Abstract
Although bacteriophages have been overshadowed as therapeutic agents by antibiotics for decades, the emergence of multidrug-resistant bacteria and a better understanding of the role of the gut microbiota in human health and disease have brought them back into focus. In this Perspective, we briefly introduce basic phage biology and summarize recent discoveries about phages in relation to their role in the gut microbiota and gastrointestinal diseases, such as inflammatory bowel disease and chronic liver disease. In addition, we review preclinical studies and clinical trials of phage therapy for enteric disease and explore current challenges and potential future directions.
Introduction
Changes in the human intestinal microbiota have been associated with gastrointestinal and liver diseases including inflammatory bowel disease, colorectal cancer (CRC), alcohol-associated liver disease and nonalcoholic fatty liver disease1–7. Although most of the changes have been described for bacteria, some studies have revealed changes in the gut virome that are associated with disease and developmental dysfunction8–14.
The human virome is dominated by bacteriophages (also known as phages), which are viruses that can infect bacteria15. After their discovery >100 years ago16–18, phages were widely used as antibacterials. However, the primitive state of microbial biology, decades before Watson and Crick, prevented meaningful scientific development of phage therapeutics, especially in the mid-century context of the discovery and rapid industrialization of small-molecule antibiotics19,20. In the past few decades, the widespread emergence of multidrug-resistant bacteria has reduced the practical utility of antibiotics21,22. Moreover, a new understanding of the close relationship between the intestinal microbiota and human health has brought into question the general applicability of broad-spectrum antibiotics23,24. Finally, modern molecular genetics, structural biology and high-throughput genomics have revealed such a high quantity and diversity of phages that most pathogenic bacteria could be targeted.
In this Perspective, we review the role of phages in maintaining human health and in disease pathogenesis, summarize advances in phage-based therapeutics including the direct use of phages in treating enteric disease, and discuss the manipulation of the gut microbiota by the targeting of specific bacterial species. Finally, we discuss the challenges to clinical application of phages and possible future directions for research.
Phage biology: structure and function
With an estimated population of more than 1031 particles, phages are the most abundant and diverse biological entities on Earth25,26. As natural predators of bacteria27,28, phages are ubiquitous in bacteria-rich environments, including soil, ocean and the human body29–36.
Generally, phages consist of a protein capsid (rarely with an internal membrane) that contains genomic nucleic acid, which can be linear double-stranded DNA (dsDNA), linear single- or double-stranded RNA, or circular single-stranded DNA (sscDNA)37,38. Phages are usually classified according to their structure, based on transmission electron microscopy and genome sequence39–41. The vast majority of DNA phages in the human gut microbiota belong to the order Caudovirales, which are dsDNA phages with genomic DNAs (gDNAs) of ~15–750 kb42. Caudovirales have protein capsids based on icosahedral symmetry and come in three general morphologies defined by a tail structure: siphophages (flexible tail), myophages (contractile tail) and podophages (short tail)43. The tails and associated tail fibres constitute an apparatus that not only defines the target specificity of the virion, but also contributes to the efficient infection44. The human gut microbiota also contains substantial numbers of much smaller (~5 kb gDNA) phages of the sscDNA family Microviridae45,46, which are isometric phages that lack tail structures and are restricted to Gram-negative bacteria such as Enterobacteria47.
In general, phages can be categorized as virulent or temperate48,49. Virulent phages (such as Escherichia coli phage T4) follow only a lytic pathway that begins with specific adsorption to a bacterial surface receptor, which can be a protein, carbohydrate, lipid or other external features such as pili, extracellular polysaccharide or flagella50–52. This adsorption is followed by injection of the gDNA into the host cytoplasm, a programme of DNA replication and gene expression, assembly of the progeny virions, and, finally, release of the progeny by lysis of the host53,54 (Supplementary Fig. 1). In contrast, temperate phages (such as E. coli λ phage) initiate infection in the same way but have the option to undergo lysogeny55,56, in which viral gene expression is shut off by a phage-encoded repressor and a dormant prophage, either integrated into the host chromosome or as a linear or circular self-replicating plasmid, is formed 57,58 (Supplementary Fig. 1). Importantly, the resultant lysogenic cell is thereby immune to further infection by the same phage because of the presence of the lysogenic repressor59.
These prophages can be passively carried by the bacterial host indefinitely; they also often carry genes that affect the bacterial host, including pathogenesis factors and defenses against other phages59,60. Moreover, either spontaneously at a low frequency or at a high frequency as a consequence of cell stress, the prophage can undergo induction and enter the lytic pathway, resulting in cell death and release of the progeny virions61–64. Even in undisturbed planktonic culture, all lysogenic strains spontaneously produce a certain concentration of free virions at a level that depends on the stability of repression, which can vary by >6 orders of magnitude65.
The host range of phages is primarily determined by the receptors on the host surfaces, the receptor recognition proteins of phages, and their interactions. In addition, there are numerous anti-phage systems that impose blocks at nearly every level of the infection process, including inhibition of DNA penetration into the cell, destruction of the phage DNA, inhibition of phage gene expression, and altruistic suicide of the infected cell66. Moreover, phages have mustered countervailing molecular and genetic strategies against these defenses67. In sum, there are many factors that define host range. Phages generally have host ranges that are restricted to one bacterial species68,69; efficient propagation of a single phage on widely different bacterial genera has not been convincingly documented. However, the methods used to isolate phages usually involve enrichment on a particular species, which probably biases searches for phages towards finding ‘specialist’ viruses. Indeed, the famous E. coli P1 phage, which was initially isolated as a prophage , is capable of injecting its DNA into Myxococcus xanthus, a bacterial species belonging to the class Deltaproteobacteria — for comparison, E. coli belongs to the class Gammaproteobacteria70.
Phages in the gastrointestinal tract
The human body contains diverse communities of microorganisms, consisting of bacteria, viruses (including phages and eukaryotic viruses), fungi and others71–75. It is estimated that there are approximately the same number of bacterial cells as human cells in the human body, with most of them in the gut 76. Phages are inherently more difficult to quantify in the diverse microenvironments of the gut, but most estimates for the phage-to-cell ratio are in the 0.1–10 range15. As noted earlier, the predominant phages in the human gut, as in all environments that are rich in bacteria, are dsDNA podophages, myophages and siphophages of the order Caudovirales77, followed by the small isometric viruses from the family Microviridae78.
Intestinal phageome of healthy individuals
Phages are hardly detectable in faecal samples of newborns79, but diverse populations can be detected within a few months79–81. In the first 2 years of life, the richness of the gut phageome decreases, which correlates with early-life bacterial colonization80–82. Although a core gut phageome has been proposed34,83, other researchers have suggested that each individual has a unique intestinal phageome45,46,84. The intestinal phageome consists of both prophages in bacterial cells and free virions or virus-like particles. Previous studies have described core bacterial members (such as species of the genera Bacteroides and Ruminococcus) in the human gut that are common among different individuals71,85, thus, intestinal phage sequences that were detected in multiple individuals might be the prophages in those core bacteria rather than free virions. Caudovirales, especially those of a temperate lifestyle, have highly mosaic genomes, meaning that different phages can have clusters of identical gene sequences86–90, making it a challenge to accurately assign a particular sequence read to a particular phage.
Different sample preparation protocols lead to variance among studies91–95. Additionally, other factors that can affect acquisition and interpretation of data include the analytical methods (metagenomic sequencing versus microscopy)34,96–99, which bioinformatics tools and databases are used100,101, as well as sampling positions and testing materials102,103. In addition, there is evidence that some phages can exist in ‘carrier’ states, in which they are dormant but not repressed or integrated into the host genome104,105. Altogether, it is therefore not unexpected to see apparently contradictory results regarding the composition and dynamics of the human intestinal phageome. Further work is needed to develop standardized protocols across the methodological spectrum, from viral DNA and RNA extraction to bioinformatic analyses.
Patients with gastrointestinal diseases
Gut bacterial dysbiosis is commonly seen in patients with gastrointestinal and liver diseases1–3, and, unsurprisingly, the intestinal phageomes of these patients differ from those of healthy individuals.
Patients with Crohn’s disease (n=27) or ulcerative colitis (n=42) have been reported to have a higher relative abundance of Caudovirales compared with Microviridae, and different compositions of Caudovirales families, compared with healthy individuals (n=61), by metagenomic sequencing of the DNA of virus-like particles from faecal samples8,9 (Fig. 1). Patients with Crohn’s disease had relatively more temperate phages, and the changes in virome composition were reflected in bacterial alterations (for example, patients with inflammatory bowel disease had reduced abundance of Firmicutes and increased levels of phages targeting Firmicutes)9. As the gut microbiota varies with the environment (including diet)106–110, researchers recruited healthy individuals from the same household for these studies, instead of using matched controls from different households8,9. Interestingly, metagenomic sequencing of faecal virus-like particles from 55 patients with irritable bowel syndrome and 51 control individuals showed that patients with irritable bowel syndrome had a less diverse faecal virome than controls, but the shift from lytic to temperate phages was not observed, which is different from patients with inflammatory bowel disease111.
Figure 1. Intestinal phageome of healthy individuals and patients with inflammatory bowel disease.
