Abstract
Clinical isolates of Staphylococcus aureus (a total of 206) and S. epidermidis (a total of 188) from various countries were tested with multiplex PCR assays to detect clinically relevant antibiotic resistance genes associated with staphylococci. The targeted genes are implicated in resistance to oxacillin (mecA), gentamicin [aac(6′)-aph(2")], and erythromycin (ermA, ermB, ermC, and msrA). We found a nearly perfect correlation between genotypic and phenotypic analysis for most of these 394 strains, showing the following correlations: 98% for oxacillin resistance, 100% for gentamicin resistance, and 98.5% for erythromycin resistance. The discrepant results were (i) eight strains found to be positive by PCR for mecA or ermC but susceptible to the corresponding antibiotic based on disk diffusion and (ii) six strains of S. aureus found to be negative by PCR for mecA or for the four erythromycin resistance genes targeted but resistant to the corresponding antibiotic. In order to demonstrate in vitro that the eight susceptible strains harboring the resistance gene may become resistant, we subcultured the susceptible strains on media with increasing gradients of the antibiotic. We were able to select cells demonstrating a resistant phenotype for all of these eight strains carrying the resistance gene based on disk diffusion and MIC determinations. The four oxacillin-resistant strains negative for mecA were PCR positive for blaZ and had the phenotype of β-lactamase hyperproducers, which could explain their borderline oxacillin resistance phenotype. The erythromycin resistance for the two strains found to be negative by PCR is probably associated with a novel mechanism. This study reiterates the usefulness of DNA-based assays for the detection of antibiotic resistance genes associated with staphylococcal infections.
Nosocomial infections caused by multiresistant staphylococci are a growing problem for many health care institutions (26, 42, 50). Of all species of staphylococci, Staphylococcus epidermidis and S. aureus have the greatest pathogenic potential. S. epidermidis is widely recognized as one of the etiologic agents of bacteremia, postoperative cardiac infections and endocarditis, osteomyelitis, urinary tract infections, and peritonitis caused by ambulatory dialysis, with a frequent association with colonization of intravascular catheters and orthopedic devices (26, 50). As for S. aureus, it is responsible for diseases caused by exotoxin production (toxic shock and staphylococcal scalded-skin syndromes) and by direct invasion and systemic dissemination (bacteremia, septic shock syndrome, skin infections, and abscesses) (7, 54).
Methicillin-resistant staphylococci (MRS) are resistant to all penicillins, including semisynthetic penicillinase-resistant congeners, penems, carbapenems, and cephalosporins. The basis of this resistance is conferred by an additional penicillin-binding protein, PBP-2′ (or PBP-2a), which is absent in methicillin-susceptible strains (11, 15). Plasmid-mediated aminoglycoside-modifying enzymes of all three classes (aminoglycoside phosphotransferases, acetyltransferases, and nucleotidyltransferases) have been found in staphylococci (53). The bifunctional enzyme AAC(6′)/APH(2"), encoded by the aac(6′)-aph(2") gene, inactivates a broad range of clinically useful aminoglycosides such as gentamicin, tobramycin, netilmicin, and amikacin (20, 49) and is the most frequently encountered aminoglycoside resistance mechanism among staphylococcal isolates (8). Resistance to erythromycin in staphylococci is usually associated with resistance to other macrolides, to the lincosamides, and to type B streptogramin (MLS). This resistance is mediated by a single alteration in the ribosome, the N6-dimethylation of an adenine residue in the 23S rRNA. This dimethylation leads to a conformational change in the ribosome, rendering the strain resistant to most antibiotic of the MLS group. Three genes (ermA, ermB, and ermC) encoding methylases have been found in staphylococci (18, 25, 55). Another mechanism of inducible resistance to erythromycin is conferred by the gene msrA, which encodes an ATP-dependent efflux pump (47, 48, 65).
The aim of this study was to develop rapid multiplex PCR assays for the detection of clinically relevant antibiotic resistance genes in staphylococci and the identification of the staphylococcal species and to compare those PCR assays with standard microbiological methods for susceptibility testing and microbial identification. In this study, a panel of 206 strains of S. aureus and 188 strains of S. epidermidis from various sources were tested by (i) conventional susceptibility testing methods and (ii) PCR for the antibiotic resistance genes mecA, aac(6′)-aph(2"), ermA, ermB, ermC, and msrA.
(This study was presented in part at the 98th General Meeting of the American Society for Microbiology, Atlanta, Ga., May 1998.)
MATERIALS AND METHODS
Bacterial strains.
A total of 206 S. aureus and 188 S. epidermidis strains were used in this study. These isolates were obtained from the American Type Culture Collection (ATCC) (10 strains), the microbiology laboratory of the Centre Hospitalier Universitaire de Québec, The Pavillon Centre Hospitalier de l'Université Laval (CHUL) (Ste-Foy, Québec, Canada) (184 strains), the Laboratoire de Santé Publique du Québec (LSPQ) (Sainte-Anne-de-Bellevue, Québec, Canada) (80 strains), the microbiology laboratory of Hôpital Laval (Ste-Foy, Québec, Canada) (91 strains), the Mount Sinai Hospital (Toronto, Ontario, Canada) (5 strains), the Huashan Hospital (Shanghaï, China) (21 strains), and the Institut Pasteur (Paris, France) (3 strains). The 184 staphylococcal strains from the Pavillon CHUL were identified by using the MicroScan Autoscan-4 system equipped with the Positive BP Combo Panel Type 6 (Dade Diagnostics, Mississauga, Ontario, Canada). A reconfirmation of the staphylococcal species identification for the 210 remaining clinical isolates was performed by using the MicroScan Autoscan-4 system. There was no known relationship between any of the patients. These isolates were implicated in a variety of classical staphylococcal diseases. Duplicate isolates from the same patients, even if the site of infection was different, were excluded from this study. Strains were cultured on sheep blood agar or in brain heart infusion (BHI). Stock cultures were stored frozen (−80°C) in BHI containing 10% glycerol.
Susceptibility testing. (i) Disk diffusion.
Disk diffusion tests were performed for each of the 394 isolates previously identified as S. aureus or S. epidermidis by following the method recommended by the National Committee for Clinical Laboratory Standards (NCCLS) (39). Disks (Becton Dickinson Microbiology Systems, Cockeysville, Md.) containing 1 μg of oxacillin, 10 μg of gentamicin, or 15 μg of erythromycin were added to inoculated Mueller-Hinton agar plates. Subsequently, they were incubated 24 h at 35°C. Any growth, including pinpoint colonies, within the 10-mm-diameter zone for oxacillin, the 12-mm-diameter zone for gentamicin, and the 13-mm-diameter zone for erythromycin was considered indicative of resistance. S. aureus ATCC 29213 was used as negative control.
(ii) MIC determinations.
MICs of oxacillin, gentamicin, and erythromycin (ranging for each antibiotic from 0 to 64 μg/ml, by serial twofold dilutions) were determined for each discordant result between genotype and phenotype by the broth microdilution methodology as recommended by the NCCLS (38). The trays were incubated at 35°C and were read for turbidity with indirect light after 24 h. S. aureus ATCC 29213 was used as negative control.
(iii) Breakpoint determination.
Breakpoints for oxacillin, gentamicin, and erythromycin were determined by using the automated microbial identification system MicroScan Autoscan-4 equipped with the Positive BP Combo Panel Type 6 (Dade Diagnostics). S. aureus ATCC 29213 was used as negative control.
(iv) Nitrocefin test.
The chromogenic cephalosporin nitrocefin disk test was used as recommended by the manufacturer (Becton Dickinson) on each of the four S. aureus strains found to be negative for mecA by PCR assays but resistant to oxacillin. Also these strains were tested for susceptibility to amoxicillin and clavulanic acid (20 and 10 μg, respectively) by disk diffusion according to NCCLS guidelines. S. aureus ATCC 43300 was used as a positive control, and S. aureus ATCC 25923 was used as a negative control.
Multiplex PCR.
S. aureus- and S. epidermidis-specific PCR assays used in this study have been previously described by us (34, 35). PCR primers were chosen from the antibiotic resistance genes mecA, aac(6′)-aph(2"), ermA, ermB, ermC, and msrA (Table 1). Primers were designed with the help of the Oligo Primer Analysis software version 4.0 (National Biosciences, Inc., Plymouth, Minn.) and synthesized by using a model 391 DNA synthesizer (Perkin-Elmer Corp./Applied Biosystems Division, Foster City, Calif.).
TABLE 1.
Antibiotic resistance gene-specific and species-specific primers used in this study
| Target gene | Accession number (GenBank) | Primer sequence | Annealing position |
|---|---|---|---|
| aac(6′)-aph(2") | M13771 | 5′-TTG GGA AGA TGA AGT TTT TAG A-3′ | 159–180 |
| 5′-CCT TTA CTC CAA TAA TTT GGC T-3′ | 311–332 | ||
| blaZ | M60253 | 5′-ACT TCA ACA CCT GCT GCT TTC-3′ | 511–531 |
| 5′-TGA CCA CTT TTA TCA GCA ACC-3′ | 663–683 | ||
| ermA | K02987 | 5′-TAT CTT ATC GTT GAG AAG GGA TT-3′ | 370–392 |
| 5′-CTA CAC TTG GCT TAG GAT GAA A-3′ | 487–508 | ||
| ermB | U35228 | 5′-CTA TCT GAT TGT TGA AGA AGG ATT-3′ | 366–389 |
| 5′-GTT TAC TCT TGG TTT AGG ATG AAA-3′ | 484–507 | ||
| ermC | M17990 | 5′-CTT GTT GAT CAC GAT AAT TTC C-3′ | 214–235 |
| 5′-ATC TTT TAG CAA ACC CGT ATT C-3′ | 382–403 | ||
| Internal control (16S rRNA gene) | J01859 | 5′-GGA GGA AGG TGG GGA TGA CG-3′ | 1244–1263 |
| 5′-ATG GTG TGA CGG GCG GTG TG-3′ | 1469–1489 | ||
| mecA | X52594 | 5′-AAC AGG TGA ATT ATT AGC ACT TGT AAG-3′ | 1059–1085 |
| X52593 | 5′-ATT GCT GTT AAT ATT TTT TGA GTT GAA-3′ | 1206–1232 | |
| msrA | X52085 | 5′-TCC AAT CAT TGC ACA AAA TC-3′ | 891–910 |
| 5′-AAT TCC CTC TAT TTG GTG GT-3′ | 1034–1053 | ||
| S. aureusa | AF033191 | 5′-AAT CTT TGT CGG TAC ACG ATA TTC TTC ACG-3′ | 5–34 |
| 5′-CGT AAT GAG ATT TCA GTA GAT AAT ACA ACA-3′ | 83–112 | ||
| S. epidermidisa | NAb | 5′-ATC AAA AAG TTG GCG AAC CTT TTC A-3′ | 21–45 |
| 5′-CAA AAG AGC GTG GAG AAA AGT ATC A-3′ | 121–145 |
Multiplex PCR assays were all performed directly from a bacterial suspension whose turbidity was adjusted to that of a 0.5 McFarland standard, which corresponds to approximately 1.5 × 108 bacteria per ml. Then, 1 μl of the standardized bacterial suspension was transferred directly to a 20-μl PCR mixture containing 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 2.5 mM MgCl2, 0.4 μM concentrations (each) of the specific primers, 200 μM concentrations (each) of the four deoxynucleoside triphosphates, and 0.5 U of Taq DNA polymerase (Promega). In order to reduce the formation of nonspecific extension products, a “hot-start” protocol was used (34). The PCR mixtures were subjected to thermal cycling (3 min at 96°C and then 30 cycles of 1 s at 95°C for the denaturation step and 30 s at 55°C for the annealing-extension step) with a PTC-200 thermal cycler (MJ Research, Inc., Watertown, Mass.). All multiplex PCR assays (Table 2) also included a primer pair specific to conserved regions of the 16S rRNA gene (241-bp amplicon) which was used to provide an internal control (34). This allowed us to control the efficiency of the quick protocol for bacterial lysis of the PCR assays and to ensure that significant PCR inhibition was absent. Of the PCR-amplified reaction mixture, 10 μl was resolved by electrophoresis through a 2% agarose gel containing 0.5 μg of ethidium bromide per ml in Tris-borate-EDTA buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA) at 12 V/cm for 30 min (34) (Fig. 1). The gels were visualized under 254-nm UV lights. The sizes of the amplification products were estimated by comparison with a 50-bp molecular size standard ladder. The total time for the PCR assays from a standardized bacterial suspension was about 1.5 h.
TABLE 2.
Multiplex PCR assays
| Multiplex no.a | PCR primer pairs combined (amplicon sizes) |
|---|---|
| 1 | S. aureus (108 bp) + S. epidermidis (125 bp) + mecA (174 bp) |
| 2 | S. aureus (108 bp) + S. epidermidis (125 bp) + aac(6′)-aph(2") (174 bp) |
| 3 | S. aureus (108 bp) + S. epidermidis (125 bp) + ermA (139 bp) |
| 4 | S. aureus (108 bp) + S. epidermidis (125 bp) + ermB (142 bp) |
| 5 | S. aureus (108 bp) + S. epidermidis (125 bp) + ermC (190 bp) |
| 6 | S. aureus (108 bp) + S. epidermidis (125 bp) + msrA (163 bp) |
| 7 | S. aureus (108 bp) + S. epidermidis (125 bp) + blaZ (173 bp) |
All multiplex PCR assays included an internal control (241 bp).
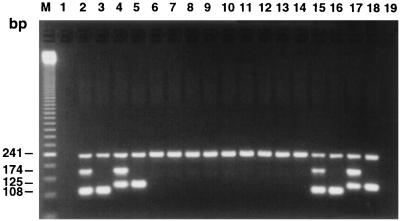
FIG. 1.
Example of multiplex PCR amplifications with the mecA-, S. aureus-, S. epidermidis-, and internal control-specific primer pairs. PCR assays were performed either from 10 pg of purified genomic DNA (lanes 2 to 14) or from 1 μl of a bacterial suspension whose turbidity was adjusted to that of a 0.5 McFarland standard (lanes 15 to 18). The content of each lane is as follows: 2, S. aureus ATCC 43300 (Oxar); 3, S. aureus ATCC 25923 (Oxas); 4, S. epidermidis ATCC 35983 (Oxar); 5, S. epidermidis ATCC 14990 (Oxas); 6, S. capitis ATCC 49326 (Oxar); 7, S. haemolyticus ATCC 29970 (Oxas); 8, S. hominis ATCC 27844 (Oxas); 9, S. saprophyticus ATCC 15305 (Oxas); 10, S. simulans ATCC 27848 (Oxas); 11, S. warneri ATCC 27836 (Oxas); 12, Enterococcus faecalis ATCC 29212; 13, Enterococcus faecium ATCC 51559; 14, Streptococcus pneumoniae ATCC 27336; 15, S. aureus ATCC 43300 (Oxar); 16, S. aureus ATCC 25923 (Oxas); 17, S. epidermidis ATCC 35983 (Oxar); and 18, S. epidermidis ATCC 14990 (Oxas). Lanes 1 and 19, controls to which no DNA was added; lane M, 50-bp ladder molecular size standard.
In vitro selection of resistant cells.
In order to select resistant cells from susceptible strains carrying the corresponding antibiotic resistance gene, we subcultured them onto medium containing increasing concentration gradients of the antibiotic of interest (9, 57). For medium preparation, 10 ml of autoclaved antibiotic medium 1 (Difco Laboratories) without antibiotic was poured into a round petri plate and allowed to cool with one edge elevated so that the agar level just reached the intersection of the bottom and side of the plate. Subsequently, the plate was leveled and an additional 10 ml of the same medium containing low (1 μg/ml), intermediate (5 μg/ml), or high (10 μg/ml) concentrations of oxacillin or erythromycin was added. After 18 h at room temperature, a gradient of antibiotic concentrations ranging from near 0 μg/ml at one edge to the maximal concentration (near the antibiotic concentration in the added medium) at the opposite edge was established. Inoculated plates were incubated for 18 to 24 h at 35°C. For subculturing, colonies were always picked in the area of highest antibiotic concentrations and streaked onto an agar plate containing the same or an increased gradient of antibiotic concentrations. The resistant cells selected from the plates with the highest concentrations of antibiotic were submitted to MIC determinations to confirm their resistance phenotype. They were also tested with the multiplex PCR assays to verify that their resistance genotype was unchanged. To verify the stability of the selected antibiotic resistance, MICs were reevaluated after three passages in medium without antibiotic. The S. aureus strain ATCC 29213, which (i) is susceptible to oxacillin and erythromycin and (ii) does not carry any of the antibiotic resistance gene tested in this study, was always used in parallel as a negative control.
RAPD assay.
Randomly amplified polymorphic DNA (RAPD) assay (66) was performed by using the Operon 10-base kit “AD” which contains 20 different oligonucleotides (Operon Technologies, Inc., Alameda, Calif.). For all bacterial species tested, amplification was performed directly from a bacterial suspension whose turbidity was adjusted to that of a 0.5 McFarland standard. The PCR mixtures (50 mM KCl, 10 mM Tris-HCl [pH 9.0], 0.1% Triton X-100, 2.5 mM MgCl2, a 1.5 μM concentration of a single 10-nucleotide primer, 200 μM concentrations (each) of the four deoxynucleoside triphosphates, and 0.5 U of Taq DNA polymerase [Promega]) were subjected to thermal cycling (3 min at 96°C and then 40 cycles of 1 min at 94°C for the denaturation step, 1 min at 32°C for the annealing step, and 2 min at 72°C for the extension step) by using a PTC-200 thermal cycler (MJ Research, Inc.). A 7-min extension step at 72°C was performed to allow the completion of DNA synthesis. Amplification products were analyzed by electrophoresis in 1.8% agarose gels containing 0.5 μg of ethidium bromide per ml.
RESULTS
Correlation between susceptibility testing and the multiplex PCR assays.
We have compared gentamicin, oxacillin, and erythromycin susceptibility results determined by the disk diffusion method for 394 staphylococcal clinical isolates with those obtained by the multiplex PCR assays for the detection of antibiotic resistance genes. The multiplex PCR assays allowed us also to verify the identity of all S. aureus and S. epidermidis strains by using the species-specific PCR assays previously described by our group (34, 35). The various multiplex PCR assays developed in this study are given in Table 2. A comparison of the multiplex PCR-based assays with conventional susceptibility testing and identification methods (disk diffusion, MIC determination, and the MicroScan identification system) showed the following correlations: 100% for gentamicin resistance (Table 3), 98% for oxacillin resistance (Table 4), 98.5% for erythromycin resistance (Table 5), and 100% for staphylococcal species identification.
TABLE 3.
Correlation between gentamicin resistance and the presence of the resistance gene aac(6′)-aph(2")
| Species | No. of strains (type) tested by disk diffusion with gentamicin | PCR results for aac(6′)-aph(2")
|
|
|---|---|---|---|
| No. positive | No. negative | ||
| S. aureus | 41 (resistant) | 41 | 0 |
| S. aureus | 165 (susceptible) | 0 | 165 |
| S. epidermidis | 131 (resistant) | 131 | 0 |
| S. epidermidis | 57 (susceptible) | 0 | 57 |
TABLE 4.
Correlation between oxacillin resistance and the presence of the resistance gene mecA
| Species | No. of strains (type) tested by disk diffusion with oxacillin | PCR results for mecA
|
|
|---|---|---|---|
| No. positive | No. negative | ||
| S. aureus | 66 (resistant) | 62 | 4 |
| S. aureus | 140 (susceptible) | 1 | 139 |
| S. epidermidis | 146 (resistant) | 146 | 0 |
| S. epidermidis | 42 (susceptible) | 3 | 39 |
TABLE 5.
Correlation between erythromycin resistance and the presence of the resistance gene ermA, ermB, ermC, or msrA
| Species | No. of strains (type) tested by disk diffusion with erythromycin | No. of strains positive by PCR for:
|
No. of PCR-negative strains | |||
|---|---|---|---|---|---|---|
| ermA | ermB | ermC | msrA | |||
| S. aureus | 73 (resistant) | 43 | 5 | 21 | 2 | 2 |
| S. aureus | 133 (susceptible) | 0 | 0 | 4 | 0 | 129 |
| S. epidermidis | 142 (resistant) | 9 | 1 | 124 | 8 | 0 |
| S. epidermidis | 46 (susceptible) | 0 | 0 | 0 | 0 | 46 |
The correlation between gentamicin resistance and the presence of aac(6′)-aph(2") is summarized in Table 3. Of 206 S. aureus strains, 165 (80%) were susceptible to gentamicin as determined by the disk diffusion method and negative by PCR for aac(6′)-aph(2"). The remaining 41 S. aureus strains were resistant to gentamicin while harboring the aac(6′)-aph(2"). Interestingly, the situation is quite different for S. epidermidis. Of 188 S. epidermidis strains, 131 (70%) were resistant to gentamicin and harbor the aac(6′)-aph(2") resistance gene. The remaining 57 strains were negative by PCR for aac(6′)-aph(2") and susceptible to gentamicin.
The correlation between oxacillin resistance and the presence of the mecA gene is summarized in Table 4. There were 139 strains (68%) of S. aureus which were both negative by PCR for mecA and susceptible to oxacillin. However, there was one S. aureus strain which was oxacillin susceptible and positive for mecA. This strain had an MIC of 1 μg/ml. Therefore, although present, the mecA gene did not confer a detectable level of oxacillin resistance. However, as described below, it was possible to select resistant cells by exposing this strain to increasing concentrations of oxacillin. Of 66 oxacillin-resistant S. aureus strains (32%), 62 were harboring mecA based on PCR analysis. Importantly, four oxacillin-resistant mecA-negative S. aureus strains showed a borderline level of resistance based on susceptibility testing by disk diffusion and MIC determination. In fact, these strains all had MICs ranging from 4 to 8 μg/ml for oxacillin. Further testing was performed on these four S. aureus strains to verify if they were β-lactamase hyperproducers, a mechanism which may mediate S. aureus resistance to methicillin (2). They were characterized by testing with (i) nitrocefin disks, (ii) a PCR assay for the detection of blaZ, and (iii) amoxicillin-clavulanic acid disks. Positive reaction to nitrocefin and the presence of the blaZ gene revealed by PCR confirmed that these strains were β-lactamase producers. In vitro testing with oxacillin and amoxicillin-clavulanic acid disks suggests that these four strains are β-lactamase hyperproducers, which could explain their borderline oxacillin-resistant phenotype (Table 6).
TABLE 6.
Characteristics of the four strains resistant to oxacillin but negative for mecA by PCR
| Strains | Oxacillin MIC (μg/ml) | Nitrocefin test result | Disk diffusion test result (mm) amoxicillin with clavulanic acida | blaZ (PCR detection) |
|---|---|---|---|---|
| S. aureus R620 | 4 | + | 24.0 | + |
| S. aureus R626 | 8 | + | 22.0 | + |
| S. aureus R627 | 4 | + | 24.5 | + |
| S. aureus STA572 | 8 | + | 23.3 | + |
| S. aureus ATCC 43300b | 64 | + | 15.2 | + |
| S. aureus ATCC 25923b | 1 | − | 28.3 | − |
Strains were considered β-lactamase hyperproducers when (i) the diameter of the inhibition zone with the amoxicillin-clavulanic acid (20 and 10 μg, respectively) disk exceeded 20 mm and (ii) they were resistant to oxacillin (2).
S. aureus ATCC 43300 is a nonhyperproducing strain carrying mecA, while S. aureus ATCC 25923 is a β-lactamase-negative strain not carrying mecA.
As expected, there was far more resistance to oxacillin in S. epidermidis than in S. aureus. There were 146 S. epidermidis strains (78%) which were both mecA positive and oxacillin resistant. Of the 42 oxacillin-susceptible strains (22%), there were 39 not harboring the mecA gene. The three S. epidermidis oxacillin-susceptible mecA-positive strains had MICs ranging from 0.25 to 1 μg/ml. Therefore, as for S. aureus, the presence of mecA was not sufficient to confer a resistance phenotype in these three S. epidermidis strains. Again, as described below, it was possible to select resistant cells from these oxacillin-susceptible strains.
Table 5 shows the correlation between erythromycin resistance and the presence of ermA, ermB, ermC, and msrA. As for oxacillin and gentamicin, there was far more resistance to erythromycin in S. epidermidis than in S. aureus. For S. aureus, of the 133 susceptible strains (64%), there were 129 not harboring any of the four genes associated with erythromycin resistance, based on multiplex PCR assays. The four erythromycin-susceptible S. aureus strains harboring an erythromycin resistance gene were all positive for ermC only (Table 7). Therefore, as observed for oxacillin resistance, the presence of ermC did not confer a detectable level of resistance. Again, it was possible to select resistant cells from these four strains. Regarding the S. aureus that was resistant to erythromycin, of 73 strains (35%) there were 71 harboring one of the four erythromycin resistance gene tested (Table 4). The incidences of the different genotypes were 21% for ermA, 2.4% for ermB, 10% for ermC, and 1% for msrA. None of the resistant strains harbored more than one resistance gene. The erythromycin resistance in the two S. aureus strains not carrying any of the four resistance genes tested is likely mediated by as-yet-unknown mechanisms. We are now investigating the mechanisms of such resistance.
TABLE 7.
S. aureus (n = 11) and S. epidermidis (n = 3) strains for which the resistance genotype was discordant with the resistance phenotype
| Group and straina | Profile | MIC (μg/ml) | MIC after selectionb (μg/ml) |
|---|---|---|---|
| Group 1 | |||
| S. aureus R632 | mecA, Oxas | 2 | >64 |
| S. epidermidis R588 | mecA, Oxas | 0.125 | >64 |
| S. epidermidis 184 | mecA, Oxas | 1 | >64 |
| S. epidermidis 3348 | mecA, Oxas | 0.25 | >64 |
| S. aureus ATCC 29213 | mecA negative, Oxas | 0.5 | 0.5 |
| S. aureus 18 | ermC, Erys | 1 | >64 |
| S. aureus 75 | ermC, Erys | 1 | >64 |
| S. aureus 93 | ermC, Erys | 1 | >64 |
| S. aureus 5919 | ermC, Erys | 1 | >64 |
| S. aureus ATCC 29213 | ermC negative, Erys | 0.5 | 0.5 |
| Group 2 | |||
| S. aureus R572 | mecA negative, Oxar | 8 | |
| S. aureus R620 | mecA negative, Oxar | 4 | |
| S. aureus R626 | mecA negative, Oxar | 8 | |
| S. aureus R627 | mecA negative, Oxar | 4 | |
| S. aureus MA-5091 | ermABC negative, msrA negative, Eryr | >32 | |
| S. aureus R566 | ermABC negative, msrA negative, Eryr | >32 |
Group 1, resistance gene present, susceptible; group 2, resistance gene absent, resistant. MIC column data for group 1 strains are the initial MICs.
The selection of resistant cells was done by plating on media with increasing gradients of the target antibiotic.
For S. epidermidis, only 46 strains (24%) were both susceptible to erythromycin and did not contain any erythromycin resistance gene. The incidences of the three erythromycin ribosomal methylase genes tested were 4.8% for ermA, 0.5% for ermB, and 66% for ermC. msrA was present in 4.3% of S. epidermidis strains. No discrepant results were found between the genotypes and the phenotypes in these 188 S. epidermidis clinical strains.
In vitro selection of resistant cells.
As shown in Table 7, there were 8 susceptible staphylococcal strains carrying the corresponding antibiotic resistance gene. Four strains (one S. aureus strain and three S. epidermidis strains) were PCR positive for mecA but susceptible for oxacillin, and four strains of S. aureus were positive for ermC but susceptible to erythromycin. These strains were all initially subcultured twice on plates containing a gradient of oxacillin or erythromycin ranging from 0 to 1 μg/ml in order to reach growth confluence. Subsequently, strains were subcultured twice on plates with gradients ranging from 0 to 5 μg/ml and then twice again on plates with gradients ranging from 0 to 10 μg/ml in order to attain growth confluence.
The culture on media with increasing gradients of the target antibiotic allowed for the selection of cells having MICs greater than 64 μg/ml for both oxacillin and erythromycin. This is a major increase, considering that the MICs for the original susceptible strains ranged from 0.5 to 1 μg/ml for these two antibiotics. Furthermore, we have confirmed by PCR testing that the genotypes of these strains harboring mecA or ermC remained unchanged after the selection process. Interestingly, even after three passages in antibiotic-free media, the MICs remained unchanged. Importantly, with the control strain ATCC 29213, it was not possible to select cells resistant to oxacillin or erythromycin, thereby confirming that the resistance gene must be present to allow for the rapid selection of resistant cells. The negative control strain always remained low, at 0.5 μg/ml. These experiments strongly suggest that susceptible strains harboring a resistance gene have the potential to develop resistance upon in vivo selection by the appropriate antimicrobial agent.
In order to further confirm that the resistant cells selected were derived from the same susceptible initial strain, RAPD-PCR was performed with genomic DNA purified from the four oxacillin-susceptible mecA-positive and the four erythromycin-susceptible ermC-positive strains as well as with the resistant cells derived from these eight strains (five S. aureus and three S. epidermidis) after in vitro selection. The RAPD assay was performed with three different oligonucleotide primers shown to be highly discriminatory for these strains. For the four staphylococcal oxacillin-susceptible mecA-positive strains (Table 7), three distinct RAPD patterns were obtained for each oligonucleotide primer tested. When used in combination, these three primers produced unique amplification patterns for each of those four strains. We found that the amplification patterns with the three selected RAPD primers were identical for both the initial susceptible strain and the selected resistant strain (data not shown). These findings strongly suggest that the selected resistant cells were derived from the corresponding original susceptible strain. Similarly, distinct RAPD patterns were obtained for each of the four S. aureus erythromycin-susceptible ermC-positive strains (Table 7). Again, identical amplification patterns were obtained with both the initial susceptible strain and the selected resistant strain.
DISCUSSION
We describe here PCR primers that can be used to survey clinically relevant antibiotic resistance genes frequently encountered in staphylococci. We have compared multiplex PCR assays for the detection of antibiotic resistance genes with classical methods for the determination of susceptibility to antibiotics. Overall, we found correlations between these two methods of 98% for oxacillin resistance, 100% for gentamicin resistance, 98.5% for erythromycin resistance, and 100% for species identification.
The mecA gene is a 2.4-kb chromosomal determinant encoding the PBP-2′ protein which is not subjected to dissemination among staphylococcal strains via plasmid spread. Expression of PBP-2′ is under control of the negative regulation elements mecI and mecRI (11, 27, 40). Five phenotypic methods are commonly used by clinical laboratories to detect MRS. For all screening methods, oxacillin is used because it is the most sensitive member of the penicilinase-resistant semisynthetic β-lactam agents for the detection of resistance. The easiest and among the most accurate test is the use of oxacillin screening agar (60). Broth dilution methods are generally accurate when cation-adjusted Mueller-Hinton broth containing 2% NaCl is used and when the results are interpreted by visual inspection of broth turbidity (21). Agar disk diffusion is the most used antibiotic susceptibility assay in clinical microbiology laboratories, but it is not particularly accurate because it may demonstrate a lack of reproducibility and sensitivity (2). The Crystal MRSA (Becton Dickinson) is a reliable and rapid method (4 to 5 h) with a fluorescent indicator to detect methicillin-resistant S. aureus (MRSA) (46). Finally, the E test is an excellent quantitative system for laboratories that do not wish to stock the more cumbersome MIC trays (41). Although these culture-based methods are generally reliable for detecting MRS, the detection of mecA is now considered the gold standard method, mainly because (i) phenotypic methods may be difficult to interpret and (ii) some isolates do not express their mecA gene unless selective pressure via antibiotic treatment is applied.
Several studies deal with the detection by PCR of the mecA gene only (36, 44, 59) or combined in multiplex with S. aureus-specific amplification assays (3, 6, 19, 51, 62). Overall, we found in this study that 30.6% of S. aureus and 79.3% of S. epidermidis strains were carrying the mecA gene. When restricted to strains isolated from Canada (obtained from CHUL, LSPQ, Laval Hospital, and Mount Sinai Hospital), the proportion of strains carrying mecA falls to 25.2% for S. aureus and 77.1% for S. epidermidis. It should be noted that this high percentage of MRSA is not representative of the true Canadian incidence because the S. aureus isolates obtained from LSPQ and Mount Sinai Hospital were selected for their phenotypic resistance to oxacillin. When considering only isolates from the CHUL and Laval Hospital, which were not selected for resistant strains, we observed an incidence of 2% of S. aureus harboring mecA. This is comparable to the study from the SENTRY antimicrobial Surveillance Program for S. aureus isolates from Canada, in which an incidence of 2.5% of oxacillin-resistant S. aureus was reported (43).
The aac(6′)-aph(2") is the gene coding for the most frequently encountered aminoglycoside modifying enzyme (AME) in gram-positive bacteria, AAC(6′)-APH(2") (61). This bifunctional enzyme inactivates a broad range of clinically useful aminoglycosides, especially gentamicin and tobramycin, because it catalyzes both acetyltransferase and phosphotransferase reactions. Two other genes associated with aminoglycoside resistance in staphylococci may also be found. These genes, aphA3 [coding for APH(3′)III enzyme], and aadC, coding for ANT(4′,4") enzyme, are much less frequently encountered than aac(6′)-aph(2") and are not clinically relevant because they mediate resistance to aminoglycosides not usually prescribed to treat staphylococcal infections (53). DNA amplification by PCR has been shown to be a reliable tool for the specific detection of AME genes in gram-negative bacteria as well as for the detection of aac(6′)-aph(2") in epidemic strains of MRSA (61). DNA dot blot analysis of AME genes has also been described (8). In this study, we found aac(6′)-aph(2") in 20% of S. aureus isolates and in 69.7% of S. epidermidis isolates, with no discrepant results with the resistance phenotypes obtained by the disk diffusion method. The absence of discrepant results between the resistance genotypes and phenotypes suggests that there were no strains harboring (or at least not expressing) aminoglycoside resistance genes other than aac(6′)-aph(2"). Therefore, all isolates harboring the aac(6′)-aph(2") gene were clearly resistant to gentamicin in susceptibility tests, and all isolates not harboring this resistance gene were susceptible to gentamicin. These findings are in agreement with other studies in which it was reported that all aminoglycoside-resistant strains were carrying aac(6′)-aph(2") (16, 31, 32, 45, 61).
Several genes are implicated in erythromycin resistance, especially in staphylococci and streptococci. Simultaneous resistance to macrolides, lincosamides, and type-B streptogramins (MLS resistance) in clinical isolates is a form of acquired resistance due to several evolutionary variants of erm genes, which encodes a 23S rRNA methylase (23, 64). The inducible gene ermA is found on the transposon Tn554 and has a single specific site for insertion into the S. aureus chromosome (37). The ermB gene is found on the transposon Tn551 of a penicillinase plasmid (25). The ermC gene is responsible for constitutive or inducible resistance to erythromycin and is generally located on small plasmids (18, 63, 64). Staphylococcal strains resistant to macrolides and type-B streptogramins also frequently harbor msrA, which encodes an ATP-dependent efflux pump and mediates the macrolide-streptogramin B (MS) resistance (18). Several studies concerning the epidemiological distribution of genes encoding erythromycin ribosomal methylases and efflux pumps have been performed by dot blot or Southern hybridization (18, 23, 65), and detection of erythromycin resistance determinants by PCR has been performed with staphylococci and streptococci (55).
In this study, the incidences of ermA in erythromycin-resistant staphylococci were 21% for S. aureus and 4.8% for S. epidermidis. These findings are in agreement with the study by Eady et al. (18) conducted with coagulase-negative staphylococci (CoNS) in the United Kingdom in which an incidence of 5.9% for ermA was reported. In a study performed in Denmark (64), 16% of S. aureus strains were carrying ermA, while only 3% of CoNS strains had this gene. These observations are also in agreement with our data. On the other hand, Thakker-Varia et al. (58) reported a higher incidence for ermA. They found that 31% of S. aureus and 19% of CoNS from three New Jersey hospitals were harboring the ermA gene. Regarding ermB, we found that this gene was less frequently encountered than ermA in erythromycin-resistant staphylococci with 1.9% of S. aureus and 0.5% of S. epidermidis strains carrying ermB. In the United Kingdom, an incidence of 7.2% for ermB in CoNS has been reported (18). Interestingly, in that study it was observed that ermB was found exclusively in animal isolates of S. intermedius, S. xylosus, and S. hyicus but was absent in CoNS of human origin. This observation could explain why our incidence for ermB in CoNS is much lower because our CoNS strains were exclusively of human origin. For ermC, we found an incidence of 10.2% for S. aureus strains and 66% for S. epidermidis strains. Eady et al. (18) have also reported a high incidence of CoNS strains carrying ermC (i.e., detected in 112 of 221 [50.6%] CoNS isolates). In New Jersey hospitals, an even higher incidence for the ermC gene (i.e., 69% for S. aureus and 81% for CoNS isolates) has been reported (58). A high incidence of S. aureus carrying ermC (i.e., 84%) has also been reported in Denmark (64). The much lower prevalence of ermC-positive strains found in Canada suggests that the selective pressure for ermC-positive S. aureus is weaker in this country. These variations may be associated with variable use of erythromycin in each country. Finally, among erythromycin-resistant strains, we found msrA in only 1% of S. aureus strains and 4.3% of S. epidermidis strains. A much higher incidence for msrA in MRS (i.e., 33%) has been reported in the United Kingdom (18). Again, these results suggest a lower selective pressure on populations of msrA-positive staphylococci in Canada.
Among the staphylococcal strains showing discrepancies between the genotype and the phenotype (four oxacillin-susceptible mecA-positive strains and four erythromycin-susceptible ermC-positive strains), we were able to select cells demonstrating a resistant phenotype from all of them. Others have also reported S. aureus strains carrying the mecA gene but susceptible to oxacillin (28, 40). The heterogeneous nature of methicillin and erythromycin resistances suggests that numerous factors could explain the sensitive phenotype in these strains (1, 10, 22, 29, 30, 40, 56). Such factors include (i) the regulation of the expression of mecA or ermC and (ii) the absence of host factors associated with the phenotypic expression of methicillin resistance. The fact that we were able to select resistant cells from originally susceptible strains demonstrates that upon in vitro selection in the presence of increasing gradients of the antimicrobial agent, it is possible to select for resistance. Furthermore, once induced, the resistance phenotype was shown to be stable. Kolbert et al. (28) were also able to select for oxacillin resistance for 6 of 10 mecA-positive and oxacillin-sensitive isolates. Our ability to select resistant cells from all mecA-positive and oxacillin-susceptible strains is probably explained by the fact that we used plates with medium containing an increasing concentration gradient of oxacillin (9, 57) as opposed to the method used by Kolbert et al., in which a series of plates containing increasing concentrations of oxacillin were inoculated. Consequently, the use of plates with an increasing concentration gradient of antibiotic appear more efficient to select for resistant cells. To our knowledge, the selection of erythromycin-resistant cells from originally susceptible bacterial strains has not been previously reported. From a clinical perspective, these findings suggest that a susceptible strain harboring but not expressing an antibiotic resistance gene should be regarded as potentially resistant to that antibiotic. Well-documented clinical observations of this phenomenon have not been demonstrated with erythromycin or a macrolide-like antibiotic. Investigators using teicoplanin have denoted a failure of treatment associated with an increase in MIC during therapy of S. aureus septicemia (12, 24, 33).
The second type of discrepancy was encountered in four mecA-negative S. aureus strains which were borderline oxacillin resistant based on MIC determinations. Since these four strains were positive for the nitrocefin test and positive by PCR for the blaZ gene and were sensitive to amoxicillin-clavulanic acid (i.e., inhibition zone of >20 mm), they appeared to be β-lactamase hyperproducers, which could explain their borderline resistance to oxacillin. Others (2, 14, 51) have also used this criteria to determine the phenotype of β-lactamase hyperproducers. However, we cannot exclude the possibility that the methicillin resistance in these strains, which is not mediated by mecA, is associated with other mechanisms of resistance (2).
We also reported two S. aureus strains fully resistant (>32 μg/ml) to erythromycin but negative for ermA, ermB, ermC, and msrA by PCR. Based on these results, we postulate that this resistance is probably associated with a novel mechanism not yet characterized in staphylococci. As reported recently by Huovinen et al. (52), a novel erythromycin methylase resistance gene, ermTR, was discovered in Streptococcus. The nucleotide sequence of ermTR is 82.5% identical to the staphylococcal ermA. Another finding concerns a novel macrolide efflux pump from Streptococcus agalactiae, called mreA (13). This putative efflux determinant is distinct from the multicomponent MsrA pump found in staphylococci. Consequently, it is possible that novel erythromycin resistance gene(s) in staphylococci will be found in the future.
This study reiterates the usefulness of DNA-based assays for the detection of antibiotic resistance genes associated with staphylococcal infections. However, the approach presented in this study would not be appropriate for detecting the clinically important resistance to fluoroquinolones because it is associated with many point mutations in two different genes (i.e., gyrA and grlA) (17). On the other hand, such complex resistance genotypes could be detected by PCR amplification of the mutated genetic target, followed by hybridization on a DNA chip or rapid automated DNA sequencing.
In the clinical setting, the simultaneous identification of the bacteria and determination of its susceptibility to antibiotics generally require 48 h (4, 5). Yet in the choice of empiric antibiotic therapy for suspected staphylococcal sepsis, the clinician must know rapidly which species is involved and its susceptibility to antibiotics. The multiplex PCR assays developed in this study could be adapted for direct detection from positive blood cultures or from clinical specimens (e.g., detection of MRSA from nasal swabs or infected wounds), thereby allowing a much faster diagnosis than conventional culture methods. We believe that a direct impact of such rapid PCR assays is that they should allow for a faster establishment of effective antibiotic therapy and a reduction of empirical treatments with broad-spectrum antibiotics, which are associated with high costs and toxicity. Utilization of the multiplex PCR technology in the clinical laboratory in combination with the recommended NCCLS guidelines would better enable physicians to prescribe appropriate antibiotic therapy, leading to therapeutic success, more prudent antibiotic usage, and conditions less conducive to staphylococcal resistance selection (5).
ACKNOWLEDGMENTS
We thank Louise Côté, director of the Microbiology Laboratory of CHUL, for free access to the laboratory and for providing the staphylococcal isolates. We also thank Louise Jetté (Laboratoire de Santé Publique du Québec), Pierre Auclair (Laval Hospital), Donald E. Low (Mount Sinai Hospital), Wang Fu (Huashan Hospital), and Nevine El Solh (Institut Pasteur) for providing staphylococcal strains. We also thank Ann Huletsky and Maurice Boissinot for helpful suggestions.
F. Martineau has a scholarship from le Fonds de Recherche en Santé du Québec. M. Ouellette is an MRC Scientist and a Burroughs-Wellcome Fund New Investigator in molecular parasitology. This research project was supported by grant PA-15586 from the Medical Research Council of Canada and Infectio Diagnostic (IDI), Inc., Sainte-Foy, Québec, Canada.
REFERENCES
- 1.Archer G L, Niemeyer D M, Thanassi J A, Pucci M J. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron E J. Genetic aspects of methicillin resistance in Staphylococcus aureus and methods used for its detection in clinical laboratories in the United States. J Chemother. 1995;7(Suppl. 3):87–92. [PubMed] [Google Scholar]
- 3.Barski P, Piechowicz L, Galinski J, Kur J. Rapid assay for detection of methicillin-resistant Staphylococcus aureus using multiplex PCR. Mol Cell Probes. 1996;10:471–475. doi: 10.1006/mcpr.1996.0066. [DOI] [PubMed] [Google Scholar]
- 4.Bergeron M G, Ouellette M. Diagnosing bacterial infectious diseases in one hour: an essential upcoming revolution. Infection. 1995;23:69–72. doi: 10.1007/BF01833867. [DOI] [PubMed] [Google Scholar]
- 5.Bergeron M G, Ouellette M. Preventing antibiotic resistance through rapid genotypic identification of bacteria and of their antibiotic resistance genes in the clinical microbiology laboratory. J Clin Microbiol. 1998;36:2169–2172. doi: 10.1128/jcm.36.8.2169-2172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brakstad O G, Maeland J A, Tveten Y. Multiplex polymerase chain reaction for detection of genes for Staphylococcus aureus thermonuclease and methicillin resistance and correlation with oxacillin resistance. APMIS. 1993;101:681–688. doi: 10.1111/j.1699-0463.1993.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 7.Brumfitt W, Hamilton-Miller J. Methicillin-resistant Staphylococcus aureus. N Engl J Med. 1989;320:1188–1196. doi: 10.1056/NEJM198905043201806. [DOI] [PubMed] [Google Scholar]
- 8.Busch-Sorensen C, Frimodt-Moller N, Miller G H, Espersen F. Aminoglycoside resistance among Danish blood culture isolates of coagulase-negative staphylococci. APMIS. 1996;104:873–880. doi: 10.1111/j.1699-0463.1996.tb04952.x. [DOI] [PubMed] [Google Scholar]
- 9.Carlton B C, Brown B J. Antibiotic resistance (gradient plate) In: Gerhardt P, Murray R, Costillow R, Nester E, Wood W, Krieg N, Briggs Phillips G, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. pp. 229–230. [Google Scholar]
- 10.Catchpole I, Thomas C, Davies A, Dykes K G H. The nucleotide sequence of Staphylococcus aureus plasmid pT48 conferring inducible resistance and comparison with similar plasmids expressing constitutive resistance. J Gen Microbiol. 1988;134:697–709. doi: 10.1099/00221287-134-3-697. [DOI] [PubMed] [Google Scholar]
- 11.Chambers H F. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomarat M, Espinouse D, Flandrois J P. Coagulase-negative staphylococci emerging during teicoplanin therapy and problems in the determination of their sensitivity. J Antimicrob Chemother. 1991;27:475–480. doi: 10.1093/jac/27.4.475. [DOI] [PubMed] [Google Scholar]
- 13.Clancy J, Dib-Hajj F, W. P J, Yuan W. Cloning and characterization of a novel macrolide efflux gene, mreA, from Streptococcus agalactiae. Antimicrob Agents Chemother. 1997;41:2719–2723. doi: 10.1128/aac.41.12.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cloney L, Marlowe C, Wong A, Chow R, Bryan R. Rapid detection of mecA in methicillin resistant Staphylococcus aureus using cycling probe technology. Mol Cell Probes. 1999;13:191–197. doi: 10.1006/mcpr.1999.0235. [DOI] [PubMed] [Google Scholar]
- 15.de Lencastre H, de Jonge B L, Matthews P R, Tomasz A. Molecular aspects of methicillin resistance in Staphylococcus aureus. J Antimicrob Chemother. 1994;33:7–24. doi: 10.1093/jac/33.1.7. [DOI] [PubMed] [Google Scholar]
- 16.Dornbusch K, Miller G H, Hare R S, Shaw K J. Resistance to aminoglycoside antibiotics in gram-negative bacilli and staphylococci isolated from blood. Report from a European collaborative study. The ESGAR study group (European Study Group on Antibiotic Resistance) J Antimicrob Chemother. 1990;26:131–144. doi: 10.1093/jac/26.1.131. [DOI] [PubMed] [Google Scholar]
- 17.Dubin D T, Fitzgibbon J E, Nahvi M D, John J F. Topoisomerases sequences of coagulase-negative staphylococcal isolates resistant to ciprofloxacin or trovafloxacin. Antimicrob Agents Chemother. 1999;43:1631–1637. doi: 10.1128/aac.43.7.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eady E A, Ross J I, Tipper J L, Walters C E, Cove J H, Noble W C. Distribution of genes encoding erythromycin ribosomal methylases and an erythromycin efflux pump in epidemiologically distinct groups of staphylococci. J Antimicrob Chemother. 1993;31:211–217. doi: 10.1093/jac/31.2.211. [DOI] [PubMed] [Google Scholar]
- 19.Geha D L, Uhl J R, Gustaferro C A, Persing D H. Multiplex detection for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol. 1994;32:1768–1772. doi: 10.1128/jcm.32.7.1768-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodel-Christian S L, Murray B E. Characterization of the gentamicin resistance transposon Tn5281 from Enterococcus faecalis and comparison to staphylococcal transposons Tn4001 and Tn4031. Antimicrob Agents Chemother. 1991;35:1147–1152. doi: 10.1128/aac.35.6.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang M B, Gay T E, Baker C N, Banerjee S N, Tenover F C. Two percent sodium chloride is required for susceptibility testing of staphylococci with oxacillin when using agar-based dilution methods. J Clin Microbiol. 1993;31:2683–2688. doi: 10.1128/jcm.31.10.2683-2688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hürlimann-Dalel R L, Ryffel C, Kayser F H, Berger-Bächi B. Survey of the methicillin resistance-associated genes mecA, mecR1-mecI, and femA-femB in clinical isolates of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:2617–2621. doi: 10.1128/aac.36.12.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenssen W D, Thakker Varia S, Dubin D T, Weinstein M P. Prevalence of macrolides-lincosamides-streptogramin B resistance and erm gene classes among clinical strains of staphylococci and streptococci. Antimicrob Agents Chemother. 1987;31:883–888. doi: 10.1128/aac.31.6.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaatz G W, Seo S M, Dorman N J, Lerner S A. Emergence of teicoplanin resistance during therapy of Staphylococcus aureus endocarditis. J Infect Dis. 1990;162:103–108. doi: 10.1093/infdis/162.1.103. [DOI] [PubMed] [Google Scholar]
- 25.Khan S A, Novick R P. Terminal nucleotide sequences of Tn551, a transposon specifying erythromycin resistance in Staphylococcus aureus: homology with Tn3. Plasmid. 1980;4:148–154. doi: 10.1016/0147-619x(80)90004-9. [DOI] [PubMed] [Google Scholar]
- 26.Kloos W E, Bannerman T L. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7:117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi N, Taniguchi K, Kojima K, Urasawa S, Uehara N, Omizu Y, Kishi Y, Yagihashi A, Kurokawa I, Watanabe N. Genomic diversity of mec regulator genes in methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Epidemiol Infect. 1996;117:289–295. doi: 10.1017/s0950268800001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolbert C P, Arruda J, Varga-Delmore P, Zheng X, Lewis M, Kolberg J, Persing D H. Branched-DNA assay for detection of the mecA gene in oxacillin-resistant and oxacillin-sensitive staphylococci. J Clin Microbiol. 1998;36:2640–2644. doi: 10.1128/jcm.36.9.2640-2644.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuwahara-Arai K, Kondo N, Hori S. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP2′ production. Antimicrob Agents Chemother. 1996;40:2680–2685. doi: 10.1128/aac.40.12.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lampson B C, Parisi J T. Naturally occurring Staphylococcus epidermidis plasmid expressing constitutive macrolides-lincosamides-streptogramin B resistance contains a deleted attenuator. J Bacteriol. 1986;166:479–483. doi: 10.1128/jb.166.2.479-483.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovering A M, Bywater M J, Holt H A, Champion H M, Reeves D S. Resistance of bacterial pathogens to four aminoglycosides and six other antibacterials and prevalence of aminoglycosides modifying enzymes. J Antimicrob Chemother. 1988;22:823–839. doi: 10.1093/jac/22.6.823. [DOI] [PubMed] [Google Scholar]
- 32.Madsen O R, Lorentzen J S, Frimodt-Moller N, Mortensen I, Rosdahl V T. Mechanisms of aminoglycoside resistance in Danish Staphylococcus aureus strains during the years 1979–1987. APMIS. 1991;99:537–540. [PubMed] [Google Scholar]
- 33.Manquat G, Croizé J, Stahl J P, Meyran M, Hirtz P, Micoud M. Failure of teicoplanin treatment associated with an increase in MIC during therapy of Staphylococcus aureus septicaemia. J Antimicrob Chemother. 1992;29:731–732. doi: 10.1093/jac/29.6.731. [DOI] [PubMed] [Google Scholar]
- 34.Martineau F, Picard F J, Roy P H, Ouellette M, Bergeron M G. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus aureus. J Clin Microbiol. 1998;36:618–623. doi: 10.1128/jcm.36.3.618-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martineau F, Picard F J, Roy P H, Ouellette M, Bergeron M G. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J Clin Microbiol. 1996;34:2888–2893. doi: 10.1128/jcm.34.12.2888-2893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy E. Nucleotide sequence of ermA, a macrolide-lincosamide-streptogramin B determinant in Staphylococcus aureus. J Bacteriol. 1985;162:633–640. doi: 10.1128/jb.162.2.633-640.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A2. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1990. [Google Scholar]
- 39.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-4A. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1990. [Google Scholar]
- 40.Niemeyer D M, Pucci M J, Thanassi J A, Sharma V K, Archer G L. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J Bacteriol. 1996;178:5464–5471. doi: 10.1128/jb.178.18.5464-5471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petersson A C, Miorner H, Kamme C. Identification of mecA-related oxacillin resistance in staphylococci by the E test and the broth microdilution method. J Antimicrob Chemother. 1996;37:445–456. doi: 10.1093/jac/37.3.445. [DOI] [PubMed] [Google Scholar]
- 42.Pfaller M A, Herwaldt L A. Laboratory, clinical and epidemiological aspects of coagulase-negative staphylococci. Clin Microbiol Rev. 1988;1:281–299. doi: 10.1128/cmr.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfaller M A, Jones R N, Doern G V, Kugler K The SENTRY Participants Group. Bacterial pathogens isolated from patients with bloodstream infection: frequencies of occurrence and antimicrobial susceptibility patterns from the SENTRY antimicrobial surveillance program (United States and Canada, 1997) Antimicrob Agents Chemother. 1998;42:1762–1770. doi: 10.1128/aac.42.7.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Predari S C, Ligozzi M, Fontana R. Genotypic identification of methicillin-resistant coagulase-negative staphylococci by polymerase chain reaction. Antimicrob Agents Chemother. 1991;35:2568–2573. doi: 10.1128/aac.35.12.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price K E, Kresel P A, Farchione L A, Siskin S B, Karpow S A. Epidemiological studies of aminoglycoside resistance in the U.S.A. J Antimicrob Chemother. 1981;8(Suppl. A):89–105. doi: 10.1093/jac/8.suppl_a.89. [DOI] [PubMed] [Google Scholar]
- 46.Qadri S M H, Ueno Y, Imambaccus H, Almodovar E. Rapid detection of methicillin-resistant Staphylococcus aureus by Crystal MRSA ID System. J Clin Microbiol. 1994;32:1830–1832. doi: 10.1128/jcm.32.7.1830-1832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross J I, Eady E A, Cove J H, Baumberg S. Identification of a chromosomally encoded ABC-transport system with which the staphylococcal erythromycin exporter MsrA may interact. Gene. 1995;153:93–98. doi: 10.1016/0378-1119(94)00833-e. [DOI] [PubMed] [Google Scholar]
- 48.Ross J I, Eady E A, Cove J H, Cunliffe W J, Baumberg S, Wootton J C. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol Microbiol. 1990;4:1207–1214. doi: 10.1111/j.1365-2958.1990.tb00696.x. [DOI] [PubMed] [Google Scholar]
- 49.Rouch D A, Byrne M E, Kong Y C, Skurray R A. The aacA-aphD gentamicin and kanamycin resistance determinant of Tn4001 from Staphylococcus aureus: expression and nucleotide sequence analysis. J Gen Microbiol. 1987;133:3039–3052. doi: 10.1099/00221287-133-11-3039. [DOI] [PubMed] [Google Scholar]
- 50.Rupp M E, Archer G L. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin Infect Dis. 1994;19:231–245. doi: 10.1093/clinids/19.2.231. [DOI] [PubMed] [Google Scholar]
- 51.Salisbury S M, Sabatini L M, Spiegel C A. Identification of methicillin-resistant staphylococci by multiplex polymerase chain reaction. Am J Clin Pathol. 1997;107:368–373. doi: 10.1093/ajcp/107.3.368. [DOI] [PubMed] [Google Scholar]
- 52.Seppälä H, Skurnik M, Soini H, Roberts M, Huovinen P. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob Agents Chemother. 1998;42:257–262. doi: 10.1128/aac.42.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaw K J, Rather P N, Hare R S, Miller G H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57:138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheagren J N. Staphylococcus aureus: the persistent pathogen. N Engl J Med. 1984;310:1368–1442. doi: 10.1056/NEJM198405243102107. [DOI] [PubMed] [Google Scholar]
- 55.Sutcliffe J, Grebe T, Taitkamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki E, Kuwahara-Arai K, Richardson J F. Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob Agents Chemother. 1993;37:1219–1226. doi: 10.1128/aac.37.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szybalski W. Microbial selection. I. Gradient plate technique for study of bacterial resistance. Science. 1952;116:46–48. [Google Scholar]
- 58.Thakker-Varia S, Jenssen W D, Moon-McDermott L, Weinstein M P, Dubin D T. Molecular epidemiology of macrolides-lincosamides-streptogramin B resistance in Staphylococcus aureus and coagulase-negative staphylococci. Antimicrob Agents Chemother. 1987;31:735–743. doi: 10.1128/aac.31.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ünal S, Hoskins J, Flokowitsch J E, Wu C Y E, Preston D A, Skatrud P L. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J Clin Microbiol. 1992;30:1685–1691. doi: 10.1128/jcm.30.7.1685-1691.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ünal S, Werner K, De Girolami P, Barsant F, Eliopoulos G. Comparison of tests for detection of methicillin-resistant Staphylococcus aureus in a clinical microbiology laboratory. Antimicrob Agents Chemother. 1994;38:345–347. doi: 10.1128/aac.38.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanhoof R, Godard C, Content J, Nyssen H J, Hannecart-Pokorni E. Detection by polymerase chain reaction of genes encoding aminoglycoside-modifying enzymes in methicillin-resistant Staphylococcus aureus isolates of epidemic phage types. J Med Microbiol. 1994;41:282–290. doi: 10.1099/00222615-41-4-282. [DOI] [PubMed] [Google Scholar]
- 62.Vannuffel P, Gigi J, Ezzedine H, Vandercam B, Delmee M, Wauters G, Gala J-L. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J Clin Microbiol. 1995;33:2864–2867. doi: 10.1128/jcm.33.11.2864-2867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Westh H, Hougaard D M, Vuust J, Rosdahl V T. Erm genes in erythromycin-resistant Staphylococcus aureus and coagulase-negative staphylococci. APMIS. 1995;103:225–232. [PubMed] [Google Scholar]
- 65.Westh H, Hougaard D M, Vuust J, Rosdahl V T. Prevalence of erm gene classes in erythromycin-resistant Staphylococcus aureus strains isolated between 1959 and 1988. Antimicrob Agents Chemother. 1995;39:369–373. doi: 10.1128/aac.39.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams J G, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;25:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]