Abstract
Introduction: This study aims to assess the utility of Boosting ensemble classification methods for increasing the diagnostic performance of multiparametric Magnetic Resonance Imaging (mpMRI) radiomic models, in differentiating benign and malignant breast lesions. Methods: The dataset includes mpMR images of 140 female patients with mass-like breast lesions (70 benign and 70 malignant), consisting of Dynamic Contrast Enhanced (DCE) and T2-weighted sequences, and the Apparent Diffusion Coefficient (ADC) calculated from the Diffusion Weighted Imaging (DWI) sequence. Tumor masks were manually defined in all consecutive slices of the respective MRI volumes and 3D radiomic features were extracted with the Pyradiomics package. Feature dimensionality reduction was based on statistical tests and the Boruta wrapper. Hierarchical Clustering on Spearman's rank correlation coefficients between features and Random Forest classification for obtaining feature importance, were implemented for selecting the final feature subset. Adaptive Boosting (AdaBoost), Gradient Boosting (GB), Extreme Gradient Boosting (XGBoost) and Light Gradient Boosting Machine (LightGBM) classifiers, were trained and tested with bootstrap validation in differentiating breast lesions. A Support Vector Machine (SVM) classifier was also exploited for comparison. The Receiver Operator Characteristic (ROC) curves and DeLong's test were utilized to evaluate the classification performances. Results: The final feature subset consisted of 5 features derived from the lesion shape and the first order histogram of DCE and ADC images volumes. XGboost and LGBM achieved statistically significantly higher average classification performances [AUC = 0.95 and 0.94 respectively], followed by Adaboost [AUC = 0.90], GB [AUC = 0.89] and SVM [AUC = 0.88]. Conclusion: Overall, the integration of Ensemble Learning methods within mpMRI radiomic analysis can improve the performance of computer-assisted diagnosis of breast cancer lesions.
Keywords: AdaBoost, Boruta, Gradient Boosting, LightGBM, Radiomics, XGBoost
Introduction
Female breast cancer was the leading cause of global cancer incidence in 2020 and the fifth in cancer mortality rates among both sexes worldwide. 1 Over the past decades effective breast cancer prognosis and patients’ survival rates have increased due to the improvements and availability of innovative screening technologies. 2 Magnetic Resonance Imaging (MRI) of the breast has emerged as an exceptionally powerful technique, with increased sensitivity in breast cancer detection, even compared to mammography and ultrasonography. 3 Additionally, the simultaneous evaluation of different MRI sequences, such as Dynamic Contrast Enhanced (DCE) and Diffusion Weighted Imaging (DWI), referred as multiparametric MRI (mpMRI), can be used to assess a multitude of morphological and functional cancer-related processes, related to breast tumor development, progression, and response to treatment. 4 Currently, mpMRI has a pivotal role in differentiating benign and malignant breast lesions that present highly overlapping enhancement patterns, non-invasively. 3 However, despite the potential to obviate unnecessary biopsies and follow-up examinations of benign tumors, mpMRI-based breast tumor differentiation still has increased false positive findings. 5
Breast MRI diagnosis has been further enriched by computer-aided image analysis, to assist the radiologists in leveraging the substantial quantitative imaging information and assessing tumor profile. 6 The spread of “-omics” strategies has provided a novel conceptual framework, termed Radiomics, aiming at the extraction of immense numbers of imaging features, that can serve as imaging biomarkers. Especially, when coupled with sophisticated supervised Machine Learning (ML) algorithms, these data can be used to construct clinically significant diagnostic and predictive models for assisting personalized care of oncologic patients.7,8
In this context, a few previous studies have developed radiomic models with Support Vector Machines (SVM) for classifying breast tumors in mpMRI datasets, demonstrating high predictive efficiencies in terms of Area Under Curve (AUC), ranging from 0.85 to 0.92.9–12 In another study, 13 four different classification algorithms, ie, SVM, Naïve Bayes, k-Nearest Neighbours, and Logistic Regression, were evaluated, demonstrating comparable performances with an average AUC = 0.93. Recently, a newly designed classification model, the difference-weighted local hyperplane has been proposed, 14 that have shown a performance of AUC = 0.90 in differentiating benign versus malignant lesions. Although very promising, the difficulty in collecting mpMRI datasets of adequate size, as well as the inherent complexity of mpMRI biomarkers, are hampering the capabilities of the proposed models, in terms of performance and generalizability.
Recently, Ensemble Learning methods that combine the predictions of multiple classifiers to reach better performance than a single estimator does, have gained interest in radiomics research. 15 These strategies have proven very useful in modelling heterogeneous datasets of any size and complexity, 7 while also excel at trading off the approximation and estimation errors compared to the more conventional ML approaches. 16 Particularly, Boosting Ensemble Classifiers have shown to outperform other classification techniques within breast mpMRI radiomics, for molecular subtypes recognition,17,18 prediction of sentinel lymph node metastasis, 19 and early prediction of treatment response and survival outcomes. 20 Additionally, their predictive efficiency for differentiating benign from malignant breast lesions has shown promise within DCE MRI radiomics alone in a recent study (AUC = 0.96), 21 but this have not yet been evaluated within mpMRI datatsets.
The current study sought to investigate the optimization of the aforementioned radiomics approaches in terms of evaluating the performance of four popular implementations of Decision Trees (DT) Boosting classifiers, namely Adaptive Boosting (AdaBoost), 22 Gradient Boosting (GB), 23 Extreme Gradient Boosting (XGBoost) 24 and Light Gradient Boosting Machine (LightGBM), 25 for breast cancer classification with mp-MRI radiomic features. A feature selection process based on the Boruta algorithm, Hierarchical Clustering (HC) on Spearman's rank correlation coefficients between features and Random Forest (RF) classification was adopted, for determining the all relevant and non-redundant feature subset, to improve the diagnostic efficiency of the classification models. For reference, an SVM classifier was also trained and evaluated on the same feature subset to allow performance comparisons, since this algorithm represents the current state-of-the-art ML method in breast mpMRI diagnostic radiomic models. To our knowledge this is the first study presenting the value of Ensemble Learning methods within multiparametric MRI radiomics for breast cancer classification.
Materials and Methods
Patient Cohort and MRI Acquisition
The reporting of this study conforms to the STROBE checklist, 26 according to the relevant Equator Network reporting guidelines (https://www.equator-network.org/reporting-guidelines/). This retrospective study was granted approval by the Internal Ethics Committee of the Department of Medicine of the University of Thessaly (approval number: 195). A sample of breast MRI data was obtained from a cohort of 293 female patients that have been consecutively examined in our institution the past five years and gave written informed consent for their participation in the study. The inclusion criteria were mass-like lesions detected on mammography and/or ultrasonography prior to any type of biopsy, with histological status verification from core needle biopsy or surgical excision, that was considered as the gold-standard of diagnosis. The exclusion criteria for this study were the receiving of neoadjuvant chemotherapy or radiation therapy, pregnancy/breastfeeding, presence of any implants or metallic clips from previous surgical procedures and generally contraindications to MRI scanning or to the administration of contrast agents. Breast lesions with a maximum diameter less than 1.0 cm in any direction were also excluded to reduce potential bias in radiomic feature measurements.
