholar]
- Coley C. W.; Green W. H.; Jensen K. F. Machine Learning in Computer-Aided Synthesis Planning. Acc. Chem. Res. 2018, 51, 1281–1289. 10.1021/acs.accounts.8b00087. [DOI] [PubMed] [Google Scholar]
- Lecun Y.; Bengio Y.; Hinton G. Deep learning. Nature 2015, 521, 436–444. 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Ching T.; Himmelstein D. S.; Beaulieu-Jones B. K.; Kalinin A. A.; Do B. T.; Way G. P.; Ferrero E.; Agapow P.-M.; Zietz M.; Hoffman M. M.; Xie W.; Rosen G. L.; Lengerich B. J.; Israeli J.; Lanchantin J.; Woloszynek S.; Carpenter A. E.; Shrikumar A.; Xu J.; Cofer E. M.; Lavender C. A.; Turaga S. C.; Alexandari A. M.; Lu Z.; Harris D. J.; DeCaprio D.; Qi Y.; Kundaje A.; Peng Y.; Wiley L. K.; Segler M. H. S.; Boca S. M.; Swamidass S. J.; Huang A.; Gitter A.; Greene C. S. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc., Interface 2018, 15, 20170387. 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaxys. https://www.reaxys.com/, 2020. (accessed 2020-12-16).
- Lowe D. Chemical Reactions from US Patents (1976–Sep2016). 2017; https://figshare.com/articles/Chemical_reactions_from_US_patents_1976-Sep2016_/5104873 (accessed 2020-12-16).
- Landrum G. RDKit: Open-Source Cheminformatics Software. 2016; https://github.com/rdkit/rdkit.
- Coley C. W.; Green W. H.; Jensen K. F. RDChiral: An RDKit Wrapper for Handling Stereochemistry in Retrosynthetic Template Extraction and Application. J. Chem. Inf. Model. 2019, 59, 2529–2537. 10.1021/acs.jcim.9b00286. [DOI] [PubMed] [Google Scholar]
- Coley C.connorcoley/ASKCOS: First public release of ASKCOS. Zenodo; 2019; 10.5281/zenodo.3261361. [DOI]
- Genheden S.; Thakkar A.; Chadimová V.; Reymond J.-L.; Engkvist O.; Bjerrum E. AiZynthFinder: a fast, robust and flexible open-source software for retrosynthetic planning. J. Cheminf. 2020, 12, 12. 10.1186/s13321-020-00472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segler M. H. S.; Waller M. P. Neural-Symbolic Machine Learning for Retrosynthesis and Reaction Prediction. Chem. - Eur. J. 2017, 23, 5966–5971. 10.1002/chem.201605499. [DOI] [PubMed] [Google Scholar]
- Schwaller P.; Laino T.; Gaudin T.; Bolgar P.; Hunter C. A.; Bekas C.; Lee A. A. Molecular Transformer: A Model for Uncertainty-Calibrated Chemical Reaction Prediction. ACS Cent. Sci. 2019, 5, 1572–1583. 10.1021/acscentsci.9b00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpov P.; Godin G.; Tetko I. V.. Artificial Neural Networks and Machine Learning - ICANN 2019: Workshop and Special Sessions; Springer International Publishing: 2019; pp 817–830. [Google Scholar]
- Ishida S.; Terayama K.; Kojima R.; Takasu K.; Okuno Y. Prediction and Interpretable Visualization of Retrosynthetic Reactions Using Graph Convolutional Networks. J. Chem. Inf. Model. 2019, 59, 5026–5033. 10.1021/acs.jcim.9b00538. [DOI] [PubMed] [Google Scholar]
- Schreck J. S.; Coley C. W.; Bishop K. J. M. Learning Retrosynthetic Planning through Simulated Experience. ACS Cent. Sci. 2019, 5, 970–981. 10.1021/acscentsci.9b00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaller P.; Petraglia R.; Zullo V.; Nair V. H.; Haeuselmann R. A.; Pisoni R.; Bekas C.; Iuliano A.; Laino T. Predicting retrosynthetic pathways using transformer-based models and a hyper-graph exploration strategy. Chem. Sci. 2020, 11, 3316–3325. 10.1039/C9SC05704H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibukawa R.; Ishida S.; Yoshizoe K.; Wasa K.; Takasu K.; Okuno Y.; Terayama K.; Tsuda K. CompRet: a comprehensive recommendation framework for chemical synthesis planning with algorithmic enumeration. J. Cheminf. 2020, 12, 52. 10.1186/s13321-020-00452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw J.; Paige B.; Kusner M.; Segler M.; Hernández-Lobato J. M.. Barking up the right tree: an approach to search over molecule synthesis DAGs. Adv. Neural Informat. Process. Syst. 33 2020. [Google Scholar]
