Abstract
Bombyx mori nucleopolyhedrovirus (BmNPV) is one of the most serious pathogens and causes serious economic losses in sericulture. At present, there is no epigenetic modification of BmNPV transcripts, especially of m6A, and this modification mediates diverse cellular and viral functions. This study showed that m6A modifications are widespread in BmNPV transcripts in virally infected cells and the identified m6A peaks with a conserved RRACH sequence. m6A sites predominantly appear in the coding sequences (CDS) and the 3′-end of CDS. About 37% of viral genes with m6A sites deleted from the viral genome did not produce any infectious virions in KOV-transfected cells. Among the viral genes related to replication and proliferation, ie-1 mRNA was identified with a higher m6A level than other viral genes. The m6A sites in the ie-1 mRNA may be negatively related to the protein expression. Viral replication was markedly inhibited in cells overexpressed with BmYTHDF3 in a dose-dependent manner, and a contrary effect was found in si-BmYTHDF3-transfected cells. Collectively, the identification of putative m6A modification in BmNPV transcripts provides a foundation for comprehensively understanding the viral infection, replication, and pathobiology in silkworms.
Keywords: BmNPV, m6A modification, ie-1, BmYTHDF3, viral replication
Introduction
Bombyx mori nucleopolyhedrovirus (BmNPV) is one of the most serious pathogens and causes serious economic losses in sericulture (1). Its genome is circular double-strand DNA with 130 kb and encodes 141 putative proteins (2). BmNPV is a classical member of baculoviruses with two types of virions in the whole life cycle. The budded virus (BV) is responsible for the horizontal transmission among the cells, and the occlusion-derived virus (ODV) is responsible for the vertical transmission (3). BmNPV belongs to the nuclear-replicated virus and is inevitably modified by a series of host modification enzymes, including m6A modification.
N6-Methyladenosine (m6A) is one of eukaryotic mRNA’s most abundant internal modifications. It is also a dynamic reversible RNA modification, which plays an important role in the structure, positioning, and function of RNA (4, 5). m6A is involved in various biological processes, including stress response, fertility, stem cell differentiation, circadian rhythm, microRNA biogenesis, and cancer occurrence development (6–9). m6A modification systems in eukaryotic cells mainly include methyltransferase (Writer), demethylases (Eraser), and m6A reader protein (Reader) (10). The motif domain of RRAmCH (R=G/A, G > A, H=A, C, U) is recognized by Writer and Reader in the cells (5). m6A modification on mRNA is catalyzed by the methyltransferase complex (Writers), including methyltransferase-like 3/14 (METTL3/METTL14) and other cofactors (11). Demethylase fat mass and obesity-associated protein (FTO) and alkB homologous protein (ALKBH) as Erasers are mainly responsible for the demethylation of m6A modification (12). The YTH domain-containing m6A reader proteins (YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2) and the homogeneous nuclear protein family (HNRNPA2B1 and HNRNPC) can recognize the m6A site or region by participating in the regulation of mRNA stability, mRNA structure, mRNA nuclear export, and mRNA splicing and translation (13–19). To date, m6A modifications appear in many DNA and RNA viral mRNAs via the host m6A modification machinery (20). m6A modifications were reported to play important roles in the viral life cycle. Both influenza A virus (IAV) mRNA and viral genome RNA are m6A modified, and the expression level of a viral gene can be upregulated by cis-regulation of this m6A modification (21). m6A modification appears in the RNA genome of hepatitis C virus (HCV), and simultaneous silencing of m6A methyltransferase-like 3/14 (METTL3/METTL14) significantly increased the expression level of the viral NS5A protein. By silencing the demethylase FTO gene, the non-structural 5A (NS5A) protein level was significantly reduced (22). Similarly, the effects of Writer and Eraser on Zika virus (ZIKV) were consistent with HCV (23). The m6A modification system inhibits HCV production by affecting viral assembly, while inhibition of ZIKV is through its effects on viral replication (23–25). After vesicular stomatitis virus (VSV) infection, the level of DEAD-box 46 (DDX46) RNA helicase and demethylase ALKBH5 in the nucleus increased significantly, which inhibited antiviral protein expression and the host innate immune response to VSV (26). When the human immunodeficiency virus (HIV) infects lymphocytes, the level of m6A methylation is elevated in both viral RNAs and host cellular RNAs. Downregulation of METTL3 and METTL14 gene expression levels in CD4 T lymphocytes inhibits HIV replication, while downregulation of ALKBH5 gene expression levels promotes HIV replication (25). METTL3 also interacted with enterovirus 71 (EV71) RNA-dependent RNA polymerase (RdRp) 3D. The 3D protein stability and transcriptional efficiency were increased by increasing 3D sumoylation/ubiquitination modification to promote viral replication (27). The m6A modification of human metapneumovirus (HMPV) can help the virus successfully evade host immune system recognition, thus promoting HMPV replication and gene expression (28). It has been reported that mRNAs of DNA viruses are modified by m6A and participate in the viral life cycle. It was found that Kaposi sarcoma-associated herpesvirus (KSHV) replication was inhibited by knockdown of the METTL3 or YTHDF1-3 gene (29). m6A modification has a positive regulatory effect in the life cycle of Simian virus 40 (SV40), and overexpression of YTHDF2 can accelerate virus replication (30). The m6A modification site located in the 3′UTRs and 3′ epsilon loop of the hepatitis B virus (HBV) genome is not conducive to the stability of HBV RNA. The m6A modification site located in the 5′ ϵ ring of the HBV genome has a positive regulatory effect on pregenomic RNA (pgRNA) reverse transcription (31). EBV virus (EBV) transcripts are many m6A modification sites. YTHDF1 can recognize m6A sites accelerating viral RNA uncapping and recruit RNA degradation complexes to inhibit viral infection and replication (32). m6A modification in adenovirus (ADV) transcripts involves regulating splicing and viral RNA expression (33). The porcine epidemic diarrhea virus (PEDV) genome contains multiple m6A modification sites and negatively regulates the virus infection (34). Our preliminary findings showed that the expression levels of writers (BmMETTL3 and BmMETTL3) and reader (BmYTHDF3) of m6A machinery in BmN cells were negative to the expression of viral structural protein VP39 in BmNPV infection (35). These studies indicate that both DNA and RNA viruses are regulated by the m6A modification system in host cells during viral infection, but the m6A modification system has different regulatory mechanisms for different viruses.