MR images were acquired on a 3.0 T MRI scanner (GE Healthcare, Signa HDx, Milwaukee, WI, USA) with patients placed in the prone position, using a dedicated phased array 8-channel breast coil. All patients underwent the same imaging protocol including conventional breast MRI with Dynamic Contrast Enhance and Diffusion Weighted Imaging. Each conventional MRI examination included scanning of the two breasts. Breast DCE-MRI protocol consisted of an axial T2-weighted Fast Spin Echo sequence (T2-FSE), (Repetition Time/Echo Time (TR/TE) = 3600/100 ms, flip angle = 90°, matrix size = 512 × 512, slice thickness = 4.0 mm), an axial Short Tau Inversion Recovery sequence (STIR), (TR/TE = 3900/90 ms, flip angle = 90°, matrix size = 512 × 512, slice thickness = 4.0 mm), and a 3D T1-weighted VIBRANT dynamic sequence with fat-suppression (TR/TE = 4.94/2.1 ms, flip angle = 10°, matrix size = 512 × 512, isotropic voxel size of 1 mm3) which was applied before and five times after the intravenous (IV) injection of a Gadolinium-based contrast agent with a 10 second timing delay, using an automatic injector system. The DWI protocol consisted of a DWI sequence which was acquired before injection of the contrast medium (TR/TE = 6000 ms/90 ms, flip angle = 90°, matrix size = 256 × 256, slice thickness = 4.0 mm, and b-values of 0 and 850 s/mm2.
Image Post-Acquisition Processing and Feature Extraction
The DCE-MRI volumes that were acquired 2-3 minutes after contrast agent administration and present the maximum enhancement between the different post-contrast time frames, were included in the analysis. Apparent Diffusion Coefficient (ADC) maps were calculated the from DWI images with two b-values (0 and 850 s/mm2) using the mono-exponential model fitting. Tumor contours in all consecutive slices in the three parametric datasets (DCE, T2-w, ADC), were manually drawn by a radiologist (20 years of experience), and the corresponding 3D volume masks of tumor masses were generated. Since the precision in tumor contouring may crucially affect the radiomic analysis, a second radiologist (23 years of experience) was recruited to provide independent segments for validation. The Dice coefficient implemented with an in-house python code was used to assess overlapping between segments. In case of segments with poor overlapping (Dice<0.85) a consensus between the two radiologists was reached for standardizing the delineation.
Radiomic feature extraction was implemented in Python 3.6 with the Pyradiomics library 27 which complies with the Imaging Biomarkers Standardization Initiative guidelines (IBSI). 28 Prior to feature extraction, outliers from pixel values distributions, as determined by the μ ± 3σ criterion, were excluded. Radiomic feature extraction was applied on the original parametric images without any filtering and 19 3D Shape-based, 16 First Order Statistics, 10 Gray Level Cooccurrence Matrix (GLCM), 24 Gray Leven Run Length Matrix (GLRLM), 16 Gray Level Size Zone Matrix (GLSZM), 5 Neighbouring Gray Tone Difference Matrix (NGTDM) and 14 Gray Level Dependence Matrix (GLDM) features were calculated, resulting in a total of 293 features for the whole imaging set of each subject. Since shape features’ calculation relies solely on imaging information of tumor margins and are independent of the whole tumor voxel intensity histogram, the extraction of shape features was applied only in DCE-MRI sequence, that presents an isotropic pixel spacing acquisition. In this way, we avoided to include redundant and misleading information regarding the tumor shape in the analysis. Additionally, the calculation of texture features utilized histogram binning with a fixed bin count of 64 bits-per-pixel for GLCM, GLDM, GLRLM and GLSZM features, and 32 bits-per-pixel for NGTDM features according to the Pyradiomics guidelines.
Statistical Analyses
A filter-based method was utilized to identify and exclude the non-informative features by assessing their individual discriminatory power. Specifically, univariate parametric (Student's t-test) or non-parametric (Mann-Whitney U-test) statistical tests (a = 0.05) were applied on each feature separately to assess statistically significantly differences between the corresponding distributions of benign and malignant cases. The selection of the appropriate statistical test was made according to the outcome of the Shapiro-Wilk test for normality (a = 0.05). With this filter-based approach we achieved to reduce the dimensionality of the initial feature space to increase the efficiency of the subsequent selection algorithm. A z-score transformation was applied to the remaining features, to standardize their values on the same scale.
Feature Selection
Feature selection processes were implemented in Python 3.6 using numpy [numpy.org], scipy [scipy.org], and scikit-learn [scikit-learn.org] libraries, and the Boruta_py package obtained from [https://github.com/scikit-learn-contrib/boruta_py]. The graphics were generated with the matplotlib library. 29
The Boruta algorithm which is a wrapper method around a Random Forest (RF) classifier was implemented for the feature selection process. 30 In principle, the Boruta method, generates artificial features by shuffling the original feature values across subjects. The original and artificial features are combined and evaluated with the RF classifier. Finally, the importance of artificial features is used as reference for selecting original features according to the RF permutation importance measure. By default, the Boruta algorithm generates two subsets of relevant features, one presenting high and the other intermediate importance, respectively. Here, only the subset of highly important features was kept for further analysis. However, as is the case with the most wrapper feature selection techniques, Boruta does not handle feature multicollinearity, thus redundant information tends to be present in the final subset, and this might compromise the subsequent classification performance. Two previous tudies have utilized additional steps of collinearity analysis to identify and exclude redundant features.31,32 In the specific implementation adopted here, Hierarchical Clustering with Ward's linkage method was applied to a cross-correlation matrix of Spearman's rank correlation coefficients, between the highly important features selected with Boruta. In each cluster of features with high dependency, defined as those having Spearman rho values above 0.6, a new RF classifier was applied to rank the within-cluster feature importance, according to the RF Gini's Index. The most important feature per cluster was retained to form the final feature subset.
Classification Modelling and Evaluation
Python implementations for XGBoost and LightGBM were obtained from their original sources [https://github.com/dmlc/xgboost], [https://github.com/microsoft/LightGBM] and used through the scikit-learn Application Programming Interface (API), which is a common framework for ML applications. 33
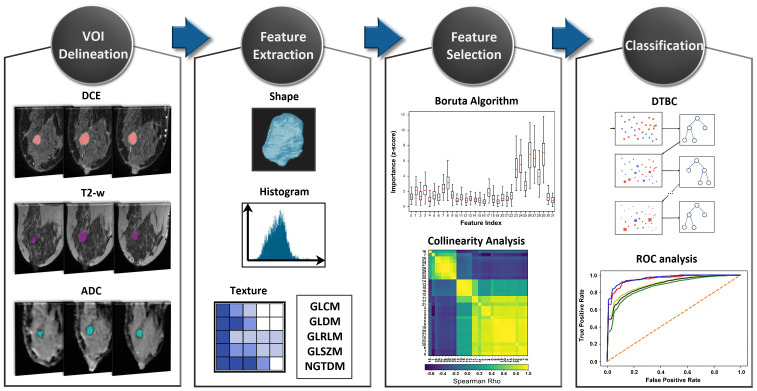
The final feature subset was used to train the GB, AdaBoost, XGBoost, LightGBM and SVM classifiers in differentiating benign from malignant breast lesions. All four boosting classification algorithms have shared the same hyperparameters, ie number of trees = 1000, max depth = 3, learning rate = 0.1 and the ‘early stopping’ option enabled. SVM classifier was built with scikit-learn library default hyperparameters, ie, Radial Basis Kernel, ’scale’ option for kernel coefficient gamma, and regularization parameter C = 1. The ‘.632 + bootstrap’ validation method 34 as implemented in mlxtend python package (http://rasbt.github.io/mlxtend/) and Receiver Operation Characteristic (ROC) analysis were employed to validate the models’ performance and obtain the Area Under the Curve (AUC) evaluation metric. The resulting classification scores of Accuracy (Acc), Sensitivity (Se), Specificity (Sp) and AUC were averaged across 300 bootstraps. Additionally, the DeLong's test was utilized to identify pairwise statistically significant differences between the AUC values of the models. 35 The complete workflow of the radiomic analysis implemented in this study is presented in Figure 1.
Figure 1.
The radiomic analysis workflow.
Results
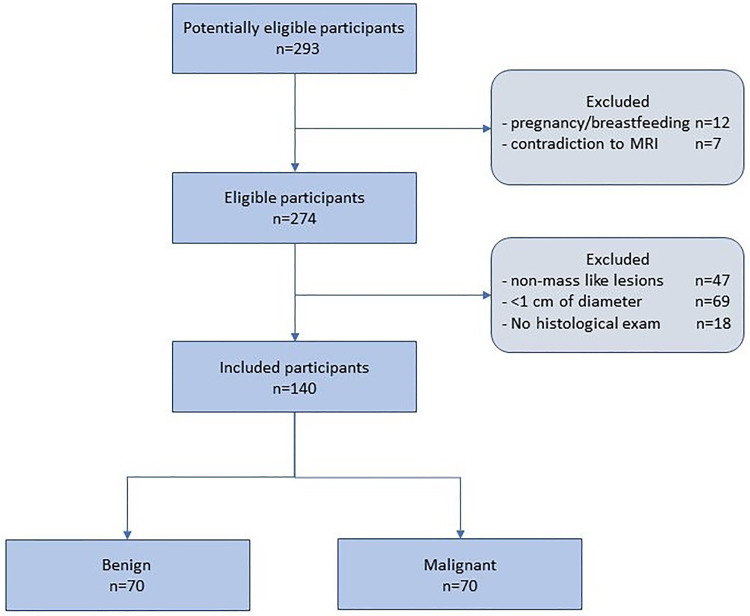
From the initially available patient cohort, a sample of 140 patients with all required data available that also met the inclusion criteria, was included, as shown in the flow diagram (Figure 2). In case where multiple lesions were present in the same or the opposite breast, only a single lesion per subject was selected, finally conforming a balanced dataset of 70 benign and 70 malignant lesions that was considered for the analysis.
Figure 2.
Flow diagram of the study participants selection.
Demographic and clinical characteristics of the sample utilized are presented in Table 1. Specifically, the mean age was 44.6 ± 11.8 for the benign and 57.4 ± 12.5 for the malignant cases. The mean volume size was 1.8 ± 1.6 cm3 and 4.0 ± 2.4 cm3 for benign and malignant lesions, respectively. Benign lesions were of type Fibroadenomas according to the histopathology examination. The radiologic assessment had assigned 48/70 lesions to type 2, 19/70 to type 3 and 3/70 to type 4 according to the MRI Breast Imaging Reporting and Data System (BI-RADS) categorization of the American College of Radiology (ACR). 36 The malignant lesion sample consisted of 53 Invasive Ductal Carcinomas (IDCs,) 12 Invasive Lobular Carcinomas (ILCs), 4 Ductal Carcinoma In-Situ (DCIS) and 1 Lobular Carcinoma In-Situ (LCIS), while 10/70 were found to be of histological grade I, 39/70 of histological grade II and 21/70 of histological grade III. The radiologic assessment had assigned 14/70 and 56/70 to MRI BI-RADS categories 4 and 5, respectively.
Table 1.
Demographic and Clinical Characteristics of the Patient Sample.
| Benign | Malignant | |
|---|---|---|
| Patients (N) | 70 females (50%) | 70 females (50%) |
| Lesions (N) | 70 (50%) | 70 (50%) |
| Age (mean ± std) | 44.6 ± 11.8 | 57.4 ± 12.5 |
| Volume in cm3 (mean ± std) | 1.8 ± 1.6 | 4.0 ± 2.4 |
| Histological type (N) | ||
| FA | 70 (100%) | |
| IDC | 53 (76%) | |
| ILC | 12 (17%) | |
| DCIS | 4 (6%) | |
| LCIS | 1 (1%) | |
| Histological grade | ||
| I | 10 (14%) | |
| II | 39 (56%) | |
| III | 21 (30%) | |
| MRI BI-RADS categories | ||
| 2 | 48 (69%) | |
| 3 | 19 (27%) | |
| 4 | 3 (4%) | 14 (20%) |
| 5 | 56 (80%) |
FA, Fibroadenoma; IDC, Invasive Ductal Carcinoma; ILC, Invasive Lobular Carcinoma; DCIS, Ductal Carcinoma In-Situ; LCIS, Lobular Carcinoma In-Situ
Manual segmentations of the lesions had presented a high overlapping among the two radiologists (raters), denoted by an average Dice coefficient of 0.88 ± 0.09.
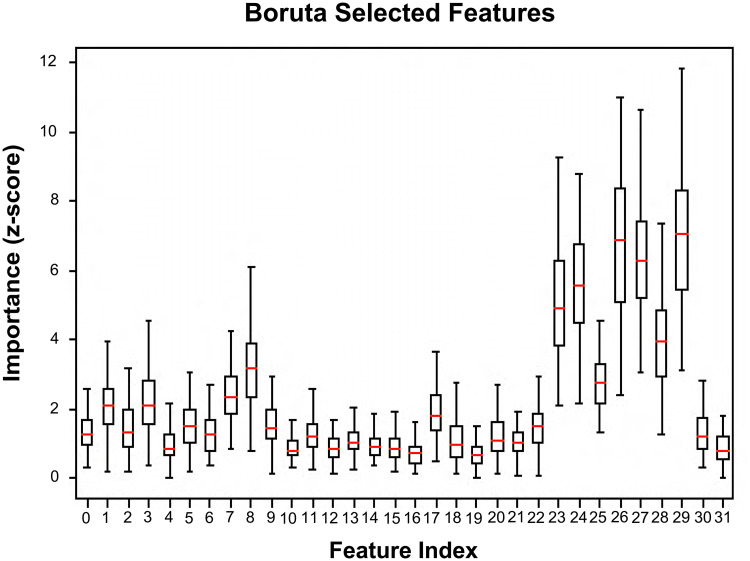
The parametric and non-parametric statistical tests have revealed 82 out of 293 total features to be non-informative (p > 0.05) and these were excluded from the analysis. Out of the remaining 211 features, the Boruta algorithm have nominated 32 highly important features, ie, 10 from shape, 8 from DCE histogram, 5 from DCE texture, 8 from ADC histogram and 1 from ADC texture presented in Table 2. Interestingly, we observed that T2-w based features were totally absent from this highly important feature subset, while ADC histogram features were the most important among the remaining features (z-scores = 1.3 ± 0.60-7.0 ± 2.00). Figure 3 presents boxplots of each feature's importance (z-score) distribution obtained from the RF permutations.
Table 2.
Boruta Selected Features.
| Index | Feature name | Importance (mean ± std) | Index | Feature name | Importance (mean ± std) |
|---|---|---|---|---|---|
| 0 | original_shape_LeastAxisLength | 1.43 ± 0.76 | 16 | DCE_original_firstorder_Skewness | 0.72 ± 0.35 |
| 1 | original_shape_Maximum2DDiameterColumn | 2.14 ± 0.80 | 17 | DCE_original_firstorder_TotalEnergy | 1.81 ± 0.70 |
| 2 | original_shape_Maximum2DDiameterRow | 1.52 ± 0.82 | 18 | DCE_original_gldm_DependenceNonUniformity | 1.06 ± 0.58 |
| 3 | original_shape_Maximum2DDiameterSlice | 2.22 ± 1.10 | 19 | DCE_original_glrlm_GrayLevelNonUniformity | 0.70 ± 0.44 |
| 4 | original_shape_Maximum3DDiameter | 0.94 ± 0.54 | 20 | DCE_original_glrlm_RunLengthNonUniformity | 1.35 ± 0.60 |
| 5 | original_shape_MeshVolume | 1.65 ± 0.63 | 21 | DCE_original_glszm_GrayLevelNonUniformity | 1.12 ± 0.48 |
| 6 | original_shape_MinorAxisLength | 1.38 ± 0.58 | 22 | DCE_original_glszm_SizeZoneNonUniformity | 1.55 ± 0.49 |
| 7 | original_shape_Sphericity | 2.46 ± 0.72 | 23 | ADC_original_firstorder_10Percentile | 4.91 ± 1.75 |
| 8 | original_shape_SurfaceArea | 3.51 ± 1.37 | 24 | ADC_original_firstorder_90Percentile | 5.36 ± 1.58 |
| 9 | original_shape_VoxelVolume | 1.50 ± 0.58 | 25 | ADC_original_firstorder_Maximum | 2.74 ± 0.87 |
| 10 | DCE_original_firstorder_90Percentile | 0.82 ± 0.36 | 26 | ADC_original_firstorder_Mean | 6.65 ± 2.18 |
| 11 | DCE_original_firstorder_Energy | 1.33 ± 0.61 | 27 | ADC_original_firstorder_Median | 6.22 ± 1.59 |
| 12 | DCE_original_firstorder_Maximum | 0.89 ± 0.40 | 28 | ADC_original_firstorder_Minimum | 3.86 ± 1.41 |
| 13 | DCE_original_firstorder_Mean | 0.94 ± 0.44 | 29 | ADC_original_firstorder_RootMeanSquared | 7.10 ± 1.98 |
| 14 | DCE_original_firstorder_Median | 0.90 ± 0.37 | 30 | ADC_original_firstorder_TotalEnergy | 1.26 ± 0.59 |
| 15 | DCE_original_firstorder_RootMeanSquared | 0.87 ± 0.43 | 31 | ADC_original_ngtdm_Busyness | 0.67 ± 0.45 |
Figure 3.
Feature importance in terms of z-score distributions of RF permutations for the highly important feature subset nominated by Boruta.
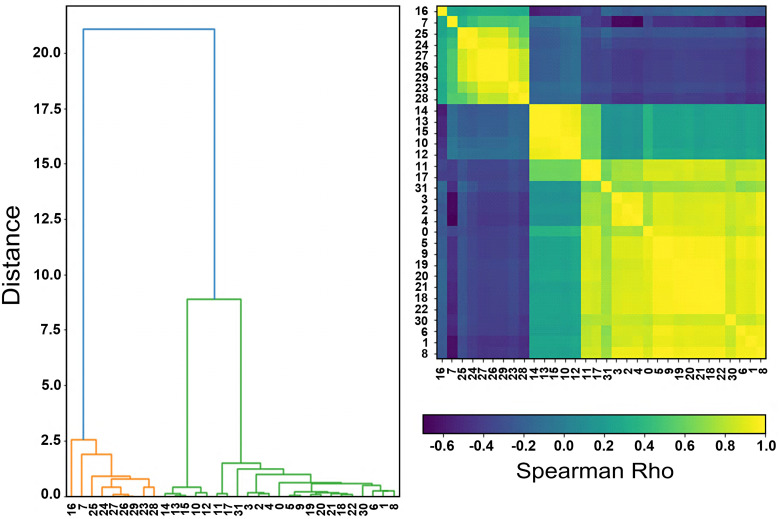
The pairwise Spearman's rank correlation coefficients have revealed several statistical dependencies (rho = 0.60 to 0.98), especially between shape and texture features of DCE and DWI (Figure 4). Histogram features have shown smaller correlation values with either shape or textural features. The second step of Boruta selected subsets elimination with HC and RF importance resulted in the determination of a minimum subset, consisting of 5 features, namely Sphericity, Surface Area, DCE_median, DCE_skewness, ADC_mean.
Figure 4.
The Hierarchical Clustering dendrogram (a) illustrating clusters arrangement as informed by the correlation plot (b) of Boruta selected features.
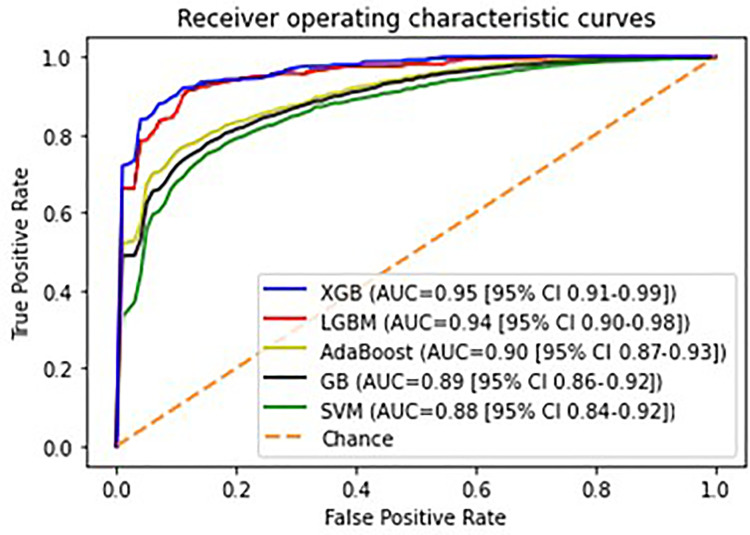
Average classification metrics of the different models, in terms of mean and 95% Confidence Intervals (CI) across the bootstrap validation subsamples, are presented in Table 3. Regarding Boosting Ensemble methods, it was observed that XGboost achieved the highest accuracy (Acc = 0.88 [95% CI 0.84-0.92]) and overall performance (AUC = 0.95 [95% CI 0.91-0.99]) followed by LGBM (Acc = 0.87 [95% CI 0.83-0.91]/ AUC = 0.94 [95% CI 0.90-0.98]), Adaboost (Acc = 0.83 [95% CI 0.80-0.86] / AUC = 0.90 [95% CI 0.87-0.93]), and GB (Acc = 0.83 [95% CI 0.80-0.86] / AUC = 0.89 [95% CI 0.86-0.92]). According to the pairwise statistical comparisons that were performed between the AUC values (Table 4), the observed interindividual differences in overall performances of XGBoost and LGBM were statistically significantly higher than AdaBoost and GB. The SVM classification model has yielded statistically significantly lower performance (Acc = 0.84 [95% CI 0.80-0.88] / AUC = 0.88 [95% CI 0.84-0.92]) than XGBoost and LGBM, but this was found statistically comparable with the performances demonstrated by AdaBoost and GB (Table 4). XGBoost has also achieved the highest sensitivity (Se = 0.91 [95% CI 0.85-0.97]) and specificity (Sp = 0.90 [95% CI 0.82-0.98]). Sensitivity and specificity metrics for the rest of the classification models were: LGBM Se = 0.90 [95% CI 0.84–0.96] / Sp = 0.89 [95% CI 0.81–0.97], AdaBoost Se = 0.83 [95% CI 0.78–0.88] / Sp = 0.82 [95% CI 0.75–0.89], GB Se = 0.82 [95% CI 0.77–0.87] / Sp = 0.80 [95% CI 0.73–0.87], SVM Se = 0.80 [95% CI 0.77–0.88] / Sp = 79 [95% CI 0.70–0.88]. Figure 5 presents the ROC curves plots of the classification models.
Table 3.
Classification Model Results (Mean [95% Confidence Intervals]).
| XGBoost | LGBM | AdaBoost | GB | SVM | |
|---|---|---|---|---|---|
| Acc | 0.88 [0.84-0.92] | 0.87 [0.83-0.91] | 0.83 [0.80-0.86] | 0.83 [0.80-0.86] | 0.84 [0.80-0.88] |
| Se | 0.91 [0.85-0.97] | 0.90 [0.84-0.96] | 0.83 [0.78-0.88] | 0.82 [0.77-0.87] | 0.80 [0.77-0.88] |
| Sp | 0.90 [0.82-0.98] | 0.89 [0.81-0.97] | 0.82 [0.75-0.89] | 0.80 [0.73-0.87] | 0.79 [0.70-0.88] |
| AUC | 0.95 [0.91-0.99] | 0.94 [0.90-0.98] | 0.90 [0.87-0.93] | 0.89 [0.86-0.92] | 0.88 [0.84-0.92] |
Table 4.
P-values of the Pairwise Statistical Comparisons of the Classification Models AUC Values Derived From DeLong's Test.
| XGBoost | LGBM | AdaBoost | GB | SVM | |
|---|---|---|---|---|---|
| XGBoost | 0.77 | 0.029 | 0.022 | 0.017 | |
| LGBM | 0.032 | 0.026 | 0.020 | ||
| AdaBoost | 0.93 | 0.71 | |||
| GB | 0.81 | ||||
| SVM |
Figure 5.
Receiver operating characteristic (ROC) curves of the classification models.
Discussion
In this study we investigate the utility of a novel radiomic analysis pipeline, based on Ensemble Learning, for increasing the mpMRI predictive capability towards differentiation of benign and malignant breast lesions. A feature selection process, based on Random Forest classification with the Boruta wrapper and hierarchical clustering on Sprearman's rank correlation coefficients has nominated 5 radiomic features from shape, DCE and DWI as being the most important for breast cancer differentiation. Four DT Boosting Ensembles were evaluated with bootstrapping validation and their performances were compared with an SVM classification model. XGBoost and LGBM achieved statistically significantly higher AUC values, compared to the performances of the rest of the methods. Overall, this study provides novel evidence regarding the robustness of the newer implementations of Boosting ensemble classification methods, which hold potential to enhance breast cancer precision medicine with minimum invasive approaches.
Differentiation of benign and malignant breast lesions is an important step for breast cancer management, since it determines the therapeutic plan that the clinicians will follow, ranging from active surveillance to chemotherapy/radiotherapy and tumor excision. 2 MRI of the breast has been increasingly recognized as a powerful diagnostic tool. The American College of Radiology has established the BI-RADS MRI lexicon that provides standardized assessment and reporting of MRI findings and a classification system to determine the probability of malignancy and biopsy recommendations. Currently, MRI BI-RADS incorporates morphological and functional descriptors for DCE-MRI and T2-w, which constitute the typical MRI protocol. 3 However, due to an overlap in imaging descriptors between benign and malignant tumors the standard MRI protocol presents moderate specificity, thus many BI-RADS 4 and 5 biopsies might be misdiagnosed. 5 Notably, the incorporation of DWI ADC mapping has been shown to significantly improve specificity which may reduce unnecessary biopsies and invasive diagnostic procedures. 37 In a recent meta-analysis of 22 studies, it was shown that the pooled specificity increased to 0.85 with the inclusion of DWI parameters compared to the pooled specificity of 0.71 for the DCE-MRI alone. 38
Previous studies have demonstrated that breast mpMRI radiomic features, representing a quantitative description of a specific geometrical or physical property of the image, hold great potential for overcoming the caveats of the subjective qualitative radiologic assessment. 39 Indeed, these features derived from tumor shape, texture, kinetics, etc, encode both simple patterns within medical images but also many higher order patterns not apparent to the human eye. 40 More importantly, radiomic features can be input to supervised machine learning models that hold diagnostic and predictive power for automating the quantitative evaluation towards further reduction of the false positive findings.12,41 However, not all of the extracted features are important, therefore a feature selection technique is needed within radiomics analysis to determine the most tumor subtype discriminative and biologically relevant features to construct robust classification models. 8
In this study we have used Boruta, which is a wrapper algorithm around a Random Forest classifier. The highly important feature subset nominated by Boruta consisted of shape, histogram and textural features from DCE and ADC. Notably, ADC features were the most important for the specific classification task, while none of T2-w features were found to participate in the selected subset. We consider this an important finding, in line with previous evidence expanding the findings of conventional evaluation to radiomic analysis, where quantitative measurements of ADC maps were reported to have more impact on classification performance than those of T2-w.12,42 Generally, Boruta has proven to be the most robust all-relevant feature selection strategy in gene selection, 43 while very recently this algorithm has been successfully used in radiomics studies,31,32 but not yet in breast MRI diagnosis. Additionally, in a recent radiogenomics study 44 it was shown to outperform other robust feature selection ensemble methods, such as the Minimum Redundancy Maximum Relevance algorithm.
Since Boruta is an all-relevant feature selection method, we additionally implemented steps of Hierarchical Clustering on Spearman's rank correlation coefficients and Random Forest classification as published elsewhere,31,32 to exclude redundant features and form the final feature subset for classification. Interestingly, shape features were found highly correlated with texture features and outperformed them, thus, only shape and first order histogram features were included in the final subset.
Boosting models’ training with the specific radiomic signature have demonstrated high performances of XGBoost (AUC = 0.95) and LGBM (AUC = 0.94) algorithms in benign versus malignant breast lesions differentiation. Of note, the AUC values between the two models were not found statistically different, however, despite the apparent algorithmic similarities, these models may present substantial variations in classification accuracy, as well as processing time, in larger datasets. 25 Adaboost and GB were found to have significantly lower AUC values of 0.90 and 0.89, respectively. Generally, these findings come in agreement with a growing body of recent literature, suggesting the unique efficiency of Boosting ensembles either for classifying conventional radiomic features,38,39 imaging features extracted from Convolutional Neural Networks (CNN),20,45 or both. 46 Additionally, they are adding more credence to the existing reports within various breast MRI radiomics paradigms.17–20 Regarding breast MRI diagnosis, in a previously proposed CADx based on ensemble methods for feature selection and classification, AdaBoost has achieved a high performance (AUC = 0.96) in differentiating malignant and benign lesions by means of DCE MRI radiomic features. 21 Beyond the exploitation of ensemble learning methods, the authors have also made use of wavelet features. As it was previously demonstrated, the wavelet transformation of medical images holds potential for capturing various spatial frequency texture patterns within heterogeneous breast lesion regions. 47 This might partially explain the high performance observed in their study, although the model was evaluated on DCE-MRI alone and utilized the AdaBoost algorithm which has achieved a significantly lower performance in our study. Besides, the additional use of the DWI sequence in our study has resulted in the adoption of a minimum set of clinically perceivable radiomic features which facilitates the enhanced interpretability of the specific diagnostic model.
Additionally, XGBoost and LGBM models were found to outperform the SVM classifier (AUC = 0.88), which has been a commonly utilized ML classification strategy in breast MRI radiomics. Specifically, in the studies of Damiel Naranjo et al., 9 Parekh et al. 10 and Hu et al., 11 SVM classifiers were utilized to differentiate breast lesions over mpMRI datasets consisting of DCE, DWI and T2-w images, with all models achieving similar performances of AUC = 0.85, AUC = 0.87 and AUC = 0.87, respectively. Cai et al. 13 have compared four different classification algorithms, ie, SVM, Naïve Bayes, k-Nearest Neighbours, and Logistic Regression. SVM has demonstrated a high performance (AUC = 0.91), although this was not found significantly different from the other classifiers utilized in their study. Further evidence regarding SVM capability for breast cancer diagnosis has been presented by Zhang et al., 12 where their model achieved a performance of 0.92. However, it is worth to mention that the authors have utilized advanced pharmacokinetic parameters of DCE MRI and Diffusion Kurtosis Imaging (DKI) in their radiomic signature, while in the present study a more conventional mpMRI protocol was available, and thus we might didn’t observed such a high performance for the SVM model. Considering the above-mentioned findings and with respect to any particular methodological differences between the studies, it is evident that ensemble learning classification holds great potential for overcoming the limited efficacy of conventional ML models towards increasing the breast mpMRI diagnostic accuracy.
Our study has some limitations. Specifically, only one mpMRI dataset of restricted size was available to train and test our classification models, while an additional external independent validation dataset would allow to evaluate their generalization on new ‘unseen’ data. Additionally, no power calculation for estimating the sample size selected for the study was done. Since this was an exploratory analysis investigating the utility of Boosting algorithms in breast cancer classification, reproducibility analysis of segmentation and feature extraction procedures was not performed. Furthermore, due to the abbreviated nature of the mpMRI protocol in our institution, the inclusion of DCE kinetic features or other MRI sequences (eg Diffusion Tensor Imaging, Diffusion Kurtosis Imaging, MR Spectroscopy, etc), previously demonstrated to have a significant impact in breast cancer diagnosis,48–50 was not feasible.
Conclusion
In conclusion, mpMRI of the breast holds potential for accurate differentiation of benign and malignant breast lesions, to reduce invasive diagnostic procedures. The integration of Ensemble Learning methods within mpMRI radiomics could provide valuable precise quantification of the diagnostic information and identify while reducing the subjective reader interpretation errors.
Acknowledgements
Professor Marianna Vlychou is highly acknowledged for contributing to the validation of the manual segmentation process.
Abbreviations
- 3D
Three-dimensional
- Acc
Accuracy
- ACR
American College of Radiology
- AdaBoost
Adaptive Boosting classifier
- ADC
Apparent Diffusion Coefficient
- API
Application Programming Interface
- AUC
Area Under Curve
- BI-RADS
Breast Imaging Reporting and Data System
- CAD
Computer Assisted Diagnosis
- CI
Confidence Interval
- CNN
Convolutional Neural Networks
- DCE
Dynamic Contrast Enhanced
- DCIS
Ductal Carcinoma In-Situ
- DKI
Diffusion Kurtosis Imaging
- DT
Decision Trees
- DWI
Diffusion Weighted Imaging
- FA
Fibroadenoma
- FSE
Fast Spin Echo
- GB
Gradient Boosting
- GLCM
Gray Level Cooccurrence Matrix
- GLDM
Gray Level Dependence Matrix
- GLRLM
Gray Level Run Length Matrix
- GLSZM
Gray Level Size Zone Matrix
- HC
Hierarchical Clustering
- IBSI
Imaging Biomarkers Standardization Initiative
- IDC
Invasive Ductal Carcinoma
- ILC
Invasive Lobular Carcinoma
- LCIS
Lobular Carcinoma In-Situ
- LightGBM
Light Gradient Boosting Machine
- ML
Machine Learning
- MRI
Magnetic Resonance Imaging
- mpMRI
multi-parametric Magnetic Resonance Imaging
- NGTDM
Neighbouring Gray Tone Difference Matrix
- RF
Random Forest
- ROC
Receiver Operation Characteristic Curve
- Se
Sensitivity
- Sp
Specificity
- STIR
Short Tau Inversion Recovery
- SVM
Support Vector Machines
- T2-w
T2 weighted imaging
- TE
Echo Time
- TR
Repetition Time
- XGBoost
Extreme Gradient Boosting
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics Statement: This study was approved by the Internal Ethics Committee of the Department of Medicine of the University of Thessaly (Approval Number: 195).
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article:This research is co-financed by Greece and the European Union (European Social Fund-ESF) through the Operational Programme (Human Resources Development, Education and Lifelong Learning 2014-2020) in the context of the project “Breast cancer assessment through advanced multiparametric imaging techniques and development of differential diagnosis software using artificial intelligence systems” [MIS5048948].
ORCID iD: Alexandros Vamvakas https://orcid.org/0000-0002-4766-8564
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Sheth D, Giger ML. Artificial intelligence in the interpretation of breast cancer on MRI. J Magn Reson Imaging. 2020;51(5):1310-1324. doi: 10.1002/jmri.26878 [DOI] [PubMed] [Google Scholar]
- 3.Zhang M, Horvat JV, Bernard-Davila B, et al. Multiparametric MRI model with dynamic contrast-enhanced and diffusion-weighted imaging enables breast cancer diagnosis with high accuracy. J Magn Reson Imaging. 2019;49(3):864-874. doi: 10.1002/jmri.26285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marino MA, Helbich T, Baltzer P, Pinker-Domenig K. Multiparametric MRI of the breast: a review. J Magn Reson Imaging. 2018;47(2):301-315. doi: 10.1002/jmri.25790 [DOI] [PubMed] [Google Scholar]
- 5.Pötsch N, Dietzel M, Kapetas P, et al. An A.I. classifier derived from 4D radiomics of dynamic contrast-enhanced breast MRI data: potential to avoid unnecessary breast biopsies. Eur Radiol. 2021;31(8):5866-5876. doi: 10.1007/s00330-021-07787-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chitalia RD, Rowland J, McDonald ES, et al. Imaging phenotypes of breast cancer heterogeneity in preoperative breast dynamic contrast enhanced magnetic resonance imaging (DCE-MRI) scans predict 10-year recurrence. Clin Cancer Res. 2020;26(4):862-869. doi: 10.1158/1078-0432.CCR-18-4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forghani R, Savadjiev P, Chatterjee A, Muthukrishnan N, Reinhold C, Forghani B. Radiomics and artificial intelligence for biomarker and prediction model development in oncology. Comput Struct Biotechnol J. 2019;17:995-1008. doi: 10.1016/j.csbj.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castiglioni I, Gallivanone F, Soda P, et al. AI-based applications in hybrid imaging: how to build smart and truly multi-parametric decision models for radiomics. Eur J Nucl Med Mol Imaging. 2019;46(13):2673-2699. doi: 10.1007/s00259-019-04414-4 [DOI] [PubMed] [Google Scholar]
- 9.Daimiel Naranjo I, Gibbs P, Reiner JSet al. Radiomics and machine learning with multiparametric breast MRI for improved diagnostic accuracy in breast cancer diagnosis. Diagnostics. 2021;11(6):919. 10.3390/diagnostics11060919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parekh VS, Jacobs MA. Multiparametric radiomics methods for breast cancer tissue characterization using radiological imaging. Breast Cancer Res Treat. 2020;180(2):407-421. doi: 10.1007/s10549-020-05533-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Q, Whitney HM, Giger ML. Radiomics methodology for breast cancer diagnosis using multiparametric magnetic resonance imaging. J Med Imaging (Bellingham). 2020;7(4):044502. doi: 10.1117/1.JMI.7.4.044502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Peng Y, Liu W, et al. Radiomics based on multimodal MRI for the differential diagnosis of benign and malignant breast lesions. J Magn Reson Imaging. 2020;52(2):596-607. doi: 10.1002/jmri.27098 [DOI] [PubMed] [Google Scholar]
- 13.Cai H, Peng Y, Ou C, Chen M, Li L. Diagnosis of breast masses from dynamic contrast-enhanced and diffusion-weighted MR: a machine learning approach. PLoS One. 2014;9(1):e87387. doi: 10.1371/journal.pone.0087387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X, Xie F, Liu L, Peng Y, Cai H, Li L. Discrimination of malignant and benign breast masses using automatic segmentation and features extracted from dynamic contrast-enhanced and diffusion-weighted MRI. Oncol Lett. 2018;16(2):1521-1528. doi: 10.3892/ol.2018.8805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunese L, Mercaldo F, Reginelli A, Santone A. An ensemble learning approach for brain cancer detection exploiting radiomic features. Comput Methods Programs Biomed. 2020 Mar;185:105134. doi: 10.1016/j.cmpb.2019.105134. Epub 2019 Oct 22. PMID: 31675644. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Liu B, Peng S, Sun J, Qiao X. Computer-aided grading of gliomas combining automatic segmentation and radiomics. Int J Biomed Imaging. 2018;2018:2512037. doi: 10.1155/2018/2512037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Yu K, Feng C, Zhao D. Molecular subtypes recognition of breast cancer in dynamic contrast-enhanced breast magnetic resonance imaging phenotypes from radiomics data. Comput Math Methods Med. 2019;2019:6978650. doi: 10.1155/2019/6978650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng W, Sun Y, Qian H, et al. Computer-aided diagnosis evaluation of the correlation between magnetic resonance imaging with molecular subtypes in breast cancer. Front Oncol. 2021;11:693339. doi: 10.3389/fonc.2021.693339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Sun D, Chen L, et al. Radiomics analysis of dynamic contrast-enhanced magnetic resonance imaging for the prediction of sentinel lymph node metastasis in breast cancer. Front Oncol. 2019;9:980. doi: 10.3389/fonc.2019.00980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tahmassebi A, Wengert GJ, Helbich TH, et al. Impact of machine learning with multiparametric magnetic resonance imaging of the breast for early prediction of response to neoadjuvant chemotherapy and survival outcomes in breast cancer patients. Invest Radiol. 2019;54(2):110-117. doi: 10.1097/RLI.0000000000000518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu W, Li Z, Chu J. A novel computer-aided diagnosis system for breast MRI based on feature selection and ensemble learning. Comput Biol Med. 2017;83:157-165. doi: 10.1016/j.compbiomed.2017.03.002 [DOI] [PubMed] [Google Scholar]
- 22.Freund Y, Schapire R. A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting. 1997;COLT 1997.
- 23.Mason L, Baxter J, Bartlett P, et al. Boosting algorithms as gradient descent in function space. 1999; In Proc. NIPS (Vol. 12, pp. 512–518).
- 24.Chen T, He T, Benesty M, et al. Xgboost: extreme gradient boosting. R package version. 0.4–2. 2015;1(4):1-4. [Google Scholar]
- 25.Ke G, Meng Q, Finley T, et al. Lightgbm: a highly efficient gradient boosting decision tree. Adv Neural Inf Process Syst. 2017;30:3146-3154. [Google Scholar]
- 26.von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med. 2007;147:573-577. [DOI] [PubMed] [Google Scholar]
- 27.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104-e107. doi: 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2):328-338. doi: 10.1148/radiol.2020191145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007;9(3):90-95. [Google Scholar]
- 30.Kursa M, Rudnicki W. Feature selection with the Boruta package. J Stat Softw. 2010;36:1-13. [Google Scholar]
- 31.Laukamp KR, Shakirin G, Baeßler B, et al. Accuracy of radiomics-based feature analysis on multiparametric magnetic resonance images for noninvasive meningioma grading. World Neurosurg. 2019;132:e366-e390. doi: 10.1016/j.wneu.2019.08.148 [DOI] [PubMed] [Google Scholar]
- 32.Kimura K, Yoshida S, Tsuchiya J, et al. Usefulness of texture features of apparent diffusion coefficient maps in predicting chemoradiotherapy response in muscle-invasive bladder cancer. Eur Radiol. 2022;32(1):671-679. doi: 10.1007/s00330-021-08110-6 [DOI] [PubMed] [Google Scholar]
- 33.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825-2830. [Google Scholar]
- 34.Efron B, Tibshirani R. Improvements on cross-validation: the .632 + bootstrap method. J Am Stat Assoc. 1997;92(438):548. doi: 10.2307/2965703 [DOI] [Google Scholar]
- 35.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837-845. [PubMed] [Google Scholar]
- 36.Morris EA, Comstock CE, Lee CH, et al. ACR BI-RADS® Magnetic Resonance Imaging. In: ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System. Reston, VA. American College of Radiology. 2013.
- 37.Clauser P, Krug B, Bickel H, et al. Diffusion-weighted imaging allows for downgrading MR BI-RADS 4 lesions in contrast-enhanced MRI of the breast to avoid unnecessary biopsy. Clin Cancer Res. 2021;27(7):1941-1948. doi: 10.1158/1078-0432.CCR-20-3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu CR, Chen KY, Li P, Xia ZY, Wang B. Accuracy of multiparametric MRI in distinguishing the breast malignant lesions from benign lesions: a meta-analysis. Acta Radiol. 2021;62(10):1290-1297. 10.1177/0284185120963900 [DOI] [PubMed] [Google Scholar]
- 39.Tagliafico AS, Piana M, Schenone D, Lai R, Massone AM, Houssami N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast. 2020;49:74-80. doi: 10.1016/j.breast.2019.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gullo R L, Eskreis-Winkler S, Morris EA, Pinker K. Machine learning with multiparametric magnetic resonance imaging of the breast for early prediction of response to neoadjuvant chemotherapy. Breast. 2020;49:115-122. doi: 10.1016/j.breast.2019.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer-Base A, Morra L, Tahmassebi A, Lobbes M, Meyer-Base U, Pinker K. AI-enhanced diagnosis of challenging lesions in breast MRI: a methodology and application primer. J Magn Reson Imaging. 2021;54(3):686-702. doi: 10.1002/jmri.27332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalmiş MU, Gubern-Mérida A, Vreemann S, et al. Artificial intelligence-based classification of breast lesions imaged with a multiparametric breast MRI protocol With ultrafast DCE-MRI, T2, and DWI. Invest Radiol. 2019;54(6):325-332. doi: 10.1097/RLI.0000000000000544 [DOI] [PubMed] [Google Scholar]
- 43.Li ZC, Zhai G, Zhang J, et al. Differentiation of clear cell and non-clear cell renal cell carcinomas by all-relevant radiomics features from multiphase CT: a VHL mutation perspective. Eur Radiol. 2019;29(8):3996-4007. doi: 10.1007/s00330-018-5872-6 [DOI] [PubMed] [Google Scholar]
- 44.Sakai Y, Yang C, Kihira S, et al. MRI Radiomic features to predict IDH1 mutation status in gliomas: a machine learning approach using gradient tree boosting. Int J Mol Sci. 2020;21(21):8004. doi: 10.3390/ijms21218004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koyasu S, Nishio M, Isoda H, Nakamoto Y, Togashi K. Usefulness of gradient tree boosting for predicting histological subtype and EGFR mutation status of non-small cell lung cancer on 18F FDG-PET/CT. Ann Nucl Med. 2020;34(1):49-57. doi: 10.1007/s12149-019-01414-0 [DOI] [PubMed] [Google Scholar]
- 46.Fangoh AM, Selim S. "Using CNN-XGBoost Deep Networks for COVID-19 Detection in Chest x-ray Images," 2020 15th International Conference on Computer Engineering and Systems (ICCES), 2020, pp. 1-7, doi: 10.1109/ICCES51560.2020.9334600. [DOI]
- 47.Chitalia RD, Kontos D. Role of texture analysis in breast MRI as a cancer biomarker: a review. J Magn Reson Imaging. 2019;49(4):927-938. doi: 10.1002/jmri.26556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang YQ, Cao SE, Cao S, et al. Preoperative identification of microvascular invasion in hepatocellular carcinoma by XGBoost and deep learning. J Cancer Res Clin Oncol. 2021;147(3):821-833. doi: 10.1007/s00432-020-03366-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsougos I, Vamvakas A, Kappas C, Fezoulidis I, Vassiou K. Application of radiomics and decision support systems for breast MR differential diagnosis. Comput Math Methods Med. 2018;2018:7417126. doi: 10.1155/2018/7417126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vamvakas A, Vassiou K, Tsivaka D, Tsougos I. Decision support systems in breast cancer. 2020. In Precision Medicine for Investigators, Practitioners and Providers (pp. 319–327). Academic Press.