Abstract
Background
Head and neck squamous cell carcinoma (HNSCC) is one of the most common malignant cancers. The treatment of HNSCC remains challenging despite recent progress in targeted therapies and immunotherapy. Research on predictive biomarkers in clinical settings is urgently needed.
Methods
Next-generation sequencing analysis was performed on tumor samples from 121 patients with recurrent or metastatic HNSCC underwent sequencing analysis. Clinicopathological information was collected, and the clinical outcomes were assessed. Progression-free survival (PFS) was estimated using the Kaplan-Meier method and cox regression model was used to conduct multivariate analysis. Fisher’s exact tests were used to calculate clinical benefit. A p value of less than 0.05 was designated as significant (p < 0.05).
Results
Chromosome 11q13 amplification (CCND1, FGF3, FGF4, and FGF19) and EGFR mutations were significantly associated with decreased PFS and no clinical benefits after treatment with a programmed death 1 (PD-1) inhibitor. The same results were found in the combined positive score (CPS) ≥ 1 subgroup. In patients who were treated with an EGFR antibody instead of a PD-1 inhibitor, a significant difference in PFS and clinical benefits was only observed between patients with CPS ≥ 1 and CPS < 1.
Conclusion
Chromosome 11q13 amplification and EGFR mutations were negatively correlated with anti-PD-1 therapy. These markers may serve as potential predictive biomarkers to identify patients for whom immunotherapy may be unsuitable.
Keywords: head and neck squamous cell carcinoma (HNSCC), immune checkpoint inhibitor (ICI), predictive biomarkers, immunotherapy, 11q13 amplification, EGFR mutation
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignant cancer worldwide (1), accounting for 450,000 deaths per year (2). Treatment for HNSCC typically includes surgery, radiotherapy, chemotherapy, or combined modalities, and more recently, targeted therapy and immunotherapy. Compared to traditional therapies, both targeted therapy and immune checkpoint inhibitor (ICI) therapy have shown significantly improved clinical benefits (CBs) in the treatment of recurrent and metastatic disease (3–6).
ICIs that target the programmed death 1 (PD-1) and PD ligand 1 (PD-L1) axis, such as nivolumab and pembrolizumab, have been widely used and have proven to be effective in solid tumors. In 2016, based on the clinical trials CheckMate141 and KEYNOTE-012, these drugs were approved for use as second-line treatments in patients with recurrent or metastatic HNSCC that was refractory to platinum-based therapy (7, 8). In 2019, pembrolizumab was approved by the US Food and Drug Administration as a first-line treatment for patients with metastatic or unresectable recurrent HNSCC with a combined positive score (CPS) ≥1, based on the clinical trial KEYNOTE-048 (3).
However, successful ICI therapy generally requires the identification of clinically effective biomarkers with which to screen potential patients for treatment sensitivity. Currently, widely accepted pancancer biomarkers such as microsatellite instability, tumor mutation burden, and PD-L1 expression are used to identify patients who could potentially benefit from ICI treatment. Nonetheless, immunotherapy is applicable in a very limited proportion of patients (approximately 18%) (7, 9), leaving the vast majority vulnerable to trial and error methods for ICI treatment. More predictive biomarkers are needed to improve the quality of care for the unresponsive majority. Furthermore, there is a need to create protocols for the stratification of patients based on predictive biomarkers in order to apply personalized treatment at a clinical level.
Recent genomic and transcriptomic investigations have substantially improved our knowledge of the molecular mechanisms underpinning HNSCC (10, 11). Genetic aberrations and the abnormal expression of certain genes, such as EGFR (12, 13) and 11q13 amplification (CCND1, FGF3, FGF4, and FGF19) (14, 15) are closely associated with prognosis and may be useful prognostic biomarkers. In 2017, Singavi et al. identified EGFR and 11q13 amplification to be potential predictive biomarkers, as these genetic alterations were associated with hyper-progression in response to ICI in five patients with lung, esophageal, and renal cancers (16). Nevertheless, research on their roles as predictive biomarkers remains limited, especially in HNSCC. To meet the urgent need for more predictive biomarkers that can guide clinical decision-making, the aim of our retrospective study was therefore to explore the predictive roles of these genetic abnormalities in identifying patients who are unsuitable for immunotherapies in the real world.