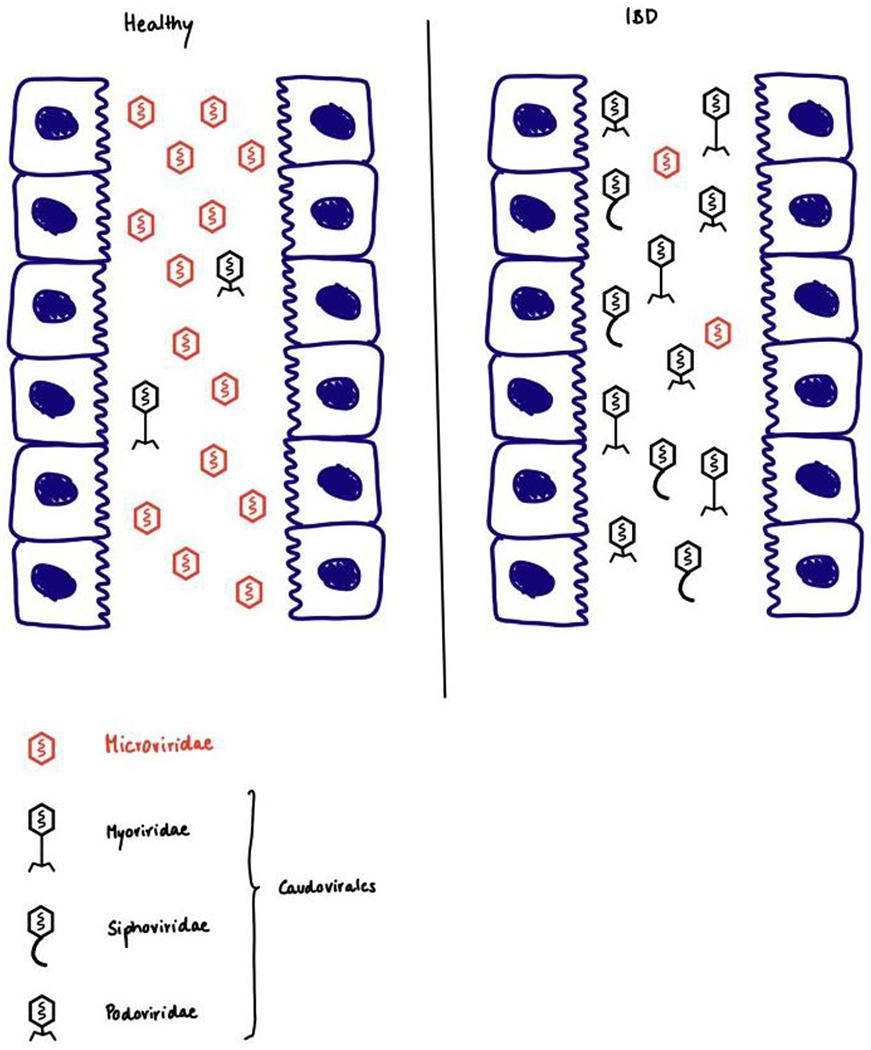
Left: healthy individual; Right: patients with inflammatory bowel disease. Compared with healthy individuals, patients with inflammatory bowel disease have a higher relative abundance of Caudovirales compared with Microviridae, and different compositions of Caudovirales families.
Viromes of colonic mucosa samples from patients with Crohn’s disease contained increased abundance of virus-like particles compared with colonic mucosa samples from healthy individuals112. Rectal mucosa viromes of patients with ulcerative colitis had a higher relative abundance but lower diversity of Caudovirales phages compared with healthy individuals10, which is consistent with results from analyses of stool samples8. Changes in the enteric virome were also observed in patients with CRC using metagenomic sequencing of faecal samples11,12. In a random forest analysis, researchers identified virome signatures that differentiated patients with CRC from healthy individuals11 and four taxonomic markers associated with patient mortality12.
Two studies have also reported virome compositions in patients with liver diseases13,14. One study included 89 patients with alcoholic hepatitis, 36 patients with alcohol use disorder, and 17 controls13, while the other study contained 73 patients with nonalcoholic fatty liver disease and 22 individuals as controls14. Compared with healthy individuals as controls, increased viral diversity was observed in faecal samples from patients with alcoholic hepatitis. Escherichia, Enterobacteria and Enterococcus phages were overrepresented in these patients, and increased abundance of Staphylococcus phages was associated with higher disease severity13. Interestingly, patients with more-severe nonalcoholic fatty liver disease had lower intestinal viral diversity, with a significant reduction in the proportion of phages compared with other intestinal viruses14. In another study, 40 control individuals and 163 patients with cirrhosis were included. The alpha diversity of the faecal virome was similar between groups, while patients with cirrhosis had more phages against Lactobacillales and Enterobacteriaceae113.
In summary, intestinal phages have been studied predominantly in patients with inflammatory bowel disease; independent cohort studies will be required to extend and validate these findings. No causative links have been built between intestinal phages and diseases, and further studies are therefore needed to determine whether intestinal phageome changes cause disease development or progression or result from disease. Moreover, findings to date have been largely limited to very broad categories of phages, rather than specific phage types or phages of particular hosts. Thus, we are still at an early stage in understanding these ‘dark matters’ of the intestine and their influences on human health and disease.
Phage-based therapy: past and present
Early history
Immediately after proposing the term ‘bacteriophage’ in 1917, Félix d’Herelle started phage treatment in patients with shigellosis114. Patients with advanced disease exhibited dramatic recoveries after being treated with oral doses of a Shigella phage114. Others also reported success using phage therapy against dysentery, including researchers from United States and Australia115,116. In the late 1920s, d’Herelle and colleagues reported that oral doses of a Vibrio cholerae phage greatly reduced mortality during cholera epidemics in Assam117. Mortality was ~6% in the treated group of patients (n=74), compared with 63% among patients who refused the phage treatment and thereby served as controls (n=124)117. There were multiple contemporaneous reports of the use of phages against other intestinal diseases such as typhoid fever, although the results were not always positive118–120. Nevertheless, phage-based therapy was widely considered a viable strategy against bacterial infections prior to the discovery of antibiotics.
However, in the 1930s, clinical reviews, especially a major comprehensive study commissioned by the American Medical Association, concluded that phage-based therapies lacked proven efficacy, specifically citing multiple reports in which phage treatment of cholera and other intestinal diseases had failed121. After that, interest in developing phages as anti-bacterials declined in the West, especially after the industrialization of small-molecule antibiotics during the World War II era20. In retrospect, the use of phages in clinical practice before the era of molecular biology might have been premature. Nevertheless, phage-based therapies are still used today in some eastern European countries/regions122; unfortunately these therapies have not been very well-documented in English language peer-reviewed literature, and we await more solid preclinical studies and better-designed clinical trials.
Current potential
Treating bacterial infection
Over the past 2 decades, some clinical trials and case studies reported the use of phages to treat gastrointestinal diseases (Table 1). The safety and efficacy of oral administration of E. coli T4-like phages have been tested in healthy individuals and patients with bacterial diarrhoea in several small-scale studies including both adults and children123–126. No severe adverse effects were reported, but no efficacy was observed either. Similar results were obtained in a large clinical trial using phages to treat bacterial diarrhoea in Bangladeshi children (n=120)127. In these studies, faecal phages against the target bacterial hosts (E. coli) were increased in treated children, but the titres did not show substantial intestinal phage replication; E. coli was low in absolute abundance, so applying higher titres of phages might have achieved better results.
Table 1.
Clinical trials and case reports of phage-based therapy against bacterial infections in gastrointestinal diseases
| Infectious agent targeting | Phage and dose | Population and treatment method | Outcome and interpretation | Reference |
|---|---|---|---|---|
| E. coli | Phage T4 (Dose A 105 PFU/ml, dose B 103 PFU/ml) | 15 healthy individuals Oral administration | Phage T4 is safe, E. coli abundance not changed | 123 |
| E. coli | 9 T4-like phages (Dose A 3x109 PFU/ml, dose B 3x107 PFU/ml) | 15 healthy individuals Oral administration | Phage cocktail is safe, gut microbiota profile not affected | 124 |
| E. coli | Commercial phage cocktail ColiProteus 20ml for adults, 10ml for children, and 10-fold dilution | 5 healthy adults, 10 healthy children Oral administration | Phage cocktail is overall safe, with occasional reported adverse effect not relevant to dosage | 125 |
| E. coli | 11 T4-like phages (3.6x108 PFU) or ColiProteus (1.4x109 PFU) | 120 male children with diarrhoea Oral administration | Safe but lack of efficacy | 127 |
| E. coli | T4-like phage cocktail (108 or 106 PFU for older children, 107 or 105 PFU for younger children) or ColiProteus (5x108 or 109 PFU) | 20 older children, 20 younger children Oral administration | Both cocktails are safe | 126 |
| Acinetobacter baumannii | 9 phages in 3 cocktails (5x109 PFU intravenous) | 68-year-old male patient with necrotizing pancreatitis complicated by pancreatic pseudocyst Intracavitary and Intravenous | Patient completely recovered | 128 |
| Klebsiella pneumoniae | 1 phage (107 PFU orally, 106 PFU intra-rectally) | 57-year-old female patient with Crohn’s disease, with multi-site infection (gastrointestinal tract, urinary tract, etc) Oral and intra-rectal | The original host (Klebsiella pneumoniae) was no longer detected | 201 |
English language publications only. E. coli, Escherichia coli.
In 2016, a 68-year-old male patient with diabetes, infected with multidrug-resistant Acinetobacter baumannii, developed necrotizing pancreatitis complicated by a pancreatic pseudocyst128. Despite multiple antibiotic courses and a percutaneous drainage of a pancreatic pseudocyst, the patient deteriorated over a four-month period. On the basis of an emergency Investigational New Drug (IND) permission from the US Food and Drug Administration, phage therapy was initiated (intracavitary and intravenous) and the patient returned to health after approximately five months128. Although this is only a case report, the obvious downward clinical course before phage treatment and the clear turning point after phage administration generated wide publicity and brought renewed hope that phage-based therapies might be used to treat bacterial infections (especially multidrug-resistant bacteria). Multiple case reports in other emergency IND situations have accumulated in the past 4 years against the multidrug-resistant bacteria Pseudomonas aeruginosa and Mycobacterium abscessus129,130. However, standardized clinical trials will be required to further determine the efficacy of phage therapies for different infectious diseases.
Phages have also been evaluated for disease prophylaxis in preclinical models. Oral administration of a three-phage cocktail to infant mice 24-hours prior to V. cholerae challenge had significant reductions in bacterial colonization of the intestine131. In addition, using an infant rabbit model, administration of phage before bacterial challenge protected them from cholera-like diarrhoea131. As cholera epidemics are seasonal and self-limiting132, phage prophylaxis might be used to control disease spread and protect high-risk individuals during outbreaks. More studies should be performed to explore the potential protective effect shortly after bacterial challenge, thereby aiming to reduce bacterial colonization and prevent disease.
Manipulating the gut microbiota
Strategies to manipulate the gut microbiota include faecal microbiota transplantation (FMT)133, use of prebiotics and probiotics134, and adjustments to diet and nutrient intake108. In the past decade, phages have also been used for precision editing of the gut microbiota (Table 2). In 2017, a United States patent was granted for PreforPro (Deerland Probiotics and Enzymes, Kennesaw, GA), a mix of phages targeting E. coli135. Two placebo-controlled trials have been conducted to determine the safety and efficacy of PreforPro in improving intestinal health by altering gut bacterial composition. One trial evaluated the effect of the phage cocktail alone136, while the other trial tested the additive effect of PreforPro on probiotics Bifidobacterium animalis subsp. lactis BL04137. Both trials included healthy individuals who reported having mild-to-moderate gastrointestinal distress but no diagnosed gastrointestinal disorders. Over the 28-day study in both trials, encapsulated PreforPro was found to be safe and tolerated, but the evidence of efficacy was not clear-cut 136,137. The connection between E. coli and abdominal symptoms has not been well established; thus, more studies are needed to better evaluate the potential efficacy.