BmNPV belongs to the nuclear-replicated virus. Whether m6A modification is introduced into the viral transcripts in the viral proliferation process, and whether this apparent modification is beneficial for the virus to evade the detection of the host’s innate immune system, is still unclear. In this study, the m6A modification was investigated by methylated RNA immunoprecipitation (MeRIP) sequencing from the BmNPV-infected midgut, and m6A sites were widespread in the viral transcripts. Furthermore, the m6A machinery negatively mediated the viral replication. The results will uncover the roles of BmNPV transcript m6A modification in the viral life cycle and provide a scientific base for anti-BmNPV.
Materials and Methods
Sample Preparation
The domesticated silkworm strain (Jingsong×Haoyue) was reared in our lab with fresh mulberry leaves at 25°C. The polyhedrin (1*106) of BmNPV stored in our lab at Soochow University was evenly coated on the mulberry leaves with 4*4 cm for the first day of 5 instar larvae. 48 h postinfection, the midguts were collected and extracted for total RNA. BmN cells were cultured in TC-100 medium with 10% fetal bovine serum (HyClone, Logan, UT, USA) in a 26°C incubator. The recombinant BmNPV-GFP was gifted by Professor Xiaofeng Wu from the Zhejiang University.
Methylated RNA Immunoprecipitation Sequencing
30 silkworm larvae were infected by BmNPV, and the midguts were used to extract the total RNA with the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The total RNA was fragmented with the corresponding buffer (10 mM ZnCl2, 10 mM Tris–HCl pH 7.0). The obtained fragments were captured with the m6A antibody, and the corresponding libraries were utilized for MeRIP sequencing (MeRIP-Seq), and further bioinformatics analysis was also conducted by Shanghai OE Biotech. Co., Ltd. (Shanghai, China). The sequencing data were deposited in the NCBI with accession numbers (SRR10141250, SRR10141249, SRR10141248, and SRR10141247) (35). BmNPV T3 (accession number: L33180.1) was used as the reference genome for mapping.
MeRIP-qPCR
The method for preparation of total RNA for MeRIP-qPCR was the same protocol as MeRIP sequencing, except that total RNA was not treated with the fragmentation buffer. Briefly, BmNPV-infected BmN cells were lysed with lysis buffer. The lysed cells were separated into parts. One part of lysate was incubated with an anti-m6A antibody, and another part was incubated with IgG control (Rabbit) conjugated to Protein A/G beads in 1 ml RNA immunoprecipitation (RIP) buffer. After incubation at 4°C overnight, the beads were washed with wash buffer 3 times, and the RIP samples were reverse transcribed into cDNA using the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China) and subjected to RT-qPCR. Translation initiation factor 4a (TIF-4A) was used as a housekeeping gene (36). The specific primers for each viral gene used in real-time PCR are listed in Supplementary Table 1 . All experiments were conducted three times.
Plasmids and siRNA
The genome loci of ie-1 in BmNPV T3 (L33180.1) are 116994.118748 and were synthesized by the Sangon Biotechnology company (Shanghai, China). The DNA sequence was inserted into the Kpn I and Xba I sites in the pIZT-V5/his vector (Invitrogen, Frederick, MD, USA) for pIZT-V5his-ie. A mutant fragment of ie-1 was mutated A to T at the potential m6A sites. This mutant DNA sequence was also synthesized by the Sangon Biotechnology company (Shanghai, China) and was inserted into the sites of Kpn I and Xba I in the pIZT-V5/his vector for pIZT-V5his-ie-1mut. pIZT-BmYTHDF3 and siRNA (si-BmYTHDF3) specifically targeting BmYTHDF3 were reported in our previous study (35).
Western Blotting
BmN cells (1*105) were transfected with 2 μg pIZT-V5/his vector and pIZT-V5his-ie-1mut with Lipofectamine (Roche, Mannheim, Germany), respectively. 48 h post-transfection, the cells were harvested and lysed in RIPA lysis buffer (Beyotime, Shanghai, China) supplemented with protease inhibitors (Beyotime, Shanghai, China). The expression levels of IE-1 and enhanced green fluorescence protein (EGFP) were detected with anti-His, anti-tubulin, and anti-EGFP antibodies (Proteintech, Rosemont, IL, USA) as the primary antibodies, and horseradish peroxidase-conjugated goat anti-mouse IgG (Proteintech, USA) was used as the secondary antibody. The reactions were visualized using an ECL reagent (Sangon, Shanghai).
Overexpression and Depletion of BmYTHDF3 in BmN Cells and BmNPV Infection
BmN cells (1*105 cells/well) were transfected with a recombinant expression plasmid (pIZT-BmYTHDF3) with the transfection reagent. 48 h post-transfection, BmN cells were infected with BmNPV-GFP (MOI = 2) and harvested for extraction of total proteins at 12 h postinfection for Western blotting. Meanwhile, BmN cells (1*105 cells/well) were transfected with specific siRNA targeting BmYTHDF3 (si-BmYTHDF3) with the transfection reagent. 48 h post-transfection, BmN cells were infected with BmNPV-GFP (MOI = 2) and harvested for extraction of total proteins at 12 h postinfection for Western blotting. The expression of the Bm59 gene of BmNPV was used as the indicator of viral replication (37).
Bioinformatics
m6A modification sites on the interested RNA sequences were carried out by the SRAMP prediction server (38). The motif discovery from the sets of sequences was investigated by HOMER software (39). The flexibility of protein local structures was studied through (i) the B-factor of the X-ray experiment and (ii) the fluctuation of residues during molecular dynamics simulations (40). The Flexibility Prediction was utilized to analyze the flexibility classes (0, 1, and 2 represent rigid, intermediate, and flexible, respectively) according to the observed dynamic properties from the target protein sequences.