- Tetko I. V.; Karpov P.; Deursen R. V.; Godin G. State-of-the-art augmented NLP transformer models for direct and single-step retrosynthesis. Nat. Commun. 2020, 11, 5575. 10.1038/s41467-020-19266-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley C. W.; Jin W.; Rogers L.; Jamison T. F.; Jaakkola T. S.; Green W. H.; Barzilay R.; Jensen K. F. A graph-convolutional neural network model for the prediction of chemical reactivity. Chem. Sci. 2019, 10, 370–377. 10.1039/C8SC04228D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakkar A.; Kogej T.; Reymond J.-L.; Engkvist O.; Bjerrum E. J. Datasets and their influence on the development of computer assisted synthesis planning tools in the pharmaceutical domain. Chem. Sci. 2020, 11, 154–168. 10.1039/C9SC04944D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Qian Y.; Gao H.; Coley C. W.; Mo Y.; Barzilay R.; Jensen K. F. Towards efficient discovery of green synthetic pathways with Monte Carlo tree search and reinforcement learning. Chem. Sci. 2020, 11, 10959–10972. 10.1039/D0SC04184J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik M.; Gothard C. M.; Drews A. M.; Gothard N. A.; Weckiewicz A.; Fuller P. E.; Grzybowski B. A.; Bishop K. J. M. Parallel Optimization of Synthetic Pathways within the Network of Organic Chemistry. Angew. Chem., Int. Ed. 2012, 51, 7928–7932. 10.1002/anie.201202209. [DOI] [PubMed] [Google Scholar]
- Kishimoto A.; Buesser B.; Chen B.; Botea A. Depth-First Proof-Number Search with Heuristic Edge Cost and Application to Chemical Synthesis Planning. Advances in Neural Information Processing Systems 2019, 7224–7234. [Google Scholar]
- Coley C. W.; Thomas D. A.; Lummiss J. A.; Jaworski J. N.; Breen C. P.; Schultz V.; Hart T.; Fishman J. S.; Rogers L.; Gao H.; Hicklin R. W.; Plehiers P. P.; Byington J.; Piotti J. S.; Green W. H.; Hart A. J.; Jamison T. F.; Jensen K. F. A robotic platform for flow synthesis of organic compounds informed by AI planning. Science 2019, 365, 6453. 10.1126/science.aax1566. [DOI] [PubMed] [Google Scholar]
- de Almeida A. F.; Moreira R.; Rodrigues T. Synthetic organic chemistry driven by artificial intelligence. Nat. Rev. Chem. 2019, 3, 589–604. 10.1038/s41570-019-0124-0. [DOI] [Google Scholar]
- Strieth-Kalthoff F.; Sandfort F.; Segler M. H. S.; Glorius F. Machine learning the ropes: principles, applications and directions in synthetic chemistry. Chem. Soc. Rev. 2020, 49, 6154–6168. 10.1039/C9CS00786E. [DOI] [PubMed] [Google Scholar]
- Pflüger P. M.; Glorius F. Molecular Machine Learning: The Future of Synthetic Chemistry?. Angew. Chem., Int. Ed. 2020, 59, 18860–18865. 10.1002/anie.202008366. [DOI] [PubMed] [Google Scholar]
- Badowski T.; Gajewska E. P.; Molga K.; Grzybowski B. A. Synergy Between Expert and Machine-Learning Approaches Allows for Improved Retrosynthetic Planning. Angew. Chem., Int. Ed. 2020, 59, 725–730. 10.1002/anie.201912083. [DOI] [PubMed] [Google Scholar]
- Molga K.; Szymkuć S.; Grzybowski B. A. Chemist Ex Machina: Advanced Synthesis Planning by Computers. Acc. Chem. Res. 2021, 54, 1094–1106. 10.1021/acs.accounts.0c00714. [DOI] [PubMed] [Google Scholar]
- Chen W.; Liu F.; Zhao Q.; Ma X.; Lu D.; Li H.; Zeng Y.; Tong X.; Zeng L.; Liu J.; Yang L.; Zuo J.; Hu Y. Discovery of Phthalazinone Derivatives as Novel Hepatitis B Virus Capsid Inhibitors. J. Med. Chem. 2020, 63, 8134–8145. 10.1021/acs.jmedchem.0c00346. [DOI] [PubMed] [Google Scholar]