Table 2.
Studies of phage-based strategies for gut microbiota modulation in gastrointestinal diseases
| Infectious agent targeting | Status | Phage and dose | Population and treatment method | Outcome and interpretation | Reference |
|---|---|---|---|---|---|
| E. coli | Clinical trial, complete | PreforPro (4 phages) 1 capsule daily for 28 days | 32 healthy individuals with mild-to-moderate gastrointestinal distress Oral administration | Phage cocktail is safe and tolerable, but no difference from placebo | 136 |
| E. coli | Clinical trial, complete | PreforPro (4 phages) together with probiotics Bifidobacterium animalis subspecies lactis strain BL04 1 capsule daily for 28 days | 68 healthy individuals with mild-to-moderate gastrointestinal distress Oral administration | Phage supplement is tolerated, but no compelling evidence of efficacy. Only marginally significant effects on self-diagnosed gastrointestinal inflammation are reported as evidence of a benefit | 137 |
| E. coli | Clinical trial, active | EcoActive (phage cocktail) Twice daily for 15 days | 30 patients with Crohn’s disease in remission Oral administration | 202 | |
| Enterococcus faecalis | Preclinical | Phage cocktail (3-4 phages) 1010 PFU, one day before sacrifice | Germ-free mice colonized with stool samples from patients with alcoholic hepatitis Oral administration | Phage cocktail is beneficial for alcohol-related liver disease | 141 |
English language publications only. E. coli, Escherichia coli.
Adherent-invasive E. coli has been implicated in the pathogenesis of inflammatory bowel disease over the past 2 decades138,139. Phages against such E. coli strains have been proposed as a treatment option. Conventional mice colonized with adherent-invasive E. coli were given drinking water with 2% dextran sodium sulfate (DSS) to induce mild symptoms of colitis. After one week, a three-phage cocktail was orally gavaged to the mice, followed by 2% DSS drinking water for another two weeks140. Mice receiving phage treatment were found to be protected from DSS-induced colitis140, and E. coli colonization was reduced140. Active phage replication was detected in ileal biopsy samples spiked with E. coli from patients with Crohn’s disease, providing additional evidence for the killing potential of phages in such environments140. A phase I/IIa randomized, double-blind, placebo-controlled clinical trial is underway to assess the safety and efficacy of oral administration of phages that target intestinal adherent-invasive E. coli in patients with Crohn’s disease in remission (NCT03808103).
Researchers have shown that faecal levels of Enterococcus faecalis, a commensal member of the human gut microbiota with low abundance, is significantly increased in patients with alcoholic hepatitis compared with patients with alcohol use disorder and non-alcoholic individuals. The presence of E. faecalis strains that produce cytolysin (a bacterial exotoxin) correlates with worse outcomes and with mortality in patients with alcoholic hepatitis141. Oral administration of cytolysin-positive E. faecalis exacerbated ethanol-induced liver disease in conventional mice. To extend these findings to humans, gnotobiotic mice were colonized with faecal samples from cytolysin-positive and cytolysin-negative patients with alcoholic hepatitis. Phages specifically targeting cytolysin-positive E. faecalis were administered by oral gavage and they reduced ethanol-induced liver disease, whereas phages against cytolysin-negative E. faecalis did not have any beneficial effect141. Besides the implications for potential treatment of ethanol-induced liver disease, this study can be regarded as one of the first documented examples of precision editing of the gut microbiota, by extirpation of a subpopulation of E. faecalis (Fig. 2). Larger studies are needed to validate these results and a clinical trial is necessary to test the therapeutic effects in patients with alcoholic hepatitis.
Figure 2. Manipulation of the gut microbiota by phages.
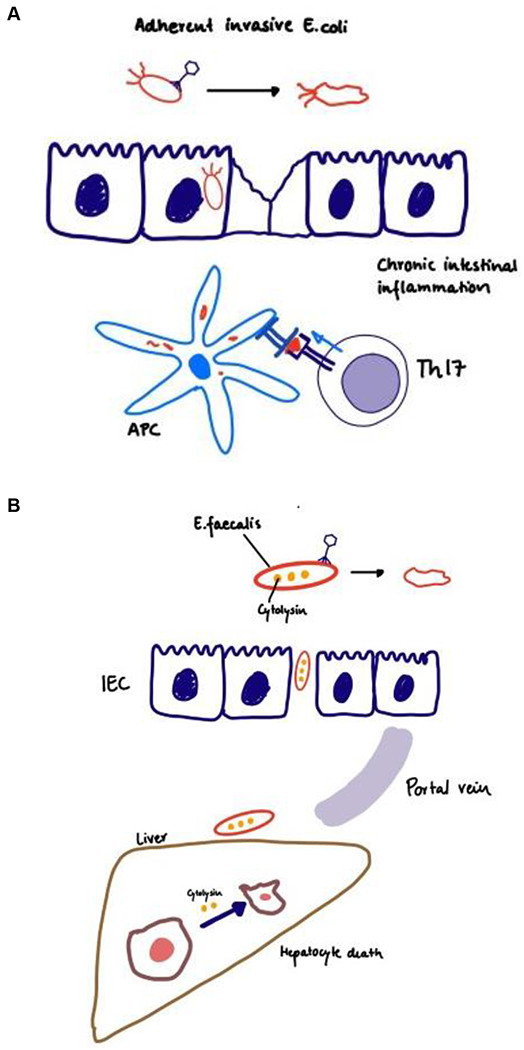
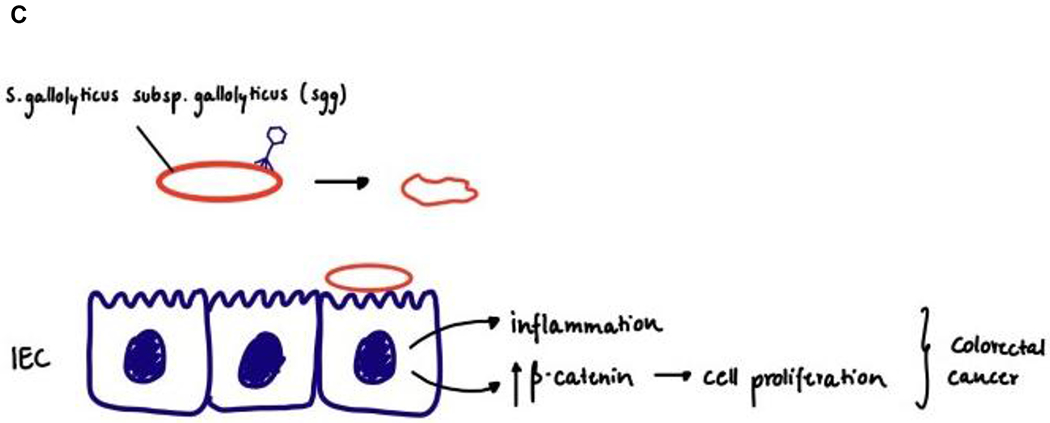
(a) Phage therapy in Crohn’s disease. Adherent-invasive E. coli stimulates antigen-presenting cell (APC) driving Th17 responses, phages targeting adherent-invasive E. coli were found to be beneficial of DSS-induced colitis. (b) Phage therapy in alcoholic liver disease. Cytolysin positive E. faecalis translocates from the gut to the liver, directly damaging hepatocytes. Phages against cytolysin positive E. faecalis could protect mice from alcoholic liver disease. (c) Phage therapy in colorectal cancer. Streptococcus gallolyticus subsp. gallolyticus (Sgg) upregulates β-catenin, stimulating cancer cell proliferation. Phage therapy might be a promising treatment option.
After first being reported by McCoy and Mason in 1951142, several studies have revealed that Streptococcus gallolyticus subsp. gallolyticus (Sgg), a cause of septicaemia and infective endocarditis, is associated with CRC (for a detailed review, see Abdulamir et al.143 and Boleij et al.144). Fusobacterium nucleatum is more abundant in faecal samples from patients with CRC than in control individuals145–147; it is also over-represented in tumours versus matched control tissue specimens from patients with CRC148. Preclinical studies have shown that a number of bacterial species of the gut microbiota, in particular F. nucleatum, Bacteroides fragilis, E. coli and E. faecalis, are also associated with CRC development and progression via different mechanisms149–155. In vitro and in vivo studies using mouse models have suggested a CRC tumour-promoting role for Sgg by upregulating β-catenin, a central signalling molecule in colon tumorigenesis156. In addition, inhibiting intestinal E. coli overgrowth by oral administration of sodium tungstate reduced gut inflammation and the incidence of colitis-associated colonic tumours in two mouse models (azoxymethane/dDSS colitis model and azoxymethane-treated Il10-deficient mouse model)157. Although more studies are required to confirm the causative link between intestinal bacteria and CRC, phage-mediated precision editing of the gut microbiota might be worth exploring as a promising treatment option (Fig. 2).
It is important to note, however, that it will be necessary to have some understanding of what determines the phage specificity in such studies in order to achieve ‘precision’. E. coli phage T4 uses different receptors for different E. coli strains158. A similar E. coli phage, Ox2, can change its receptor from one outer membrane protein (the usual receptor of phage Ox2 is OmpA) to another (OmpC and/or OmpX) or to different carbohydrate residues in the lipopolysaccharide as a result of single mutations in the tail fibre158. Moreover, some phage genomes encode arrays of tail fibre genes that can be switched in and out by high-frequency recombination processes159,160. Amazingly, phages against Bordetella spp. that encode an error-prone reverse transcriptase that causes extreme hypermutation of the receptor-binding domain of the tail fibre have been isolated161. It is therefore important to identify the receptors of phages to precisely target the host bacteria.
Phage therapy as a precision medicine approach
Phages can not only precisely edit the gut microbiota, they can also deliver drugs to a specific location. As the natural predators of bacteria, phages propagate in environments where their hosts reside. With the development of more powerful tools to engineer phages, drugs can be attached to the phage surface to be released when phages reach their destinations162. Thus, site-specific administration of high doses might be possible, enabling reduced concentrations of drugs in the circulation and decreased toxic effects on non-target tissues163,164.
Several preclinical studies have tested this approach. In an in vitro study, thousands of molecules of the antibiotic chloramphenicol were attached to a phage surface via ester linkage, enabling it to be slowly released by serum esterases165. The phages were able to target Staphylococcus aureus, providing local high concentrations of chloramphenicol that were sufficient to inhibit the growth of previously resistant S. aureus cells165. A similar idea has also been applied in vivo in a mouse model166. . F. nucleatum was largely found in CRC tumours and promoted CRC resistance to chemotherapy in mice167. F. nucleatum targets Toll-like receptor 4 and specific microRNAs to activate the autophagy pathway, thus altering the CRC chemotherapeutic response167. In a study using a CRC mouse model, Zheng et al. coated F. nucleatum phages with irinotecan, a first-line treatment for CRC166. These phages target F. nucleatum, which reside in CRC tumour tissues. Phages therefore accumulated in CRC tumours and precisely delivered the drug to its destination, with minimal adverse effects to non-tumour tissues166. Oral administration of these irinotecan-coated phages also decreased the abundance of F. nucleatum, thus re-sensitizing tumour cells to chemotherapy166. A similar approach has been used in another study in which the researchers assembled anti-bacterial silver nanoparticles on the phage surface. Those phages specifically targeted F. nucleatum and accumulated in CRC tumour cells. Compared with the mice receiving only chemotherapy, mice receiving a combination of both phage therapy and chemotherapy showed less tumour growth and a longer survival time 168. Thus, the idea of phage-mediated drug delivery has promise for wide application in clinical practice.
Gut microbial metabolites have important roles in human health and also contribute to diseases169,170. Short-chain fatty acids (such as butyrate) produced by gut bacteria inhibit tumour growth and stimulate antitumour immune responses in mouse models of CRC171,172. Zheng et al. showed that phages can be covalently linked with dextran nanoparticles that promote proliferation of Clostridium butyricum, which increased faecal levels of short-chain fatty acids in mice and inhibited tumour growth166. These phages targeted pathogenic F. nucleatum without affecting C. butyricum. Multi-functional phage particles can be administered orally, and might be used not only to deliver drugs to specific locations, but also to increase treatment efficacy by modulating the intestinal microbiota (that is, reducing the amount of pathogenic bacteria and promoting the growth of beneficial bacteria). This novel and convenient route of administration (that is, oral administration of chemically-coated phages) deserves more attention. Studies are needed in different models of diseases and to assess long-term effects.
Challenges and future directions
A century after their discovery, phages are the subjects of renewed interest for treatment of bacterial infections, especially for gastrointestinal diseases, in which systemic introduction of phages into the bloodstream would not be required. These bacterial predators have broad applications, but there are many challenges to overcome.
Most studies have reported phage-based therapies to be safe124,136,173, as phages only propagate in bacteria. However, using a mouse model, one study showed that filamentous Pseudomonas phages can directly interact with human leukocytes, with phage RNA being produced and stimulating interferon production174. This observation indicates that filamentous phages might interact with the human immune system and have direct effects on human health. Several preclinical studies have also assessed the immune response induced by phages. Some reported that orally administrated phages could stimulate inflammatory cytokine production and induce inflammation, mostly in mouse models of intestinal inflammation and dysbiosis175–177. On the other hand, phage administration in vitro either had no effect on the inflammatory response or exerted an anti-inflammatory response on mammalian cells, as measured by the levels of inflammatory cytokines176,178–180. Given the long-term presence of bacteria and phages in the mammalian gut, it will not be surprising to find out that phages are capable of many interactions with the human immune system and other diverse cell types.
The narrow host range is another apparent limitation of phage-based treatment. Therefore, the superb specificity of phages, which enables precise targeting of bacteria, is also a potential problem, because the narrow host range could limit wide therapeutic utility. One option is to create a phage cocktail, comprised of multiple phages that each target a different receptor. However, this increases the complexity and safety risk of the treatment, because under current guidelines, the safety of each individual phage, as well as each different combination of phages, would need to be tested. Many factors therefore must be considered in developing a therapeutic phage cocktail, including the host range, the receptors, and the infectious efficiency (for detailed guidelines, see Merabishvili et al.181). Another possible strategy involves ‘phage training’, or phage adaptation. This process selects for evolved phages with broader host ranges or that can overcome bacterial resistance through experimental procedures performed in the laboratory. Phages that can target multiple hosts can be obtained via multiple rounds of selection using either different bacterial isolations or resistant mutants (for detailed protocols, see Betts et al.182 and Friman et al.183). This approach has the additional attraction of being ‘natural’ by avoiding recombinant DNA methods and the complications of genetically modified organism (GMO) classification. On the other hand, rapid development of synthetic biology has made engineering phages attractive, albeit subject to GMO regulation. By identifying phage proteins that are responsible for host recognition, genetic modifications could be made to broaden host ranges184 or reduce the potential for the emergence of phage resistance185. To achieve this goal, cutting-edge genomic editing tools and more knowledge about host determination will be indispensable.
The research field of phage therapy is still at an early stage, with many scientific questions remaining to be answered. In addition, to obtain a better result, many factors should be considered and carefully evaluated before phage administration186,187. One of the most important is to screen all the individuals for the presence of the target bacterial host in their gut. The bacterial host should also be tested in vitro to confirm its sensitivity to the selected phages. Another critical point is to determine the dose of phages being applied. Multiple studies have reported the safety of using a relatively high dose of phages (for example, 109 PFU, both orally and intravenously)124,127,188. Applying phages in high concentrations might be necessary, especially for oral administration as gastric acid could decrease the amount of surviving phages189. Concurrent administration of an acid-neutralizing reagent and phage encapsulation methods might be helpful in this regard126,190,191. Additionally, the pharmacokinetic and pharmacodynamic properties of phages also need to be evaluated192. Multiple studies found phages being cleared in the circulation system in a few hours, in both mice and humans188,193, which might pose a problem in maintaining sufficient number of phages for therapeutic purposes. However, some researchers have reported that orally administrated phages could still be detected in the gut after several days124,126. Modifications of the phage capsid proteins might improve its half-life, as such changes might help phages better evade phagocytosis194–196.
Although there are still many concerns and challenges (Box 1), phage therapy has great potential for use in clinical practice (Fig. 3). Well-designed, placebo-controlled clinical trials showing safety and efficacy could help the field and attract more scientists and physicians. However, the development of this field not only depends on the ‘researchers’ (scientists, physicians and even patients) but also on the regulatory context. Phages are considered as medicinal products in the US and the European Union197, which are under very strict constraints related to their production and marketing authorization, such as compliance to Good Manufacturing Practice (GMP)197. As a customized treatment, therapeutic phages need to be selected and produced ad hoc, making it impractical to be an immutable pre-defined medicinal product198. Manufacturing GMP-certified medicinal products is overall costly and time-consuming, making it even harder to initiate a phage-based clinical trial199. Recently, the Belgian government classified therapeutic phages as magistral preparations, providing more flexibilities to phage treatments200. In European law, magistral preparation (compounded prescription drug product in the US) is defined as “any medicinal product prepared in a pharmacy in accordance with a medical prescription for an individual patient” (Article 3 of Directive 2001/83 and Article 6 quater, § 3 of the Law of 25 March 1964). Although this might be less likely to be approved by the US Food and Drug Administration (FDA) due to stricter rules and more concerns, phage therapeutics would require some specific rules and regulations that are different from other standardized medicines. Phages might be considered for GRAS (generally regarded as safe) materials by the US FDA, given their ubiquity in the human body and environment and their fundamental inability to attack human tissues. This designation might set the stage for conducting a few set-piece clinical trials of phage cocktails; assuming positive results in terms of safety and efficacy, regulatory approval might then be advanced for other combinations of phages prepared and formulated in the same way, much like the way new flu vaccines are approved each season. In any case, it is clear that more government-funded, phage-based clinical trials are required to better explore the therapeutic potential of phages in a broad range of gastrointestinal diseases.
Box 1. Important questions for further studies of phage-based therapies.
Although phages were discovered a century ago, phage therapy is still a relatively new research area, with many challenges and problems as well as open questions and opportunities.
Is phage-based therapy safe for clinical practice? What will the regulations and rules be?
Will phage-based therapy replace antibiotic treatment? If not, when to choose which option? Or both simultaneously?
How to decide the best administration route and the dose for each phage therapy?
Which is better: a single phage or phage cocktail? How to decide which one to use in clinical practice?
Biofilm is a big challenge for antibiotic treatment; can phages be found that will propagate efficiently in biofilms?
Could phages be used against intracellular bacterial infections?
More phages can always be found. Is it possible to quickly and easily ascertain whether new isolates are going to be helpful, adding power to the therapy? Would bioinformatic tools help?
What are the long-term effects of phage-based treatments on the intestinal microbiota and humans?
Figure 3. Potential applications of phages.
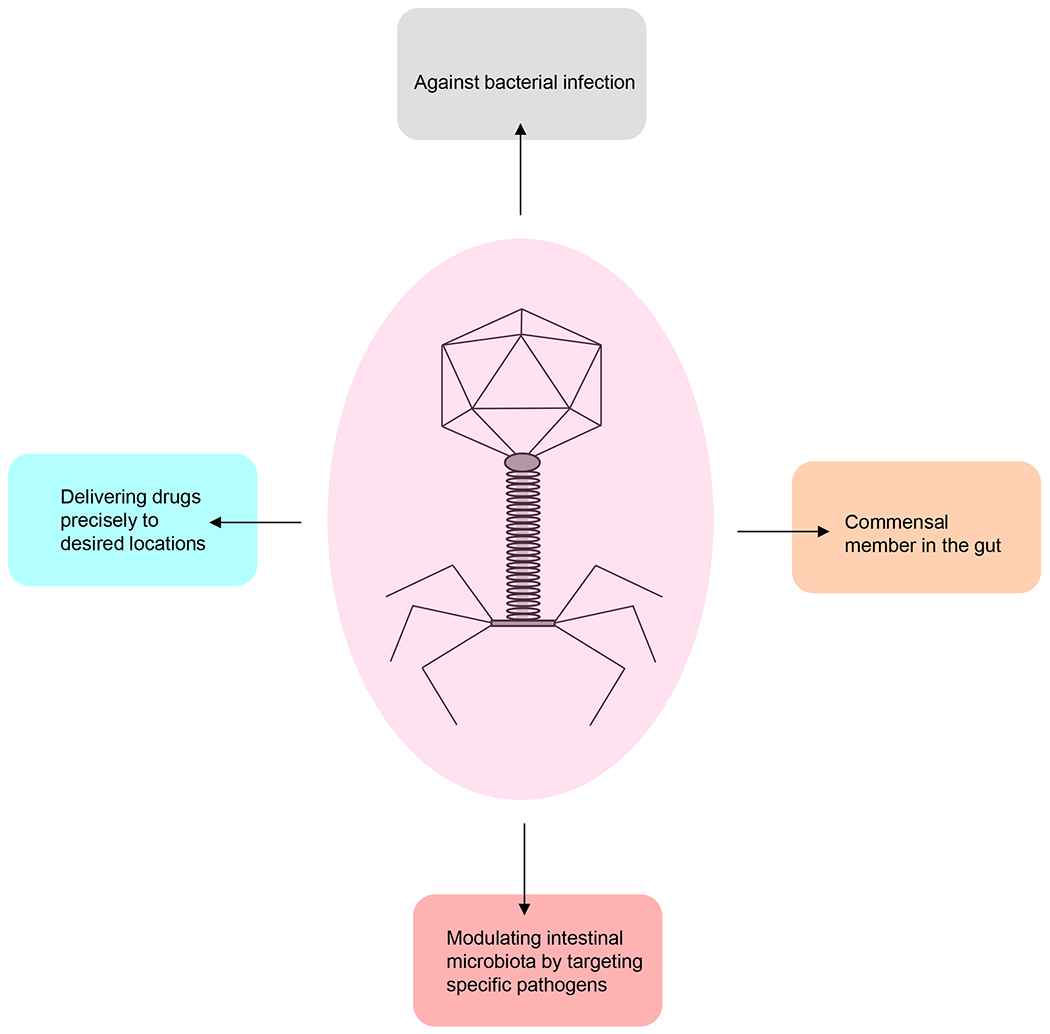
Apart from targeting their bacterial hosts, phages might also interact with the human body and could thereby have multiple effects on human health and diseases.
Conclusion
Bacteriophages have been used to combat bacterial infections for over a century. Discoveries of the association between the gut microbiota and human diseases have prompted renewed attention to this research area. Phages are powerful weapons not only against pathogenic bacterial infections, but they can also precisely edit the intestinal microbiota and harbour promising therapeutic effects for many different gastrointestinal diseases. Multiple therapeutic possibilities have been proposed, but more basic and preclinical studies, as well as properly designed randomized, double-blind, placebo-controlled trials are required to help the field move forward. Still at the early stage, the field has significant problems and challenges to be solved, such as the beneficial or harmful effects of potential phage–human interactions, the evolving nature of phages as vital biological entities, and the long-term effects of a phage-modulated gut microbiota on human health. Overall, phage-based therapies could become promising and powerful approaches to treat many gastrointestinal and possibly extra-intestinal diseases, and are deserving of greater attention and further exploration.
Supplementary Material
In this Perspective, Duan, Young and Schnabl explore the effects of bacteriophages on the gut microbiota and the potential applications of phage therapy for treatment of gastrointestinal diseases. Limitations and challenges of phage therapy for gastrointestinal diseases are also discussed.
Acknowledgements
The authors were supported in part by a Biocodex Microbiota Foundation Grant, NIH grants R01 AA024726, U01 AA026939, by Award Number BX004594 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development (to B.S.) and services provided by P30 DK120515 and P50 AA011999.
Competing interests
B.S. has been consulting for Ferring Research Institute, Intercept Pharmaceuticals, HOST Therabiomics, Mabwell Therapeutics, Patara Pharmaceuticals and Takeda. B.S.’s institution UC San Diego has received grant support from BiomX, NGM Biopharmaceuticals, CymaBay Therapeutics, Synlogic Operating Company, Prodigy Biotech and Axial Biotherapeutics. R.Y. was formerly involved with GangaGen (Bangalore India) as a member of its scientific advisory board.
Footnotes
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s415XX-XXX-XXXX-X
References
- 1.Ni J, Wu GD, Albenberg L & Tomov VT Gut microbiota and IBD: causation or correlation? Nature Reviews Gastroenterology & Hepatology 14, 573–584, doi: 10.1038/nrgastro.2017.88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tripathi A et al. The gut–liver axis and the intersection with the microbiome. Nature Reviews Gastroenterology & Hepatology 15, 397–411, doi: 10.1038/s41575-018-0011-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong SH & Yu J Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nature Reviews Gastroenterology & Hepatology 16, 690–704, doi: 10.1038/s41575-019-0209-8 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Young VB The role of the microbiome in human health and disease: an introduction for clinicians. The BMJ 356, j831, doi: 10.1136/bmj.j831 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Lurie-Weinberger MN & Gophna U Archaea in and on the Human Body: Health Implications and Future Directions. PLOS Pathogens 11, e1004833, doi: 10.1371/journal.ppat.1004833 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu H et al. The Candida albicans exotoxin candidalysin promotes alcohol-associated liver disease. Journal of Hepatology 72, 391–400, doi: 10.1016/j.jhep.2019.09.029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lang S et al. Intestinal Fungal Dysbiosis and Systemic Immune Response to Fungi in Patients With Alcoholic Hepatitis. Hepatology 71, 522–538, doi: 10.1002/hep.30832 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norman JM et al. Disease-Specific Alterations in the Enteric Virome in Inflammatory Bowel Disease. Cell 160, 447–460, doi: 10.1016/j.cell.2015.01.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clooney AG et al. Whole-Virome Analysis Sheds Light on Viral Dark Matter in Inflammatory Bowel Disease. Cell Host & Microbe 26, 764–778, doi: 10.1016/j.chom.2019.10.009 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Zuo T et al. Gut mucosal virome alterations in ulcerative colitis. Gut 68, 1169–1179, doi: 10.1136/gutjnl-2018-318131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannigan GD, Duhaime MB, Ruffin MT, Koumpouras CC & Schloss PD Diagnostic Potential and Interactive Dynamics of the Colorectal Cancer Virome. mBio 9, e02248–02218, doi: 10.1128/mBio.02248-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakatsu G et al. Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology 155, 529–541, doi: 10.1053/j.gastro.2018.04.018 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Jiang L et al. Intestinal virome in patients with alcoholic hepatitis. Hepatology 72, 2182–2196, doi: 10.1002/hep.31459 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang S et al. Intestinal Virome Signature Associated With Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology 159, 1839–1852, doi: 10.1053/j.gastro.2020.07.005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shkoporov AN & Hill C Bacteriophages of the Human Gut: The “Known Unknown” of the Microbiome. Cell Host & Microbe 25, 195–209, doi: 10.1016/j.chom.2019.01.017 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Twort FW AN INVESTIGATION ON THE NATURE OF ULTRA-MICROSCOPIC VIRUSES. The Lancet 186, 1241–1243, doi: 10.1016/S0140-6736(01)20383-3 (1915). [DOI] [Google Scholar]
- 17.d’Herelle F Sur un microbe invisible antagoniste des bacilles dysenteriques. Comptes Rendus Academie des Sciences (Paris) 165, 373–375 (1917). [Google Scholar]
- 18.Summers WC. Bacteriophage therapy. Annual Review of Microbiology 55, 437–451, doi: 10.1146/annurev.micro.55.1.437 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Fleming A On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to their Use in the Isolation of B. influenza. British Journal of Experimental Pathology 10, 226–236 (1929). [PubMed] [Google Scholar]
- 20.Aminov R History of antimicrobial drug discovery: Major classes and health impact. Biochemical Pharmacology 133, 4–19, doi: 10.1016/j.bcp.2016.10.001 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Goossens H, Ferech M, Vander Stichele R & Elseviers M Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. The Lancet 365, 579–587, doi: 10.1016/S0140-6736(05)17907-0 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Spellberg B, Bartlett JG & Gilbert DN The Future of Antibiotics and Resistance. New England Journal of Medicine 368, 299–302, doi: 10.1056/NEJMp1215093 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becattini S, Taur Y & Pamer EG Antibiotic-Induced Changes in the Intestinal Microbiota and Disease. Trends in Molecular Medicine 22, 458–478, doi: 10.1016/j.molmed.2016.04.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vila AV et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nature Communications 11, 362, doi: 10.1038/s41467-019-14177-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bobay LM & Ochman H Biological species in the viral world. Proceedings of the National Academy of Sciences 115, 6040–6045, doi: 10.1073/pnas.1717593115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mushegian AR & Margolin W Are There 1031 Virus Particles on Earth, or More, or Fewer? Journal of Bacteriology 202, e00052–00020, doi: 10.1128/JB.00052-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohwer F Global Phage Diversity. Cell 113, 141, doi: 10.1016/S0092-8674(03)00276-9 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Clokie MRJ, Millard AD, Letarov AV & Heaphy S Phages in nature. Bacteriophage 1, 31–45, doi: 10.4161/bact.1.1.14942 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashelford KE, Day MJ & Fry JC Elevated Abundance of Bacteriophage Infecting Bacteria in Soil. Applied and Environmental Microbiology 69, 285–289, doi: 10.1128/AEM.69.1.285-289.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pratama AA & van Elsas JD The ‘Neglected’ Soil Virome – Potential Role and Impact. Trends in Microbiology 26, 649–662, doi: 10.1016/j.tim.2017.12.004 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Suttle CA Viruses in the sea. Nature 437, 356–361, doi: 10.1038/nature04160 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Suttle CA Marine viruses — major players in the global ecosystem. Nature Reviews Microbiology 5, 801–812, doi: 10.1038/nrmicro1750 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Paez-Espino D et al. Uncovering Earth’s virome. Nature 536, 425–430, doi: 10.1038/nature19094 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Manrique P et al. Healthy human gut phageome. Proceedings of the National Academy of Sciences 113, 10400–10405, doi: 10.1073/pnas.1601060113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reyes A et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338, doi: 10.1038/nature09199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reyes A, Semenkovich NP, Whiteson K, Rohwer F & Gordon JI Going viral: next-generation sequencing applied to phage populations in the human gut. Nature Reviews Microbiology 10, 607–617, doi: 10.1038/nrmicro2853 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hendrix RW Bacteriophage genomics. Current Opinion in Microbiology 6, 506–511, doi: 10.1016/j.mib.2003.09.004 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Hatfull GF Bacteriophage genomics. Current Opinion in Microbiology 11, 447–453, doi: 10.1016/j.mib.2008.09.004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ackermann HW & Prangishvili D Prokaryote viruses studied by electron microscopy. Archives of Virology 157, 1843–1849, doi: 10.1007/s00705-012-1383-y (2012). [DOI] [PubMed] [Google Scholar]
- 40.Dion MB, Oechslin F & Moineau S Phage diversity, genomics and phylogeny. Nature Reviews Microbiology 18, 125–138, doi: 10.1038/s41579-019-0311-5 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Koonin EV et al. Global Organization and Proposed Megataxonomy of the Virus World. Microbiology and Molecular Biology Reviews 84, e00061–00019, doi: 10.1128/MMBR.00061-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Shayeb B et al. Clades of huge phages from across Earth’s ecosystems. Nature 578, 425–431, doi: 10.1038/s41586-020-2007-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ackermann HW Classification of bacteriophages. The Bacteriophages, 8–16 (Oxford, 2006). [Google Scholar]
- 44.Goldberg EB Recognition, attachment and injection. Bacteriophage T4, 32–39 (American Society for Microbiology, 1983). [Google Scholar]
- 45.Shkoporov AN et al. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell Host & Microbe 26, 527–541, doi: 10.1016/j.chom.2019.09.009 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Gregory AC et al. The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut. Cell Host & Microbe 28, 724–740, doi: 10.1016/j.chom.2020.08.003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldberg EB Bacteriophage nuclear acid penetration. Receptors and Recognition (Series B, Volume 7): Virus Receptors (Part 1: Bacterial Viruses), 115–141 (Chapman & Hall, Ltd., 1980). [Google Scholar]
- 48.Campbell A The future of bacteriophage biology. Nature Reviews Genetics 4, 471–477, doi: 10.1038/nrg1089 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ofir G & Sorek R Contemporary Phage Biology: From Classic Models to New Insights. Cell 172, 1260–1270, doi: 10.1016/j.cell.2017.10.045 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Lindberg AA Bacteriophage Receptors. Annual Review of Microbiology 27, 205–241, doi: 10.1146/annurev.mi.27.100173.001225 (1973). [DOI] [PubMed] [Google Scholar]
- 51.Ge H et al. The “fighting wisdom and bravery” of tailed phage and host in the process of adsorption. Microbiological Research 230, 126344, doi: 10.1016/j.micres.2019.126344 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Silva JB, Storms Z & Sauvageau D Host receptors for bacteriophage adsorption. FEMS Microbiology Letters 363, fnw002, doi: 10.1093/femsle/fnw002 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Young R Bacteriophage lysis: mechanism and regulation. Microbiological Reviews 56, 430–481 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cahill J & Young R Chapter Two - Phage Lysis: Multiple Genes for Multiple Barriers. Advances in Virus Research 103, 33–70 (Academic Press, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng L et al. Decision Making at a Subcellular Level Determines the Outcome of Bacteriophage Infection. Cell 141, 682–691, doi: 10.1016/j.cell.2010.03.034 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dou C et al. Structural and functional insights into the regulation of the lysis–lysogeny decision in viral communities. Nature Microbiology 3, 1285–1294, doi: 10.1038/s41564-018-0259-7 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Feiner R et al. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nature Reviews Microbiology 13, 641–650, doi: 10.1038/nrmicro3527 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Howard-Varona C, Hargreaves KR, Abedon ST & Sullivan MB Lysogeny in nature: mechanisms, impact and ecology of temperate phages. The ISME Journal 11, 1511–1520, doi: 10.1038/ismej.2017.16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bondy-Denomy J et al. Prophages mediate defense against phage infection through diverse mechanisms. The ISME Journal 10, 2854–2866, doi: 10.1038/ismej.2016.79 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fortier LC & Sekulovic O Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 4, 354–365, doi: 10.4161/viru.24498 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banks DJ, Lei B & Musser JM Prophage Induction and Expression of Prophage-Encoded Virulence Factors in Group A Streptococcus Serotype M3 Strain MGAS315. Infection and Immunity 71, 7079–7086, doi: 10.1128/IAI.71.12.7079-7086.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goerke C, Köller J & Wolz C Ciprofloxacin and Trimethoprim Cause Phage Induction and Virulence Modulation in Staphylococcus aureus. Antimicrobial Agents and Chemotherapy 50, 171–177, doi: 10.1128/AAC.50.1.171-177.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi J, Kotay SM & Goel R Various physico-chemical stress factors cause prophage induction in Nitrosospira multiformis 25196- an ammonia oxidizing bacteria. Water Research 44, 4550–4558, doi: 10.1016/j.watres.2010.04.040 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Alexeeva S, Guerra Martínez JA, Spus M & Smid EJ Spontaneously induced prophages are abundant in a naturally evolved bacterial starter culture and deliver competitive advantage to the host. BMC Microbiology 18, 120, doi: 10.1186/s12866-018-1229-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nanda AM, Thormann K & Frunzke J Impact of Spontaneous Prophage Induction on the Fitness of Bacterial Populations and Host-Microbe Interactions. Journal of Bacteriology 197, 410–419, doi: 10.1128/JB.02230-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Labrie SJ, Samson JE & Moineau S Bacteriophage resistance mechanisms. Nature Reviews Microbiology 8, 317–327, doi: 10.1038/nrmicro2315 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Samson JE, Magadán AH, Sabri M & Moineau S Revenge of the phages: defeating bacterial defences. Nature Reviews Microbiology 11, 675–687, doi: 10.1038/nrmicro3096 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Ackermann HW & Dubow MS Viruses of Prokaryotes: General Properties of Bacteriophages 1, 49–85 (CRC Press Inc., 1987). [Google Scholar]
- 69.Dufour N et al. Bacteriophage LM33_P1, a fast-acting weapon against the pandemic ST131-O25b:H4 Escherichia coli clonal complex. Journal of Antimicrobial Chemotherapy 71, 3072–3080, doi: 10.1093/jac/dkw253 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Kaiser D & Dworkin M Gene transfer to myxobacterium by Escherichia coli phage P1. Science 187, 653–654, doi: 10.1126/science.803710 (1975). [DOI] [PubMed] [Google Scholar]
- 71.Qin J et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65, doi: 10.1038/nature08821 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huttenhower C et al. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214, doi: 10.1038/nature11234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lloyd-Price J et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66, doi: 10.1038/nature23889 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arumugam M et al. Enterotypes of the human gut microbiome. Nature 473, 174–180, doi: 10.1038/nature09944 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lourenço M et al. The Spatial Heterogeneity of the Gut Limits Predation and Fosters Coexistence of Bacteria and Bacteriophages. Cell Host & Microbe 28, 390–401, doi: 10.1016/j.chom.2020.06.002 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Sender R, Fuchs S & Milo R Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 164, 337–340, doi: 10.1016/j.cell.2016.01.013 (2016). [DOI] [PubMed] [Google Scholar]
- 77.Breitbart M et al. Metagenomic Analyses of an Uncultured Viral Community from Human Feces. Journal of Bacteriology 185, 6220–6223, doi: 10.1128/JB.185.20.6220-6223.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim MS, Park EJ, Roh SW & Bae JW Diversity and Abundance of Single-Stranded DNA Viruses in Human Feces. Applied and Environmental Microbiology 77, 8062–8070, doi: 10.1128/AEM.06331-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liang G et al. The stepwise assembly of the neonatal virome is modulated by breastfeeding. Nature 581, 470–474, doi: 10.1038/s41586-020-2192-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reyes A et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proceedings of the National Academy of Sciences 112, 11941–11946, doi: 10.1073/pnas.1514285112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim ES et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nature Medicine 21, 1228–1234, doi: 10.1038/nm.3950 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lim ES, Wang D & Holtz LR The Bacterial Microbiome and Virome Milestones of Infant Development. Trends in Microbiology 24, 801–810, doi: 10.1016/j.tim.2016.06.001 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Edwards RA et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nature Microbiology 4, 1727–1736, doi: 10.1038/s41564-019-0494-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moreno-Gallego JL et al. Virome Diversity Correlates with Intestinal Microbiome Diversity in Adult Monozygotic Twins. Cell Host & Microbe 25, 261–272, doi: 10.1016/j.chom.2019.01.019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tap J et al. Towards the human intestinal microbiota phylogenetic core. Environmental Microbiology 11, 2574–2584, doi: 10.1111/j.1462-2920.2009.01982.x (2009). [DOI] [PubMed] [Google Scholar]
- 86.Pedulla ML et al. Origins of Highly Mosaic Mycobacteriophage Genomes. Cell 113, 171–182, doi: 10.1016/S0092-8674(03)00233-2 (2003). [DOI] [PubMed] [Google Scholar]
- 87.Deng L et al. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 513, 242–245, doi: 10.1038/nature13459 (2014). [DOI] [PubMed] [Google Scholar]
- 88.Mavrich TN & Hatfull GF Bacteriophage evolution differs by host, lifestyle and genome. Nature Microbiology 2, 17112, doi: 10.1038/nmicrobiol.2017.112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hendrix RW, Smith MCM, Burns RN, Ford ME & Hatfull GF Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proceedings of the National Academy of Sciences 96, 2192–2197, doi: 10.1073/pnas.96.5.2192 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shapiro JW & Putonti C Gene Co-occurrence Networks Reflect Bacteriophage Ecology and Evolution. mBio 9, e01870–01817, doi: 10.1128/mBio.01870-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shkoporov AN et al. Reproducible protocols for metagenomic analysis of human faecal phageomes. Microbiome 6, 68, doi: 10.1186/s40168-018-0446-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Conceição-Neto N et al. Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis. Scientific Reports 5, 16532, doi: 10.1038/srep16532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Waller AS et al. Classification and quantification of bacteriophage taxa in human gut metagenomes. The ISME Journal 8, 1391–1402, doi: 10.1038/ismej.2014.30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma Y, You X, Mai G, Tokuyasu T & Liu C A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome 6, 24, doi: 10.1186/s40168-018-0410-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kleiner M, Hooper LV & Duerkop BA Evaluation of methods to purify virus-like particles for metagenomic sequencing of intestinal viromes. BMC Genomics 16, 7, doi: 10.1186/s12864-014-1207-4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Flewett TH, Bryden AS & Davies H Diagnostic electron microscopy of faeces. Journal of Clinical Pathology 27, 603–608, doi: 10.1136/jcp.27.8.603 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim KH & Bae JW Amplification Methods Bias Metagenomic Libraries of Uncultured Single-Stranded and Double-Stranded DNA Viruses. Applied and Environmental Microbiology 77, 7663–7668, doi: 10.1128/AEM.00289-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yilmaz S, Allgaier M & Hugenholtz P Multiple displacement amplification compromises quantitative analysis of metagenomes. Nature Methods 7, 943–944, doi: 10.1038/nmeth1210-943 (2010). [DOI] [PubMed] [Google Scholar]
- 99.Roux S et al. Towards quantitative viromics for both double-stranded and single-stranded DNA viruses. PeerJ 4, e2777, doi: 10.7717/peerj.2777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Krishnamurthy SR & Wang D Origins and challenges of viral dark matter. Virus Research 239, 136–142, doi: 10.1016/j.virusres.2017.02.002 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Santiago-Rodriguez MT & Hollister BE Human Virome and Disease: High-Throughput Sequencing for Virus Discovery, Identification of Phage-Bacteria Dysbiosis and Development of Therapeutic Approaches with Emphasis on the Human Gut. Viruses 11, 656, doi: 10.3390/v11070656 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barr JJ et al. Bacteriophage adhering to mucus provide a non–host-derived immunity. Proceedings of the National Academy of Sciences 110, 10771–10776, doi: 10.1073/pnas.1305923110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wagner J et al. Bacteriophages in Gut Samples From Pediatric Crohn’s Disease Patients: Metagenomic Analysis Using 454 Pyrosequencing. Inflammatory Bowel Diseases 19, 1598–1608, doi: 10.1097/MIB.0b013e318292477c (2013). [DOI] [PubMed] [Google Scholar]
- 104.Siringan P, Connerton PL, Cummings NJ & Connerton IF Alternative bacteriophage life cycles: the carrier state of Campylobacter jejuni. Open Biology 4, 130200, doi: 10.1098/rsob.130200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pourcel C, Midoux C, Vergnaud G & Latino L A carrier state is established in Pseudomonas aeruginosa by phage LeviOr01, a newly isolated ssRNA levivirus. Journal of General Virology 98, 2181–2189, doi: 10.1099/jgv.0.000883 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Joossens M et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 60, 631–637, doi: 10.1136/gut.2010.223263 (2011). [DOI] [PubMed] [Google Scholar]
- 107.Lax S et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052, doi: 10.1126/science.1254529 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zmora N, Suez J & Elinav E You are what you eat: diet, health and the gut microbiota. Nature Reviews Gastroenterology & Hepatology 16, 35–56, doi: 10.1038/s41575-018-0061-2 (2019). [DOI] [PubMed] [Google Scholar]
- 109.Minot S et al. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Research 21, 1616–1625, doi: 10.1101/gr.122705.111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zuo T et al. Human-Gut-DNA Virome Variations across Geography, Ethnicity, and Urbanization. Cell Host & Microbe 28, 741–751, doi: 10.1016/j.chom.2020.08.005 (2020). [DOI] [PubMed] [Google Scholar]
- 111.Coughlan S et al. The gut virome in Irritable Bowel Syndrome differs from that of controls. Gut Microbes 13, 1887719, doi: 10.1080/19490976.2021.1887719 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lepage P et al. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut 57, 424–425, doi: 10.1136/gut.2007.134668 (2008). [DOI] [PubMed] [Google Scholar]
- 113.Bajaj JS et al. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut 70, 1162–1173, doi: 10.1136/gutjnl-2020-322470 (2021). [DOI] [PubMed] [Google Scholar]
- 114.d’Herelle F & Smith GH The bacteriophage and its behavior, 490–497 (Williams & Wilkins, 1926). [Google Scholar]
- 115.Spence RC & B. ME The therapeutic value of the bacteriophage in treatment of bacillary dysentery. Southern Medical Journal 17, 563–571, doi: 10.1097/00007611-192408000-00005 (1924). [DOI] [Google Scholar]
- 116.Burnet FM, McKie M & Wood IJ INVESTIGATIONS ON BACILLARY DYSENTERY IN INFANTS, WITH SPECIAL REFERENCE TO BACTERIOPHAGE PHENOMENA. Medical Journal of Australia 2, 71–78, doi: 10.5694/j.1326-5377.1930.tb41310.x (1930). [DOI] [Google Scholar]
- 117.D’Herelle F Studies Upon Asiatic Cholera. Yale Journal of Biology and Medicine 1, 195–219 (1929). [PMC free article] [PubMed] [Google Scholar]
- 118.Becherish A. a. H., P. Le bacteriophage dans le traitement de la fievre typhoid. Comptes Rendus Hebdomadaires des Séances et Mémoires de la Société de Biologie et des ses Filiales 86, 168 (1923). [Google Scholar]
- 119.Smith J THE BACTERIOPHAGE IN THE TREATMENT OF TYPHOID FEVER. British Medical Journal 2, 47–49, doi: 10.1136/bmj.2.3315.47 (1924). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hadley P The Twort-D’Herelle Phenomenon: A Critical Review and Presentation of a New Conception (Homogamic Theory) Of Bacteriophage Action. The Journal of Infectious Diseases 42, 263–434, doi: 10.1093/infdis/42.4.263 (1928). [DOI] [Google Scholar]
- 121.Eaton MD & Bayne-Jones S BACTERIOPHAGE THERAPY: REVIEW OF THE PRINCIPLES AND RESULTS OF THE USE OF BACTERIOPHAGE IN THE TREATMENT OF INFECTIONS. Journal of the American Medical Association 103, 1769–1776, doi: 10.1001/jama.1934.72750490003007 (1934). [DOI] [Google Scholar]
- 122.Merabishvili M et al. Quality-Controlled Small-Scale Production of a Well-Defined Bacteriophage Cocktail for Use in Human Clinical Trials. PLoS One 4, e4944, doi: 10.1371/journal.pone.0004944 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bruttin A & Brüssow H Human Volunteers Receiving Escherichia coli Phage T4 Orally: a Safety Test of Phage Therapy. Antimicrobial Agents and Chemotherapy 49, 2874–2878, doi: 10.1128/AAC.49.7.2874-2878.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sarker SA et al. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology 434, 222–232, doi: 10.1016/j.virol.2012.09.002 (2012). [DOI] [PubMed] [Google Scholar]
- 125.McCallin S et al. Safety analysis of a Russian phage cocktail: From MetaGenomic analysis to oral application in healthy human subjects. Virology 443, 187–196, doi: 10.1016/j.virol.2013.05.022 (2013). [DOI] [PubMed] [Google Scholar]
- 126.Sarker SA et al. Oral application of Escherichia coli bacteriophage: safety tests in healthy and diarrheal children from Bangladesh. Environmental Microbiology 19, 237–250, doi: 10.1111/1462-2920.13574 (2017). [DOI] [PubMed] [Google Scholar]
- 127.Sarker SA et al. Oral Phage Therapy of Acute Bacterial Diarrhea With Two Coliphage Preparations: A Randomized Trial in Children From Bangladesh. EBioMedicine 4, 124–137, doi: 10.1016/j.ebiom.2015.12.023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schooley RT et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrobial Agents and Chemotherapy 61, e00954–00917, doi: 10.1128/AAC.00954-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jennes S et al. Use of bacteriophages in the treatment of colistin-only-sensitive Pseudomonas aeruginosa septicaemia in a patient with acute kidney injury—a case report. Critical Care 21, 129, doi: 10.1186/s13054-017-1709-y (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dedrick RM et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nature Medicine 25, 730–733, doi: 10.1038/s41591-019-0437-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yen M, Cairns LS & Camilli A A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nature Communications 8, 14187, doi: 10.1038/ncomms14187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Faruque SM et al. Self-limiting nature of seasonal cholera epidemics: Role of host-mediated amplification of phage. Proceedings of the National Academy of Sciences 102, 6119–6124, doi: 10.1073/pnas.0502069102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Khoruts A & Sadowsky MJ Understanding the mechanisms of faecal microbiota transplantation. Nature Reviews Gastroenterology & Hepatology 13, 508–516, doi: 10.1038/nrgastro.2016.98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sanders ME, Merenstein DJ, Reid G, Gibson GR & Rastall RA Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nature Reviews Gastroenterology & Hepatology 16, 605–616, doi: 10.1038/s41575-019-0173-3 (2019). [DOI] [PubMed] [Google Scholar]
- 135.Deaton J, Ertle E & Dawson HGUS Patent Number: 9,839,657; Assignee: Dearland Enzymes, Inc. (Kennesaw, GA). United States Patent and Trademark Office; (2017). [Google Scholar]
- 136.Gindin M, Febvre HP, Rao S, Wallace TC & Weir TL Bacteriophage for Gastrointestinal Health (PHAGE) Study: Evaluating the Safety and Tolerability of Supplemental Bacteriophage Consumption. Journal of American College of Nutrition 38, 68–75, doi: 10.1080/07315724.2018.1483783 (2019). [DOI] [PubMed] [Google Scholar]
- 137.Grubb DS et al. PHAGE-2 Study: Supplemental Bacteriophages Extend Bifidobacterium animalis subsp. lactis BL04 Benefits on Gut Health and Microbiota in Healthy Adults. Nutrients 12, 2474, doi: 10.3390/nu12082474 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rolhion N & Darfeuille-Michaud A Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflammatory Bowel Diseases 13, 1277–1283, doi: 10.1002/ibd.20176 (2007). [DOI] [PubMed] [Google Scholar]
- 139.Palmela C et al. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 67, 574–587, doi: 10.1136/gutjnl-2017-314903 (2018). [DOI] [PubMed] [Google Scholar]
- 140.Galtier M et al. Bacteriophages Targeting Adherent Invasive Escherichia coli Strains as a Promising New Treatment for Crohn’s Disease. Journal of Crohn’s and Colitis 11, 840–847, doi: 10.1093/ecco-jcc/jjw224 (2017). [DOI] [PubMed] [Google Scholar]
- 141.Duan Y et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511, doi: 10.1038/s41586-019-1742-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McCoy WC & Mason JM 3rd. Enterococcal endocarditis associated with carcinoma of the sigmoid; report of a case. Journal of the Medical Association of the State of Alabama 21, 162–166 (1951). [PubMed] [Google Scholar]
- 143.Abdulamir AS, Hafidh RR & Bakar FA The association of Streptococcus bovis/gallolyticus with colorectal tumors: The nature and the underlying mechanisms of its etiological role. Journal of Experimental & Clinical Cancer Research 30, 11, doi: 10.1186/1756-9966-30-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Boleij A, van Gelder MMHJ, Swinkels DW & Tjalsma H Clinical Importance of Streptococcus gallolyticus Infection Among Colorectal Cancer Patients: Systematic Review and Meta-analysis. Clinical Infectious Diseases 53, 870–878, doi: 10.1093/cid/cir609 (2011). [DOI] [PubMed] [Google Scholar]
- 145.Wirbel J et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nature Medicine 25, 679–689, doi: 10.1038/s41591-019-0406-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yachida S et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nature Medicine 25, 968–976, doi: 10.1038/s41591-019-0458-7 (2019). [DOI] [PubMed] [Google Scholar]
- 147.Wong SH et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 66, 1441–1448, doi: 10.1136/gutjnl-2016-312766 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Castellarin M et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Research 22, 299–306, doi: 10.1101/gr.126516.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kostic AD et al. Fusobacterium nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host & Microbe 14, 207–215, doi: 10.1016/j.chom.2013.07.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bullman S et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448, doi: 10.1126/science.aal5240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wu S et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nature Medicine 15, 1016–1022, doi: 10.1038/nm.2015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Arthur JC et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science 338, 120–123, doi: 10.1126/science.1224820 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dejea CM et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597, doi: 10.1126/science.aah3648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Huycke MM, Abrams V & Moore DR Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 23, 529–536, doi: 10.1093/carcin/23.3.529 (2002). [DOI] [PubMed] [Google Scholar]