Materials and Methods
Patients
One hundred and twenty-one patients were enrolled in the study from January 9, 2019, to November 10, 2020. The main inclusion criteria were as follows: (a) Pathology confirmed recurrent or metastatic HNSCC treated with anti-PD-1 antibody or EGFR antibody; (b) an age of 18 to 80 years; (c) no serious comorbidities (e.g., had suffered from other malignant tumors); (d) complete and usable follow-up data. Two cohorts of patients were enrolled: a PD-1 cohort that included 98 patients who were treated with anti-PD-1 antibody (PD-1 group), and a non-PD-1 cohort (NPD-1 group) that included 32 patients who were treated with non-PD-1 inhibitor (EGFR antibody). This study was approved by the ethics committee of Shanghai Ninth People’s Hospital (No. SH9H-2020-T257-1). Clinicopathological data were collected from patients during treatment and follow-up visits.
Assessment of Clinical Benefit
In Assessment of clinical benefit, we referred to Yu et al. (17). Clinical efficacy was evaluated per RECIST 1.1 every 8 weeks. CB was defined as a patient exhibiting a complete response (CR) or partial response (PR) according to RECIST 1.1 (i.e., tumor shrinkage > 30% from baseline) or stable disease (SD) if they had any objective reduction in tumor burden lasting at least 6 months. No clinical benefit (NCB) were defined as those experiencing progressive disease according to RECIST 1.1 or SD lasting <6 months and were discontinued from immunotherapy within 3 months (18, 19).
Targeted Sequencing of Clinical Samples
Tumor genomic DNA was isolated from formalin-fixed paraffin-embedded (FFPE) tissue sections using the QIAgen DNA FFPE tissue kit (Germantown, MD, USA), and was used for targeted sequencing with a cancer-related-gene panel (Genecast Biotech., Wuxi, China). Using Qiagen DNA blood mini kit, genomic DNA was extracted from peripheral blood collected from participants, and was used as a matched control. The library was constructed with 300 ng of genomic DNA from each participant. Fragment libraries were prepared from samples sheared by sonication, and target regions were enriched using customized IDT library prep kits (Integrated DNA Technologies, Coralville, IA, USA). The captured DNA was amplified, and the paired-end library was sequenced using the NovaSeq 6000 platform (Illumina, San Diego, CA, USA). Bioinformatics analysis was performed using an in-house program (Genecast Biotech.).
Statistical Analysis
Statistical analyses were performed using MedCalc (version 19.0.4) (MedCalc Software Ltd, Ostend, Belgium). Progression-free survival (PFS) was estimated using the Kaplan−Meier method, and between-group differences in PFS were tested using the log-rank test. Cox regression model was used for multivariate analysis of PFS. Fisher exact tests were used to analyze the association between genetic aberrations with CBs. Odds ratios (ORs) and their associated 95% confidence intervals (CIs), and multivariate analysis of CB/NCB were estimated using the logistic regression model. A P value of less than 0.05 was considered statistically significant (p < 0.05).
Results
Patient Characteristics
Of the 121 patients enrolled in the study, 92 were male and 29 were female. The median age of the patients was 57 years. Ninety-one patients had oral squamous cell carcinoma, 23 had oropharyngeal carcinoma, and 7 had other cancer types. Assessment of PD-L1 status revealed that 84 patients had a CPS ≥ 1, 30 patients had a CPS < 1, and 7 patients had an unknown CPS status. The clinicopathological data are summarized in Table 1. The detailed clinicopathological data for each patient is shown in Supplemental Table S1.
Table 1.
Summary of clinical information.
| Number | % | |
|---|---|---|
| Sex | ||
| Male | 92 | 76.0% |
| Female | 29 | 24.0% |
| Age (years) | ||
| Median | 57 (27–79) | |
| Cancer type | ||
| Oral squamous cell carcinoma | 91 | 75.2% |
| Oropharyngeal carcinoma | 23 | 19.0% |
| P16 positive | 5 | 4.1% |
| P16 negative | 18 | 14.9% |
| Other cancer type | 7 | 5.8% |
| PD-L1 | ||
| CPS < 1 | 30 | 25.0% |
| CPS ≥ 1 | 84 | 69.4% |
| Unknown | 7 | 6.0% |
| Number of prior lines of therapy* | ||
| 0 | 70 | NA |
| ≥1 | 60 | NA |
*9 patients were treated with both PD-1 antibody and EGFR antibody at different lines. NA, not applicable.