- Jian Y.; Merceron R.; De Munck S.; Forbes H. E.; Hulpia F.; Risseeuw M. D.P.; Van Hecke K.; Savvides S. N.; Munier-Lehmann H.; Boshoff H.I.M.; Van Calenbergh S. Endeavors towards transformation of M. tuberculosis thymidylate kinase (MtbTMPK) inhibitors into potential antimycobacterial agents. Eur. J. Med. Chem. 2020, 206, 112659. 10.1016/j.ejmech.2020.112659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji G.; Yusa M.; Masada S.; Yokoo H.; Hosoe J.; Hakamatsuka T.; Demizu Y.; Uchiyama N. Facile Synthesis of Kwakhurin, a Marker Compound of Pueraria mirifica and Its Quantitative NMR Analysis for Standardization as a Reagent. Chem. Pharm. Bull. 2020, 68, 797–801. 10.1248/cpb.c20-00346. [DOI] [PubMed] [Google Scholar]
- Pismataro M. C.; Horenstein N. A.; Stokes C.; Quadri M.; De Amici M.; Papke R. L.; Dallanoce C. Design, synthesis, and electrophysiological evaluation of NS6740 derivatives: Exploration of the structure-activity relationship for alpha7 nicotinic acetylcholine receptor silent activation. Eur. J. Med. Chem. 2020, 205, 112669. 10.1016/j.ejmech.2020.112669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzato T. P.; Gubiani J. R.; Bernardi D. I.; Nogueira C. R.; Monteiro A. F.; Juliano F. F.; de Alencar S. M.; Pilli R. A.; de Lima C. A.; Longato G. B.; Ferreira A. G.; Foglio M. A.; de Carvalho J. E.; Vendramini-Costa D. B.; Berlinck R. G. S. Antiproliferative Flavanoid Dimers Isolated from Brazilian Red Propolis. J. Nat. Prod. 2020, 83, 1784–1793. 10.1021/acs.jnatprod.9b01136. [DOI] [PubMed] [Google Scholar]
- Su Z.; Yang T.; Wang J.; Lai M.; Tong L.; Wumaier G.; Chen Z.; Li S.; Li H.; Xie H.; Zhao Z. Design, synthesis and biological evaluation of potent EGFR kinase inhibitors against 19D/T790M/C797S mutation. Bioorg. Med. Chem. Lett. 2020, 30, 127327. 10.1016/j.bmcl.2020.127327. [DOI] [PubMed] [Google Scholar]
- Mendez D.; Gaulton A.; Bento A P.; Chambers J.; De Veij M.; Felix E.; Magarinos M. P.; Mosquera J. F; Mutowo P.; Nowotka M.ł; Gordillo-Maranon M.; Hunter F.; Junco L.; Mugumbate G.; Rodriguez-Lopez M.; Atkinson F.; Bosc N.; Radoux C. J; Segura-Cabrera A.; Hersey A.; Leach A. R ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis C. L.; Wills M.. Organic synthesis; Oxford Univ. Press, 1995. [Google Scholar]
- ChemAxon. http://www.chemaxon.com, 2020. (accessed 2020-12-16).
- Sterling T.; Irwin J. J. ZINC 15 - Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewska E. P.; Szymkuć S.; Dittwald P.; Startek M.; Popik O.; Mlynarski J.; Grzybowski B. A. Algorithmic Discovery of Tactical Combinations for Advanced Organic Syntheses. Chem. 2020, 6, 280–293. 10.1016/j.chempr.2019.11.016. [DOI] [Google Scholar]
- Lin Y.; Zhang Z.; Mahjour B.; Wang D.; Zhang R.; Shim E.; McGrath A.; Shen Y.; Brugger N.; Turnbull R.; Trice S.; Jasty S.; Cernak T. Reinforcing the supply chain of umifenovir and other antiviral drugs with retrosynthetic software. Nat. Commun. 2021, 12, 7327. 10.1038/s41467-021-27547-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan R. P. Time-Split Cross-Validation as a Method for Estimating the Goodness of Prospective Prediction. J. Chem. Inf. Model. 2013, 53, 783–790. 10.1021/ci400084k. [DOI] [PubMed] [Google Scholar]
- Browne C. B.; Powley E.; Whitehouse D.; Lucas S. M.; Cowling P. I.; Rohlfshagen P.; Tavener S.; Perez D.; Samothrakis S.; Colton S. A survey of Monte Carlo tree search methods. IEEE Trans. Comput. Intell. AI in Games 2012, 4, 1–43. 10.1109/TCIAIG.2012.2186810. [DOI] [Google Scholar]
- Kojima R.; Ishida S.; Ohta M.; Iwata H.; Honma T.; Okuno Y. KGCN: A graph-based deep learning framework for chemical structures. J. Cheminf. 2020, 12, 32. 10.1186/s13321-020-00435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.