- 155.Wang X et al. 4-Hydroxy-2-Nonenal Mediates Genotoxicity and Bystander Effects Caused by Enterococcus faecalis–Infected Macrophages. Gastroenterology 142, 543–551, doi: 10.1053/j.gastro.2011.11.020 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kumar R et al. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLOS Pathogens 13, e1006440, doi: 10.1371/journal.ppat.1006440 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhu W et al. Editing of the gut microbiota reduces carcinogenesis in mouse models of colitis-associated colorectal cancer. Journal of Experimental Medicine 216, 2378–2393, doi: 10.1084/jem.20181939 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Henning U et al. Chapter 23: Receptor recognition by T-even type coliphages. Molecular Biology of Bacteriophage T4 1, 291–298 (ASM Press, 1994). [Google Scholar]
- 159.Iida S Bacteriophage P1 carries two related sets of genes determining its host range in the invertible C segment of its genome. Virology 134, 421–434, doi: 10.1016/0042-6822(84)90309-x (1984). [DOI] [PubMed] [Google Scholar]
- 160.Summer EJ et al. Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. Journal of Molelcular Biology 340, 49–65, doi: 10.1016/j.jmb.2004.04.053 (2004). [DOI] [PubMed] [Google Scholar]
- 161.Liu M et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295, 2091–2094, doi: 10.1126/science.1067467 (2002). [DOI] [PubMed] [Google Scholar]
- 162.Bar H, Yacoby I & Benhar I Killing cancer cells by targeted drug-carrying phage nanomedicines. BMC Biotechnology 8, 37, doi: 10.1186/1472-6750-8-37 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yacoby I, Shamis M, Bar H, Shabat D & Benhar I Targeting Antibacterial Agents by Using Drug-Carrying Filamentous Bacteriophages. Antimicrobial Agents and Chemotherapy 50, 2087, doi: 10.1128/AAC.00169-06 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yacoby I, Bar H & Benhar I Targeted Drug-Carrying Bacteriophages as Antibacterial Nanomedicines. Antimicrobial Agents and Chemotherapy 51, 2156, doi: 10.1128/AAC.00163-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vaks L & Benhar I Antibacterial Application of Engineered Bacteriophage Nanomedicines: Antibody-Targeted, Chloramphenicol Prodrug Loaded Bacteriophages for Inhibiting the Growth of Staphylococcus aureus Bacteria. Biomedical Nanotechnology: Methods and Protocols, 187–206 (Humana Press, 2011). [DOI] [PubMed] [Google Scholar]
- 166.Zheng D-W et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nature Biomedical Engineering 3, 717–728, doi: 10.1038/s41551-019-0423-2 (2019). [DOI] [PubMed] [Google Scholar]
- 167.Yu T et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 170, 548–563, doi: 10.1016/j.cell.2017.07.008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dong X et al. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Science Advances 6, eaba1590, doi: 10.1126/sciadv.aba1590 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Devkota S et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487, 104–108, doi: 10.1038/nature11225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hendrikx T et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 68, 1504–1515, doi: 10.1136/gutjnl-2018-317232 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Singh N et al. Activation of Gpr109a, Receptor for Niacin and the Commensal Metabolite Butyrate, Suppresses Colonic Inflammation and Carcinogenesis. Immunity 40, 128–139, doi: 10.1016/j.immuni.2013.12.007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cani PD & Jordan BF Gut microbiota-mediated inflammation in obesity: a link with gastrointestinal cancer. Nature Reviews Gastroenterology & Hepatology 15, 671–682, doi: 10.1038/s41575-018-0025-6 (2018). [DOI] [PubMed] [Google Scholar]
- 173.Febvre PH et al. PHAGE Study: Effects of Supplemental Bacteriophage Intake on Inflammation and Gut Microbiota in Healthy Adults. Nutrients 11, 666, doi: 10.3390/nu11030666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sweere JM et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 363, eaat9691, doi: 10.1126/science.aat9691 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tetz GV et al. Bacteriophages as potential new mammalian pathogens. Scientific Reports 7, 7043, doi: 10.1038/s41598-017-07278-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Van Belleghem JD, Clement F, Merabishvili M, Lavigne R & Vaneechoutte M Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Scientific Reports 7, 8004, doi: 10.1038/s41598-017-08336-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gogokhia L et al. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host & Microbe 25, 285–299, doi: 10.1016/j.chom.2019.01.008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang L et al. Staphylococcus aureus Bacteriophage Suppresses LPS-Induced Inflammation in MAC-T Bovine Mammary Epithelial Cells. Frontiers in Microbiology 9, 1614, doi: 10.3389/fmicb.2018.01614 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hong Y et al. The impact of orally administered phages on host immune response and surrounding microbial communities. Bacteriophage 6, e1211066, doi: 10.1080/21597081.2016.1211066 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Miernikiewicz P et al. T4 Phage and Its Head Surface Proteins Do Not Stimulate Inflammatory Mediator Production. PLoS One 8, e71036, doi: 10.1371/journal.pone.0071036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Merabishvili M, Pirnay JP & De Vos D Guidelines to Compose an Ideal Bacteriophage Cocktail. Bacteriophage Therapy: From Lab to Clinical Practice, 99–110 (Springer; New York, 2018). [Google Scholar]
- 182.Betts A, Vasse M, Kaltz O & Hochberg ME Back to the future: evolving bacteriophages to increase their effectiveness against the pathogen Pseudomonas aeruginosa PAO1. Evolutionary Applications 6, 1054–1063, doi: 10.1111/eva.12085 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Friman VP et al. Pre-adapting parasitic phages to a pathogen leads to increased pathogen clearance and lowered resistance evolution with Pseudomonas aeruginosa cystic fibrosis bacterial isolates. Journal of Evolutionary Biology 29, 188–198, doi: 10.1111/jeb.12774 (2016). [DOI] [PubMed] [Google Scholar]
- 184.Dunne M et al. Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins. Cell Reports 29, 1336–1350, doi: 10.1016/j.celrep.2019.09.062 (2019). [DOI] [PubMed] [Google Scholar]
- 185.Yehl K et al. Engineering Phage Host-Range and Suppressing Bacterial Resistance through Phage Tail Fiber Mutagenesis. Cell 179, 459–469, doi: 10.1016/j.cell.2019.09.015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chatain-Ly MH The factors affecting effectiveness of treatment in phages therapy. Frontiers in Microbiology 5, 51, doi: 10.3389/fmicb.2014.00051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cui Z, Guo X, Feng T & Li L Exploring the whole standard operating procedure for phage therapy in clinical practice. Journal of Translational Medicine 17, 373, doi: 10.1186/s12967-019-2120-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Fabijan AP et al. Safety of bacteriophage therapy in severe Staphylococcus aureus infection. Nature Microbiology 5, 465–472, doi: 10.1038/s41564-019-0634-z (2020). [DOI] [PubMed] [Google Scholar]
- 189.Jończyk E, Kłak M, Międzybrodzki R & Górski A The influence of external factors on bacteriophages—review. Folia Microbiologica 56, 191–200, doi: 10.1007/s12223-011-0039-8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Colom J et al. Microencapsulation with alginate/CaCO3: A strategy for improved phage therapy. Scientific Reports 7, 41441, doi: 10.1038/srep41441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hsu BB et al. In situ reprogramming of gut bacteria by oral delivery. Nature Communications 11, 5030, doi: 10.1038/s41467-020-18614-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dąbrowska K Phage therapy: What factors shape phage pharmacokinetics and bioavailability? Systematic and critical review. Medicinal Research Reviews 39, 2000–2025, doi: 10.1002/med.21572 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Srivastava AS, Kaido T & Carrier E Immunological factors that affect the in vivo fate of T7 phage in the mouse. Journal of Virological Methods 115, 99–104, doi: 10.1016/j.jviromet.2003.09.009 (2004). [DOI] [PubMed] [Google Scholar]
- 194.Merril CR et al. Long-circulating bacteriophage as antibacterial agents. Proceedings of the National Academy of Sciences 93, 3188–3192, doi: 10.1073/pnas.93.8.3188 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Capparelli R, Parlato M, Borriello G, Salvatore P & Iannelli D Experimental Phage Therapy against Staphylococcus aureus in Mice. Antimicrobial Agents and Chemotherapy 51, 2765, doi: 10.1128/AAC.01513-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Capparelli R, Ventimiglia I, Roperto S, Fenizia D & Iannelli D Selection of an Escherichia coli O157:H7 bacteriophage for persistence in the circulatory system of mice infected experimentally. Clinical Microbiology and Infection 12, 248–253, doi: 10.1111/j.1469-0691.2005.01340.x (2006). [DOI] [PubMed] [Google Scholar]
- 197.Fauconnier A Guidelines for Bacteriophage Product Certification. Bacteriophage Therapy: From Lab to Clinical Practice 1693, 253–268 (Springer; New York, 2018). [DOI] [PubMed] [Google Scholar]
- 198.Pirnay JP et al. The Phage Therapy Paradigm: Prêt-à-Porter or Sur-mesure? Pharmaceutical Research 28, 934–937, doi: 10.1007/s11095-010-0313-5 (2011). [DOI] [PubMed] [Google Scholar]
- 199.Servick K Beleaguered phage therapy trial presses on. Science 352, 1506, doi: 10.1126/science.352.6293.1506 (2016). [DOI] [PubMed] [Google Scholar]
- 200.Pirnay JP et al. The Magistral Phage. Viruses 10, 64, doi: 10.3390/v10020064 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Corbellino M et al. Eradication of a Multidrug-Resistant, Carbapenemase-Producing Klebsiella pneumoniae Isolate Following Oral and Intra-rectal Therapy With a Custom Made, Lytic Bacteriophages Preparation. Clinical Infectious Diseases 70, 1998–2001, doi: 10.1093/cid/ciz782 (2020). [DOI] [PubMed] [Google Scholar]
- 202.Intralytix Inc. and Mount Sinai Hospital. Safety and Efficacy of EcoActive on Intestinal Adherent Invasive E. coli in Patients With Inactive Crohn’s Disease. ClinicalTrials.gov (NCT03808103). (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