Overall Analysis of Patients Treated With PD-1 Antibody
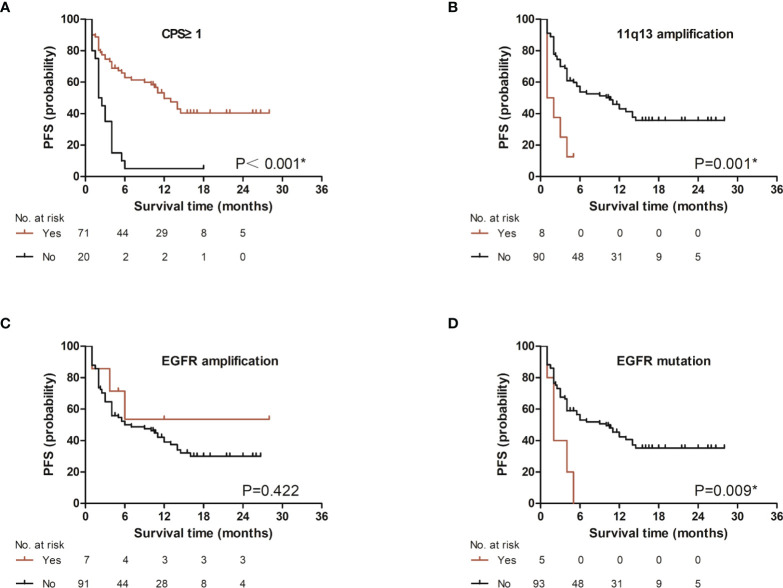
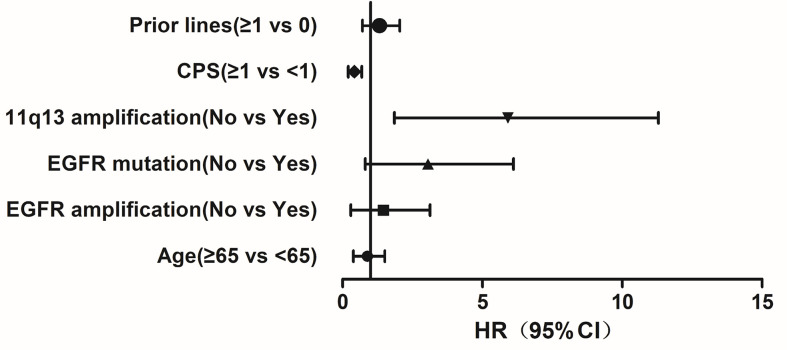
We first analyzed the PFS based on CPS level (CPS ≥ 1 and CPS < 1), 11q13 status, EGFR amplification status and EGFR mutation status, we found that PFS was significantly different between the groups with different CPS (p < 0.001, Figure 1A). A significant difference in PFS was observed between the groups with different 11q13 amplification status (p = 0.001, Figure 1B). The PFS was not significantly different between the groups with different EGFR amplification status (p = 0.422, Figure 1C). A significant difference was observed between the groups with different EGFR mutation status (p = 0.009, Figure 1D). Multivariate analyses of treatment lines, CPS level, 11q13 amplification, EGFR mutation, EGFR amplification, and age showed that CPS and 11q13 amplification are independent factors affecting PFS with HR = 0.37 and 4.58, respectively (Figure 2).
Figure 1.
Association of several factors with progression-free survival in patients treated with PD-1 inhibitor. Association of (A) CPS level. (CPS, combined positive score), (B) 11q13 amplification status. (11q13: CCND1_FGF3_FGF4_FGF19 or any one of them), (C) EGFR amplification status, or (D) EGFR mutation status, with progression-free survival. *p<0.05.
Figure 2.
Multivariate analyses of treatment lines, CPS level, 11q13 amplification, EGFR mutation, EGFR amplification, and age with progression-free survival. CPS and 11q13 amplification are independent factor affecting progression-free survival.
