Abstract
We previously demonstrated (M. M. Exner, P. Doig, T. J. Trust, and R. E. W. Hancock, Infect. Immun. 63:1567–1572, 1995) that Helicobacter pylori has at least one nonspecific porin, HopE, which has a low abundance in the outer membrane but forms large channels. H. pylori is relatively susceptible to most antimicrobial agents but less susceptible to the polycationic antibiotic polymyxin B. We demonstrate here that H. pylori is able to take up higher basal levels of the hydrophobic fluorescent probe 1-N-phenylnaphthylamine (NPN) than Pseudomonas aeruginosa or Escherichia coli, consistent with its enhanced susceptibility to hydrophobic agents. Addition of polymyxin B led to a further increase in NPN uptake, indicative of a self-promoted uptake pathway, but it required a much higher amount of polymyxin B to yield a 50% increase in NPN uptake in H. pylori (6 to 8 μg/ml) than in P. aeruginosa or E. coli (0.3 to 0.5 μg/ml), suggesting that H. pylori has a less efficient self-promoted uptake pathway. Since intrinsic resistance involves the collaboration of restricted outer membrane permeability and secondary defense mechanisms, such as periplasmic β-lactamase (which H. pylori lacks) or efflux, we examined the possible role of efflux in antibiotic susceptibility. We had previously identified in H. pylori 11637 the presence of portions of three genes with homology to potential restriction-nodulation-division (RND) efflux systems. It was confirmed that H. pylori contained only these three putative RND efflux systems, named here hefABC, hefDEF, and hefGHI, and that the hefGHI system was expressed only in vivo while the two other RND systems were expressed both in vivo and in vitro. In uptake studies, there was no observable energy-dependent tetracycline, chloramphenicol, or NPN efflux activity in H. pylori. Independent mutagenesis of the three putative RND efflux operons in the chromosome of H. pylori had no effect on the in vitro susceptibility of H. pylori to 19 antibiotics. These results, in contrast to what is observed in E. coli, P. aeruginosa, and other clinically important gram-negative bacteria, suggest that active efflux does not play a role in the intrinsic resistance of H. pylori to antibiotics.
Helicobacter pylori is an important human pathogen that is estimated to infect 50% of the world's population (7, 31). H. pylori infection in humans is associated with the development of numerous gastric pathologies, including gastritis, gastric ulcers, and gastric cancers. Once H. pylori is established in the gastric submucosa, infected individuals usually carry it for life unless treated.
Although many H. pylori strains are quite susceptible to most antibiotics in vitro, treatment of H. pylori-infected individuals has proven difficult (7, 31). Single-antibiotic therapy has not been very effective, and successful treatment usually requires two or three antibiotics given in combination with a proton pump inhibitor (31). Several hypotheses have been proposed in the literature to explain this seemingly elevated in vivo antibiotic resistance, including acid inactivation of the drug, inappropriate formulation, variable compliance, and inability of the drug to accumulate in the gastric mucosa. However, such speculation on the basis for in vivo resistance is premature, since there have been no fundamental studies of antibiotic uptake and efflux mechanisms in H. pylori and the basis for the normal high susceptibility of this bacterium to antibiotics is not known.
There are three basic uptake systems across the outer membrane (10), namely, the uptake of hydrophilic substances through the water-filled channels of porins, uptake of polycations via self-promoted uptake at divalent cation binding sites on lipopolysaccharide, and uptake of hydrophobic substances through the outer membrane bilayer. We previously demonstrated that Helicobacter contains at least five porins (part of a 32-member family of outer membrane proteins) (6, 8). These porins are expressed relatively weakly compared to those of other bacteria, although the main candidate nonspecific porin of H. pylori, HopE, formed large channels (6). Here we have examined the other uptake pathways.
Intrinsic antibiotic resistance in gram-negative bacteria involves the collaboration of restricted outer membrane permeability with secondary defense mechanisms, such as periplasmic β-lactamase or antibiotic efflux (23). In Pseudomonas aeruginosa, for example, intrinsic resistance can be overcome by increasing the outer membrane permeability (e.g., by expressing in the outer membrane a large-channel porin [11] or by utilizing a permeabilizing agent, such as a chelator or polycation [28]) and also by mutating the MexA-MexB-OprM efflux system (17) or, for specific β-lactams, by mutationally preventing chromosomal β-lactamase induction (5). The genomes of two strains of H. pylori have been sequenced (1, 30), and there are no β-lactamase homolog genes in either strain.
There are at least four conserved families of efflux systems associated with bacterial resistance to antibiotics (25). One of these families, which is widespread among gram-negative bacteria, is the resistance-nodulation-division (RND) family of efflux systems (22, 26). The RND family efflux systems are dependent on the proton motive force. Such systems, including the AcrAB-TolC system in Escherichia coli (19), the AcrAB system in Haemophilus influenzae (27), the MexAB-OprM system in P. aeruginosa (17), and the MtrCDE system in Neisseria gonorrhoeae (9), are responsible for the intrinsic resistance to a wide variety of structurally and chemically unrelated antibiotics and other antimicrobial compounds (22, 25, 26). In each of these cases, inactivation of the RND systems by mutation renders the bacteria hypersusceptible to a wide variety of antimicrobials (22, 25, 26), whereas upregulation of these systems leads to multiple antibiotic resistance (14, 17).
We previously reported the presence of three gene fragments encoding components of putative RND efflux systems in H. pylori 11637 (4). In this study, we have examined the determinants of the intrinsic susceptibility of H. pylori to antibiotics and whether efflux mediated by any of the three putative efflux systems is a determinant.