Results
m6A Modifications Are Widespread in BmNPV Transcripts in Virally Infected Cells
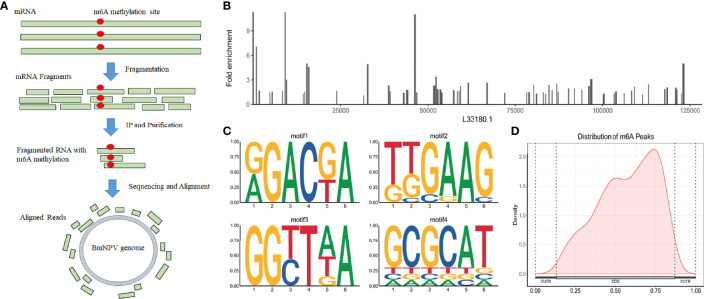
BmNPV-infected silkworm midguts were collected and sequenced with methylated RNA immunoprecipitation combined with high-throughput sequencing (MeRIP-Seq) to understand the m6A modification in mRNAs, including host mRNAs and viral mRNAs ( Figure 1A ). Our previous study found that thousands of m6A peaks were identified from BmNPV-infected tissues, and the host m6A modification system was related to the expression of the viral gene. However, this modification is still a puzzle whether it appears in the BmNPV mRNAs. To better understand the post-transcription modification, especially m6A sites on the viral genes and the potential roles in the viral life cycle, we compared the MeRIP-Seq data with the BmNPV T3 (L33180.1) reference genome and found that 74 regions with high potential m6A methylation (m6A peak) were identified in 59 mRNA transcripts of BmNPV ( Figure 1B and Table 1 ). Among these m6A peaks, the mean size of the m6A peak is 262 bp, while the maximum and minimum peaks are 599 and 135 bp, respectively. HOMER motif enrichment analysis was utilized to analyze the RRACH motif conservation in 74 identified m6A peaks. The results showed that four conserved RRACH motifs were identified from the alignments between the de novo motif and known motifs and met the requirement of the motif domain of RRAmCH (R=G/A, G > A; H=A, C, U) ( Figure 1C ). The positions of m6A sites were analyzed to understand the appearance of m6A sites in viral mRNAs. The results indicated that m6A modification mainly appeared in coding sequences (CDS) and the 3′-end of CDS ( Figure 1D ). These results suggested a potential biofunctional significance for m6A modification in viral mRNA.
Figure 1.
BmNPV transcripts are m6A modified. (A) The workflow of MeRIP is for the detection of m6A modification in BmNPV transcripts. (B) The distribution of m6A peaks on the genome of BmNPV T3 (L33180.1). (C) The conserveness of the RRACH motifs within the identified m6A peaks using HOMER. (D) Distribution of m6A peaks on viral transcripts including 5′-UTR, CDS, and 3′-UTR.
Table 1.
The information of m6A peaks on BmNPV transcripts from the MeRIP sequencing.
| Chromosome/genome | ChromStart | ChromEnd | Name | Score | Strand | Sizes |
|---|---|---|---|---|---|---|
| L33180.1 | 57541 | 57675 | Orf_61 | 0.022 | – | 134 |
| L33180.1 | 91856 | 91999 | Orf_95 | 0.0018 | – | 143 |
| L33180.1 | 85439 | 85584 | he65 | 6.60E-05 | – | 145 |
| L33180.1 | 78238 | 78383 | bro-c | 0.00025 | + | 145 |
| L33180.1 | 82971 | 83117 | Orf_87 | 5.60E-07 | – | 146 |
| L33180.1 | 9016 | 9163 | Orf_9 | 0.00095 | + | 147 |
| L33180.1 | 71601 | 71748 | Orf_76 | 0.11 | + | 147 |
| L33180.1 | 127894 | 128041 | lef-2 | 0.00018 | + | 147 |
| L33180.1 | 72394 | 72394 | odv-e25 | 0.49 | + | 148 |
| L33180.1 | 122464 | 122612 | pe38 | 1.30E-66 | + | 148 |
| L33180.1 | 11024 | 11172 | arif-1 | 0.0036 | – | 148 |
| L33180.1 | 25369 | 25517 | 39k | 0.0098 | – | 148 |
| L33180.1 | 26006 | 26154 | 39k | 0.97 | – | 148 |
| L33180.1 | 121197 | 121345 | ie-2 | 2.20E-07 | – | 148 |
| L33180.1 | 121636 | 121784 | ie-2 | 2.20E-07 | – | 148 |
| L33180.1 | 14226 | 14375 | Orf_14 | 0.00052 | + | 149 |
| L33180.1 | 30886 | 31035 | gta | 0.033 | + | 149 |
| L33180.1 | 52237 | 52386 | Orf_54 | 0.037 | + | 149 |
| L33180.1 | 83635 | 83784 | vp80 | 0.018 | + | 149 |
| L33180.1 | 93966 | 94115 | Orf_97 | 1.00E-05 | + | 149 |
| L33180.1 | 80205 | 80355 | lef-5 | 7.90E-10 | + | 150 |
| L33180.1 | 36040 | 36190 | lef-8 | 1.80E-05 | – | 150 |
| L33180.1 | 125383 | 125533 | bro-d | 0.00071 | – | 150 |
| L33180.1 | 108994 | 109144 | p74 | 7.90E-06 | – | 150 |
| L33180.1 | 106072 | 106223 | p35 | 0.00098 | + | 151 |
| L33180.1 | 4703 | 4854 | Orf_5 | 0.0035 | – | 151 |
| L33180.1 | 61422 | 61610 | Orf_67 | 0.0017 | – | 188 |
| L33180.1 | 56767 | 56957 | Orf_60 | 2.90E-05 | – | 190 |
| L33180.1 | 16947 | 17139 | dbp | 0.001 | – | 192 |
| L33180.1 | 108182 | 108376 | p26 | 2.10E-05 | + | 194 |