We next evaluated whether CB was associated with CPS, 11q13 amplification, EGFR amplification, or EGFR mutations in patients who received anti-PD-1 treatment. Of the 71 patients with CPS ≥ 1, 46 (64.8%) benefited from anti-PD-1 treatment, while only three of the 20 patients with CPS < 1 obtained CB (p = 0.001; OR = 10.43; 95% CI: 2.78 - 39.05). Of the 90 patients without an 11q13 amplification, 51 (56.7%) obtained CB, while one of the 8 patients with an 11q13 amplification exhibited CB (p = 0.024; OR = 9.15; 95% CI: 1.08 - 77.52). Of the 93 patients with no EGFR mutations, 52 (55.9%) obtained CB, whereas none of the 5 patients with EGFR mutations obtained CB (p = 0.020; OR = 13.92; 95% CI: 0.75 – 258.94). Details are shown in Table 2. Multivariate analysis including treatment lines, CPS level, 11q13 amplification, EGFR mutation, EGFR amplification, and age showed only CPS level significantly associated with CB (p = 0.001).
Table 2.
Logistic analysis of factors that affect clinical benefit.
| Characteristic | Clinical benefit (n, %) | p value (Fisher exact test) | Odds ratio (95% CI) | |
|---|---|---|---|---|
| CB | NCB | |||
| CPS | ||||
| ≥1 | 46 (64.8) | 25 (35.2) | <0.001 | 10.43 (2.78, 39.05) |
| <1 | 3 (15.0) | 17 (85.0) | ||
| 11q13 amplification | ||||
| No | 51 (56.7) | 39 (43.3) | 0.024 | 9.15 (1.08, 77.52) |
| Yes | 1 (12.5) | 7 (87.5) | ||
| EGFR amplification | ||||
| No | 48 (52.7) | 43 (47.3) | 1 | 0.84 (0.18, 3.95) |
| Yes | 4 (57.1) | 3 (42.9) | ||
| EGFR mutation | ||||
| No | 52 (55.9) | 41 (44.1) | 0.020 | 13.92 (0.75, 258.94) |
| Yes | 0 (0) | 5 (100) | ||
CB, clinical benefit; NCB, non-clinical benefit; CPS, combined positive score; 11q13: CCND1_FGF3_FGF4_FGF19 or any one of them.
Analysis of Predictive Biomarkers in Patients From the CPS ≥ 1 Subgroup Treated With PD-1 Inhibitor
Focusing further on patients with CPS ≥1 who received anti-PD-1 treatment, analysis of PFS between groups with or without 11q13 amplification, EGFR amplification, EGFR mutations revealed that PFS was significantly different between the groups with or without 11q13 amplification (p < 0.001) and EGFR mutation (p = 0.022) (Figure S1). PFS was not significantly different between the groups with or without EGFR amplification (p = 0.990). Multivariate analyses of treatment lines, 11q13 amplification, EGFR mutation, EGFR amplification, and age showed that 11q13 amplification and EGFR mutation are independent factors affecting PFS with HR = 8.62 and 4.78, respectively (Figure S2).
We next evaluated whether 11q13 amplification, EGFR amplification, or EGFR mutation with CB. Our analysis indicated that 46 of 66 patients lacking an 11q13 amplification exhibited CB (69.7%), while none of the five patients with an 11q13 amplification demonstrated CB (p = 0.004; OR = 24.95; 95% CI: 1.32 - 472.62). Of the 65 patients with no EGFR amplification, 42 were evaluated as CB (64.6%), while four of the six patients with EGFR amplification exhibited CB (66.7%) (p=1.000; OR = 0.91; 95% CI: 0.16 - 5.37). The analysis of patients with no EGFR mutations showed that 42 of 69 exhibited CB (64.6%), whereas neither of the 2 patients with EGFR mutations demonstrated CB (0.00%) (p = 0.121; OR = 9.89; 95% CI: 0.46 - 214.55). EGFR mutations were not markedly associated with NCB in patients with CPS ≥1. Detailed information is shown in Table 3. Multivariate analyses show no factors are significantly associated with CB.
Table 3.