MATERIALS AND METHODS
Bacteria, plasmids, and culture conditions.
H. pylori strains were grown as previously described (8). H. pylori 26695 was provided by K. Eaton (Ohio State University, Columbus); H. pylori 11637, SS1 (16), J99, ArHp64, ArHp12, ArHp18, ArHp25, ArHp210, and UA861 (13) were obtained from the culture collection of Astra Research Center Boston (Cambridge, Mass.). These strains represent isolates from Australia (SS1 and ArHp25), Sweden (ArHp210), Argentina (ArHp64), the United States (J99 and ArHp12), and Canada (ArHp18 and UA861). An H. pylori 11637 genomic library constructed in the cosmid pLAFR3 (29) and maintained in E. coli HB101 was used for the cloning of the H. pylori hefABC RND operon. P. aeruginosa H103 and K372 and its oprM::Ω mutant K613 were previously described (16). The vector pBluescript was from Stratagene (La Jolla, Calif.). E. coli DH5α was used for subcloning the hefABC operon. E. coli W4680 and its ΔacrAB::Tn903 Kanr mutant WZM120 (18) were kindly provided by Keith Poole (Queens University, Ontario, Canada). E. coli and P. aeruginosa strains were grown in Luria broth. For cloning in E. coli, ampicillin was used at 100 μg/ml and tetracycline was used at 18 μg/ml.
Reagents.
Chemical reagents were purchased from Sigma Chemical (St. Louis, Mo.). Restriction enzymes, polymerases, and other molecular biology reagents were purchased from Gibco BRL (Burlington, Ontario) and used as described in the manufacturer's instructions. [3H]chloramphenicol (1 mCi/ml; 30 to 60 Ci/mmol) and [3H]tetracycline (1 mCi/ml; 0.77 Ci/mmol) were purchased from Du Pont, NEN (Boston, Mass.).
MIC determination.
The MICs in Table 1 were determined by the broth microdilution method (3). Disk diffusion assays to assess the knockout mutants were done as follows. A sterile cotton-tipped swab was dipped into the H. pylori suspension (diluted to twice the McFarland standard) and streaked in three directions across a tryptic soy agar (BBL; Becton Dickinson and Company, Cockeysville, Md.) supplemented with 5% sheep blood (Remel, Lenexa, Kans.). The plates were dried for 5 min, and then disks containing the antibiotics (Remel) were placed on the agar surface. The plates were incubated for 3 days in a microaerophilic atmosphere (5% O2, 10% CO2, and 85% N2); (Cellstar Trigas incubator; Queue Systems, Asheville, N.C.) at 37°C. At the end of the incubation period, the diameters of the zones of growth inhibition were measured.
TABLE 1.
MICs of different classes of antibiotics
| Antibiotic | MIC (μg/ml) for:
|
||||
|---|---|---|---|---|---|
| H. pylori 11637 | E. coli K4401 | E. coli KZM120 ΔacrAB | P. aeruginosa K372 | P. aeruginosa K613 oprM::Ω | |
| Ampicillin | 0.06 | 4 | 2 | >128 | >128 |
| Penicillin | 0.12 | 32 | 16 | >1,024 | 512 |
| Cefotaxime | 0.02 | 0.5 | NDa | 4 | 1 |
| Ciprofloxacin | 0.12 | 0.12 | ND | 2 | 0.12 |
| Novobiocin | 0.1 | 32 | 0.5 | 64 | 4 |
| Erythromycin | 0.5 | 32 | 2 | 128 | 8 |
| Tetracycline | 0.03 | 2 | 0.25 | 4 | 0.12 |
| Chloramphenicol | 0.5 | 4 | 0.5 | 16 | 1 |
| Polymyxin B | 5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Colistin | 8 | 0.5 | 0.5 | 1.0 | 1.0 |
ND, not done.
Cloning and DNA sequencing of the hefABC operon.
Molecular biology protocols were performed according to standard methods (20). The DNA sequence of the hefABC operon of H. pylori 11637 was generated by shotgun sequencing of random subclones of cosmid clone 200:4, which contained the hefABC putative efflux system (4). Briefly, p200:4 was partially restricted with Sau3A1 restriction enzyme, and the resulting DNA was size fractionated on an agarose gel. Restriction fragments approximately 1 to 3 kb long were purified and ligated into BamHI-digested pBluescript. The resultant recombinant clones were screened in hybridization experiments with the DNA insert from the H. pylori alkaline phosphatase clone 200 (4) as a probe. The clones that reacted positively with the probe were sequenced with the universal reverse and forward primers. The remaining DNA sequence gaps were filled in by primer walking with p200:4 as a template. Sequence assembly was facilitated by the use of the Contig Alignment Program (12). The resulting DNA sequences covered the hefABC operon at least twice on each DNA strand. All DNA sequencing was done on an ABI 373 automated DNA sequencer with the ABI Prism dye terminator sequencing kit.
Presence of putative efflux operons in other H. pylori strains.
The linkage of the hefABC, hefDEF, and hefGHI genes was examined in H. pylori SS1, ArHp25, ArHp210, ArHp64, ArHp12, ArHp18, and UA861 by using PCR primers specific for each gene in the particular cluster with a “reverse” primer specific for the gene encoding the RND pump protein (hefC, hefF, or hefI) in the respective cluster. The primers used were hefA, 5′CTTCAAAGGCTAGTTTGGATGCGGC; hefB, 5′CAGCGATACAGCAAAATTGGGGGCGC; hefC, 5′TTACTACGACTTATCCTGGGGCTAGCGCTG; hefD, 5′GACACGATGCTAGCGATTGTGGAGCC; hefE, 5′GGCGAGCAGGTGAAGGCTGGAGATGC; hefF, Hel1 (see below); hefG, 5′GGCGATTACAACGCTTATTACAACGC; hefH, 5′ATGGCTATGTGGAAAAGCTTTATGC; and hefI, 5′GAAATTGTGGTTAAAAAAGAAGGGGC. The reverse RND pump gene-specific primers were hefC, 5′CCCCATCAATTTCCACATTCTCCGC; hefF, Hel2 (see below), and hefI, 5′GCAAAAAATATCGCGCCCCCCACATGC. PCRs were performed in a Perkin-Elmer 9600 PCR machine. The reactions consisted of 35 cycles, with each cycle composed of 15 s at 94°C (denaturation), 30 s at 50°C (annealing), and 150 s at 72°C (extension). After a final extension of 10 min at 72°C, the amplicons were electrophoresed on a 1.2% agarose gel with 0.04 M Tris-acetate–0.001 M EDTA, pH 8.0 (TAE).
Gene expression analysis of the hef operons by RT PCR.
RNA was isolated from H. pylori SS1 that had been grown in vitro (on chocolate blood agar plates) or directly from the stomachs of C57BL/6 mice that were colonized with H. pylori SS1 (in vivo). The RNA was prepared with the S.N.A.P. total RNA isolation kit from Invitrogen (Carlsbad, Calif.). The RNA was analyzed by reverse transcriptase (RT) PCR with the SuperScript RT-PCR kit from Gibco BRL (Gaithersburg, Md.). The gene-specific primers for the efflux pumps used were designed from the sequence alignments of the genes from H. pylori 26695 and J99. For hefF, the Hel1 (5′CTAGCCCTGAAGAAATGGAAAACAACATCG) and Hel2 (5′CCTTGCCATGTCATTAAAATCCG) PCR primers were predicted to amplify a 418-bp amplicon, whereas for hefI, the Hel3 (5′TCAAATCCAGCGTGATGTAGTAGCCG) and Hel4 (5′ATTTATCCCCCAAATCAATGAAGGGG) PCR primers were predicted to produce a 515-bp amplicon. Three independent reactions were used for each analysis. Total nucleic acid (before DNA removal) was used as a positive control template, whereas RNA was used as a template in two identical reactions where the only difference was the addition of the RT enzyme. The reactions were held at 37°C for 30 min prior to 35 cycles of PCR with parameters of 15 s at 94°C, 15 s at 55°C, and 1 min at 72°C. The reaction mixtures were then electrophoresed on a 1% TAE agarose gel.
Uptake of [3H]tetracycline, [3H]chloramphenicol, and 1-N-phenylnapthylamine (NPN) by H. pylori.
Two-day cultures of H. pylori 26695, grown on chocolate blood agar plates, were resuspended in phosphate-buffered saline, washed once in 5 mM HEPES buffer (pH 7.2) containing 10 μM MgCl2, and resuspended at an optical density at 600 nm of 0.5 in 5 mM glucose–5 mM HEPES buffer at either pH 4, pH 7.2, or pH 9; the cell suspension was aliquoted and used immediately in the uptake assays.
Carbon sources were added when needed at a final concentration of 0.4% (acetate, citrate, ethanol, fructose, gluconate, glucose, glycerol, lactose, maltose, mannitol, mannose, propionate, succinate, and sucrose) or 0.2% (alanine, glycine, histidine, and proline), and the resulting cell suspension was incubated at 37°C for 20 min prior to assay initiation. Carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to a final concentration of 40 μM 15 min prior to assay initiation.
The antibiotic uptake assays were initiated by the addition of approximately 3 μCi of radiolabeled antibiotic to a 500-μl suspension of the cells. At various time points, 100-μl aliquots were removed from the reaction mixture and added to 2 ml of 100 mM LiCl2 and the cells were vacuum filtered onto a 0.2-μm-pore-size filter. The filter was washed twice with 2 ml of 100 mM LiCl2 and dried, and the amount of radioactivity on the filter was determined by scintillation counting. Antibiotic uptake assays were performed three times at 25°C.
The NPN uptake assays were performed as described previously (18). Briefly, 1 ml of the H. pylori, P. aeruginosa, or E. coli cell suspension was placed in a quartz cuvette, NPN was added to a final concentration of 10 μM, and when stated, 1 to 250 μM CCCP was added. The cell suspension was briefly mixed by inversion and placed into a spectrofluorimeter to determine the background level of fluorescence. Subsequently, polymyxin B was added to the cuvette, the cuvette was briefly mixed and placed back into the fluorimeter, and the change in fluorescence over time was recorded on a chart recorder. The fluorescence measurements were done on a Perkin-Elmer 650-10S spectrofluorimeter with an excitation wavelength of 350 nm, an emission wavelength of 420 nm, and a slit width of 5 nm. The NPN assays were performed at room temperature.
DNA and protein sequence analysis.
Pairwise sequence alignments were performed with the ALIGN program at the Genestream Search network server (http://genome.eerie.fr/bin/align-guess.cgi), Montpellier, France. Phylogenetic analysis was done with the PHYLIP phylogeny inference package (8a). Phylogenetic trees were constructed with the nearest-neighbor distance matrix. DNA similarity searches were performed with the BLAST algorithm (2).
Mutagenesis of the putative efflux operons.
Mutagenesis of hefL was performed as follows. The internal portion (bp 724 to 2152) of the hefI gene (JHP1249) was amplified from H. pylori J99 as a 1.43-kb fragment and cloned into pGEM-T (Gibco BRL). Inverse PCR was then used to inactivate the gene by the insertion of the Kmr cassette from pILL600 (15) and a concomitant deletion between positions 1325 and 1647 of the hefI gene. This suicide plasmid construct was then introduced into H. pylori J99 by natural transformation, and transformants were selected by plating on selective media containing 25 μg of kanamycin/ml. Chromosomal DNA was prepared from several transformants and analyzed by PCR to ensure that the double crossover had occurred in the correct position. Mutagenesis of hefC and hefF were carried out in a similar fashion. Specifically, an internal fragment of the hefC gene (JHP554) was cloned into pGEM-T (bp 134 to 2729), and after inverse PCR, the Kmr cassette was inserted between positions 640 and 2219 of hefC, resulting in a 1,579-bp deletion. An internal portion of the hefF gene (JHP903) was cloned into pGEM-T (bp167 to 1429), and after inverse PCR, the Kmr cassette was inserted between positions 585 and 1019 of hefF, resulting in a 434-bp deletion.
Nucleotide sequence accession number.
The DNA sequence of the hefABC operon was submitted to the GenBank database (accession no. AF059041).
RESULTS
Hydrophobic and self-promoted uptake.
Consistent with the data from other laboratories (21), H. pylori demonstrated high susceptibility to β-lactams, quinolones, and other hydrophilic agents and to chloramphenicol and other hydrophobic agents but relatively low susceptibility to the polycations polymyxin B and cationic antimicrobial peptides (Table 1).
In contrast, wild-type E. coli bacteria were relatively resistant to hydrophobic agents but susceptible to polycations, while P. aeruginosa was also relatively resistant to the hydrophilic antibiotics. Mutants deficient in the major efflux systems of P. aeruginosa and E. coli showed high susceptibility to chloramphenicol, tetracycline, and, in part, novobiocin, ciprofloxacin, and erythromycin, analagous to the susceptibilities observed for H. pylori (Table 1). H. pylori was more susceptible to β-lactams than even the efflux mutants of E. coli and P. aeruginosa (Table 1), probably due in part to the complete absence of a β-lactamase in this organism (1, 30).
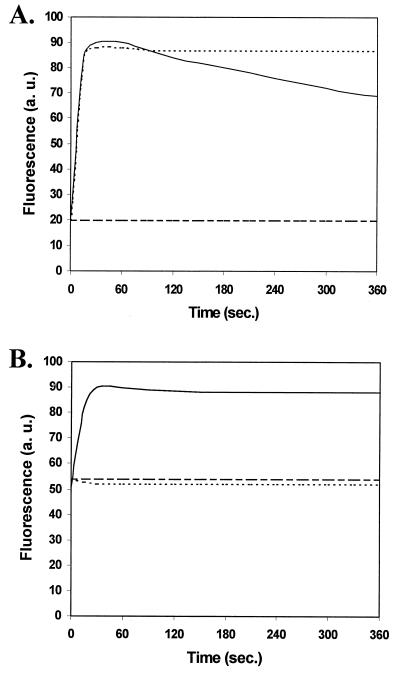
NPN is a small hydrophobic fluorescent probe which has been used to define the hydrophobic and self-promoted uptake pathways across the outer membranes of gram-negative bacteria (17). NPN fluoresces weakly in aqueous environments but strongly in hydrophobic environments, such as the membrane interior. It is usually excluded from the outer membrane by the outer monolayer of the outer membrane, which comprises the polyanion lipopolysaccharide, stabilized by divalent cation cross bridging. In most bacteria, a destabilizing polycation, such as polymyxin B, is required to permeabilize the outer membrane to NPN, a result consistent with and diagnostic of self-promoted uptake of the polycation. H. pylori (Fig. 1) demonstrated relatively high intrinsic uptake (50 arbitrary units) of NPN compared to E. coli (10 to 20 arbitrary units) or P. aeruginosa (10 arbitrary units), a result consistent with its relatively high susceptibility to hydrophobic agents. The higher background level of NPN fluorescence indicated that either the outer membrane of in vitro-grown H. pylori was permeable to hydrophobic compounds or there was a lack of active efflux in in vitro-grown H. pylori.
FIG. 1.
Uptake of NPN by E. coli (A) and H. pylori (B) in the presence of NPN alone (dashed lines), after the addition of NPN and polymyxin B (solid lines), and after incubation of E. coli with CCCP prior to the addition of NPN and polymyxin B (dotted lines). a. u., arbitrary unit.
To probe the self-promoted uptake pathway, we added the polycationic antibiotic polymyxin B to cells preincubated with NPN. Although, as mentioned above, incubation of H. pylori in the presence of NPN produced a significantly higher level of fluorescence than with E. coli (Fig. 1B), the addition of 6.4 μg of polymyxin B/ml resulted in further rapid uptake of NPN, similar to that seen for E. coli in the presence of the same concentration of polymyxin B.
The addition of graded concentrations of polymyxin B to H. pylori indicated that half-maximal uptake of NPN was achieved at 6 to 8 μg/ml, whereas only 0.3 μg of NPN/ml was required for half-maximal uptake in E. coli and 0.5 μg/ml was required in P. aeruginosa, numbers that correlated well with the MICs of polymyxin B for those bacteria. This was consistent with the hypothesis that self-promoted uptake is less effective in H. pylori and with the relatively low susceptibility of this bacterium to polycations.
H. pylori putative RND efflux operons.
We considered whether a lack of efflux was responsible for the high intrinsic antibiotic susceptibility of H. pylori. The H. pylori 26695 genomic sequence was reported to contain only six open reading frames (ORFs) with homology to RND efflux components (ORFs 606, 607, 969, 970, 1328, and 1329) (1, 30), and these pairs of genes correlated to the DNA inserts from H. pylori alkaline phosphatase fusion clones 200, 61, and 12, identified by us as portions of efflux operons (4). We identified three additional linked genes. There were no other RND efflux systems predicted in either strain of H. pylori for which the genomic sequence has been elucidated.
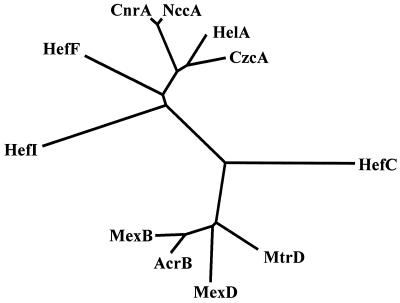
The operon containing hefABC from H. pylori 11637 was cloned, with the insert from fusion clone 200 (4) as a probe, and sequenced, and was found to be homologous to H. pylori 26695 ORFs 605, 606, and 607 (30) and H. pylori J99 ORFs 552, 553, and 554 (1). Phylogenetic analysis showed that this operon was most similar to the branch of the RND family proteins that are known to be involved in multiple drug efflux (Fig. 2), e.g., the AcrAB-TolC RND efflux system, which mediates the resistance of E. coli to a variety of different antibiotics, detergents, and dyes (22). The hefA gene encoded a homolog of E. coli TolC outer membrane protein, and the hefB and hefC genes were homologs of the E. coli acrA and acrB genes, which encode membrane fusion and RND cytoplasmic pump proteins, respectively. The hefABC operon was highly conserved among the three H. pylori strains (11637, 26695, and J99); each individual protein was more than 97% identical to each of its corresponding proteins in the other two H. pylori strains. In addition, and E. coli hemE homolog was present in the upstream region of the hefABC operon in all three of the strains examined (J99, 11637, and 26695).
FIG. 2.
Phylogenetic tree showing the relationship of the H. pylori RND pump proteins to other RND pump proteins. The three putative H. pylori RND efflux pump proteins (HefC, HefF, and HefI) are divergent from the clusters of RND efflux pump proteins involved in the efflux of divalent cations (CnrA [Ralstonia eutropha], NccA [Alcaligenes denitrificans], HelA [Legionella pneumophila], and CzcA [R. eutropha]) and those pumps involved in multiple-drug efflux (AcrA [E. coli], MexB [P. aeruginosa], MexD [P. aeruginosa], and MtrD [N. gonorrhoeae]).
A second H. pylori putative RND efflux operon contained three ORFs, named here hefDEF, which corresponded to H. pylori 26695 ORFs 971, 970, and 969 and J99 ORFs 905, 904, and 903. The hefD gene possessed similarity to tolC, and its product was identified as a putative outer membrane protein: the product of hefE (equivalent to clone 61 [4]) was a membrane fusion protein homolog, and hefF encoded an RND pump protein homolog (Fig. 2). The hefD and hefE genes each contained a typical signal sequence.
A third H. pylori RND putative operon contained four ORFs, named here orf1326-hefGHI, which corresponded to the H. pylori 26695 ORFs 1326, 1327, 1328, and 1329 and strain J99 ORFs 1246, 1247, 1248, and 1249. The hefGHI genes encoded the putative outer membrane protein, membrane fusion protein, and RND pump proteins (hefI being equivalent to clone 61 [4]), respectively. The orf1326 gene encoded a 125-amino-acid protein of unknown function. The orf1326, hefG, and hefH genes all contained typical signal sequences. The hefDEF and hefGHI operons were highly conserved between H. pylori 26695 and H. pylori J99, being greater than 97 and 91% identical at the amino acid level. Phylogenetic analysis of the H. pylori efflux homologs with other characterized RND efflux systems revealed that the HefDEF and HefGHI systems were most similar to those systems involved in the efflux of divalent cations, while HefABC was most similar to those systems characterized as multiple-drug efflux pumps (Fig. 2). However, it was apparent that, e.g., the H. pylori RND pump proteins are divergent (i.e., branch earlier on the phylogenetic tree) from the other bacterial sequences.
The presence and genetic linkages of the three putative efflux operons were examined in a panel of seven other diverse H. pylori isolates (see Materials and Methods). In all seven additional strains, PCR amplicons were obtained for hefA-hefC, hefB-hefC, hefD-hefF, hefE-hefF, and hefG-hefI, and the sizes of the amplicons were indistinguishable from those obtained from H. pylori J99 and predicted from H. pylori 26695 and 11637. Furthermore, amplicons indistinguishable in size from that of J99 representing internal portions of hefC, hefF, and hefI were generated from all seven additional strains, with the exception of hefC from ArHp18. This is likely due to some minor difference in the primer sequence, as the positive results from the hefA-hefC and hefB-hefC reactions clearly indicate that the hefC gene is present in ArHp18.
In contrast to what is seen in other bacteria, the individual efflux operons within H. pylori 26695 or H. pylori J99 were not highly similar. The pump proteins (HefC, -F, and -I) were 23.8 to 28.4% identical and 44 to 59% similar to each other. The membrane fusion proteins (HefB, -E, and -H) were 16 to 22% identical (42 to 53% similar), while the outer membrane proteins (HefA, -D, and -F) were 18 to 22% identical (49 to 45% similar) to each other.
In vitro and in vivo expression of putative efflux operons.
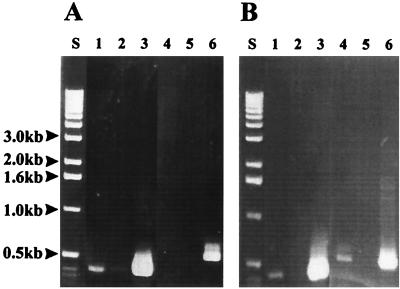
RT-PCR was used to analyze the in vitro and in vivo expression of the H. pylori putative efflux systems. The results of these experiments are shown in Fig. 3. Using PCR primers specific for hefF, a PCR amplicon of the expected size (418 bp) was visible both in vitro and in vivo and was absent in the negative control (Fig. 3A), suggesting that hefF was expressed both in vivo and in vitro. The PCR primers specific for hefI failed to amplify a product in vitro (even when the autoradiograms were overexposed or when larger amounts of template were used) but did produce an amplicon of the expected size (515 bp) in vivo (Fig. 3B, lane 4), and the amplicon was absent in the negative control (Fig. 3B, lane 5). Analysis of hefC revealed that it was also expressed both in vitro and in vivo (data not shown).
FIG. 3.
In vitro and in vivo expression of the H. pylori hef operons; RT-PCR of H. pylori grown in vitro (A) and in vivo (B). Lanes 1 to 3 were done with hefF-specific PCR primers, while lanes 4 to 6 were done with the hefI-specific PCR primers. The RT-PCR mixtures in lanes 1 and 4 contained reverse transcriptase, Taq DNA polymerase, and RNA; lanes 2 and 5 are the negative controls and contained Taq DNA polymerase and RNA; lanes 3 and 6 are the positive controls and contained all the nucleic acids (prior to DNase treatment) plus Taq DNA polymerase. Lanes S, size standards.
NPN efflux.
The addition of 6.4 μg of polymyxin B/ml to E. coli resulted in the rapid uptake of the hydrophobic fluorescent dye NPN followed by a decrease in fluorescence over time, in the presence of a proton motive force, due to active efflux of NPN by the bacterial cells (Fig. 1A). Dissipation of the proton motive force by preincubation of the cells with the proton translocator CCCP completely abolished the extrusion of NPN by the cells, as previously shown for P. aeruginosa (18). In contrast, in H. pylori, once NPN fluorescence reached its maximum level after polymyxin B treatment, it did not diminish over the time course of these experiments, suggesting that H. pylori did not efflux NPN (Fig. 1B). The lack of any detectable efflux activity may have been caused by the cells becoming deenergized over the course of the experiments. However, examination of the cells by phase-contrast microscopy showed that the cells were actively motile, indicating that the cells maintained a proton motive force throughout the time course of these experiments. To see if the addition of a carbon source could energize active efflux of NPN, the H. pylori cells were preincubated with 18 different potential carbon sources. The experiments were also repeated at pH 4 and pH 9 in an attempt to induce an artificial proton motive force. However, the addition of these carbon sources and the alteration of the assay medium pH were not able to promote the extrusion of NPN by H. pylori, suggesting that NPN was not a substrate for active efflux by in vitro-grown H. pylori.
Accumulation of [3H]tetracycline and [3H]chloramphenicol by H. pylori.
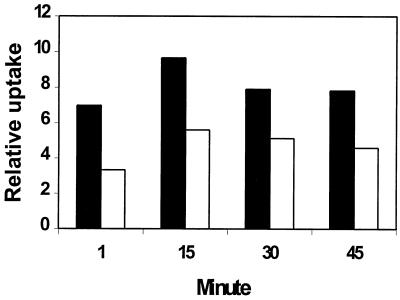
Since NPN was not a substrate for active efflux by H. pylori, we examined the accumulation of [3H]tetracycline and [3H]chloramphenicol, which are substrates for active efflux by many RND multiple-drug efflux systems. If these compounds were effluxed by H. pylori, the addition of CCCP would result in increased uptake, as observed in other bacteria (14, 19). Incubation of H. pylori 11637 with either [3H]tetracycline or [3H]chloramphenicol resulted in the accumulation of radiolabel by the cells. Pretreatment of H. pylori with CCCP, however, resulted in a reduction rather than an increase in the accumulation of both [3H]chloramphenicol (Fig. 4) and [3H]tetracycline (not shown) relative to the untreated control. These results are consistent with an influence of the membrane potential on the partitioning of these antibiotics across the cell membrane.
FIG. 4.
CCCP inhibition of [3H]chloramphenicol uptake by H. pylori (solid bars) and H. pylori pretreated with 40 μM CCCP (open bars). The cells were incubated separately in the presence of [3H]chloramphenicol. Aliquots were removed at 1, 15, 30, and 45 min following the addition of [3H]chloramphenicol and processed as stated in Materials and Methods. The amount of radioactivity on the filter was determined by scintillation counting. The relative uptake is defined as the percentage of radioactivity (counts per minute) accumulated by the cells divided by the total radioactivity (counts per minute) present. Representative results from one of three experiments are shown.
Mutagenesis of the H. pylori putative RND efflux operons.
The three efflux pump proteins were individually mutagenized in the chromosome of H. pylori J99 by marker exchange mutagenesis. The abilities of the resulting mutants to grow or divide in vitro were not affected. When analyzed by disk diffusion, the susceptibilities of the mutants to 19 antibiotics (amoxicillin-clavulanic acid, amikacin, aztreonam, chloramphenicol, ceftazidime, cefaclor, ciprofloxacin, cephalothin, clarithromycin, clindamycin, cefotaxime, cefoxitin, gentamicin, imipenem, kanamycin, nalidixic acid, methicillin, tetracycline, and vancomycin), many of which were known substrates for active efflux systems, were unchanged.
DISCUSSION
H. pylori is an enigma, in that it is quite sensitive to most antimicrobial agents in the laboratory, with the exception of polycations, but very difficult to treat in patients. This paper describes data providing reasons for this observation. We demonstrate here that H. pylori apparently lacks an effective RND efflux system for antibiotics. Studies of uptake of tetracycline, chloramphenicol, and NPN (Fig. 1 and 4), as well as metronidazole (21), indicated that these compounds, which are typical substrates for RND efflux systems, are not effluxed by H. pylori in an energy-dependent fashion. While only RND efflux systems are known to have a wide variety of types of antibiotic substrates, other, simpler efflux systems, usually with a single cytoplasmic pump protein, have been reported to influence passage of a specific class of antibiotics. For example, tetracycline and chloramphenicol pumps (22) have been reported to be involved in resistance in some bacteria (although usually in mutational or plasmid-borne rather than intrinsic resistance). However, only three gene products with any homology to this class of major facilitator efflux proteins were observed, namely, HP1165, HP1182, and HP1185 of strain 26695. However, there was no compelling evidence based on either the neighboring genes or the highest number of hits (which tended to be to genes obtained as part of genome-sequencing projects) that these genes were really involved in antibiotic resistance. In any case, these proteins were clearly not involved in the energy-dependent efflux of NPN, tetracycline, or chloramphenicol in vitro (since no such efflux was observed in H. pylori [Fig. 4]). This was also true of HP1082 and HP1206, which showed some homology to mammalian multiple-drug efflux proteins and to other members of the ATP-binding cassette family of uptake and export proteins. Consistent with the above analysis, the MICs for H. pylori of antibiotics like chloramphenicol, tetracycline, and, in part, novobiocin, ciprofloxacin, and erythromycin, were more reminiscent of those for mutants of P. aeruginosa and E. coli lacking the major RND systems involved in intrinsic antibiotic resistance rather than the MICs for wild-type gram-negative bacteria. Of the three putative efflux systems identified in H. pylori, only one, HefABC, showed any homology to the RND efflux systems involved in multidrug resistance in other bacteria, but it was clearly a somewhat distant relative (Fig. 2). It was apparently expressed under laboratory conditions, as was HefDEF (but not HefGHI), a relative of the cation efflux subfamily of the RND efflux systems (26). However, knockouts in any of these three operons failed to influence susceptibility to 19 different antibiotics. While it remains marginally possible that HefABC and HefDEF contribute equally to the efflux of multiple antibiotics, this would be unprecedented in any bacterium studied to date and seems unlikely based on the high susceptibility of H. pylori to most antibiotics and on our tetracycline, NPN, and chloramphenicol transport data, which argue against the existence of any functional efflux operon under laboratory conditions.
In marked contrast, the constitutive expression of specific RND efflux systems is a significant factor in the intrinsic resistance of all gram-negative bacteria studied to date to multiple classes of antibiotics (22, 25, 26). The role of RND multiple-drug efflux systems in intrinsic antibiotic resistance has been demonstrated in several human pathogens, including E. coli (20), H. influenzae (27), N. gonorrhoeae (9), and P. aeruginosa (17).
The expression of RND efflux systems in many bacteria is regulated by environmental stimuli, including the presence of antibiotics and antimicrobial compounds, the growth stage, and stress factors (22, 25, 26). The RT-PCR data presented here suggest the presence of a regulatory mechanism controlling the in vivo expression of the hefGHI operon. The H. pylori hefI-specific mRNA could only be detected in vivo (Fig. 4), suggesting that the entire hefGHI operon was expressed under these conditions. However, detection of hefC and hefF mRNAs (Fig. 4), both in vivo and in vitro, suggested that these operons were expressed constitutively. While our RT-PCR data did not quantitate the levels at which these two operons were expressed, transport and susceptibility data confirmed that in vitro-grown H. pylori did not express any efflux systems at a level that influenced antibiotic susceptibility. Collectively, these results indicate that active efflux does not play a role in the in vitro intrinsic resistance of H. pylori to many antibiotics. However, we cannot rule out the potential role of efflux in the in vivo intrinsic resistance of H. pylori to antibiotics.
The other part of the intrinsic resistance equation is uptake. We previously demonstrated that H. pylori, unlike other gram-negative bacteria, contained no predominant porin species. However, it did contain approximately 2,000 copies of a nonspecific porin, HopE, with a large channel (1.5 nS for the monomer in 1 M KCl; cf. E. coli OmpF, which has a monomer single-channel conductance of 0.57 nS), and we anticipate that most antibiotics would equilibrate reasonably rapidly across the outer membrane of H. pylori (cf. P. aeruginosa, which has only 200 copies or so of large-channel porins with a monomer single-channel conductance of 1.7 nS in 1 M KCl). Thus, given the lack of secondary defense mechanisms (10), such as β-lactamase (not present in H. pylori) and efflux (not functional for at least some antibiotics under laboratory conditions), we would anticipate a relatively modest influence of porin-mediated permeation on intrinsic resistance. Similarly, reasonable uptake of hydrophobic substances, like NPN, novobiocin, and chloramphenicol, coupled with nonfunctioning efflux systems, as observed in efflux-defective P. aeruginosa (24) and E. coli (19), would explain the relatively high susceptibility of H. pylori to hydrophobic antibiotics. Conversely, a poorly functioning self-promoted uptake system, as observed here, is consistent with the low sensitivity of H. pylori to the polycationic lipopeptide polymyxin B (Table 1) and the observed resistance to antimicrobial peptides. We presume that this is due to the unique structure of H. pylori lipopolysaccharide, since this molecule is very influential in self-promoted uptake.
ACKNOWLEDGMENTS
This work was partly supported by a grant to R.E.W.H. from the Medical Research Council of Canada. R.E.W.H. is a recipient of the MRC Distinguished Scientist award.
Zita Kabok is thanked for assistance in in vivo colonization and sampling.
REFERENCES
- 1.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;14:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Amsterdam D. Susceptibility testing of antimicrobials in liquid media. In: Lorian V, editor. Antibiotics in laboratory medicine. 4th ed. Baltimore, Md: Williams and Wilkins; 1996. pp. 52–111. [Google Scholar]
- 4.Bina J E, Nano F, Hancock R E W. Utilization of alkaline phosphatase fusions to identify secreted proteins, including potential efflux proteins and virulence factors from Helicobacter pylori. FEMS Microbiol Lett. 1997;148:63–68. doi: 10.1111/j.1574-6968.1997.tb10268.x. [DOI] [PubMed] [Google Scholar]
- 5.Curtiss N A C, Eisenstadt R L, Rudd C, White A J. Inducible type I β-lactamases of Gram negative bacteria and resistance to β-lactam antibiotics. J Antimicrob Chemother. 1986;17:51–61. doi: 10.1093/jac/17.1.51. [DOI] [PubMed] [Google Scholar]
- 6.Doig P, Exner M M, Hancock R E W, Trust T J. Isolation and characterization of a conserved porin protein from Helicobacter pylori. J Bacteriol. 1995;177:5447–5452. doi: 10.1128/jb.177.19.5447-5452.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn B C, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Exner M M, Doig P, Trust T J, Hancock R E W. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect Immun. 1995;63:1567–1572. doi: 10.1128/iai.63.4.1567-1572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Felgenstein J. PHYLIP (phylogeny inference package) version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 9.Haman K E, Pan W, Spratt B G, Balthazar J T, Judd R C, Shafer W M. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 10.Hancock R E W. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 11.Huang H, Hancock R E W. The role of specific surface loop regions in determining the function of the imipenem-specific pore protein OprD of Pseudomonas aeruginosa. J Bacteriol. 1996;178:3085–3090. doi: 10.1128/jb.178.11.3085-3090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X. A contig assembly program based on sensitive detection of fragment overlaps. Genomics. 1992;14:18–25. doi: 10.1016/s0888-7543(05)80277-0. [DOI] [PubMed] [Google Scholar]
- 13.Jiang Q, Hiratsuka K, Taylor D E. Variability of gene order in different Helicobacter pylori strains contributes to genome diversity. Mol Microbiol. 1996;20:833–842. doi: 10.1111/j.1365-2958.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 14.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 15.Labigne A, Courcoux P, Tompkins L. Cloning of Campylobacter jejuni genes required for leucine biosynthesis, and construction of a leu-negative mutant of C. jejuni by shuttle transposon mutagenesis. Res Microbiol. 1992;143:15–26. doi: 10.1016/0923-2508(92)90030-r. [DOI] [PubMed] [Google Scholar]
- 16.Lee A, O'Rourke J, De U M, Robertson B, Daskalopoulos G, Dixon M F. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 17.Li X Z, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh B, Grant C, Hancock R E W. Use of the fluorescent probe 1-N-phenylnapthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1984;26:546–551. doi: 10.1128/aac.26.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 21.McNulty C A, Dent J, Wise R. Susceptibility of clinical isolates of Campylobacter pyloridis to 11 antimicrobial agents. Antimicrob Agents Chemother. 1985;28:837–838. doi: 10.1128/aac.28.6.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore R A, Beckthold B, Bryan L E. Metronidazole uptake in Helicobacter pylori. Can J Microbiol. 1995;41:746–749. doi: 10.1139/m95-102. [DOI] [PubMed] [Google Scholar]
- 23.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ocaktan A, Yoneyama H, Nakae T. Use of fluorescence probes to monitor function of the subunit proteins of the MexA-MexB-OprM drug extrusion machinery in Pseudomonas aeruginosa. J Biol Chem. 1997;272:21964–21969. doi: 10.1074/jbc.272.35.21964. [DOI] [PubMed] [Google Scholar]
- 25.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saier M H M, Tam R, Reizer A, Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol. 1994;11:841–847. doi: 10.1111/j.1365-2958.1994.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez L, Pan W, Vinas M, Nikaido H. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J Bacteriol. 1997;179:6855–6857. doi: 10.1128/jb.179.21.6855-6857.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott M, Yan H, Hancock R E W. Biological properties of structurally related cationic antimicrobial peptides. Infect Immun. 1999;67:2006–2009. doi: 10.1128/iai.67.4.2005-2009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 31.van der Hulst R W, Keller J J, Rauws E A, Tytgat G N. Treatment of Helicobacter pylori infection: a review of the world literature. Helicobacter. 1996;1:6–19. doi: 10.1111/j.1523-5378.1996.tb00003.x. [DOI] [PubMed] [Google Scholar]