| L33180.1 | 53267 | 53461 | lef-3 | 0.004 | – | 194 |
| L33180.1 | 53797 | 53991 | lef-3 | 0.01 | – | 194 |
| L33180.1 | 78756 | 78950 | 38k | 0.0021 | – | 194 |
| L33180.1 | 19447 | 19643 | Orf_21 | 6.60E-05 | – | 196 |
| L33180.1 | 31475 | 31672 | gta | 0.0012 | + | 197 |
| L33180.1 | 71903 | 72100 | odv-e25 | 0.0093 | + | 197 |
| L33180.1 | 90186 | 90383 | Orf_94 | 0.0043 | – | 197 |
| L33180.1 | 8263 | 8461 | bv/odv-e26 | 0.0021 | + | 198 |
| L33180.1 | 23755 | 23955 | fgf | 6.30E-18 | + | 200 |
| L33180.1 | 86801 | 87002 | Orf_90 | 6.30E-19 | + | 201 |
| L33180.1 | 111302 | 111545 | me53 | 0.00025 | – | 243 |
| L33180.1 | 9435 | 9679 | Orf_10 | 0.0034 | – | 244 |
| L33180.1 | 112889 | 113136 | ie-0 | 0.03 | + | 247 |
| L33180.1 | 866 | 1113 | orf1629 | 2.00E-05 | – | 247 |
| L33180.1 | 46527 | 46774 | gp37 | 0.00019 | – | 247 |
| L33180.1 | 5301 | 5550 | Orf_5 | 0.003 | – | 249 |
| L33180.1 | 70170 | 70449 | Orf_74 | 3.20E-07 | – | 279 |
| L33180.1 | 17751 | 18045 | iap1 | 0.00055 | + | 294 |
| L33180.1 | 84525 | 84823 | vp80 | 0.0017 | + | 298 |
| L33180.1 | 80936 | 81234 | Orf_85 | 0.00048 | – | 298 |
| L33180.1 | 87150 | 87451 | Orf_90 | 6.30E-19 | + | 301 |
| L33180.1 | 15729 | 16053 | pkip | 0.00013 | – | 324 |
| L33180.1 | 38589 | 38925 | Orf_40 | 2.00E-96 | + | 336 |
| L33180.1 | 32541 | 32881 | Orf_36 | 0.0021 | + | 340 |
| L33180.1 | 115589 | 115929 | odv-ec27 | 0.002 | + | 340 |
| L33180.1 | 120710 | 121052 | ie-2 | 2.20E-07 | – | 342 |
| L33180.1 | 101807 | 102150 | p24 | 0.0017 | + | 343 |
| L33180.1 | 42816 | 43191 | Orf_47 | 0.021 | – | 375 |
| L33180.1 | 57051 | 57432 | Orf_60 | 2.90E-05 | – | 381 |
| L33180.1 | 118211 | 118602 | ie-1 | 0.0024 | + | 391 |
| L33180.1 | 52483 | 52879 | Orf_54 | 1.10E-05 | + | 396 |
| L33180.1 | 58288 | 58685 | vlf-1 | 0.0018 | – | 397 |
| L33180.1 | 45981 | 46402 | Orf_51 | 6.30E-08 | + | 421 |
| L33180.1 | 117432 | 117871 | ie-1 | 0.0029 | + | 439 |
| L33180.1 | 1458 | 1903 | orf1629 | 1.40E-05 | – | 445 |
| L33180.1 | 43802 | 44248 | fp | 0.0019 | – | 446 |
| L33180.1 | 103057 | 103507 | pp34 | 0.00022 | + | 450 |
| L33180.1 | 103556 | 104030 | Orf_109 | 0.01 | + | 474 |
| L33180.1 | 66807 | 67281 | p30 | 4.00E-04 | – | 474 |
| L33180.1 | 122855 | 123345 | pe38 | 1.30E-66 | + | 490 |
| L33180.1 | 51448 | 51942 | Orf_54 | 1.10E-05 | + | 494 |
| L33180.1 | 95815 | 96354 | Orf_101 | 9.10E-08 | + | 539 |
| L33180.1 | 96473 | 97059 | lef-7 | 2.50E-17 | – | 586 |
| L33180.1 | 100092 | 100690 | gp64/67 | 4.20E-05 | – | 598 |
Viral Genes With m6A Sites Were Classified Into Four Phenotypes
The KOV (bacmid)s were subdivided into four phenotypes (A to D) according to the results from the knockout BmNPVs (KOVs) for each viral gene using the lambda red recombination system (41). The defined phenotypes of type-A and -B KOVs could produce infectious viruses, but types C and D did not produce any infectious virions in KOV-transfected cells (2). According to the appearance of m6A in the viral genes, we found 33, 4, 17, and 5 viral genes in types A, B, C, and D, respectively ( Table 2 ).
Table 2.
The KOV (bacmid)s were subdivided into four phenotypes (A to D) according to the results from the knockout BmNPVs (KOVs) for each viral gene using the lambda red recombination system.
| A | B | C | D |
|---|---|---|---|
| pe38 | bro-d | lef-2 | lef-5 |
| arif-1 | me53 | odv-e25 | lef-8 |
| 39k | ie-0 | vp80 | lef-3 |
| ie-2 | pkip | 38k | ie-1 |
| gta | gp64/67 | orf1629 | |
| he65 | odv-ec27 | ||
| bro-c | vlf-1 | ||
| p74 | dbp | ||
| p35 | p45 | ||
| p26 | bv/odv-c42 | ||
| bv/odv-e26 | dnaJ domain | ||
| fgf | Orf_61 | ||
| p30 | Orf_76 | ||
| gp37 | Orf_54 | ||
| iap1 | Orf_67 | ||
| fp | Orf_90 | ||
| p24 | Orf_109 | ||
| pp34 | |||
| pif-3 | |||
| pif-1 | |||
| chaB-like | |||
| lef-7 | |||
| Orf_9 | |||
| Orf_14 | |||
| Orf_5 | |||
| Orf_60 | |||
| Orf_21 | |||
| Orf_94 | |||
| Orf_10 | |||
| Orf_74 | |||
| Orf_36 | |||
| Orf_51 | |||
| Orf_101 |
A and -B KOVs could produce infectious viruses, but type-C and -D did not produce any infectious virions in KOV-transfected cells.