Subgroup analysis of patients with CPS ≥ 1.
| Characteristic | Clinical benefit (n, row%) | p value (Fisher exact test) | Odds ratio (95% CI) | |
|---|---|---|---|---|
| CB | NCB | |||
| 11q13 amplification | ||||
| No | 46 (69.7) | 20 (30.3) | 0.004 | 24.95 (1.32, 472.62) |
| Yes | 0 (0) | 5 (100) | ||
| EGFR amplification | ||||
| No | 42 (64.6) | 23 (35.4) | 1.000 | 0.91 (0.16, 5.37) |
| Yes | 4 (66.7) | 2 (33.3) | ||
| EGFR mutation | ||||
| No | 46 (66.7) | 23 (33.3) | 0.121 | 9.89 (0.46, 214.55) |
| Yes | 0 (0) | 2 (100) | ||
CB, clinical benefit; NCB, non-clinical benefit; CPS, combined positive score; 11q13: CCND1_FGF3_FGF4_FGF19 or any one of them.
Analysis of Predictive Biomarkers in Patients Treated With EGFR Antibody
To assess whether these predictive biomarkers were associated with PFS in patients treated with non-PD-1 inhibitor (EGFR antibody), we found only CPS level were associate with PFS in both univariate analysis (Figure S3, p = 0.030) and multivariate analysis (HR = 0.29). To assess whether these predictive biomarkers were associated with CB in non-PD-1 inhibitor treatment (EGFR antibody), we analyzed the non-PD-1 inhibitor group. In the subgroup analyses based on 11q13 amplification, EGFR amplification, and EGFR mutation status, no significant differences were observed between patients with and without mutations. However, significant differences were observed between patients with CPS ≥1 compared to those with CPS <1, indicating that CPS could predict CB even in patients who did not receive a PD-1 inhibitor. Detailed information is shown in Supplemental Table S2. Multivariate analyses show no factors are significantly associated with CB.
Discussion
HNSCC is notorious for its high mortality rate, and frequent recurrence, and metastasis. While ICIs have broadened the clinical options for treating HNSCC, the successful application of these inhibitors requires the identification of predictive biomarkers that can help select for patients who are likely to exhibit CB. Here, we report the results of a retrospective, real-world study to determine the association between genetic aberrations and clinical outcomes.
The amplification of EGFR is not uncommon in HNSCC patients. EGFR has been identified as a predictive biomarker for chemotherapy or radiation/chemotherapy benefits and survival in oropharyngeal cancer and for targeted therapy benefit in HNSCC (20, 21). Prior to this study, little was known about the potential for EGFR amplification to predict immunotherapy benefit in HNSCC. We found that EGFR mutations, not EGFR amplification, adversely affected clinical outcomes when patients were treated with a PD-1 inhibitor ta. EGFR mutations were originally reported to affect the clinical outcomes of ICI treatment in TKI naïve, PD-L1-positive, and EGFR-mutant patients with advanced non-small-cell lung cancer (22). However, recent studies have indicated that the clinical outcomes of non-small-cell lung cancer patients with EGFR mutations are promising and that patients with different EGFR mutations may experience different outcomes of ICI therapy despite their generally low response to ICIs (23, 24). We observed that patients with EGFR mutations had a significantly decreased PFS and NCB to PD-1 inhibitors, which is in keeping with results from previous reports. Interestingly, when we further analyzed patients with CPS ≥ 1, we observed the same outcome in PFS, but not in CB., which may be due to the rare mutation rate in HNSCC. However, this observation requires further study because EGFR mutations are uncommon and the sample size in our study was small. EGFR overexpression was previously reported to be associated with poor PFS in HNSCC (13). In our study, however, EGFR amplification did not significantly affect clinical outcomes under any circumstances. This result is also different from that of a previous study which reported that 1 out of 26 patients with EGFR amplification who received ICI therapy experienced hyper-progression in different types of cancer (16). Further studies with large sample size are needed to validate the role of EGFR amplification in HNSCC.