Validation of m6A Modification in Viral Genes
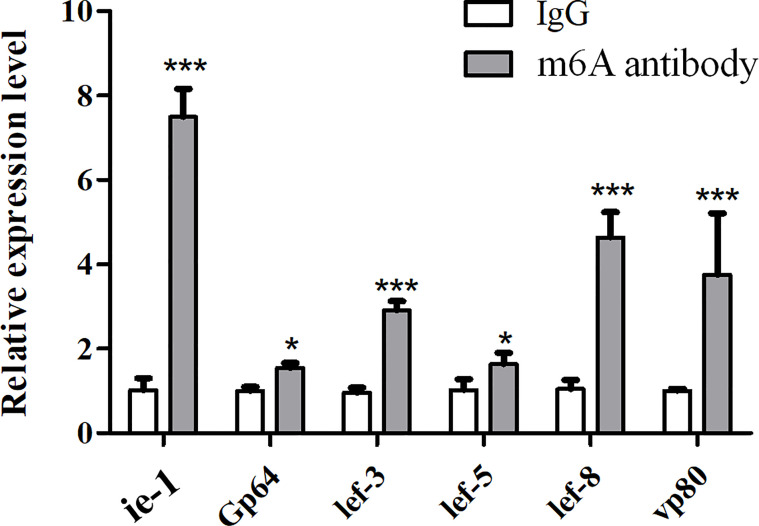
To know the existence of the m6A site in viral genes, multiple genes related to viral replication (lef-8, lef-5, lef-3, VP80, GP64/78, and ie-1) were selected for validation using m6A antibody immunoprecipitation combined with qPCR (MeRIP-qPCR). The results showed that all these viral genes could be enriched by the m6A antibody and detected by qPCR (real-time PCR) using the corresponding primers of each gene ( Supplementary Table 1 ). Compared to the relative expression levels of viral genes in the complex of the m6A antibody and viral mRNAs with m6A modification, the results showed that ie-1 mRNA with a higher m6A level than other viral genes was identified from the complex ( Figure 2 ), indicating that enriched m6A modification in the ie-1 gene may be related to the transcription or translation.
Figure 2.
Validation of m6A modification in viral replication-related genes using m6A antibody immunoprecipitation combined with qPCR (MeRIP-qPCR). The IgG or anti-m6A antibody enrichment of the viral mRNAs was measured by MeRIP-qPCR in BmNPV-infected BmN cells. *P < 0.05 and ***P < 0.001.
The Expression Level of IE-1 Was Related to the m6A Sites
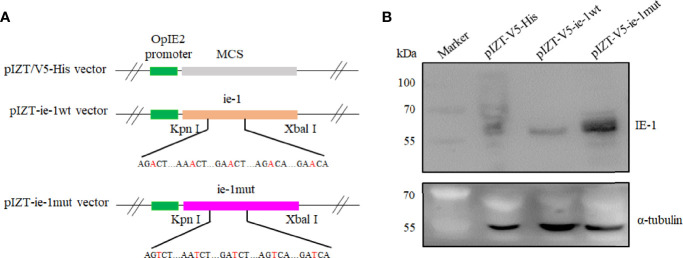
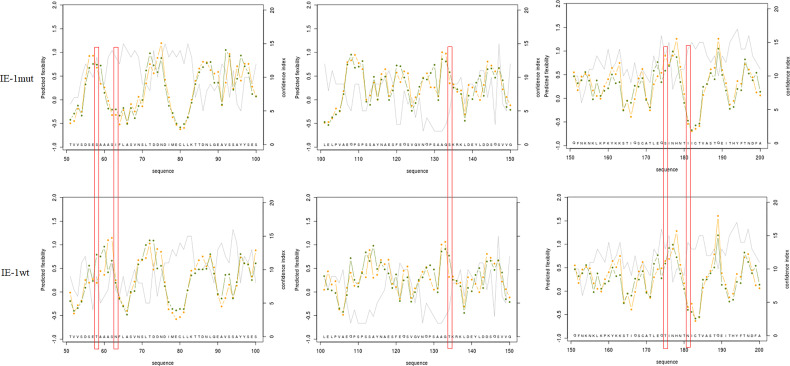
To further validate the function of m6A sites in the ie-1 mRNA, the wild-type and mutant ie-1 plasmids were constructed by cloning a wild-type 1,752-nucleotide (nt) length fragment (remove the TAA) encompassing the m6A peak region in the ie-1 mRNA and a mutant fragment with A > T mutations at the potential m6A sites, respectively ( Figure 3A ). The plasmids were transfected into BmN cells for Western blotting assay, which exhibited a higher IE-1 expression level on the mutant plasmid than wild-type ie-1 ( Figure 3B ), confirming that the presence of m6A sites in the ie-1 mRNA may be related to its expression. To exclude the influence of codon on protein expression, the flexibility of protein local structures was studied through (i) the B-factor of the X-ray experiment and (ii) the fluctuation of residues during molecular dynamics simulations (40). The prediction results showed that these 5 amino acid mutants in the IE-1 had not altered the protein flexibility ( Figure 4 ).
Figure 3.
Construction of the ie-1wt and ie-1mut plasmids and detection of the expression level of IE-1 in BmN cells transfected with plasmids. (A) The wild-type and mutant ie-1 plasmids were constructed by cloning a 1,752-nt length fragment (remove the TAA) encompassing the m6A peak region in the ie-1 mRNA and a mutant fragment with A > T mutations at the potential m6A sites, respectively. (B) IE-1 expression was detected from the plasmid-transfected BmN cells with Western blotting assay. pIZT-V5-His plasmid as the control.
Figure 4.
The flexibility of protein local structures was studied through (i) the B-factor of the X-ray experiment and (ii) the fluctuation of residues during molecular dynamic simulations. IE-1wt (58T, 63N, 134T, 175T, and 181N) > IE-1mut (58S, 63I, 134S, 175S, and 181I).
m6A Reader (BmYTHDF3) Negatively Regulated the Viral Replication
Our previous study concluded that cytoplasmic YTH-domain family 3 (BmYTHDF3) was indicated as the m6A reader of the m6A machinery in silkworm, Bombyx mori, and the m6A modification system was predicted to play important roles in the BmNPV infection (35). The appearance of m6A modification in the ie-1 gene means that BmYTHDF3 could recognize these m6A sites. However, the exact roles of BmYTHDF3 in the replication of BmNPV are still not clear, and we used the recombinant expression plasmid (pIZT-BmYTHDF3) and specific siRNA targeting BmYTHDF3 transfected into BmN cells followed by BmNPV-GFP infection for examining the roles of BmYTHDF3 in the viral replication. The Western blotting assay showed that the EGFP was markedly inhibited in cells overexpressed with BmYTHDF3 in a dose-dependent manner, and a contrary effect was found in si-BmYTHDF3-transfected cells ( Figures 5A, B ). Meanwhile, a real-time PCR assay obtained similar results via detecting the Bm59 gene expression level in BmN cells overexpressed with BmYTHDF3 and transfected with si-BmYTHDF3 ( Figures 5C, D ), suggesting that BmYTHDF3 could inhibit the BmNPV infection in BmN cells via recognizing the m6A sites in the viral gene.
Figure 5.
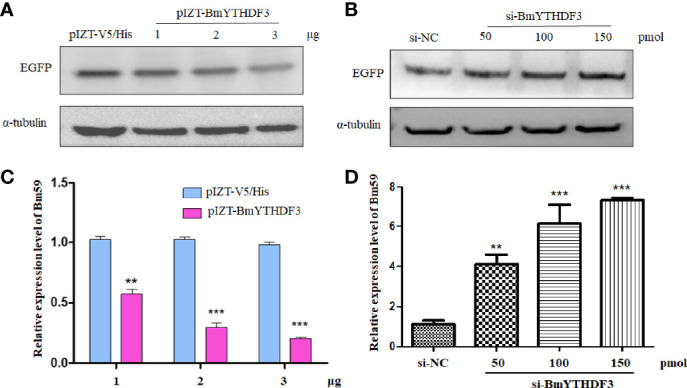
Knockdown of BmYTHDF3 suppressed BmNPV infection and replication. (A) The infection of BmNPV in BmN cells transfected with pIZT-BmYTHDF3 (1, 2, and 3 μg) or pIZT-V5/His as control following BmNPV-GFP infection was detected by Western blotting. (B) The infection of BmNPV in BmN cells transfected with si-BmYTHDF3 (50, 100, and 150 pmol) or si-NC as control following BmNPV-GFP infection was detected by Western blotting. (C) The relative expression level of Bm59 in BmN cells transfected with pIZT-BmYTHDF3 (1, 2, and 3 μg) or pIZT-V5/His as control following BmNPV-GFP infection was detected by real-time PCR. (D) The relative expression level of Bm59 in BmN cells transfected with si-BmYTHDF3 (50, 100, and 150 pmol) or si-NC as control following BmNPV-GFP infection was detected by real-time PCR. **p < 0.05 and ***p < 0.001.
Discussion
Increasing data indicated that m6A modification not only exists in the cellular mRNAs but also is widespread in the viral transcripts, including DNA and RNA viruses. Viral m6A modifications have been reported as pro-viral or antiviral functions depending on the virus species, cell type, and viral RNA location (20). We showed that m6A modification is widespread in BmNPV transcripts in virally infected cells. Viral genes with m6A sites were classified into four phenotypes according to the results from the knockout BmNPVs (KOVs) for each viral gene using the lambda red recombination system (41). We demonstrated that ie-1 mRNA with a higher m6A level than other viral genes was examined, and the expression level of IE-1 was related to the m6A sites in the viral ie-1 gene. Furthermore, m6A reader protein BmYTHDF3 suppressed the BmNPV infection in BmN cells, maybe recognizing the m6A sites in the viral gene.
m6A machinery regulates cellular RNAs of viral RNAs by writers, erasers, and readers in cells (42). The appearance of m6A modification in viral RNAs and cellular m6A modification systems played vital roles in the viral life cycle. Our previous study found that the cellular mRNAs with m6A modification were altered following BmNPV infection and the expression levels of writers and readers negatively mediated BmNPV infection (35). However, the precise regulation of m6A modification on viral infection was vague. Therefore, we made assumptions about widespread m6A sites in the BmNPV transcripts like other viruses, and the viral RNA with m6A sites could be regulated by cellular m6A reader proteins. Presently, any reports showed that BmNPV transcripts could be modified by the m6A machinery. MeRIP-Seq data were aligned with the BmNPV T3 (L33180.1) reference genome, and 74 regions with high potential m6A methylation (m6A peak) were distributed in 59 viral mRNA transcripts. Among these m6A peaks, four conserved RRACH motifs were identified as other species. The enrichment analysis of m6A peaks on viral mRNAs showed that most were distributed on the CDS and 3′-CDS of viral mRNAs. It is known that m6A modification was found near the 3′ UTR and stop codon in mammals and mice (43). The methylation landscape across human adult tissues showed m6A sites preferentially around stop codons, essential for maintaining basic cellular function. Most of the m6A in the genes with tissue specificity were found in the 5′-UTR (44), suggesting that these viral genes with m6A sites may be essential for distinct regulatory effects in the aspect of host range, viral replication, assembly, and host–virus interaction. 22/59 viral genes were demonstrated to be required to produce infectious viruses, and the m6A site appearance in these genes may be related to the viral replication. Furthermore, several viral genes related to viral replication were selected to be validated by MeRIP-qPCR, and the results also showed that m6A modifications were enriched in these selected viral genes. Among them, the modification level of m6A on the ie-1 gene was higher than other genes, indicating that these m6A sites might regulate their transcription or translation.