The amplification of 11q13 has been observed in over 30% of patients with HNSCC and its prognostic value has also been reported (15, 25, 26). Their study indicated that higher expression of 11q13 amplification was indicative of worse prognosis. Chromosome 11q13 amplification was found to correlate significantly with carcinogenesis and the attenuation of effector immune cells in the tumor microenvironment (14). CCND1 has been associated with resistance to PD-1-targeted therapy in a Chinese population with non-cutaneous melanoma due to its effects on innate immunity (17). In a retrospective study carried out at four French institutions, patients with recurrent and/or metastatic HNSCC experienced accelerated tumor growth and had shorter PFS after anti-PD-1 and anti-PD-L1 treatment, with three of the patients harboring a CCND1 amplification (27). The role of CCND1 amplification as a predictive biomarker for the efficacy of ICI therapy was also suggested in an in silico analysis performed by Chen et al. (14). In their study, the authors analyzed three large cohorts of solid tumors from The Cancer Genome Atlas (TCGA) database, the Memorial Sloan Kettering Cancer Center (MSKCC) archive, and a local database, and they concluded that CCND1 amplification correlated with shorter overall survival and poorer outcomes after ICI therapy. Singavi et al. reported that three patients in lung cancer and esophageal cancer harboring an 11q13 amplification experienced hyperprogression after treatment with ICIs (16). In our study, 11q13 amplification significantly affected ICI treatment benefits, regardless of CPS level. Our results are similar to previous observations that have implicated the immune-negative role of 11q13 amplification as a predictive biomarker.
We enrolled 121 patients in this study, which represents one of the largest HNSCC cohorts studied thus far. However, our study was a retrospective analysis, which limited the power of our findings. We are thus enrolling more patients for further studies to consolidate our conclusions. A prospective study is also planned to further examine the associations of certain gene aberrations with clinical efficacy after treatment with ICIs.
In conclusion, we showed that 11q13 amplification and EGFR mutations negatively correlated with PD-1 inhibitor therapy in a retrospectively analyzed clinical cohort. These genetic aberrations may serve as potential predictive biomarkers to identify patients who are unsuitable for immunotherapies, which will aid physicians in clinical decision-making. Therefore, further studies are required to confirm the utility of these biomarkers as predictors of treatment outcome in HNSCC.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethics committee of Shanghai Ninth People’s Hospital (No. SH9H-2020-T257-1). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
GZ, RL, SD, and LZ conceived and supervised the study. SD, LZ, CW, and YY drafted the manuscript. WJ, LY, and JL collected samples and clinical information. SW, DS, and XG analyzed the data. All authors have revised the manuscript and provided helpful advice. All authors read and approved the final manuscript.
Funding
This study was supported by the Shanghai Anti-Cancer Research Foundation (H8001-004) and Clinical Research Plan of SHDC (No. SHDC2020CR4012) and WU JIEPING MEDICAL FOUNDATION (No. 320.6750.2021-01-34). The study sponsors had no role in the study design, data collection, analysis and interpretation of data or preparation of the manuscript.
Conflict of Interest
Authors DS and XG are employed by Genecast Biotechnology.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.813732/full#supplementary-material
References