In our previous study, we found that the expression level of VP39 was decreased in BmN cells overexpressed with BmMETTL3 or BmYTHDF3, while the reverse results were obtained in the BmN cell depletion of BmMETTL3 or BmYTHDF3 using corresponding specific siRNAs (35). These suggested that the m6A modification system plays an important role in BmNPV infection with an unknown mechanism. The role of the YTHDF readers on viral replication was extensively examined by depletion and overexpression studies in numerous DNA and RNA viruses (29, 32, 45). Increasing data showed that YTHDF readers regulate the mRNA fate of cellular or viral m6A RNAs. It is described to regulate splicing, nuclear export, cap-independent translation, and decay (46). To better understand the regulatory effects of m6A machinery on the expression of viral genes or viral replication, the expression level of IE-1 was detected from BmN cells transfected with the wild-type and mutant ie-1 plasmids, and Western blotting assay exhibited a higher IE-1 expression level on the mutant plasmid than wild-type ie-1, confirming that the presence of m6A sites in the ie-1 mRNA may be related to its expression. When these mutant sites were introduced into ie-1, any adverse effect on protein flexibility was detected. Using the BmNPV-GFP-infected BmN cells overexpressed with BmYTHDF3 and transfected with si-YTHDF3, we found that the EGFP and viral gene Bm59 were significantly decreased and increased, respectively. These results suggest that BmYTHDF3 suppressed the BmNPV infection in BmN cells via recognizing the m6A sites in viral genes. The m6A modification system has different regulatory mechanisms for different viruses (45, 47). The appearance of m6A modification on viral RNA will be used as a molecular marker for evading host innate immune recognition of non-self RNA (28). Altogether, many BmNPV transcripts have been modified by the m6A machinery, including many genes related to viral infectivity. BmYTHDF3 suppressed the BmNPV infection by recognizing the m6A sites in viral genes, resulting in impairment of viral gene expression, avoiding viral overproliferation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Author Contributions
XH and CG contributed to the conception and design of the study. XZ, YZ, and JP performed the detection and analysis. XZ and XH wrote the first draft of the manuscript. CG wrote sections of the manuscript. All authors contributed to the manuscript revision and read and approved the submitted version.
Funding
This study was supported by the National Key R&D Program of China (2019YFA0905200), the China Postdoctoral Science Foundation (2019M651952), and the Priority Academic Program of Development of Jiangsu Higher Education Institutions.
Conflict of Interest
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.869313/full#supplementary-material
References
- 1. Yu H, Wang X, Xu J, Ma Y, Zhang S, Yu D, et al. iTRAQ-Based Quantitative Proteomics Analysis of Molecular Mechanisms Associated With Bombyx Mori (Lepidoptera) Larval Midgut Response to BmNPV in Susceptible and Near-Isogenic Strains. J Proteomics (2017) 165:35–50. doi: 10.1016/j.jprot.2017.06.007 [DOI] [PubMed] [Google Scholar]
- 2. Ono C, Kamagata T, Taka H, Sahara K, Asano S, Bando H. Phenotypic Grouping of 141 BmNPVs Lacking Viral Gene Sequences. Virus Res (2012) 165:197–206. doi: 10.1016/j.virusres.2012.02.016 [DOI] [PubMed] [Google Scholar]
- 3. Kelly BJ, King LA, Possee RD. Introduction to Baculovirus Molecular Biology. Methods Mol Biol (2016) 1350:25–50. doi: 10.1007/978-1-4939-3043-2_2 [DOI] [PubMed] [Google Scholar]
- 4. Meyer KD, Jaffrey SR. The Dynamic Epitranscriptome: N6-Methyladenosine and Gene Expression Control. Nat Rev Mol Cell Biol (2014) 15:313–26. doi: 10.1038/nrm3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fu Y, Dominissini D, Rechavi G, He C. Gene Expression Regulation Mediated Through Reversible M(6)A RNA Methylation. Nat Rev Genet (2014) 15:293–306. doi: 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 6. Li S, Mason CE. The Pivotal Regulatory Landscape of RNA Modifications. Annu Rev Genomics Hum Genet (2014) 15:127–50. doi: 10.1146/annurev-genom-090413-025405 [DOI] [PubMed] [Google Scholar]
- 7. Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S, Mason CE. The Birth of the Epitranscriptome: Deciphering the Function of RNA Modifications. Genome Biol (2012) 13:175. doi: 10.1186/gb-2012-13-10-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yue Y, Liu J, He C. RNA N6-Methyladenosine Methylation in Post-Transcriptional Gene Expression Regulation. Genes Dev (2015) 29:1343–55. doi: 10.1101/gad.262766.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic M(6)A mRNA Methylation Directs Translational Control of Heat Shock Response. Nature (2015) 526:591–4. doi: 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao BS, Roundtree IA, He C. Post-Transcriptional Gene Regulation by mRNA Modifications. Nat Rev Mol Cell Biol (2017) 18:31–42. doi: 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zaccara S, Ries RJ, Jaffrey SR. Reading, Writing and Erasing mRNA Methylation. Nat Rev Mol Cell Biol (2019) 20:608–24. doi: 10.1038/s41580-019-0168-5 [DOI] [PubMed] [Google Scholar]
- 12. Yang Y, Hsu PJ, Chen Y-S, Yang Y-G. Dynamic Transcriptomic M6a Decoration: Writers, Erasers, Readers and Functions in RNA Metabolism. Cell Res (2018) 28:616–24. doi: 10.1038/s41422-018-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem Cells. M6a mRNA Methylation Facilitates Resolution of Naive Pluripotency Toward Differentiation. Science (2015) 347:1002–6. doi: 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- 14. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-Methyladenosine-Dependent RNA Structural Switches Regulate RNA-Protein Interactions. Nature (2015) 518:560–4. doi: 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5' UTR M(6)A Promotes Cap-Independent Translation. Cell (2015) 163:999–1010. doi: 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-Methyladenosine-Dependent Regulation of Messenger RNA Stability. Nature (2014) 505:117–20. doi: 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-Methyladenosine Modulates Messenger RNA Translation Efficiency. Cell (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, et al. Structural Basis for Selective Binding of M6a RNA by the YTHDC1 YTH Domain. Nat Chem Biol (2014) 10:927–9. doi: 10.1038/nchembio.1654 [DOI] [PubMed] [Google Scholar]