- 1. Shield KD, Ferlay J, Jemal A, Sankaranarayanan R, Chaturvedi AK, Bray F, et al. The Global Incidence of Lip, Oral Cavity, and Pharyngeal Cancers by Subsite in 2012. CA Cancer J Clin (2017) 67(1):51–64. doi: 10.3322/caac.21384 [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3. Burtness B, Harrington KJ, Greil R, Soulieres D, Tahara M, de Castro G, Jr, et al. Pembrolizumab Alone or With Chemotherapy Versus Cetuximab With Chemotherapy for Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck (KEYNOTE-048): A Randomised, Open-Label, Phase 3 Study. Lancet (2019) 394(10212):1915–28. doi: 10.1016/S0140-6736(19)32591-7 [DOI] [PubMed] [Google Scholar]
- 4. Ferris RL, Blumenschein G, Jr, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med (2016) 375(19):1856–67. doi: 10.1056/NEJMoa1602252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guigay J, Auperin A, Fayette J, Saada-Bouzid E, Lafond C, Taberna M, et al. Cetuximab, Docetaxel, and Cisplatin Versus Platinum, Fluorouracil, and Cetuximab as First-Line Treatment in Patients With Recurrent or Metastatic Head and Neck Squamous-Cell Carcinoma (GORTEC 2014-01 TPExtreme): A Multicentre, Open-Label, Randomised, Phase 2 Trial. Lancet Oncol (2021) 22(4):463–75. doi: 10.1016/S1470-2045(20)30755-5 [DOI] [PubMed] [Google Scholar]
- 6. Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, et al. Platinum-Based Chemotherapy Plus Cetuximab in Head and Neck Cancer. N Engl J Med (2008) 359(11):1116–27. doi: 10.1056/NEJMoa0802656 [DOI] [PubMed] [Google Scholar]
- 7. Harrington KJ, Ferris RL, Blumenschein G, Jr, Colevas AD, Fayette J, Licitra L, et al. Nivolumab Versus Standard, Single-Agent Therapy of Investigator’s Choice in Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck (CheckMate 141): Health-Related Quality-of-Life Results From a Randomised, Phase 3 Trial. Lancet Oncol (2017) 18(8):1104–15. doi: 10.1016/S1470-2045(17)30421-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Plimack ER, Bellmunt J, Gupta S, Berger R, Chow LQ, Juco J, et al. Safety and Activity of Pembrolizumab in Patients With Locally Advanced or Metastatic Urothelial Cancer (KEYNOTE-012): A Non-Randomised, Open-Label, Phase 1b Study. Lancet Oncol (2017) 18(2):212–20. doi: 10.1016/S1470-2045(17)30007-4 [DOI] [PubMed] [Google Scholar]
- 9. Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, et al. Safety and Clinical Activity of Pembrolizumab for Treatment of Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck (KEYNOTE-012): An Open-Label, Multicentre, Phase 1b Trial. Lancet Oncol (2016) 17(7):956–65. doi: 10.1016/S1470-2045(16)30066-3 [DOI] [PubMed] [Google Scholar]
- 10. Chung CH, Guthrie VB, Masica DL, Tokheim C, Kang H, Richmon J, et al. Genomic Alterations in Head and Neck Squamous Cell Carcinoma Determined by Cancer Gene-Targeted Sequencing. Ann Oncol (2015) 26(6):1216–23. doi: 10.1093/annonc/mdv109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dubot C, Bernard V, Sablin MP, Vacher S, Chemlali W, Schnitzler A, et al. Comprehensive Genomic Profiling of Head and Neck Squamous Cell Carcinoma Reveals FGFR1 Amplifications and Tumour Genomic Alterations Burden as Prognostic Biomarkers of Survival. Eur J Cancer (2018) 91:47–55. doi: 10.1016/j.ejca.2017.12.016 [DOI] [PubMed] [Google Scholar]
- 12. Chung CH, Ely K, McGavran L, Varella-Garcia M, Parker J, Parker N, et al. Increased Epidermal Growth Factor Receptor Gene Copy Number Is Associated With Poor Prognosis in Head and Neck Squamous Cell Carcinomas. J Clin Oncol (2006) 24(25):4170–6. doi: 10.1200/JCO.2006.07.2587 [DOI] [PubMed] [Google Scholar]
- 13. Zhu X, Zhang F, Zhang W, He J, Zhao Y, Chen X. Prognostic Role of Epidermal Growth Factor Receptor in Head and Neck Cancer: A Meta-Analysis. J Surg Oncol (2013) 108(6):387–97. doi: 10.1002/jso.23406 [DOI] [PubMed] [Google Scholar]
- 14. Chen Y, Huang Y, Gao X, Li Y, Lin J, Chen L, et al. CCND1 Amplification Contributes to Immunosuppression and Is Associated With a Poor Prognosis to Immune Checkpoint Inhibitors in Solid Tumors. Front Immunol (2020) 11:1620. doi: 10.3389/fimmu.2020.01620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanken H, Grobe A, Cachovan G, Smeets R, Simon R, Sauter G, et al. CCND1 Amplification and Cyclin D1 Immunohistochemical Expression in Head and Neck Squamous Cell Carcinomas. Clin Oral Investig (2014) 18(1):269–76. doi: 10.1007/s00784-013-0967-6 [DOI] [PubMed] [Google Scholar]
- 16. Singavi AK, Menon S, Kilari D, Alqwasmi A, Ritch PS, Thomas JP. Predictive Biomarkers for 368 Hyper-Progression (HP) in Response to Immune Checkpoint Inhibitors (ICI) – Analysis of 369 Somatic Alterations (SAs). Ann Oncol (2017) 28(Suppl_5):v403–27.370. doi: 10.1093/annonc/mdx376.006 [DOI] [Google Scholar]
- 17. Yu J, Yan J, Guo Q, Chi Z, Tang B, Zheng B, et al. Genetic Aberrations in the CDK4 Pathway Are Associated With Innate Resistance to PD-1 Blockade in Chinese Patients With Non-Cutaneous Melanoma. Clin Cancer Res (2019) 25(21):6511–23. doi: 10.1158/1078-0432.CCR-19-0475 [DOI] [PubMed] [Google Scholar]
- 18. Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, et al. Genomic Correlates of Response to Immune Checkpoint Therapies in Clear Cell Renal Cell Carcinoma. Science (2018) 359(6377):801–6. doi: 10.1126/science.aan5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roh W, Chen PL, Reuben A, Spencer CN, Prieto PA, Miller JP, et al. Integrated Molecular Analysis of Tumor Biopsies on Sequential CTLA-4 and PD-1 Blockade Reveals Markers of Response and Resistance. Sci Transl Med (2017) 9(379). doi: 10.1126/scitranslmed.aah3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kumar B, Cordell KG, Lee JS, Worden FP, Prince ME, Tran HH, et al. EGFR, P16, HPV Titer, Bcl-xL and P53, Sex, and Smoking as Indicators of Response to Therapy and Survival in Oropharyngeal Cancer. J Clin Oncol (2008) 26(19):3128–37. doi: 10.1200/JCO.2007.12.7662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lim SM, Cho SH, Hwang IG, Choi JW, Chang H, Ahn MJ, et al. Investigating the Feasibility of Targeted Next-Generation Sequencing to Guide the Treatment of Head and Neck Squamous Cell Carcinoma. Cancer Res Treat (2019) 51(1):300–12. doi: 10.4143/crt.2018.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lisberg A, Cummings A, Goldman JW, Bornazyan K, Reese N, Wang T, et al. A Phase II Study of Pembrolizumab in EGFR-Mutant, PD-L1+, Tyrosine Kinase Inhibitor Naive Patients With Advanced NSCLC. J Thorac Oncol (2018) 13(8):1138–45. doi: 10.1016/j.jtho.2018.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garassino MC, Cho BC, Kim JH, Mazieres J, Vansteenkiste J, Lena H, et al. Durvalumab as Third-Line or Later Treatment for Advanced Non-Small-Cell Lung Cancer (ATLANTIC): An Open-Label, Single-Arm, Phase 2 Study. Lancet Oncol (2018) 19(4):521–36. doi: 10.1016/S1470-2045(18)30144-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hastings K, Yu HA, Wei W, Sanchez-Vega F, DeVeaux M, Choi J, et al. EGFR Mutation Subtypes and Response to Immune Checkpoint Blockade Treatment in Non-Small-Cell Lung Cancer. Ann Oncol (2019) 30(8):1311–20. doi: 10.1093/annonc/mdz141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Freier K, Sticht C, Hofele C, Flechtenmacher C, Stange D, Puccio L, et al. Recurrent Coamplification of Cytoskeleton-Associated Genes EMS1 and SHANK2 With CCND1 in Oral Squamous Cell Carcinoma. Genes Chromosomes Cancer (2006) 45(2):118–25. doi: 10.1002/gcc.20270 [DOI] [PubMed] [Google Scholar]
- 26. Fujii M, Ishiguro R, Yamashita T, Tashiro M. Cyclin D1 Amplification Correlates With Early Recurrence of Squamous Cell Carcinoma of the Tongue. Cancer Lett (2001) 172(2):187–92. doi: 10.1016/s0304-3835(01)00651-6 [DOI] [PubMed] [Google Scholar]
- 27. Saada-Bouzid E, Defaucheux C, Karabajakian A, Coloma VP, Servois V, Paoletti X, et al. Hyperprogression During Anti-PD-1/PD-L1 Therapy in Patients With Recurrent and/or Metastatic Head and Neck Squamous Cell Carcinoma. Ann Oncol (2017) 28(7):1605–11. doi: 10.1093/annonc/mdx178 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.