- 19. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a Mammalian RNA Demethylase That Impacts RNA Metabolism and Mouse Fertility. Mol Cell (2013) 49:18–29. doi: 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baquero-Perez B, Geers D, Díez J. From A to M6a: The Emerging Viral Epitranscriptome. Viruses (2021) 13:1049. doi: 10.3390/v13061049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, et al. Epitranscriptomic Enhancement of Influenza A Virus Gene Expression and Replication. Cell Host Microbe (2017) 22:377–86.e375. doi: 10.1016/j.chom.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe (2016) 20:654–65. doi: 10.1016/j.chom.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, et al. Dynamics of Human and Viral RNA Methylation During Zika Virus Infection. Cell Host Microbe (2016) 20:666–73. doi: 10.1016/j.chom.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dang W, Xie Y, Cao P, Xin S, Wang J, Li S, et al. N(6)-Methyladenosine and Viral Infection. Front Microbiol (2019) 10:417. doi: 10.3389/fmicb.2019.00417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, et al. Dynamics of the Human and Viral M(6)A RNA Methylomes During HIV-1 Infection of T Cells. Nat Microbiol (2016) 1:16011. doi: 10.1038/nmicrobiol.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng Q, Hou J, Zhou Y, Li Z, Cao X. The RNA Helicase DDX46 Inhibits Innate Immunity by Entrapping M(6)A-Demethylated Antiviral Transcripts in the Nucleus. Nat Immunol (2017) 18:1094–103. doi: 10.1038/ni.3830 [DOI] [PubMed] [Google Scholar]
- 27. Hao H, Hao S, Chen H, Chen Z, Zhang Y, Wang J, et al. N6-Methyladenosine Modification and METTL3 Modulate Enterovirus 71 Replication. Nucleic Acids Res (2019) 47:362–74. doi: 10.1093/nar/gky1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu M, Zhang Z, Xue M, Zhao BS, Harder O, Li A, et al. N(6)-Methyladenosine Modification Enables Viral RNA to Escape Recognition by RNA Sensor RIG-I. Nat Microbiol (2020) 5:584–98. doi: 10.1038/s41564-019-0653-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hesser CR, Karijolich J, Dominissini D, He C, Glaunsinger BA. N6-Methyladenosine Modification and the YTHDF2 Reader Protein Play Cell Type Specific Roles in Lytic Viral Gene Expression During Kaposi's Sarcoma-Associated Herpesvirus Infection. PloS Pathog (2018) 14:e1006995. doi: 10.1371/journal.ppat.1006995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsai K, Courtney DG, Cullen BR. Addition of M6a to SV40 Late mRNAs Enhances Viral Structural Gene Expression and Replication. PloS Pathog (2018) 14:e1006919. doi: 10.1371/journal.ppat.1006919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim GW, Jang JY, et al. N6-Methyladenosine Modification of Hepatitis B Virus RNA Differentially Regulates the Viral Life Cycle. Proc Natl Acad Sci USA (2018) 115:8829–34. doi: 10.1073/pnas.1808319115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xia TL, Li X, Wang X, Zhu YJ, Zhang H, Cheng W, et al. N(6)-Methyladenosine-Binding Protein YTHDF1 Suppresses EBV Replication and Promotes EBV RNA Decay. EMBO Rep (2021) 4:e50128. doi: 10.15252/embr.202050128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Price AM, Hayer KE, McIntyre ABR, Gokhale NS, Abebe JS, Della Fera AN, et al. Direct RNA Sequencing Reveals M(6)A Modifications on Adenovirus RNA Are Necessary for Efficient Splicing. Nat Commun (2020) 11:6016. doi: 10.1038/s41467-020-19787-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen J, Jin L, Wang Z, Wang L, Chen Q, Cui Y, et al. N6-Methyladenosine Regulates PEDV Replication and Host Gene Expression. Virology (2020) 548:59–72. doi: 10.1016/j.virol.2020.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang X, Zhang Y, Dai K, Liang Z, Zhu M, Pan J, et al. N (6)-Methyladenosine Level in Silkworm Midgut/Ovary Cell Line Is Associated With Bombyx Mori Nucleopolyhedrovirus Infection. Front Microbiol (2019) 10:2988. doi: 10.3389/fmicb.2019.02988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo H, Jiang L, Xia Q. Selection of Reference Genes for Analysis of Stress-Responsive Genes After Challenge With Viruses and Temperature Changes in the Silkworm Bombyx Mori . Mol Genet Genomics (2016) 291:999–1004. doi: 10.1007/s00438-015-1125-4 [DOI] [PubMed] [Google Scholar]
- 37. Hu XL, Shen YW, Zheng Q, Wang GB, Wu XF, Gong CL. Bm59 is an Early Gene, But is Unessential for the Propagation and Assembly of Bombyx Mori Nucleopolyhedrovirus. Mol Genet Genomics (2016) 291:145–54. doi: 10.1007/s00438-015-1094-7 [DOI] [PubMed] [Google Scholar]
- 38. Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: Prediction of Mammalian N6-Methyladenosine (M6a) Sites Based on Sequence-Derived Features. Nucleic Acids Res (2016) 44:e91. doi: 10.1093/nar/gkw104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime Cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell (2010) 38:576–89. doi: 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bornot A, Etchebest C, Brevern A. Predicting Protein Flexibility Through the Prediction of Local Structures. Proteins-structure Funct Bioinf (2011) 79:839–52. doi: 10.1002/prot.22922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Datsenko KA, Wanner BL. One-Step Inactivation of Chromosomal Genes in Escherichia Coli K-12 Using PCR Products. Proc Natl Acad Sci USA (2000) 97:6640–5. doi: 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xue M, Zhao BS, Zhang Z, Lu M, Harder O, Chen P, et al. Viral N(6)-Methyladenosine Upregulates Replication and Pathogenesis of Human Respiratory Syncytial Virus. Nat Commun (2019) 10(1):4595. doi: 10.1038/s41467-019-12504-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA Mediates Preferential M(6)A mRNA Methylation in 3'UTR and Near Stop Codon and Associates With Alternative Polyadenylation. Cell Discov (2018) 4:10. doi: 10.1038/s41421-018-0019-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N, et al. Dynamic Landscape and Evolution of M6a Methylation in Human. Nucleic Acids Res (2020) 48:6251–64. doi: 10.1093/nar/gkaa347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan B, Gao SJ. RNA Epitranscriptomics: Regulation of Infection of RNA and DNA Viruses by N-6-Methyladenosine (M(6)a). Rev Med Virol (2018) 28:e1983. doi: 10.1002/rmv.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao B, Wang W, Zhao Y, Qiao H, Gao Z, Chuai X. Regulation of Antiviral Immune Response by N (6)-Methyladenosine of mRNA. Front Microbiol (2021) 12:789605. doi: 10.3389/fmicb.2021.789605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu PL, Cao SJ, Wu R, Zhao Q, Yan QG. Regulatory Effect of M(6) A Modification on Different Viruses. J Med Virol (2021) 93:6100–15. doi: 10.1002/jmv.27246 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .