Abstract
Diabetic nephropathy (DN) is the main leading cause of chronic kidney disease worldwide. Although remarkable therapeutic advances have been made during the last few years, there still exists a high residual risk of disease progression to end-stage renal failure. To further understand the pathogenesis of tissue injury in this disease, by means of the Next-Generation Sequencing, we have studied the microRNA (miRNA) differential expression pattern in kidneys of Black and Tan Brachyury (BTBR) ob/ob (leptin deficiency mutation) mouse. This experimental model of type 2 diabetes and obesity recapitulates the key histopathological features described in advanced human DN and therefore can provide potential useful translational information. The miRNA-seq analysis, performed in the renal cortex of 22-week-old BTBR ob/ob mice, pointed out a set of 99 miRNAs significantly increased compared to non-diabetic, non-obese control mice of the same age, whereas no miRNAs were significantly decreased. Among them, miR-802, miR-34a, miR-132, miR-101a, and mir-379 were the most upregulated ones in diabetic kidneys. The in silico prediction of potential targets for the 99 miRNAs highlighted inflammatory and immune processes, as the most relevant pathways, emphasizing the importance of inflammation in the pathogenesis of kidney damage associated to diabetes. Other identified top canonical pathways were adipogenesis (related with ectopic fatty accumulation), necroptosis (an inflammatory and regulated form of cell death), and epithelial-to-mesenchymal transition, the latter supporting the importance of tubular cell phenotype changes in the pathogenesis of DN. These findings could facilitate a better understanding of this complex disease and potentially open new avenues for the design of novel therapeutic approaches to DN.
Keywords: miRNA, inflammation, diabetes, type 2 diabetes, diabetic nephropaty, chronic kidney disease, BTBR ob/ob mice
Introduction
Diabetic nephropathy (DN) represents the leading cause of chronic kidney disease (CKD), being the most frequent origin of end-stage renal disease (ESRD) and the main reason for renal replacement therapy in Western countries (Cheng et al., 2021; Deng et al., 2021). Nowadays, the incidence of DN continues to grow due to the increasing prevalence of type 2 diabetes (T2D) linked to metabolic syndrome, obesity, and dyslipidemia (Navarro-González et al., 2011; Alicic et al., 2017; Sharma et al., 2017). At present, the main therapeutic targets comprise blood pressure and hyperglycemia control, including the inhibition of the renin-angiotensin-aldosterone system (RAAS) as a nephroprotective strategy (Ruiz-Ortega et al., 2020). During the last few years, sodium-glucose cotransporter-2 inhibitors (SGLT2i) or glucagon-like peptide-1 receptor agonists have been used as the first and second therapeutic options in specific CKD stages. They have shown noticeable beneficial effects on cardiovascular and renal outcomes with a significant decrease in mortality (Neumiller et al., 2017). Apart from these mentioned drugs, the dipeptidyl peptidase 4 inhibitors are also a therapeutic approach currently recommended in the new KDIGO guidelines (Kidney Disease: Improving Global Outcomes Diabetes Work Group, 2020). In addition, new anti-inflammatory therapies and metabolic modulators, such as JAK/STAT, Rho-kinase and Sirtuin-3 inhibitors, peptide N-Acetyl-Seryl-Aspartyl-Proline, glycolysis inhibitors, and mineralocorticoid receptor antagonists, have been postulated as potential drugs in DN due to the preclinical evidence, showing their renoprotective role or even their ability to reverse established renal damage (Hashimoto et al., 2010; Bakris et al., 2020; Locatelli et al., 2020; Opazo-Ríos et al., 2020b; Srivastava et al., 2020; Matoba et al., 2021). Growing evidence in all these mentioned studies has shown that the reno-protective effects of these drugs, at least, in part, may be due to the regulation of epithelial and endothelial-to-mesenchymal transition (EMT and EndoMT) in the progression of DN. Unfortunately, at present, there are no interventions capable of fully prevent the progression to advanced kidney disease in people with T2D, being therefore necessary to find out new therapeutic options that limit the natural history of the disease.
Many studies have investigated the molecular mechanism involved in the onset and progression of DN. Hyperglycemia has been considered the driving and triggering force in the DN development, but recent data have also pointed out an important role of the immune system and inflammation, as well as oxidative stress, lipotoxicity, and uremic toxins (Moreno et al., 2018; Opazo-Ríos et al., 2020a; Lavoz et al., 2020b; Rayego-Mateos et al., 2020). At the molecular level, the importance of signaling pathways related to renal inflammation and fibrosis such as the loss of endothelial glucocorticoid receptor (Srivastava et al., 2021b), endothelial FGFR1 and SIRT3 signaling (Li J. et al., 2017; Srivastava et al., 2018), TGFβ/SMAD, Notch, WNT/β-Catenin, and Sonic-Hedgehog pathways (Marquez-Exposito et al., 2018; Zhao et al., 2018; Wang et al., 2022) have also demonstrated to play a relevant role in DN. Interestingly, despite good glycemic control, the inability to fully avoid chronic meta-inflammation (microinflammatory milieu caused by metabolic factors) could contribute to ESRD progression in diabetic patients. This phenomenon can be explained by the concept of hyperglycemic “metabolic memory” (Bheda, 2020). Recent evidence suggests epigenetic regulation mechanisms, including DNA methylation and the histone post-translational modifications, as drivers of metabolic memory, suggesting that epigenetic regulation may result very relevant in DN (Keating and El-Osta, 2013; Zhao et al., 2016; Morgado-Pascual et al., 2018; Martinez-Moreno et al., 2020a; Martinez-Moreno et al., 2020b). Epigenetic regulation of gene expression is a dynamic process that may be modified in response to the environment or therapeutic modulation. In particular, lysine histone methylation, acetylation, and crotonylation have been involved in kidney diseases (Martinez-Moreno et al., 2020a). In addition, epigenetic readers that identify and interpret epigenetic signals are key components of the system (Morgado-Pascual et al., 2019).
Some authors also include microRNAs (miRNAs) in the epigenetic regulation of gene expression. miRNAs are evolutionarily conserved small (20–24 nt) non-coding RNAs related to both stability and translation of target mRNAs. Since their discovery, miRNAs have progressively turned out to central stage in the understanding of the post-transcriptional gene expression regulation. Interestingly, epigenetic regulation modulates several miRNAs expression, and, conversely, some of them participate in the expression of relevant epigenetic regulators (Sato et al., 2011). The importance of miRNAs function has been demonstrated in a wide variety of processes including, cancer, cardiovascular and renal diseases, peritoneal fibrosis, and diabetes (Adams et al., 2014; Morishita et al., 2016; Prattichizzo et al., 2016; Colpaert and Calore, 2019; Ruiz-Ortega et al., 2020; Jankauskas et al., 2021). Regarding DN, some authors have suggested a potential role of miRNAs in the development and progression of the pathology, featuring some of them (miR-21, miR-200b/c, and mir-29c) (Long et al., 2011; Park et al., 2013; Zhong et al., 2013) as promising therapeutic targets, but some others (e.g., miR-146a) (Bhatt et al., 2016a) as protectors of DN progression and fibrosis. In this sense, several miRNAs have been found to modulate EMT and EndoMT and, in consequence, to prevent fibrosis in DN and other kidney diseases by targeting several genes related to these processes (Srivastava et al., 2013; Giordo et al., 2021). Thus, miR-30a, miR-30c, miR-26a, miR-130, miR-23b, Let-7days, and miR-98 have shown to modulate EMT in experimental DN (Bai et al., 2016; Liu et al., 2016; Zheng et al., 2016; Zhu et al., 2019; Gao et al., 2020; Wang Y. et al., 2021). Moreover, the anti-fibrotic effects of other miRNAs, including miR-200a, miR-455-3p, miR-92days-3p, miR-130b, miR-26a, and miR-29a, have been also described in DN (Du et al., 2010; Wang et al., 2011; Lin et al., 2014; Koga et al., 2015; Bai et al., 2016; Wu et al., 2018; Zhang, 2021). Apart from miRNAs, other non-coding RNAs have been postulated to regulate gene expression in DN. In this way, long-noncoding RNAs (lncRNAs) and circular RNAs have been described to act as miRNA sponges, regulating their actions in DN (Srivastava et al., 2021a; Patil et al., 2021). For instance, lncRNAs NR_033515, NEAT1, OIP5-AS1, and MALAT1 promote EMT and fibrosis by sponging anti-fibrotic miRNAs (Gao et al., 2018; Liu et al., 2019; Wang et al., 2019; Li N. et al., 2020; Fu et al., 2020; Meng et al., 2020), whereas other lncRNAs, such as ZEB1-AS1, prevent EMT and fibrosis in DN (Meng et al., 2020). In consequence, different non-coding RNAs and other epigenetic mechanisms can interact to regulate gene expression and, therefore, DN progression. These data, as well as their higher stability and the possibility of measuring them in different fluids, have pointed out their utility as biomarkers for early detection and progression of DN, suggesting their potential use as therapeutic agents (Simpson et al., 2016).
However, an important handicap to be dodged in the study of DN and its complications is the lack of robust animal models that replicate the key features of human diabetes to test novel therapeutic tools (Brosius and Alpers, 2013). In this regard, Black and Tan BRachyury (BTBR) ob/ob (leptin deficiency mutation) mouse has recently turned out as an excellent preclinical model for DN study, because it replicates the key histopathological features observed inT2D (Hudkins et al., 2010; Alpers and Hudkins, 2011), despite the absence of tubulointerstitial fibrosis observed in advanced human DN (PMID: 24711709). Therefore, to further advance in the better knowledge on the mechanisms involved in the genesis and progression of DN, the aim of this study was to characterize by the Next-Generation Sequencing (NGS) the miRNA differential expression pattern in an established model of advanced DN in the BTBR ob/ob mice.
Materials and Methods
Ethics Statement
All animal procedures were performed according to the guidelines of animal research in the European Community and with prior approval by the Ethics Committee of the Health Research of the IIS-Fundación Jiménez Díaz and by the Madrid regional government (Ref. PROEX 079/18). All animal procedures conformed to EU Directive 2010/63EU and the national rule 53/2013 regarding protection of animals used for experimental and other scientific purposes. The establishment and care of BTBR ob/ob diabetic and obese mice colony (referred to here as “diabetic mice”) and their corresponding controls [BTBR wild type (WT)] have been previously described (Clee et al., 2005). These mice were originally obtained from JAX™ Mice (Charles River Europe laboratory), and then, the mouse colony breeding was maintained in the Fundación Jiménez Díaz Animal facilities, following the JAX™ recommendations (Jackson Laboratory, 2007). Animals were housed at a density of four animals per cage in a temperature-controlled room (20°C–22°C) with 12-h light–dark cycles and feeding with standard chow and water ad libitum provided by the animal facilities.
Design of the Experimental Model of Diabetic Nephropathy and Characterization
Male BTBR ob/ob diabetic mice and their corresponding non-diabetic and non-obese littermates (BTBR WT) were studied over 22 weeks of age (n = 6 for each group). These mice rapidly develop morphologic renal lesions characteristic of both early and advanced human DN (Hudkins et al., 2010). Body weight and blood glucose levels were measured by Fisherbrand Precision balance and NovaPro Glu/Ket system (Nova Biomedical, Waltham, MA, USA) after clipping the distal 2–3 mm of the tail-tip. Spot urine samples were collected once a week from all mice and analyzed for albumin and creatinine by the ELISA Kit (cat. nos. ab108792 and ab65340, respectively, Abcam) to obtain the urine albumin/creatinine ratio (ACR). Animals were euthanatized by intraperitoneal anesthetic induction of ketamine (100 mg/kg) and xylazine (10 mg/kg). After anesthetic assessment, the kidneys were perfused in situ with saline before removal, and one half of each kidney was fixed in 4% formaldehyde, embedded in paraffin, and used for histological studies, whereas the other half remaining was snap-frozen in liquid nitrogen for renal cortex miRNA studies. Metabolic and renal profile were assessed in serum including: glucose, creatinine, urea, blood urea nitrogen, albumin, triglycerides (TG), total cholesterol (TC), high-density lipoprotein (HDL), and low-density lipoprotein (LDL); aspartate transaminase, alanine aminotransferase, and alkaline phosphatase. The measurement was performed at our institution's central laboratory using Roche Cobas autoanalyzer. Over the disease outcome, glycosuria was measured periodically by test strips (range 50–1,000 mg/dl) and subsequently quantified by urine dilution in institutional autoanalyzer.
Morphological and Immunohistochemical Studies
A quarter piece of each kidney sample fixed in 4% formaldehyde were embedded in paraffin and cut in serial sections (4–5 μm thickness) for further histological [periodic acid–Schiff (PAS)/Sirius red] and immunohistochemistry (IHC) studies.
Periodic Acid–Schiff Staining
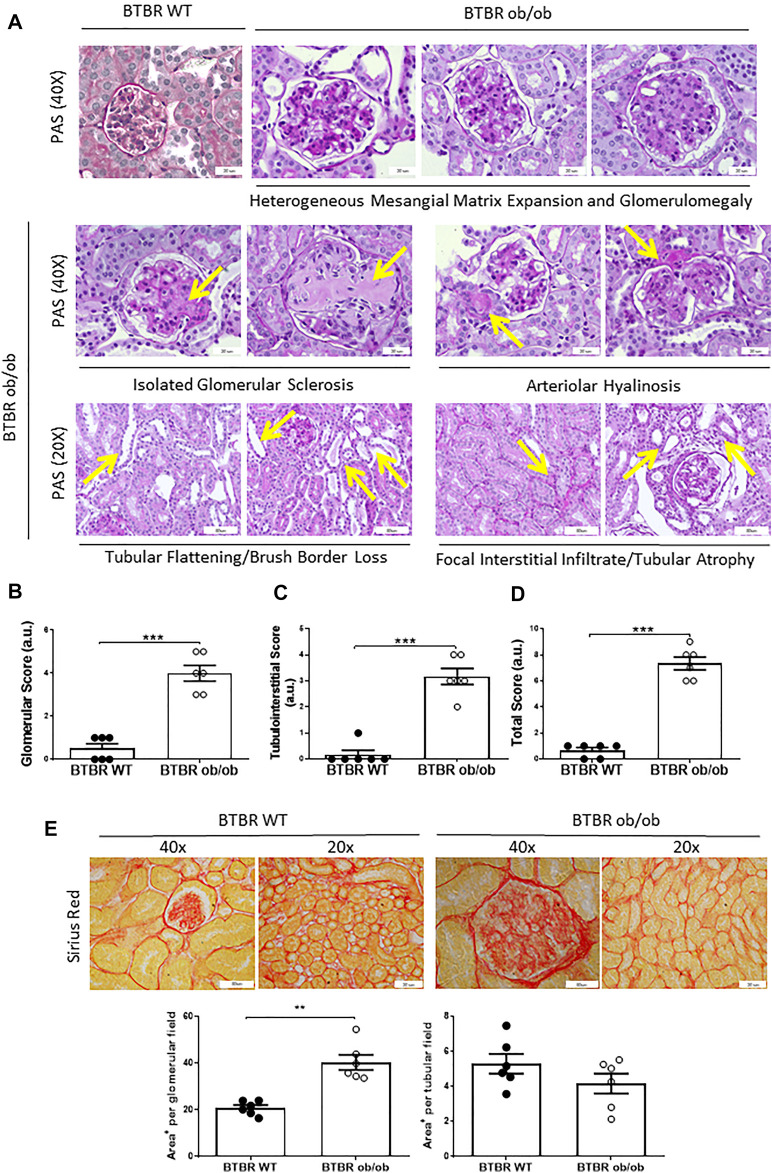
PAS staining was performed using 0.5% PAS’s Reagent (Sigma-Aldrich) and Hematoxylin (Thermo Scientific) for histopathological assessment in 4 μm sections, as previously described (Varga and Brenner, 2005). The following lesions were evaluated: the degree of mesangial matrix expansion, glomerular hypertrophy (glomerulomegaly), glomerular sclerosis, arteriolar hyalinosis, tubular casts, acute tubular damage, and tubular atrophy, as well as the presence of interstitial inflammatory cells and fibrosis. A semi-quantitative assessment of renal damage at glomerular (glomerulomegaly, mesangial matrix expansion, presence of nodular sclerosis, and arteriolar hyalinosis) and tubulointerstitial (casts, tubular flattening, tubular atrophy, inflammatory infiltrates, and fibrosis) levels as well as the total (sum of both scores) lesions were graded according to their histopathological score (from 0 to 4) as previously described (Zoja et al., 2002). The histological assessment and representative lesions were obtained by epifluorescence optical microscope (Axioscope 5 Workstation compact, Zeiss).
Sirius Red Staining
Sirius red staining was performed in 5-µm kidney sections using a picrosirius solution [10% of Sirius red (1%) in picric acid], as previously described (Varga and Brenner, 2005). Collagen deposition levels were evaluated by using the Image-Pro Plus software (Bio-Rad) determining the red positive staining per glomerular and interstitial areas in eight randomly chosen fields (×40 and ×20 magnification, respectively) acquired by epifluorescence optical microscope (Axioscope 5Workstation compact, Zeiss).
Immunohistological Studies
For immunostaining procedure, 4-µm tissue sections were deparaffinized through xylene and hydrated through graded ethanol (100%, 96%, 90%, and 70%) ending in distilled water. Antigens were restored by using PTLink system (DAKO Diagnostic), blocking endogenous peroxidase afterward. Commercial casein solution (DAKO Diagnostic) was used to release non-specific protein bindings (1 h at room temperature), and tissue sections were incubated overnight at 4°C withp-SMAD3 (1/200; ab52903, Abcam) antibody diluted in antibody solution (DAKO Diagnostics). Kidney sections were incubated with the specific HRP secondary anti-body (GENA934, Sigma Chemical) for 1 h followed by Avidin-Biotin Complex incubation (Vector laboratories) for 30 min. To develop signal, samples were incubated with substrate solution and 3,3-diaminobenzidine as a chromogen (Abcam) and counterstained with Carazzi’s hematoxylin (Thermo Fisher Scientific). Specificity was checked by omission of primary antibody (data not shown). Quantification was made by using the Image-Pro Plus software (Bio-Rad) determining the positive relative staining area per total area in five to 10 randomly chosen fields (×40 magnification) acquired by epifluorescence optical microscope (Axioscope 5Workstation compact, Zeiss).
Total RNA Extraction
Cortex kidney sections were lysed using Tissue Lyser for disaggregation of tissues, and total RNA was obtained using the KingFisher Flex station (Thermo Fisher) and a commercial kit (MagMAX™ mirVana™), following the manufacturer’s specifications; total RNA was quantified using a 2100 Bioanalyzer (Agilent) to test size profiling.
miRNA Sequencing and Bioinformatic Analysis
For sequencing of small RNAs, 12 libraries (six BTBR WT and six BTBR ob/ob) were prepared according to the instructions of the “NEBNext Multiplex Small RNA Library Prep Set for Illumina” kit (New England Biolabs). The input amount of total RNA to start the protocol was 800 ng of each sample, according to the bioanalyzer measurements in RNA 6000 Nano Chips. The library preparation procedure included a PCR step, which was adjusted to 13 cycles. The libraries obtained were run in 6% PolyAcrylamide Gel, and the region of 135–325 pb (corresponding to small RNA) was selected and assessed using an Agilent 2100 Bioanalyzer with High-Sensitivity DNA chips. The final pool of libraries was denatured prior to being seeded on a flow-cell at a density of 1.38 pM, where clusters were formed and sequenced in a NovaSeq 6000 sequencer using a flow cell SP v1.5 (Illumina). An amount of 18–37 million of single-end reads (mean 24.4 × 106) was obtained per sample. The quality of sequences was assessed using the program FastQC-0.11.7 (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). Sequences were then filtered according to size (minimum length of 16) and ambiguities (removing reads with more than 10 Ns within the sequence). To that purpose, the program Prinseq (Schmieder and Edwards, 2011) was used. Next, sequences were mapped against Mus musculus genome (release GRCh38) using TopHat (Trapnell et al., 2012; Kim et al., 2013); reads were annotated using the corresponding MmGRCm38.95-gtf file. Numbers of sequences per sample are summarized in the Supplementary Table S1. The whole bioinformatic protocol was executed using the RNAseq pipeline app of the GPRO-suite (Llorens et al., 2011). Total samples (six BTBR WT and six BTBR ob/ob) were used for differential expression analysis, made using Cufflinks/CuffDiff (Trapnell et al., 2012; Kim et al., 2013) to compare the expression of BTBR ob/ob vs. BTBR WT. We filtered out those entries whose detection levels were lower than 40 counts (sum of averages from BTBR W and BTBR ob/ob groups) and finally found a number of 191 microRNA entries which can be considered as positively detected. The bioinformatic study about the possible outcome of miRNA-based regulation was performed using the Ingenuity Pathway Analysis software (IPA, Qiagen).
RT-qPCR miRNA Validation and mRNA Levels Evaluation
Cortex kidney mRNA from BTBR WT and BTBR ob/ob mice, used to perform the miRNA-seq study, were analyzed by multiple RT-qPCR. For miRNA evaluation, a miRCURY LNA RT kit (Qiagen) was use to obtain cDNA. To confirm miRNA-seq results, qPCR was performed miRCURY LNA miRNA PCR Assays (Qiagen) following the manufacturer’s instructions: miR-802 (YP00205002), miR-34a (YP00204486), and miR-375 (YP00204362); normalized by 5S rRNA (hsa) (YP00203906). For mRNA evaluation, cDNA was obtained by using the reverse transcription kit (Applied Biosystems). Then, a multiplex RT-qPCR was performed using fluorogenic primers design by assay on demand mouse expression products (Applied Biosystems): Ngal: Mm01324470_m1, Mcp1: Mm00441242_m1, Acta2: Mm01546133_m1, and Tgf-β: Mm01178820_m1. As endogen control to normalize, Gapdh: Mm99999915_g1 was used. miRNA and mRNA copy number were calculated for each sample by the instrument software (ABIPrism 7500 Fast sequence detection PCR system software; Applied Biosystems) using Ct value (“arithmetic fit point analysis for the light cycler”), and the results were expressed in n-fold calculated vs. BTBR WT group.
Statistical Analysis
Data are expressed as mean ± standard error of the mean (SEM) of each group (n = 6 mice/group). Normality distribution was tested by using Shapiro–Wilk test. If the samples followed a Gaussian distribution or not, then means were compared by Student t-test or Mann–Whitney statistical test, respectively. Statistical significance was assumed when a null hypothesis could be rejected at p < 0.05. The statistical analysis was performed using the GraphPad Prism software (GrahPad Software).
Results
Renal Lesions in the Experimental Model of Advanced Diabetic Nephropathy
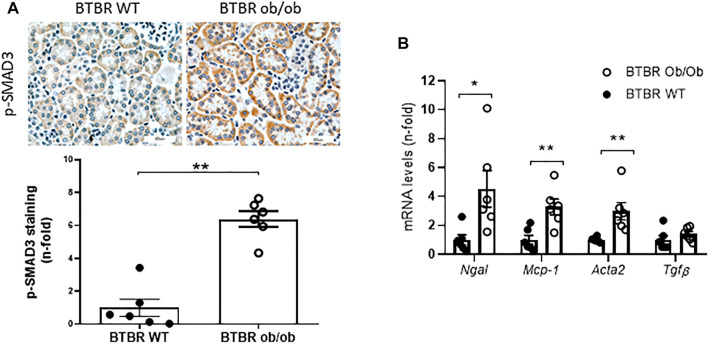
The BTBR ob/ob mice model at 22 weeks of age resembles the kidney damage observed in patients with advanced DN, as previously described (Hudkins et al., 2010; Alpers and Hudkins, 2011). The key features observed after histopathological PAS assessment are shown in the Figure 1A. At the glomerular level, the main hallmarks of kidney damage in diabetic mice are the glomerulomegaly and increased mesangial matrix. Isolated nodular glomerulosclerosis could be detected once as a blue moon in one or two glomeruli throughout the entire renal tissue assessment. Arteriolar hyalinosis was noted by thickening of the arteriolar walls with PAS-positive material at the vascular pole. At the tubulointerstitial level, the presence of cast (isolated PAS-positive), tubular flattening manifested by loss of the tubular cells brush border, focal and/or diffuse inflammatory infiltrate, and tubular atrophy were found. The quantification of kidney damage in BTBR ob/ob mice compared with non-obese non-diabetic BTRB WT mice of the same age used as controls is shown at glomerular (Figure 1B), tubulointerstitial (Figure 1C), and total areas (Figure 1D). According to previous results (Lavoz et al., 2020b), the studied kidneys did not develop tubulointerstitial fibrosis, which was also confirmed by Sirius red staining (Figure 1E). On the other hand, a high activation of the fibrotic-related pathway SMAD, assessed by increased SMAD3 phosphorylated levels (p-SMAD3), was found in BTBR ob/ob mice compared to control mice (Figure 2A). At mRNA level, an increased expression of Acta2were observed in the BTBR ob/ob mice kidneys compared with control mice, whereas no differences were found in the mRNA levels of Tgfβ (Figure 2B). In addition, mRNA levels of several inflammatory-related factors (Ngal and Mcp-1) were upregulated in the BTBR ob/ob mice kidneys compared to controls (Figure 2B).
FIGURE 1.
The glomerular and tubulointerstitial features observed in BTBR ob/ob of 22 weeks old and histopathological assessment. (A) Representative images of the kidney PAS staining at different magnifications showing the main glomerular (yellow arrows) and tubulointerstitial (green arrows) changes detected. The histopathological assessment was evaluated in glomerular (B), tubulointerstitial (C), and total score (D). (E) Representative images of the kidney Sirius red staining at different magnifications (upper) and their quantification (lower). Data are shown as mean ± SEM and graphs bar with scatter dot plots of each group (n = 6 mice/group); ***p < 0.001vs. BTBR WT. **p < 0.005 vs. BTBR WT. a.u., arbitrary units.
FIGURE 2.
Evaluation of the SMAD pathway activation and inflammatory and fibrotic markers. (A) Increased kidney levels of phosphorylated SMAD3 (p-SMAD3) were observed in BTBR ob/ob of 22 weeks old compared to BTBR WT. (B) Increased levels of Ngal, Mcp-1, and Acta2 were observed in the BTBR ob/ob group compared to BTBR WT mice, whereas no differences were detected in Tgf-β expression levels. Data are shown as mean ± SEM and graphs bar with scatter dot plots of each group (n = 6 mice/group). ***p < 0.001 vs. BTBR WT; **p < 0.005 vs. BTBR WT; *p < 0.05 vs. BTBR WT.
To further evaluate changes in renal function, both serum and urinary markers were assessed. As previously described, the BTBR ob/ob mice showed hyperglycemia and increase in body weight at early stages, which were maintained over time (22 weeks) with changes in serum lipids, such as TC, TGs, and HDL in comparison with BTBR WT (Table 1). The BTBR ob/ob mice at 22nd week presented discrete elevation of serum creatinine, severe albuminuria (measured by urinary ACR), and high glycosuria levels as compared to control littermates, revealing kidney dysfunction in diabetic mice (Table 1).
TABLE 1.
Metabolic and renal parameters in BTBR WT and BTBR ob/ob mice. *p < 0.05 vs. BTBR WT; UD, undetectable.
| General Parameters | BTBR WT | BTBR ob/ob |
|---|---|---|
| Body weight at 22th week (g) | 38 ± 0.8 | 72 ± 1.1* |
| Glycaemia at 22th week (mg/dl) | 151 ± 4 | 531 ± 24* |
| Glycosuria (mg/dl) | UD | 2190 ± 278 |
| Urinary albumin/creatinine ratio—uACR (mg/g) | 50 ± 7.2 | 850 ± 99* |
| Serum creatinine (mg/dl) | 0.20 ± 0.01 | 0.27 ± 0.01* |
| Urea (mg/dl) | 43 ± 2.9 | 42 ± 4.2 |
| Blood urea nitrogen—BUN (mg/dl) | 20 ± 1.4 | 20 ± 1.9 |
| Aspartate transaminase—AST (IU/L) | 67 ± 4.8 | 85 ± 9.2 |
| Alanine transaminase—ALT (IU/L) | 12 ± 0.8 | 44 ± 7.0* |
| Alkaline phosphatase—AP (IU/L) | 34 ± 1.6 | 89 ± 8.7* |
| Serum albumin (g/dl) | 3.5 ± 0.1 | 4.3 ± 0.3* |
| Total cholesterol (mg/dl) | 126 ± 4.8 | 203 ± 19.1* |
| Triglycerides (mg/dl) | 69 ± 8.4 | 96 ± 11.3* |
| Low-density lipid—LDL (mg/dl) | 6.8 ± 0.8 | 37 ± 7.4* |
| High-density lipid—HDL (mg/dl) | 107 ± 3.8 | 147 ± 12.8* |
Renal Cortex of BTBR Ob/Ob Mice Displays a Different miRNA Pattern Expression in Relation to BTBR WT Mice
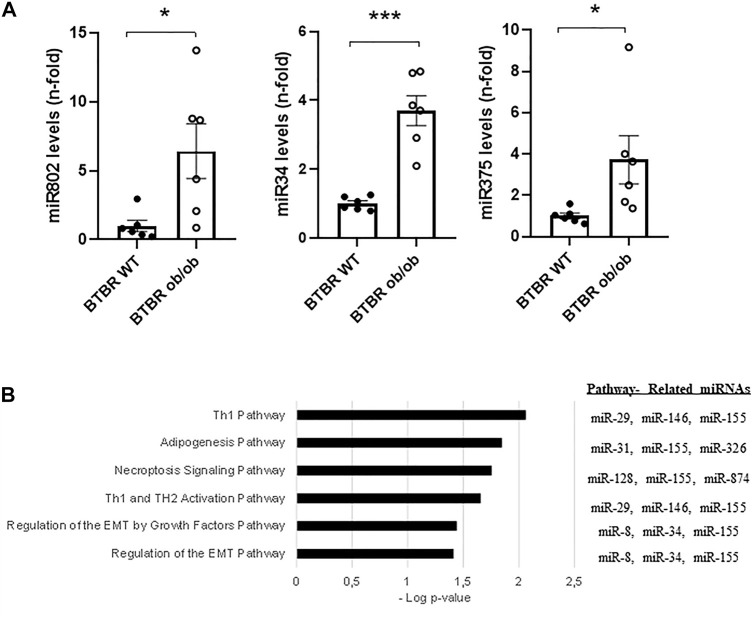
To evaluate the miRNAs potentially involved in the genesis of advanced DN in the BTBR ob/ob mice (22-weeks-old), a miRNA-seq study was performed. The results showed 198 miRNAs whose expression was dysregulated (194 upregulated vs. four downregulated) in the BTBR ob/ob group vs. WT (Supplementary Table S2). Among them, 99 upregulated miRNAs presented q-values < 0.05, whereas none deregulated miRNAs showed significant differences between groups (Table 2). According to the n-fold changes, miR-802 (44.55-fold), miR-34a (13.04-fold), miR-132 (9.68-fold), miR-101a (8.52-fold), and mir-379 (7.89-fold) were some of the most upregulated miRNAs in the BTBR ob/ob group. The elevated expression of several miRNAs (miR-802, miR-34, and miR375) was also confirmed by RT-qPCR (Figure 3A).
TABLE 2.
List of the 99 miRNAs presenting q-values < 0.05 in the comparative analysis of BTBR ob/ob vs. BTBR WT group.
| Ensembl Gene_ID | miRNA | Fold Change | q_value | Ensembl Gene_ID2 | miRNA | Fold Change | q_value |
|---|---|---|---|---|---|---|---|
| ENSMUSG00000076457 | miR-802 | 44.55 | 0.0180327 | ENSMUSG00000077962 | miR-874 | 6.69 | 0.0336 |
| ENSMUSG00000065493 | miR-34a | 13.04 | 0.00063579 | ENSMUSG00000076066 | miR-223 | 3.64 | 0.00431 |
| ENSMUSG00000065537 | miR-132 | 9.68 | 0.00063579 | ENSMUSG00000065610 | miR-29a | 3.63 | 0.0281 |
| ENSMUSG00000065451 | miR-101a | 8.52 | 0.0467852 | ENSMUSG00000077042 | miR-574 | 3.61 | 0.000636 |
| ENSMUSG00000065498 | miR-379 | 7.89 | 0.00063579 | ENSMUSG00000065611 | miR-23a | 3.54 | 0.000636 |
| ENSMUSG00000065507 | miR-204 | 7.84 | 0.00063579 | ENSMUSG00000065439 | miR-140 | 3.54 | 0.000636 |
| ENSMUSG00000092830 | miR-3963 | 7.56 | 0.00063579 | ENSMUSG00000104618 | miR-1839 | 3.49 | 0.000636 |
| ENSMUSG00000065477 | miR-411 | 7.24 | 0.00063579 | ENSMUSG00000099036 | miR-378c | 3.46 | 0.000636 |
| ENSMUSG00000098973 | miR-6236 | 6.87 | 0.00063579 | ENSMUSG00000076338 | miR-181d | 3.44 | 0.000636 |
| ENSMUSG00000065551 | miR-210 | 6.79 | 0.00063579 | ENSMUSG00000065586 | miR-96 | 3.41 | 0.000636 |
| ENSMUSG00000065616 | miR-375 | 6.18 | 0.00063579 | ENSMUSG00000076255 | miR-92b | 3.40 | 0.000636 |
| ENSMUSG00000070102 | miR-455 | 5.93 | 0.00063579 | ENSMUSG00000065418 | miR-322 | 3.36 | 0.000636 |
| ENSMUSG00000065546 | miR-196a-1 | 5.91 | 0.00063579 | ENSMUSG00000076010 | miR-615 | 3.31 | 0.000636 |
| ENSMUSG00000065443 | miR-196b | 5.90 | 0.00063579 | ENSMUSG00000065601 | miR-146 | 3.28 | 0.0271 |
| ENSMUSG00000065607 | miR-331 | 5.83 | 0.0293941 | ENSMUSG00000065484 | miR-130a | 3.25 | 0.000636 |
| ENSMUSG00000065592 | miR-145a | 5.69 | 0.00063579 | ENSMUSG00000070074 | miR-484 | 3.25 | 0.0168 |
| ENSMUSG00000076011 | miR-652 | 5.52 | 0.00063579 | ENSMUSG00000065470 | miR-149 | 3.25 | 0.00325 |
| ENSMUSG00000065402 | miR-122 | 5.37 | 0.00063579 | ENSMUSG00000076062 | miR-92–1 | 3.21 | 0.0117 |
| ENSMUSG00000076357 | miR-653 | 5.26 | 0.0266784 | ENSMUSG00000065408 | miR-31 | 3.13 | 0.00569 |
| ENSMUSG00000070076 | miR-127 | 5.18 | 0.00063579 | ENSMUSG00000105972 | miR-1843a | 3.09 | 0.00809 |
| ENSMUSG00000092998 | miR-5099 | 5.14 | 0.00063579 | ENSMUSG00000065422 | miR-221 | 3.06 | 0.000636 |
| ENSMUSG00000065417 | miR-340 | 5.06 | 0.00063579 | ENSMUSG00000065397 | miR-155 | 3.06 | 0.000636 |
| ENSMUSG00000070139 | miR-532 | 5.05 | 0.0237531 | ENSMUSG00000065580 | miR-15b | 3.03 | 0.00364 |
| ENSMUSG00000106465 | miR-374c | 5.01 | 0.00114916 | ENSMUSG00000065510 | miR-361 | 3.03 | 0.000636 |
| ENSMUSG00000065556 | miR-101b | 5.00 | 0.00063579 | ENSMUSG00000065471 | miR-222 | 3.00 | 0.000636 |
| ENSMUSG00000065500 | miR-10b | 4.84 | 0.00364284 | ENSMUSG00000105497 | miR-191 | 3.00 | 0.00204 |
| ENSMUSG00000105200 | miR-378a | 4.68 | 0.00431115 | ENSMUSG00000065528 | miR-320 | 3.00 | 0.000636 |
| ENSMUSG00000065431 | miR-186 | 4.52 | 0.00063579 | ENSMUSG00000065464 | miR-185 | 2.94 | 0.00325 |
| ENSMUSG00000076049 | miR-598 | 4.50 | 0.0420373 | ENSMUSG00000065587 | miR-34c | 2.92 | 0.000636 |
| ENSMUSG00000065593 | miR-339 | 4.44 | 0.00063579 | ENSMUSG00000076361 | miR-182 | 2.89 | 0.000636 |
| ENSMUSG00000065503 | miR-351 | 4.43 | 0.00063579 | ENSMUSG00000065462 | miR-200c | 2.86 | 0.0196 |
| ENSMUSG00000065446 | miR-139 | 4.39 | 0.00063579 | ENSMUSG00000076122 | miR-503 | 2.83 | 0.000636 |
| ENSMUSG00000065520 | miR-128-1 | 4.37 | 0.00697744 | ENSMUSG00000070130 | miR-328 | 2.83 | 0.000636 |
| ENSMUSG00000105458 | miR-3074-2 | 4.23 | 0.00063579 | ENSMUSG00000065542 | miR-224 | 2.80 | 0.00502 |
| ENSMUSG00000065396 | miR-99b | 4.22 | 0.0175788 | ENSMUSG00000098756 | miR-378d | 2.71 | 0.000636 |
| ENSMUSG00000065476 | miR-30b | 4.01 | 0.00114916 | ENSMUSG00000065619 | miR-183 | 2.68 | 0.000636 |
| ENSMUSG00000065489 | miR-365-2 | 4.01 | 0.00063579 | ENSMUSG00000076460 | miR-744 | 2.64 | 0.016 |
| ENSMUSG00000065429 | miR-345 | 3.99 | 0.00063579 | ENSMUSG00000098343 | miR-6240 | 2.63 | 0.000636 |
| ENSMUSG00000076398 | miR-676 | 3.95 | 0.00063579 | ENSMUSG00000065411 | miR-195a | 2.56 | 0.000636 |
| ENSMUSG00000065612 | miR-151 | 3.91 | 0.00063579 | ENSMUSG00000065582 | miR-194–2 | 2.49 | 0.00536 |
| ENSMUSG00000099169 | miR-7j | 3.90 | 0.00063579 | ENSMUSG00000065518 | miR-423 | 2.37 | 0.00974 |
| ENSMUSG00000065543 | miR-330 | 3.89 | 0.00063579 | ENSMUSG00000065574 | miR-203 | 2.26 | 0.01 |
| ENSMUSG00000065548 | miR-29c | 3.84 | 0.0161905 | ENSMUSG00000070106 | miR-363 | 2.13 | 0.00431 |
| ENSMUSG00000065515 | miR-152 | 3.77 | 0.00063579 | ENSMUSG00000093011 | miR-100 | 2.07 | 0.0476 |
| ENSMUSG00000080645 | miR-1198 | 3.77 | 0.00063579 | ENSMUSG00000105220 | miR-497 | 2.0 | 0.00754 |
| ENSMUSG00000076376 | miR-674 | 3.77 | 0.00063579 | ENSMUSG00000065495 | miR-150 | 1.93 | 0.00864 |
| ENSMUSG00000076140 | miR-542 | 3.76 | 0.00063579 | ENSMUSG00000065613 | miR-92–2 | 1.85 | 0.0168 |
| ENSMUSG00000105196 | miR-142 | 3.75 | 0.00063579 | ENSMUSG00000065395 | miR-193a | 1.80 | 0.0271 |
| ENSMUSG00000065571 | miR-326 | 3.71 | 0.0325777 | ENSMUSG00000065532 | miR-187 | 1.73 | 0.0301 |
| ENSMUSG00000065560 | miR-148b | 3.71 | 0.00063579 | — | — | — | — |
FIGURE 3.
miRNA RT-qPCR validation and assessment of key pathways and miRNAs. (A) miRNA relative expression (n-fold vs. BTBR WT) of miR802, miR34a, and miR375. (B) Left panel shows a graphic with the top canonical pathways related to the 99 miRNAs that were significantly predicted in the comparative analysis of the cortex kidney of BTBR ob/ob mice vs. BTBR WT, omitting cancer related pathways. Right panel indicates the most related miRNAs to each pathway. Data are shown as mean ± SEM and graphs bar with scatter dot plots of each group (n = 6 mice/group). ***p < 0.001vs. BTBR WT; *p < 0.05 vs. BTBR WT.
Functional Analysis of the miRNA-Seq Results
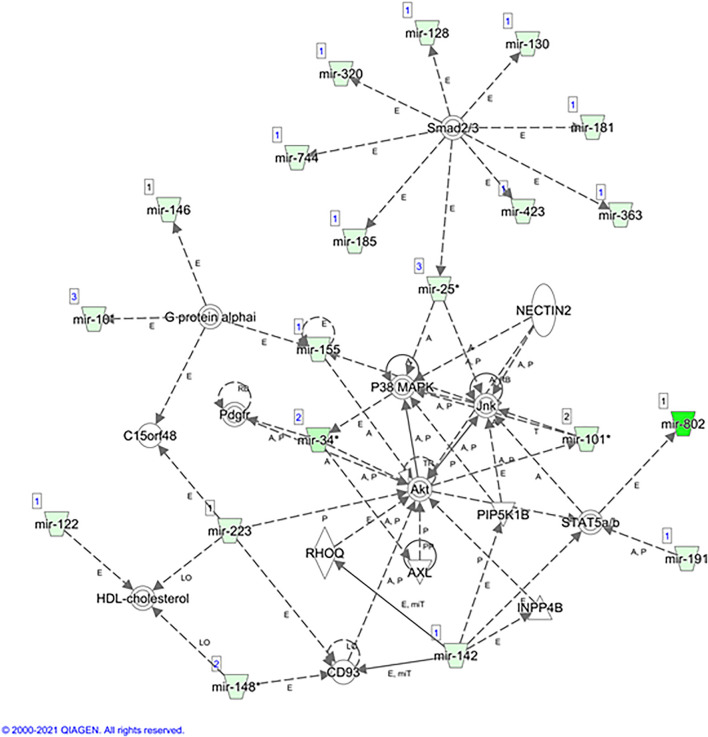
A subsequent Ingenuity Pathway Analysis (IPA) performed with the IPA software and using the 99 miRNAs (q-values < 0.05) displayed a set of canonical pathways related to these miRNAs (Supplementary Table S3). Although cancer-related pathways were the most highlighted, we decided to focus on other potential biological processes that could be deregulated in our study. This new analysis identified T helper (Th) immune responses (Th1 and Th2), adipogenesis, necroptosis, and EMT pathways as the top canonical pathways related to these miRNAs (Figure 3B). The main predicted network obtained was related to connective tissue disorders, gene expression, and inflammatory diseases (Figure 4). The top upstream regulator analysis showed the miRNA biogenesis components AGO2 and DICER1 as the most predicted factors to be regulated by the studied miRNAs, with HNF4a, SMAD2/3, and TNFRSF1B being the next factors of the list (Table 3).
FIGURE 4.
Predicted network. Interaction network of miRNAs and genes related to connective tissue disorders, gene expression, and inflammatory diseases, obtained with the IPA bioinformatic analysis for the 99 miRNAs significantly upregulated in the BTBR ob/ob mice group.
TABLE 3.
Top upstream regulator predicted molecules for the 99 miRNAs significantly upregulated in the kidney cortex of BTBR mice.
| Upstream regulator | Molecule type | p-value of overlap | Target molecules in dataset |
|---|---|---|---|
| AGO2 | Translation regulator | 6.88E-68 | mir-10, mir-101, mir-122, mir-128, mir-130, mir-139, mir-148, mir-15, mir-181, mir-182, mir-185, mir-186, mir-187, mir-188, mir-193, mir-194, mir-203, mir-204, mir-221, mir-23, mir-25, mir-28, mir-29, mir-320, mir-328, mir-330, mir-339, mir-34, mir-340, mir-361, mir-365, mir-379, mir-423, mir-484, mir-598, mir-652, mir-676, mir-8, mir-802 |
| DICER1 | Enzyme | 9.67E-39 | mir-10, mir-101, mir-122, mir-132, mir-139, mir-142, mir-145, mir-146, mir-148, mir-15, mir-150, mir-155, mir-181, mir-182, mir-188, mir-193, mir-194, mir-196, mir-204, mir-23, mir-25, mir-29, mir-30, mir-331, mir-34, mir-361, mir-365, mir-378, mir-497, mir-8, Mir7j |
| HNF4A | Transcription regulator | 9.48E-20 | mir-10, mir-101, mir-130, mir-140, mir-142, mir-148, mir-15, mir-181, mir-193, mir-194, mir-203, mir-28, mir-31, mir-34, mir-365, mir-455, mir-484, mir-497, mir-574, mir-8, mir-802 |
| Smad2/3 | Group | 0.0000000000000039 | mir-128, mir-130, mir-181, mir-185, mir-25, mir-320, mir-363, mir-423, mir-744 |
| TNFRSF1B | Transmembrane receptor | 0.000000000000014 | mir-10, mir-101, mir-130, mir-221, mir-28, mir-30, mir-320, mir-322, mir-326, mir-34, mir-96 |
| NF2 | Other | 0.00000000000225 | mir-10, mir-101, mir-15, mir-188, mir-28, mir-29 |
| MTDH | Transcription regulator | 0.0000000036 | mir-101, mir-15, mir-182, mir-28, mir-340 |
| E2F3 | Transcription regulator | 0.00000000873 | mir-10, mir-145, mir-15, mir-221, mir-23, mir-25 |
| KHDRBS1 | Transcription regulator | 0.0000000379 | mir-101, mir-142, mir-29, mir-339 |
| PPARA | Ligand-dependent nuclear receptor | 0.0000000764 | mir-10, mir-101, mir-130, mir-146, mir-148, mir-182, mir-203, mir-25, mir-30, mir-34, mir-378, mir-8 |
| SPI1 | Transcription regulator | 0.00000129 | mir-142, mir-223, mir-322, mir-351, mir-503, mir-542, mir-8 |
| ETS2 | Transcription regulator | 0.00000173 | mir-142, mir-155, mir-223, mir-29 |
| PIK3CA | Kinase | 0.00000258 | mir-210, mir-221, mir-34 |
| HMGA1 | Transcription regulator | 0.00000426 | mir-101, mir-196, mir-29, mir-331 |
| Gulo | Enzyme | 0.000012 | mir-15, mir-30, mir-322 |
| SMARCA4 | Transcription regulator | 0.0000181 | mir-145, mir-28, mir-30, mir-34, mir-674 |
| E2F1 | Transcription regulator | 0.0000241 | mir-10, mir-145, mir-15, mir-221, mir-23, mir-25 |
| IGF1R | Transmembrane receptor | 0.0000451 | mir-10, mir-127, mir-132, mir-139, mir-196, mir-34, mir-379 |
| REST | Transcription regulator | 0.0000607 | mir-139, mir-148, mir-203, mir-204 |
| MYC | Transcription regulator | 0.000124 | mir-145, mir-188, mir-29, mir-339, mir-34, mir-365, mir-378 |
| LTB4R | G protein–coupled receptor | 0.00016 | mir-10, mir-146, mir-155 |
| AGO1 | Translation regulator | 0.000178 | mir-203, mir-8 |
| SLC9A3R1 | Other | 0.000226 | mir-145, mir-155, mir-221 |
| PKD1 | Ion channel | 0.000236 | mir-10, mir-182, mir-204, mir-30, mir-8, mir-96 |
| TNFSF12 | Cytokine | 0.000327 | mir-146, mir-23, mir-322, mir-455 |
| INSR | Kinase | 0.000835 | mir-10, mir-127, mir-132, mir-139, mir-196, mir-34, mir-379 |
Discussion
In the present study, miRNA-seq analysis performed in the renal cortex of an advanced model of DN in 22-week-old BTBR ob/ob mice pointed out a set of 99 miRNA significantly increased compared to non-diabetic non-obese controls of the same age. These miRNAs could potentially be involved in the genesis and progression of DN and, therefore, used as potential biomarkers and/or therapeutic targets in this clinical condition. Our results disclosed increased levels not only of several miRNAs previously described in diabetic complications but also of other miRNAs related to different pathological processes. Interestingly, the functional predictive analysis highlighted inflammatory and immune processes as one of the most relevant pathways related to these miRNAs, remarking the importance of inflammation in the pathogenesis of kidney damage associated to diabetes.
Among the most deregulated miRNAs, miR-802 was, by far, the most upregulated in BTBR ob/ob mice. miR-802 is located on the 21st chromosome, and it has widely been associated with cancer in recent years, although showing opposite effects depending on the organ involved (Zhang et al., 2017; Feng et al., 2020; Ni et al., 2020; Wang et al., 2020; Wu et al., 2020). Interestingly, high levels of miR-802 have also been described in diabetic patients and in obesity-induced experimental nephropathy (Sun et al., 2019; Zhang et al., 2020). In addition, miR-101 and miR-375, both elevated in our miRNA-seq study, have also been involved in T1D and T2Donset and progression (Patoulias, 2018; Santos et al., 2019). Thus, increased levels of circulating miR-802, miR-101, and miR-375 have been proposed as potential biomarkers for T2D in humans (Higuchi et al., 2015). Our results point out the potential direct participation of these miRNAs in the DN progression. Mechanistically, an increase in the hepatic oxidative stress induced by miR-802 was proposed as a potential mechanism implicated in the insulin resistance observed in high-fat diet (HFD)–fed mice (Yang et al., 2019). Regarding the kidney, a recent study demonstrated that miR-802 causes nephropathy in HFD mice by suppressing NF-κB–repressing factor (Sun et al., 2019). Other authors demonstrated that miR-802 overexpression in two obese murine models, HFD and Lepr (db/db) mice, impairs glucose metabolism by silencing Hnf1b (Kornfeld et al., 2013). Our study provides further support for the direct involvement of mir-802 in DN and extends these findings to another experimental model that closely replicates the key histopathological features observed in human DN.
Our results additionally highlighted another set of miRNAs previously described to participate in the pathogenesis of DN. Thus, several authors suggested miR-200b/c and mir-29c as key therapeutic targets in experimental DN in db/db mice (Long et al., 2011; Park et al., 2013). Accordingly, we observed a significant increase in miR-200c and miR-29c levels in BTBR ob/ob mice. On the contrary, although previous studies demonstrated that miR-146 deletion accelerate DN progression in a murine streptozotocin (STZ)–induced diabetes model (Bhatt et al., 2016b; Sankrityayan et al., 2019), we found out an increase in miR-146 levels in the cortex kidney of BTBR ob/ob mice. This disparity could be explained by the differences between the experimental diabetes model used, one induced by STZ resembling T1D and the other by DN generated in BTBR ob/ob mice that resembles advanced human DN lesions in a T2D and metabolic disorder milieu (King, 2012). Increased miR-132 circulating levels have been associated with T2D and nephropathy and approaches to targeting it did impair blood glucose and improved insulin secretion (Bijkerk et al., 2015; Bijkerk et al., 2019; Florijn et al., 2019). Accordingly, we found out increased miR-132 expression in the kidney of BTBR ob/ob mice. In a recent study, a protective role of miR-379 deletion was described in STZ-induced DN in mice (Kato et al., 2021), with this miRNA being one of the most upregulated in our study. Another example of diabetes-related miRNAs in the present report is miR-204, whose genetic deletion improved glycemic control despite obesity in db/db mice (Gaddam et al., 2020). On the other hand, some miRNAs described to play a protective role in DN progression are miR-29b (Chen et al., 2014), miR-34c (Liu et al., 2015), and miR-26a (Koga et al., 2015). In the present study we did not observe differences in miR-29b andmiR-26a levels, whereas miR-34c was increased in the BTBR ob/ob. Whereas the mentioned miR-34c (Liu et al., 2015) protective effects were described in cultured podocytes, our study was performed in the renal cortex of diabetic mice, which could explain the apparent discrepancies among both studies.
Besides the potential individual effects exerted by the above-described miRNAs, our bioinformatic analysis pointed out Th1 and Th2 responses as the most common associated pathways to our miRNA-list, immediately after cancer related pathways that were not considered here as commented in the results section. The intensive research in the immunology field during the last years have markedly contributed to unravel the role of immune cells in many diseases, remarking the important role of Th subtypes, including Th1, Th2, Th17, and T regulatory (Treg) in many human diseases (Raphael et al., 2015). Importantly, Th subtype differentiation is a tightly regulated process, and mixed phenotypes can be found depending on the pathological conditions and can be modified by therapeutic interventions (DuPage and Bluestone, 2016; Gagliani and Huber, 2017). In this sense, Th1 and Th17 cells are characteristic of proinflammatory conditions, as described in immune mediated-disorders and chronic inflammatory diseases, whereas Treg cells exert protective anti-inflammatory actions (Harrington et al., 2005; Saigusa et al., 2020). Many experimental studies targeting Th17 immune response by different approaches, including neutralizing antibodies against the effector cytokine IL-17A or its soluble receptor, pharmacological inhibitors of RORγt, the main transcription factor driver of Th17 differentiation or drugs modulating Th17/Treg balance, like vitamin D agonist, as well as studies using genetically modified mice, have remarked the importance of Th17/IL-17A in the pathogenesis of chronic inflammatory diseases, including immune and non-immune renal damage (Orejudo et al., 2019; Lavoz et al., 2020a; Marchant et al., 2020; Rodrigues-Diez et al., 2021). Among the wide range of pathways implicated in DN generation and progression, we recently proposed that Th17 immune response could be playing a main role in DN in humans and BTBR ob/ob diabetic model (Lavoz et al., 2020a). In addition, we have previously demonstrated that administration of an IL-17A neutralizing antibody in BTBR ob/ob mice, starting when renal dysfunction and structural alterations were already present, caused a beneficial effect, restoring renal damage parameters, mainly due to inhibition of NF-κB/inflammation in the diabetic kidney (Lavoz et al., 2019). Other studies support the beneficial effects of IL-17A reduction in experimental DN in other mice models, as STZ-induced diabetes and autoimmune diabetes in NOD mice (Emamaullee et al., 2009; Kuriya et al., 2013; Kim et al., 2015; Tong et al., 2015).
Regarding the role of miRNAs in the regulation of the Th17 response, several authors described their participation in different pathologies including autoimmune diseases. In this sense, miR-20b and miR-30a suppress Th17 differentiation in experimental autoimmune diseases by targeting RORγt and STAT3 and IL-21R, respectively (Zhu et al., 2014; Qu et al., 2016). Similarly, in another autoimmune disease, miR-155-3p targets two genes (Dnaja2 and Dnajb1) that negatively regulated Th17 differentiation contributing to potentiate the Th17 response (Mycko et al., 2015). miR-146a elicits a different modulation of the Th17 response depending on the context. In experimental autoimmune encephalomyelitis, miR-146a reduced Th17 differentiation by targeting IL-6 and IL-2 (Li B. et al., 2017). However, a more recent study suggested that miR-146a-5p promotes Th17 cell differentiation by targeting the metalloprotease ADAM17 in the primary Sjögren’s syndrome (Wang X. et al., 2021). Increased levels of mir-34a, the second miRNA mostly upregulated in our miRNA-seq, have been described as an inductor of Th17 response by targeting FOXP3 in rheumatoid arthritis and systemic lupus erythematosus patients (Xie et al., 2019). Moreover, miR-34a may induce increased level of IL-17 and other proinflammatory cytokines via SIRT1 direct targeting and subsequent induction of NF-κB and the downstream pathway (Karbasforooshan and Karimi, 2018). Remarkably, miR-802 increases Th17 immune response by targeting the suppressor of cytokine signaling (SOCS5) in inflammatory bowel disease (Yao et al., 2020). Apart from immune diseases, the relation between miRNAs and Th17 has been demonstrated in other pathologies. In hepatocellular carcinoma–derived Th17 cells, a recent article showed increased miR-132 levels, another upregulated miRNA in our study, and demonstrated that the use of a miR-132 mimic accelerates Th17 differentiation in vitro (Feng et al., 2021). At the kidney level, there are few studies on the role of miRNAs in Th17 differentiation (Lavoz et al., 2020a). In this sense, in experimental crescentic glomerulonephritis, miR-155 participated in Th17 immune response and tissue injury (Krebs et al., 2013), and their deficiency attenuated the renal damage in hyperglycemia-induced nephropathy by promoting nephrin acetylation (Lin et al., 2015).
Interestingly, some of the miRNAs related to Th cell responses also participate in the diabetic pathology and were modified in our miRNA-seq study. Thereby, as mentioned above, miR-146a was proposed to be related to T2D susceptibility after a meta-analysis performed in a total of 12 studies that reveal a downregulation of miR-146a circulating levels in diabetic subjects in comparison to normal ones (Alipoor et al., 2017). In a similar manner, miR-155 was found downregulated in PBMC obtained from type 2 diabetic patients (Corral-Fernández et al., 2013). However, at the kidney level, both miR-155 and miR-146a were increased more than five-fold in DN patients compared with the controls as well as in experimental type 2 DN rat models (Huang et al., 2014). In the present study, both miR-155 and miR-146 were increased in the cortex kidney of BTBR ob/ob mice and were also related to the Th1 and Th2 responses. All the DN-related miRNAs that increased as found in our experimental study, combined with the fact that some of them have also been described to participate in the inflammatory/immune responses, including Th17, support our previous findings on the role of Th17 in DN (Lavoz et al., 2019) and pave the way to future studies modulating miRNAs in this clinical condition.
Other canonical pathway highlighted in our miRNA-seq studies was adipogenesis. Although there are a number of publications relating miRNAs in adipose tissue (Heyn et al., 2020), limited articles relating miRNA and adipogenesis in kidney tissue have been published so far. Recently, we demonstrated the expression of fatty acid influx/efflux markers and the presence of lipid droplets expressing perilipin-1 at both glomerular and tubulointerstitial levels in the BTBR ob/ob model (Opazo-Ríos et al., 2020b). Therefore, the present results, showing miRNAs expression related to adipocyte differentiation and lipid storage in renal tissue, constitute a novel hallmark and opens new ways to investigate the progression of type 2 DN in future studies.
Necroptosis, a form of cell death (Linkermann and Green, 2014; Newton and Manning, 2016), was another pathway remarked in our miRNA-seq studies. Necroptosis is a type of programmed necrosis characterized by the activation of receptor-interacting protein (RIP) 1 and 3 and by damage-associated molecular pattern–induced inflammation (Linkermann and Green, 2014; Newton and Manning, 2016). In kidney diseases, necroptosis has been proposed as a key mechanism involved in cell death in acute kidney damage (AKI), as described in the experimental models of renal ischemia/reperfusion injury, folic acid-induced AKI, and cisplatin nephropathy (Linkermann et al., 2013; Linkermann and Green, 2014; Xu et al., 2015; Martin-Sanchez et al., 2017; Martin-Sanchez et al., 2018). However, data about necroptosis in diabetes and experimental DN, in particular, are very limited. In this sense, in cultured podocytes, high-glucose concentrations elicited necroptosis (Xu et al., 2019). The inhibitor of necroptosis necrostatin-1 (nec-1) reduced visceral fat deposition and restored cognitive function and brain damage, but it did not result in any improvement on insulin sensitivity in prediabetic HFD rats (Jinawong et al., 2020). RIPK3 deficiency alleviated myocardial injury, improved cardiac function, and attenuated necroptosis in mice with STZ-induced diabetic cardiomyopathy (Chen et al., 2021). Our results suggest that necroptosis could be another pathway involved in kidney damage in diabetes to be explored.
In response to an insult, tubular epithelial cells undergo several changes, including loss of cell-to-cell contact and the polarized epithelial phenotype, leading to tubular dysfunction (Ruiz-Ortega et al., 2020). These changes in tubular cell phenotype are described as partial EMT and can contribute to renal damage progression, including in DN (Ruiz-Ortega et al., 2020). Recent studies have pointed out the importance of tubular damage, besides glomerular damage, in the genesis and progression of DN (Rayego-Mateos et al., 2021). In this sense, SGLT2i, besides acting as antihyperglycemic drugs, exert kidney protective effects in type 2 diabetic patients by acting in the proximal tubule reducing sodium reabsorption, probably by improving mitochondrial function (Shirakawa and Sano, 2020) and restoring tubular cell phenotype (Li J. et al., 2020; Das et al., 2020). Many data suggest the involvement of miRNAs in phenotype changes associated to diabetes and fibrotic conditions (Giordo et al., 2021). Our miRNA-seq data, showing that EMT related-processes are also connected to the upregulated miRNAs in advanced experimental DN, support the importance of tubular cells in the progression of kidney damage in DN and suggest that strategies targeting tubular damage should be investigated in DN.
Our bioinformatic analysis has also revealed that the most predictive factors to be regulated by the studied miRNAs are AGO2 and DICER1, both involved in miRNAs regulation, with other targets HNF4α, SMAD2/3 (a key pathway in fibrotic processes) (Tuleta and Frangogiannis, 2021), and TNFRSF1B being a member of the TNF Receptor (TNFR) Superfamily, also known as TNFR2 (So and Ishii, 2019). HNF4α dysfunction has been associated with metabolic disorders including diabetes (Niehof and Borlak, 2008; David-Silva et al., 2013). Mutations in HNF4α and HNF1α cause an autosomal dominant form of DM and tubular dysfunction (Terryn et al., 2016). Developmental studies have unveiled that HNF4α regulates the expression of key genes involved in differentiation and reabsorption in proximal tubules (Marable et al., 2020). TNFRSF1B binds to TNF-α and plays a critical role in immune regulation (So and Ishii, 2019) and kidney damage (Speeckaert et al., 2012). Remarkably, circulating TNFR2 levels are robust predictors of early and late renal function decline leading to ESRD in type 1 and type 2 DN patients (Carlsson et al., 2016; Niewczas et al., 2019).
To sum up, our results unveil a battery of miRNAs controlling key genes involved in adipogenesis, inflammation, immune response, necroptosis, and EMT, which constitute key mechanisms involved in the genesis and progression of DN. These data could potentially be relevant for the design of therapeutic approaches to this dreadful disease.
Acknowledgments
We want to thank Irene Rubio Soto for her technical assistance with the histological studies.
Data Availability Statement
The datasets generated for this study can be found in the NCBI SRA archive with BioProject record PRJNA759746 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA759746/) and BioSample records SRX11993569, SRX11993570, SRX11993571, SRX11993572, SRX11993573, SRX11993574, SRX11993575, SRX11993576, SRX11993577, SRX11993578, SRX11993579, and SRX11993580.
Ethics Statement
The animal study was reviewed and approved by Ethics Committee of the Health Research of the IIS-Fundación Jiménez Díaz and by the Madrid regional government (Ref. PROEX 079/18). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
All the authors have reviewed the manuscript and approved the final version. LO-R, AT-M, and RR-D contributed to the design of the experiments as wells as the acquisition, analysis, and interpretation of all data and drafted the manuscript. RR, BS-A, and MF-F have participated in RNA-seq data acquisition and analysis. MS, VM, and CL have participated in the development of mouse models, histology, IHC experiments, and analysis of data. SM, RR-D, and MR-O have evaluated all histological samples in a blinded manner. CL-L, JM, MR-O, JE, and RR-D contributed to the critical review of the manuscript results and to the financial support of the work.
Funding
This work was supported by grants from the Instituto de Salud Carlos III (ISCIII) and Fondos FEDER European Union (PI17/00119, PI17/01495, PI17/00130, PI20/00140, PI19/00184, PI20/00639, PI20/00487, PI20/00375, DTS20/00083, and DTS19/00093), Red de Investigación Renal REDINREN: RD16/0009 to MR-O (RD16/0009/0003) and to CL-L (RD16/0009/0020), Sociedad Española de Nefrología, “NOVELREN-CM: Enfermedad renal crónica: nuevas Estrategias para la prevención, Diagnóstico y tratamiento” (B2017/BMD-3751 to MR-O), “Convocatoria Dinamización Europa Investigación 2019” MINECO (EIN 2019-103294 to MR-O), Innovation programme under the Marie Skłodowska-Curie grant of the European Union’s Horizon 2020 (IMProve-PD ID: 812699 to MR-O), Spanish Biomedical Research Centre in Cardiovascular Diseases (CIBERCV), Spanish Ministry of Science and Innovation (RYC-2017-22369), Consejería de Salud y Familias-FEDER, Junta de Andalucía (PIGE-0052-2020 to JM), and Gobierno de Chile (S-2020-05 VIDCA-UACh to CL). RICORS program to RICORS2040 (KIDNEY DISEASE) RD21/0005/0002 to MR-O; RD21/0005/0017 to CL-L.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.778776/full#supplementary-material
References
- Adams B. D., Kasinski A. L., Slack F. J. (2014). Aberrant Regulation and Function of microRNAs in Cancer. Curr. Biol. 24, R762–R776. 10.1016/j.cub.2014.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alicic R. Z., Rooney M. T., Tuttle K. R. (2017). Diabetic Kidney Disease: Challenges, Progress, and Possibilities. Clin. J. Am. Soc. Nephrol. 12, 2032–2045. 10.2215/CJN.11491116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipoor B., Ghaedi H., Meshkani R., Torkamandi S., Saffari S., Iranpour M., et al. (2017). Association of MiR-146a Expression and Type 2 Diabetes Mellitus: A Meta-Analysis. Int. J. Mol. Cel. Med. 6, 156–163. 10.22088/acadpub.BUMS.6.3.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpers C. E., Hudkins K. L. (2011). Mouse Models of Diabetic Nephropathy. Curr. Opin. Nephrol. Hypertens. 20, 278–284. 10.1097/MNH.0b013e3283451901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Geng J., Zhou Z., Tian J., Li X. (2016). MicroRNA-130b Improves Renal Tubulointerstitial Fibrosis via Repression of Snail-Induced Epithelial-Mesenchymal Transition in Diabetic Nephropathy. Sci. Rep. 6, 20475. 10.1038/SREP20475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakris G. L., Agarwal R., Anker S. D., Pitt B., Ruilope L. M., Rossing P., et al. (2020). Effect of Finerenone on Chronic Kidney Disease Outcomes in Type 2 Diabetes. N. Engl. J. Med. 383, 2219–2229. 10.1056/NEJMOA2025845 [DOI] [PubMed] [Google Scholar]
- Bhatt K., Lanting L. L., Jia Y., Yadav S., Reddy M. A., Magilnick N., et al. (2016a). Anti-Inflammatory Role of MicroRNA-146a in the Pathogenesis of Diabetic Nephropathy. J. Am. Soc. Nephrol. 27, 2277–2288. 10.1681/ASN.2015010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt K., Lanting L. L., Jia Y., Yadav S., Reddy M. A., Magilnick N., et al. (2016b). Anti-Inflammatory Role of Microrna-146a in the Pathogenesis of Diabetic Nephropathy. J. Am. Soc. Nephrol. 27, 2277–2288. 10.1681/ASN.2015010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bheda P. (2020). Metabolic Transcriptional Memory. Mol. Metab. 38, 100955. 10.1016/j.molmet.2020.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijkerk R., Duijs J. M., Khairoun M., Ter Horst C. J., van der Pol P., Mallat M. J., et al. (2015). Circulating microRNAs Associate with Diabetic Nephropathy and Systemic Microvascular Damage and Normalize after Simultaneous Pancreas-Kidney Transplantation. Am. J. Transpl. 15, 1081–1090. 10.1111/ajt.13072 [DOI] [PubMed] [Google Scholar]
- Bijkerk R., Esguerra J. L. S., Ellenbroek J. H., Au Y. W., Hanegraaf M. A. J., de Koning E. J., et al. (2019). In Vivo Silencing of MicroRNA-132 Reduces Blood Glucose and Improves Insulin Secretion. Nucleic Acid Ther. 29, 67–72. 10.1089/nat.2018.0763 [DOI] [PubMed] [Google Scholar]
- Brosius F. C., Alpers C. E. (2013). New Targets for Treatment of Diabetic Nephropathy: What We Have Learned from Animal Models. Curr. Opin. Nephrol. Hypertens. 22, 17–25. 10.1097/MNH.0b013e32835b3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson A. C., Östgren C. J., Nystrom F. H., Länne T., Jennersjö P., Larsson A., et al. (2016). Association of Soluble Tumor Necrosis Factor Receptors 1 and 2 with Nephropathy, Cardiovascular Events, and Total Mortality in Type 2 Diabetes. Cardiovasc. Diabetol. 15, 40. 10.1186/S12933-016-0359-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Y., Zhong X., Huang X. R., Meng X. M., You Y., Chung A. C., et al. (2014). MicroRNA-29b Inhibits Diabetic Nephropathy in Db/db Mice. Mol. Ther. 22, 842–853. 10.1038/mt.2013.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Li X., Hua Y., Ding Y., Meng G., Zhang W. (2021). RIPK3-Mediated Necroptosis in Diabetic Cardiomyopathy Requires CaMKII Activation. Oxid. Med. Cel. Longev. 2021, 6617816. 10.1155/2021/6617816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. T., Xu X., Lim P. S., Hung K. Y. (2021). Worldwide Epidemiology of Diabetes-Related End-Stage Renal Disease, 2000-2015. Diabetes Care 44, 89–97. 10.2337/dc20-1913 [DOI] [PubMed] [Google Scholar]
- Clee S. M., Nadler S. T., Attie A. D. (2005). Genetic and Genomic Studies of the BTBR Ob/ob Mouse Model of Type 2 Diabetes. Am. J. Ther. 12, 491–498. 10.1097/01.mjt.0000178781.89789.25 [DOI] [PubMed] [Google Scholar]
- Colpaert R. M. W., Calore M. (2019). MicroRNAs in Cardiac Diseases. Cells 8, 737. 10.3390/cells8070737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Fernández N. E., Salgado-Bustamante M., Martínez-Leija M. E., Cortez-Espinosa N., García-Hernández M. H., Reynaga-Hernández E., et al. (2013). Dysregulated miR-155 Expression in Peripheral Blood Mononuclear Cells from Patients with Type 2 Diabetes. Exp. Clin. Endocrinol. Diabetes 121, 347–353. 10.1055/s-0033-1341516 [DOI] [PubMed] [Google Scholar]
- Das N. A., Carpenter A. J., Belenchia A., Aroor A. R., Noda M., Siebenlist U., et al. (2020). Empagliflozin Reduces High Glucose-Induced Oxidative Stress and miR-21-dependent TRAF3IP2 Induction and RECK Suppression, and Inhibits Human Renal Proximal Tubular Epithelial Cell Migration and Epithelial-To-Mesenchymal Transition. Cell. Signal. 68, 109506. 10.1016/j.cellsig.2019.109506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Silva A., Freitas H. S., Okamoto M. M., Sabino-Silva R., Schaan B. D., Machado U. F. (2013). Hepatocyte Nuclear Factors 1α/4α and Forkhead Box A2 Regulate the Solute Carrier 2A2 (Slc2a2) Gene Expression in the Liver and Kidney of Diabetic Rats. Life Sci. 93, 805–813. 10.1016/j.lfs.2013.10.011 [DOI] [PubMed] [Google Scholar]
- Deng Y., Li N., Wu Y., Wang M., Yang S., Zheng Y., et al. (2021). Global, Regional, and National Burden of Diabetes-Related Chronic Kidney Disease from 1990 to 2019. Front. Endocrinol. (Lausanne) 12, 672350. 10.3389/fendo.2021.672350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du B., Ma L. M., Huang M. B., Zhou H., Huang H. L., Shao P., et al. (2010). High Glucose Down-Regulates miR-29a to Increase Collagen IV Production in HK-2 Cells. FEBS Lett. 584, 811–816. 10.1016/J.FEBSLET.2009.12.053 [DOI] [PubMed] [Google Scholar]
- DuPage M., Bluestone J. A. (2016). Harnessing the Plasticity of CD4(+) T Cells to Treat Immune-Mediated Disease. Nat. Rev. Immunol. 16, 149–163. 10.1038/nri.2015.18 [DOI] [PubMed] [Google Scholar]
- Emamaullee J. A., Davis J., Merani S., Toso C., Elliott J. F., Thiesen A., et al. (2009). Inhibition of Th17 Cells Regulates Autoimmune Diabetes in NOD Mice. Diabetes 58, 1302–1311. 10.2337/db08-1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Liu L., Xu L., Wang H., Hua Q., He P. (2020). MiR-802 Suppresses Colorectal Cancer Cell Viability, Migration and Invasion by Targeting RAN. Cancer Manag. Res. 12, 2291–2300. 10.2147/CMAR.S231709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R., Cui Z., Liu Z., Zhang Y. (2021). Upregulated microRNA-132 in T Helper 17 Cells Activates Hepatic Stellate Cells to Promote Hepatocellular Carcinoma Cell Migration In Vitro . Scand. J. Immunol. 93, e13007. 10.1111/sji.13007 [DOI] [PubMed] [Google Scholar]
- Florijn B. W., Duijs J. M. G. J., Levels J. H., Dallinga-Thie G. M., Wang Y., Boing A. N., et al. (2019). Diabetic Nephropathy Alters the Distribution of Circulating Angiogenic MicroRNAs Among Extracellular Vesicles, HDL, and Ago-2. Diabetes 68, 2287–2300. 10.2337/db18-1360 [DOI] [PubMed] [Google Scholar]
- Fu J. X., Sun G. Q., Wang H. L., Jiang H. X. (2020). LncRNA OIP5-AS1 Induces Epithelial-To-Mesenchymal Transition and Renal Fibrosis in Diabetic Nephropathy via Binding to miR-30c-5p. J. Biol. Regul. Homeost. Agents 34, 961–968. 10.23812/20-199-A-68 [DOI] [PubMed] [Google Scholar]
- Gaddam R. R., Kim Y. R., Li Q., Jacobs J. S., Gabani M., Mishra A., et al. (2020). Genetic Deletion of miR-204 Improves Glycemic Control Despite Obesity in Db/db Mice. Biochem. Biophys. Res. Commun. 532, 167–172. 10.1016/j.bbrc.2020.08.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliani N., Huber S. (2017). Basic Aspects of T Helper Cell Differentiation. Methods Mol. Biol. 1514, 19–30. 10.1007/978-1-4939-6548-9_2 [DOI] [PubMed] [Google Scholar]
- Gao B. H., Wu H., Wang X., Ji L. L., Chen C. (2020). MiR-30c-5p Inhibits High Glucose-Induced EMT and Renal Fibrogenesis by Down-Regulation of JAK1 in Diabetic Nephropathy. Eur. Rev. Med. Pharmacol. Sci. 24, 1338–1349. 10.26355/EURREV_202002_20191 [DOI] [PubMed] [Google Scholar]
- Gao J., Wang W., Wang F., Guo C. (2018). LncRNA-NR_033515 Promotes Proliferation, Fibrogenesis and Epithelial-To-Mesenchymal Transition by Targeting miR-743b-5p in Diabetic Nephropathy. Biomed. Pharmacother. 106, 543–552. 10.1016/J.BIOPHA.2018.06.104 [DOI] [PubMed] [Google Scholar]
- Giordo R., Ahmed Y. M. A., Allam H., Abusnana S., Pappalardo L., Nasrallah G. K., et al. (2021). EndMT Regulation by Small RNAs in Diabetes-Associated Fibrotic Conditions: Potential Link with Oxidative Stress. Front. Cell Dev. Biol. 9, 683594. 10.3389/fcell.2021.683594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L. E., Hatton R. D., Mangan P. R., Turner H., Murphy T. L., Murphy K. M., et al. (2005). Interleukin 17-producing CD4+ Effector T Cells Develop via a Lineage Distinct from the T Helper Type 1 and 2 Lineages. Nat. Immunol. 6, 1123–1132. 10.1038/ni1254 [DOI] [PubMed] [Google Scholar]
- Hashimoto N., Phan S. H., Imaizumi K., Matsuo M., Nakashima H., Kawabe T., et al. (2010). Endothelial-Mesenchymal Transition in Bleomycin-Induced Pulmonary Fibrosis. Am. J. Respir. Cel Mol. Biol. 43, 161–172. 10.1165/rcmb.2009-0031OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn G. S., Corrêa L. H., Magalhães K. G. (2020). The Impact of Adipose Tissue-Derived miRNAs in Metabolic Syndrome, Obesity, and Cancer. Front. Endocrinol. (Lausanne) 11, 563816. 10.3389/fendo.2020.563816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi C., Nakatsuka A., Eguchi J., Teshigawara S., Kanzaki M., Katayama A., et al. (2015). Identification of Circulating miR-101, miR-375 and miR-802 as Biomarkers for Type 2 Diabetes. Metabolism 64, 489–497. 10.1016/j.metabol.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Huang Y., Liu Y., Li L., Su B., Yang L., Fan W., et al. (2014). Involvement of Inflammation-Related miR-155 and miR-146a in Diabetic Nephropathy: Implications for Glomerular Endothelial Injury. BMC Nephrol. 15, 142. 10.1186/1471-2369-15-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudkins K. L., Pichaiwong W., Wietecha T., Kowalewska J., Banas M. C., Spencer M. W., et al. (2010). BTBR Ob/Ob Mutant Mice Model Progressive Diabetic Nephropathy. J. Am. Soc. Nephrol. 21, 1533–1542. 10.1681/ASN.2009121290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson Laboratory (2007). Breeding Strategies for Maintaining Colonies of Laboratory Mice, 29. Bar Harbor, MA. [Google Scholar]
- Jankauskas S. S., Gambardella J., Sardu C., Lombardi A., Santulli G. (2021). Functional Role of miR-155 in the Pathogenesis of Diabetes Mellitus and its Complications. ncRNA 7, 39. 10.3390/ncrna7030039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinawong K., Apaijai N., Wongsuchai S., Pratchayasakul W., Chattipakorn N., Chattipakorn S. C. (2020). Necrostatin-1 Mitigates Cognitive Dysfunction in Prediabetic Rats with No Alteration in Insulin Sensitivity. Diabetes 69, 1411–1423. 10.2337/db19-1128 [DOI] [PubMed] [Google Scholar]
- Karbasforooshan H., Karimi G. (2018). The Role of SIRT1 in Diabetic Retinopathy. Biomed. Pharmacother. 97, 190–194. 10.1016/j.biopha.2017.10.075 [DOI] [PubMed] [Google Scholar]
- Kato M., Abdollahi M., Tunduguru R., Tsark W., Chen Z., Wu X., et al. (2021). miR-379 Deletion Ameliorates Features of Diabetic Kidney Disease by Enhancing Adaptive Mitophagy via FIS1. Commun. Biol. 4, 30. 10.1038/s42003-020-01516-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating S. T., El-Osta A. (2013). Glycemic Memories and the Epigenetic Component of Diabetic Nephropathy. Curr. Diab. Rep. 13, 574–581. 10.1007/s11892-013-0383-y [DOI] [PubMed] [Google Scholar]
- Kidney Disease: Improving Global Outcomes (KDIGO) Diabetes Work Group (2020). KDIGO 2020 Clinical Practice Guideline for Diabetes Management in Chronic Kidney Disease. Kidney Int. 98, S1–S115. 10.1016/J.KINT.2020.06.019 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013). TopHat2: Accurate Alignment of Transcriptomes in the Presence of Insertions, Deletions and Gene Fusions. Genome Biol. 14, R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. M., Lee S. H., Lee A., Kim D. J., Kim Y. G., Kim S. Y., et al. (2015). Targeting T Helper 17 by Mycophenolate Mofetil Attenuates Diabetic Nephropathy Progression. Transl. Res. 166, 375–383. 10.1016/j.trsl.2015.04.013 [DOI] [PubMed] [Google Scholar]
- King A. J. (2012). The Use of Animal Models in Diabetes Research. Br. J. Pharmacol. 166, 877–894. 10.1111/j.1476-5381.2012.01911.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga K., Yokoi H., Mori K., Kasahara M., Kuwabara T., Imamaki H., et al. (2015). MicroRNA-26a Inhibits TGF-β-Induced Extracellular Matrix Protein Expression in Podocytes by Targeting CTGF and Is Downregulated in Diabetic Nephropathy. Diabetologia 58, 2169–2180. 10.1007/S00125-015-3642-4 [DOI] [PubMed] [Google Scholar]
- Kornfeld J. W., Baitzel C., Könner A. C., Nicholls H. T., Vogt M. C., Herrmanns K., et al. (2013). Obesity-Induced Overexpression of miR-802 Impairs Glucose Metabolism through Silencing of Hnf1b. Nature 494, 111–115. 10.1038/nature11793 [DOI] [PubMed] [Google Scholar]
- Krebs C. F., Kapffer S., Paust H. J., Schmidt T., Bennstein S. B., Peters A., et al. (2013). MicroRNA-155 Drives TH17 Immune Response and Tissue Injury in Experimental Crescentic GN. J. Am. Soc. Nephrol. 24, 1955–1965. 10.1681/ASN.2013020130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriya G., Uchida T., Akazawa S., Kobayashi M., Nakamura K., Satoh T., et al. (2013). Double Deficiency in IL-17 and IFN-γ Signalling Significantly Suppresses the Development of Diabetes in the NOD Mouse. Diabetologia 56, 1773–1780. 10.1007/s00125-013-2935-8 [DOI] [PubMed] [Google Scholar]
- Lavoz C., Matus Y. S., Orejudo M., Carpio J. D., Droguett A., Egido J., et al. (2019). Interleukin-17A Blockade Reduces Albuminuria and Kidney Injury in an Accelerated Model of Diabetic Nephropathy. Kidney Int. 95, 1418–1432. 10.1016/j.kint.2018.12.031 [DOI] [PubMed] [Google Scholar]
- Lavoz C., Rayego-Mateos S., Orejudo M., Opazo-Ríos L., Marchant V., Marquez-Exposito L., et al. (2020a). Could IL-17A Be a Novel Therapeutic Target in Diabetic Nephropathy? J. Clin. Med. 9, 272. 10.3390/jcm9010272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoz C., Rodrigues-Diez R. R., Plaza A., Carpio D., Egido J., Ruiz-Ortega M., et al. (2020b). VEGFR2 Blockade Improves Renal Damage in an Experimental Model of Type 2 Diabetic Nephropathy. J. Clin. Med. 9, 302. 10.3390/jcm9020302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Wang X., Choi I. Y., Wang Y. C., Liu S., Pham A. T., et al. (2017a). miR-146a Modulates Autoreactive Th17 Cell Differentiation and Regulates Organ-Specific Autoimmunity. J. Clin. Invest. 127, 3702–3716. 10.1172/JCI94012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu H., Takagi S., Nitta K., Kitada M., Srivastava S. P., et al. (2020a). Renal Protective Effects of Empagliflozin via Inhibition of EMT and Aberrant Glycolysis in Proximal Tubules. JCI insight 5, e129034. 10.1172/jci.insight.129034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Shi S., Srivastava S. P., Kitada M., Nagai T., Nitta K., et al. (2017b). FGFR1 Is Critical for the Anti-Endothelial Mesenchymal Transition Effect of N-Acetyl-Seryl-Aspartyl-Lysyl-Proline via Induction of the MAP4K4 Pathway. Cell Death Dis 8, e2965. 10.1038/CDDIS.2017.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Jia T., Li Y. R. (2020b). LncRNA NEAT1 Accelerates the Occurrence and Development of Diabetic Nephropathy by Sponging miR-23c. Eur. Rev. Med. Pharmacol. Sci. 24, 1325–1337. 10.26355/EURREV_202002_20190 [DOI] [PubMed] [Google Scholar]
- Lin C. L., Lee P. H., Hsu Y. C., Lei C. C., Ko J. Y., Chuang P. C., et al. (2014). MicroRNA-29a Promotion of Nephrin Acetylation Ameliorates Hyperglycemia-Induced Podocyte Dysfunction. J. Am. Soc. Nephrol. 25, 1698–1709. 10.1681/ASN.2013050527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., You Y., Wang J., Qin Y., Huang P., Yang F. (2015). MicroRNA-155 Deficiency Promotes Nephrin Acetylation and Attenuates Renal Damage in Hyperglycemia-Induced Nephropathy. Inflammation 38, 546–554. 10.1007/s10753-014-9961-7 [DOI] [PubMed] [Google Scholar]
- Linkermann A., Bräsen J. H., Darding M., Jin M. K., Sanz A. B., Heller J. O., et al. (2013). Two Independent Pathways of Regulated Necrosis Mediate Ischemia-Reperfusion Injury. Proc. Natl. Acad. Sci. U. S. A. 110, 12024–12029. 10.1073/pnas.1305538110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkermann A., Green D. R. (2014). Necroptosis. N. Engl. J. Med. 370, 455–465. 10.1056/nejmra1310050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Qiang L., Wang G. D., Duan Q., Liu J. (2019). LncRNA MALAT1 Facilities High Glucose Induced Endothelial to Mesenchymal Transition and Fibrosis via Targeting miR-145/ZEB2 axis. Eur. Rev. Med. Pharmacol. Sci. 23, 3478–3486. 10.26355/EURREV_201904_17713 [DOI] [PubMed] [Google Scholar]
- Liu H., Wang X., Liu S., Li H., Yuan X., Feng B., et al. (2016). Effects and Mechanism of miR-23b on Glucose-Mediated Epithelial-To-Mesenchymal Transition in Diabetic Nephropathy. Int. J. Biochem. Cel Biol. 70, 149–160. 10.1016/J.BIOCEL.2015.11.016 [DOI] [PubMed] [Google Scholar]
- Liu X. D., Zhang L. Y., Zhu T. C., Zhang R. F., Wang S. L., Bao Y. (2015). Overexpression of miR-34c Inhibits High Glucose-Induced Apoptosis in Podocytes by Targeting Notch Signaling Pathways. Int. J. Clin. Exp. Pathol. 8, 4525–4534. [PMC free article] [PubMed] [Google Scholar]
- Llorens C., Futami R., Covelli L., Domínguez-Escribá L., Viu J. M., Tamarit D., et al. (2011). The Gypsy Database (GyDB) of Mobile Genetic Elements: Release 2.0. Nucleic Acids Res. 39, D70–D74. 10.1093/nar/gkq1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli M., Zoja C., Zanchi C., Corna D., Villa S., Bolognini S., et al. (2020). Manipulating Sirtuin 3 Pathway Ameliorates Renal Damage in Experimental Diabetes. Sci. Rep. 10, 8418. 10.1038/S41598-020-65423-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Wang Y., Wang W., Chang B. H., Danesh F. R. (2011). MicroRNA-29c Is a Signature microRNA under High Glucose Conditions that Targets Sprouty Homolog 1, and its In Vivo Knockdown Prevents Progression of Diabetic Nephropathy. J. Biol. Chem. 286, 11837–11848. 10.1074/jbc.M110.194969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marable S. S., Chung E., Park J. S. (2020). Hnf4a Is Required for the Development of Cdh6-Expressing Progenitors into Proximal Tubules in the Mouse Kidney. J. Am. Soc. Nephrol. 31, 2543–2558. 10.1681/ASN.2020020184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant V., Tejera-Muñoz A., Marquez-Expósito L., Rayego-Mateos S., Rodrigues-Diez R. R., Tejedor L., et al. (2020). IL-17A as a Potential Therapeutic Target for Patients on Peritoneal Dialysis. Biomolecules 10 (10), 1361. 10.3390/biom10101361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez-Exposito L., Cantero-Navarro E., Lavoz C., Fierro-Fernández M., Poveda J., Rayego-Mateos S., et al. (2018). Could Notch Signaling Pathway Be a Potential Therapeutic Option in Renal Diseases? Nefrologia 38, 466–475. 10.1016/j.nefro.2017.11.027 [DOI] [PubMed] [Google Scholar]
- Martin-Sanchez D., Fontecha-Barriuso M., Carrasco S., Sanchez-Niño M. D., Mässenhausen A. V., Linkermann A., et al. (2018). TWEAK and RIPK1 Mediate a Second Wave of Cell Death during AKI. Proc. Natl. Acad. Sci. U. S. A. 115, 4182–4187. 10.1073/pnas.1716578115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Sanchez D., Ruiz-Andres O., Poveda J., Carrasco S., Cannata-Ortiz P., Sanchez-Niño M. D., et al. (2017). Ferroptosis, but Not Necroptosis, Is Important in Nephrotoxic Folic Acid-Induced AKI. J. Am. Soc. Nephrol. 28, 218–229. 10.1681/ASN.2015121376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Moreno J. M., Fontecha-Barriuso M., Martin-Sanchez D., Guerrero-Mauvecin J., Goma-Garces E., Fernandez-Fernandez B., et al. (2020a). Epigenetic Modifiers as Potential Therapeutic Targets in Diabetic Kidney Disease. Int. J. Mol. Sci. 21 (11), 4113. 10.3390/ijms21114113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Moreno J. M., Fontecha-Barriuso M., Martín-Sánchez D., Sánchez-Niño M. D., Ruiz-Ortega M., Sanz A. B., et al. (2020b). The Contribution of Histone Crotonylation to Tissue Health and Disease: Focus on Kidney Health. Front. Pharmacol. 11, 393. 10.3389/fphar.2020.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba K., Sekiguchi K., Nagai Y., Takeda Y., Takahashi H., Yokota T., et al. (2021). Renal ROCK Activation and its Pharmacological Inhibition in Patients with Diabetes. Front. Pharmacol. 12, 738121. 10.3389/FPHAR.2021.738121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q., Zhai X., Yuan Y., Ji Q., Zhang P. (2020). lncRNA ZEB1-AS1 Inhibits High Glucose-Induced EMT and Fibrogenesis by Regulating the miR-216a-5p/BMP7 axis in Diabetic Nephropathy. Braz. J. Med. Biol. Res. 53, e9288. 10.1590/1414-431X20209288 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Moreno J. A., Gomez-Guerrero C., Mas S., Sanz A. B., Lorenzo O., Ruiz-Ortega M., et al. (2018). Targeting Inflammation in Diabetic Nephropathy: A Tale of hope. Expert Opin. Investig. Drugs 27, 917–930. 10.1080/13543784.2018.1538352 [DOI] [PubMed] [Google Scholar]
- Morgado-Pascual J. L., Marchant V., Rodrigues-Diez R., Dolade N., Suarez-Alvarez B., Kerr B., et al. (2018). Epigenetic Modification Mechanisms Involved in Inflammation and Fibrosis in Renal Pathology. Mediators Inflamm. 2018, 2931049. 10.1155/2018/2931049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgado-Pascual J. L., Rayego-Mateos S., Tejedor L., Suarez-Alvarez B., Ruiz-Ortega M. (2019). Bromodomain and Extraterminal Proteins as Novel Epigenetic Targets for Renal Diseases. Front. Pharmacol. 10, 1315. 10.3389/fphar.2019.01315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita Y., Yoshizawa H., Watanabe M., Imai R., Imai T., Hirahara I., et al. (2016). MicroRNA Expression Profiling in Peritoneal Fibrosis. Transl. Res. 169, 47–66. 10.1016/j.trsl.2015.10.009 [DOI] [PubMed] [Google Scholar]
- Mycko M. P., Cichalewska M., Cwiklinska H., Selmaj K. W. (2015). miR-155-3p Drives the Development of Autoimmune Demyelination by Regulation of Heat Shock Protein 40. J. Neurosci. 35, 16504–16515. 10.1523/JNEUROSCI.2830-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-González J. F., Mora-Fernández C., Muros de Fuentes M., García-Pérez J. (2011). Inflammatory Molecules and Pathways in the Pathogenesis of Diabetic Nephropathy. Nat. Rev. Nephrol. 7, 327–340. 10.1038/nrneph.2011.51 [DOI] [PubMed] [Google Scholar]
- Neumiller J. J., Alicic R. Z., Tuttle K. R. (2017). Therapeutic Considerations for Antihyperglycemic Agents in Diabetic Kidney Disease. J. Am. Soc. Nephrol. 28, 2263–2274. 10.1681/ASN.2016121372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K., Manning G. (2016). Necroptosis and Inflammation. Annu. Rev. Biochem. 85, 743–763. 10.1146/annurev-biochem-060815-014830 [DOI] [PubMed] [Google Scholar]
- Ni M., Zhao Y., Zhang W. J., Jiang Y. J., Fu H., Huang F., et al. (2020). microRNA-802 Accelerates Hepatocellular Carcinoma Growth by Targeting RUNX3. J. Cel. Physiol. 235, 7128–7135. 10.1002/jcp.29611 [DOI] [PubMed] [Google Scholar]
- Niehof M., Borlak J. (2008). HNF4 Alpha and the Ca-Channel TRPC1 Are Novel Disease Candidate Genes in Diabetic Nephropathy. Diabetes 57, 1069–1077. 10.2337/db07-1065 [DOI] [PubMed] [Google Scholar]
- Niewczas M. A., Pavkov M. E., Skupien J., Smiles A., Md Dom Z. I., Wilson J. M., et al. (2019). A Signature of Circulating Inflammatory Proteins and Development of End-Stage Renal Disease in Diabetes. Nat. Med. 25, 805–813. 10.1038/s41591-019-0415-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo-Ríos L., Mas S., Marín-Royo G., Mezzano S., Gómez-Guerrero C., Moreno J. A., et al. (2020a). Lipotoxicity and Diabetic Nephropathy: Novel Mechanistic Insights and Therapeutic Opportunities. Int. J. Mol. Sci. 21 (7), 2632. 10.3390/ijms21072632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo-Ríos L., Sanchez Matus Y., Rodrigues-Díez R. R., Carpio D., Droguett A., Egido J., et al. (2020b). Anti-Inflammatory, Antioxidant and Renoprotective Effects of SOCS1 Mimetic Peptide in the BTBR Ob/ob Mouse Model of Type 2 Diabetes. BMJ Open Diabetes Res. Care 8 (1), e001242. 10.1136/bmjdrc-2020-001242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orejudo M., Rodrigues-Diez R. R., Rodrigues-Diez R., Garcia-Redondo A., Santos-Sánchez L., Rández-Garbayo J., et al. (2019). Interleukin 17A Participates in Renal Inflammation Associated to Experimental and Human Hypertension. Front. Pharmacol. 10, 1015. 10.3389/fphar.2019.01015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. T., Kato M., Yuan H., Castro N., Lanting L., Wang M., et al. (2013). FOG2 Protein Down-Regulation by Transforming Growth Factor-β1-Induced microRNA-200b/c Leads to Akt Kinase Activation and Glomerular Mesangial Hypertrophy Related to Diabetic Nephropathy. J. Biol. Chem. 288, 22469–22480. 10.1074/jbc.M113.453043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil N. S., Feng B., Su Z., Castellani C. A., Chakrabarti S. (2021). Circular RNA Mediated Gene Regulation in Chronic Diabetic Complications. Sci. Rep. 11, 23766. 10.1038/S41598-021-02980-Y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patoulias D. I. (2018). Is miRNA-375 a Promising Biomarker for Early Detection and Monitoring of Patients with Type 2 Diabetes? Arch. Med. Sci. Atheroscler. Dis. 3, e119–e122. 10.5114/amsad.2018.78775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prattichizzo F., Giuliani A., De Nigris V., Pujadas G., Ceka A., La Sala L., et al. (2016). Extracellular microRNAs and Endothelial Hyperglycaemic Memory: A Therapeutic Opportunity? Diabetes Obes. Metab. 18, 855–867. 10.1111/dom.12688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Zhou J., Wang T., Han J., Ma L., Yu H., et al. (2016). MiR-30a Inhibits Th17 Differentiation and Demyelination of EAE Mice by Targeting the IL-21R. Brain Behav. Immun. 57, 193–199. 10.1016/j.bbi.2016.03.016 [DOI] [PubMed] [Google Scholar]
- Raphael I., Nalawade S., Eagar T. N., Forsthuber T. G. (2015). T Cell Subsets and Their Signature Cytokines in Autoimmune and Inflammatory Diseases. Cytokine 74, 5–17. 10.1016/j.cyto.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayego-Mateos S., Campillo S., Rodrigues-Diez R. R., Tejera-Muñoz A., Marquez-Exposito L., Goldschmeding R., et al. (2021). Interplay between Extracellular Matrix Components and Cellular and Molecular Mechanisms in Kidney Fibrosis. Clin. Sci. (Lond) 135, 1999–2029. 10.1042/CS20201016 [DOI] [PubMed] [Google Scholar]
- Rayego-Mateos S., Morgado-Pascual J. L., Opazo-Ríos L., Guerrero-Hue M., García-Caballero C., Vázquez-Carballo C., et al. (2020). Pathogenic Pathways and Therapeutic Approaches Targeting Inflammation in Diabetic Nephropathy. Int. J. Mol. Sci. 21 (11), 3798. 10.3390/ijms21113798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Diez R. R., Tejera-Muñoz A., Orejudo M., Marquez-Exposito L., Santos-Sanchez L., Rayego-Mateos S., et al. (2021). Interleukin-17A: Potential Mediator and Therapeutic Target in Hypertension. Nefrología (English Edition) 41, 244–257. 10.1016/j.nefroe.2021.06.003 [DOI] [PubMed] [Google Scholar]
- Ruiz-Ortega M., Rayego-Mateos S., Lamas S., Ortiz A., Rodrigues-Diez R. R. (2020). Targeting the Progression of Chronic Kidney Disease. Nat. Rev. Nephrol. 16, 269–288. 10.1038/s41581-019-0248-y [DOI] [PubMed] [Google Scholar]
- Saigusa R., Winkels H., Ley K. (2020). T Cell Subsets and Functions in Atherosclerosis. Nat. Rev. Cardiol. 17, 387–401. 10.1038/s41569-020-0352-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankrityayan H., Kulkarni Y. A., Gaikwad A. B. (2019). Diabetic Nephropathy: The Regulatory Interplay between Epigenetics and microRNAs. Pharmacol. Res. 141, 574–585. 10.1016/j.phrs.2019.01.043 [DOI] [PubMed] [Google Scholar]
- Santos A. S., Cunha Neto E., Fukui R. T., Ferreira L. R. P., Silva M. E. R. (2019). Increased Expression of Circulating microRNA 101-3p in Type 1 Diabetes Patients: New Insights into miRNA-Regulated Pathophysiological Pathways for Type 1 Diabetes. Front. Immunol. 10, 1637. 10.3389/fimmu.2019.01637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F., Tsuchiya S., Meltzer S. J., Shimizu K. (2011). MicroRNAs and Epigenetics. FEBS J. 278, 1598–1609. 10.1111/j.1742-4658.2011.08089.x [DOI] [PubMed] [Google Scholar]
- Schmieder R., Edwards R. (2011). Quality Control and Preprocessing of Metagenomic Datasets. Bioinformatics 27, 863–864. 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Bhattacharya P., Kalia K., Tiwari V. (2017). Diabetic Nephropathy: New Insights into Established Therapeutic Paradigms and Novel Molecular Targets. Diabetes Res. Clin. Pract. 128, 91–108. 10.1016/j.diabres.2017.04.010 [DOI] [PubMed] [Google Scholar]
- Shirakawa K., Sano M. (2020). Sodium-Glucose Co-Transporter 2 Inhibitors Correct Metabolic Maladaptation of Proximal Tubular Epithelial Cells in High-Glucose Conditions. Int. J. Mol. Sci. 21, 1–13. 10.3390/IJMS21207676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson K., Wonnacott A., Fraser D. J., Bowen T. (2016). MicroRNAs in Diabetic Nephropathy: From Biomarkers to Therapy. Curr. Diab. Rep. 16, 35. 10.1007/s11892-016-0724-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So T., Ishii N. (2019). The TNF-TNFR Family of Co-signal Molecules. Adv. Exp. Med. Biol. 1189, 53–84. 10.1007/978-981-32-9717-3_3 [DOI] [PubMed] [Google Scholar]
- Speeckaert M. M., Speeckaert R., Laute M., Vanholder R., Delanghe J. R. (2012). Tumor Necrosis Factor Receptors: Biology and Therapeutic Potential in Kidney Diseases. Am. J. Nephrol. 36, 261–270. 10.1159/000342333 [DOI] [PubMed] [Google Scholar]
- Srivastava S. P., Goodwin J. E., Kanasaki K., Koya D. (2020). Metabolic Reprogramming by N-Acetyl-Seryl-Aspartyl-Lysyl-Proline Protects against Diabetic Kidney Disease. Br. J. Pharmacol. 177, 3691–3711. 10.1111/BPH.15087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S. P., Goodwin J. E., Tripathi P., Kanasaki K., Koya D. (2021a). Interactions Among Long Non-Coding RNAs and microRNAs Influence Disease Phenotype in Diabetes and Diabetic Kidney Disease. Int. J. Mol. Sci. 22 (11), 6027. 10.3390/IJMS22116027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S. P., Li J., Kitada M., Fujita H., Yamada Y., Goodwin J. E., et al. (2018). SIRT3 Deficiency Leads to Induction of Abnormal Glycolysis in Diabetic Kidney with Fibrosis. Cel Death Dis 9, 997. 10.1038/S41419-018-1057-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S. P., Zhou H., Setia O., Liu B., Kanasaki K., Koya D., et al. (2021b). Loss of Endothelial Glucocorticoid Receptor Accelerates Diabetic Nephropathy. Nat. Commun. 12, 2368. 10.1038/S41467-021-22617-Y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S. P., Koya D., Kanasaki K. (2013). MicroRNAs in Kidney Fibrosis and Diabetic Nephropathy: Roles on EMT and EndMT. Biomed. Res. Int. 2013, 1–10. 10.1155/2013/125469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Chen J., Wu W., Tang J., Luo L., Zhang K., et al. (2019). MiR-802 Causes Nephropathy by Suppressing NF-κB-Repressing Factor in Obese Mice and Human. J. Cel. Mol. Med. 23, 2863–2871. 10.1111/jcmm.14193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terryn S., Tanaka K., Lengelé J. P., Olinger E., Dubois-Laforgue D., Garbay S., et al. (2016). Tubular Proteinuria in Patients with HNF1α Mutations: HNF1α Drives Endocytosis in the Proximal Tubule. Kidney Int. 89, 1075–1089. 10.1016/j.kint.2016.01.027 [DOI] [PubMed] [Google Scholar]
- Tong Z., Liu W., Yan H., Dong C. (2015). Interleukin-17A Deficiency Ameliorates Streptozotocin-Induced Diabetes. Immunology 146, 339–346. 10.1111/imm.12512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., et al. (2012). Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuleta I., Frangogiannis N. G. (2021). Diabetic Fibrosis. Biochim. Biophys. Acta. Mol. Basis Dis. 1867, 166044. 10.1016/j.bbadis.2020.166044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Brenner D. P. S. (2005). Fibrosis Research — Methods and Protocols. Totowa, NJ: Humana Press. [Google Scholar]
- Wang B., Koh P., Winbanks C., Coughlan M. T., McClelland A., Watson A., et al. (2011). miR-200a Prevents Renal Fibrogenesis through Repression of TGF-β2 Expression. Diabetes 60, 280–287. 10.2337/DB10-0892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang R., Wu X., Chen Y., Ji W., Wang J., et al. (2022). The Wnt Signaling Pathway in Diabetic Nephropathy. Front. Cel Dev. Biol. 9, 701547. 10.3389/FCELL.2021.701547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li D., Sun L., Shen G., Liu H., Guo H., et al. (2020). Regulation of the Small GTPase Ran by miR-802 Modulates Proliferation and Metastasis in Colorectal Cancer Cells. Br. J. Cancer 122, 1695–1706. 10.1038/s41416-020-0809-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xin S., Wang Y., Ju D., Wu Q., Qiu Y., et al. (2021a). MicroRNA-146a-5p Enhances T Helper 17 Cell Differentiation via Decreasing a Disintegrin and Metalloprotease 17 Level in Primary Sjögren's Syndrome. Bioengineered 12, 310–324. 10.1080/21655979.2020.1870321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xu Y., Zhu Y. C., Wang Y. K., Li J., Li X. Y., et al. (2019). LncRNA NEAT1 Promotes Extracellular Matrix Accumulation and Epithelial-To-Mesenchymal Transition by Targeting miR-27b-3p and ZEB1 in Diabetic Nephropathy. J. Cel. Physiol. 234, 12926–12933. 10.1002/JCP.27959 [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu Y., Zhang L., Bai L., Chen S., Wu H., et al. (2021b). miR-30b-5p Modulate Renal Epithelial-Mesenchymal Transition in Diabetic Nephropathy by Directly Targeting SNAI1. Biochem. Biophys. Res. Commun. 535, 12–18. 10.1016/J.BBRC.2020.10.096 [DOI] [PubMed] [Google Scholar]
- Wu J., Liu J., Ding Y., Zhu M., Lu K., Zhou J., et al. (2018). MiR-455-3p Suppresses Renal Fibrosis through Repression of ROCK2 Expression in Diabetic Nephropathy. Biochem. Biophys. Res. Commun. 503, 977–983. 10.1016/J.BBRC.2018.06.105 [DOI] [PubMed] [Google Scholar]
- Wu X., Liu L., Zhang H. (2020). miR-802 Inhibits the Epithelial-Mesenchymal Transition, Migration and Invasion of Cervical Cancer by Regulating BTF3. Mol. Med. Rep. 22, 1883–1891. 10.3892/mmr.2020.11267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Wang J., Gong W., Xu H., Pan X., Chen Y., et al. (2019). NF-κB-Driven miR-34a Impairs Treg/Th17 Balance via Targeting Foxp3. J. Autoimmun. 102, 96–113. 10.1016/j.jaut.2019.04.018 [DOI] [PubMed] [Google Scholar]
- Xu Y., Gao H., Hu Y., Fang Y., Qi C., Huang J., et al. (2019). High Glucose-Induced Apoptosis and Necroptosis in Podocytes Is Regulated by UCHL1 via RIPK1/RIPK3 Pathway. Exp. Cel Res. 382, 111463. 10.1016/j.yexcr.2019.06.008 [DOI] [PubMed] [Google Scholar]
- Xu Y., Ma H., Shao J., Wu J., Zhou L., Zhang Z., et al. (2015). A Role for Tubular Necroptosis in Cisplatin-Induced AKI. J. Am. Soc. Nephrol. 26, 2647–2658. 10.1681/ASN.2014080741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Xing H., Liu J., Yang L., Ma H., Ma H. (2019). MicroRNA-802 Increases Hepatic Oxidative Stress and Induces Insulin Resistance in High-Fat Fed Mice. Mol. Med. Rep. 20, 1230–1240. 10.3892/mmr.2019.10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Gao R., Luo M., Li D., Guo L., Yu Z., et al. (2020). miR-802 Participates in the Inflammatory Process of Inflammatory Bowel Disease by Suppressing SOCS5. Biosci. Rep. 40 (4), BSR20192257. 10.1042/BSR20192257 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang F., Ma D., Zhao W., Wang D., Liu T., Liu Y., et al. (2020). Obesity-Induced Overexpression of miR-802 Impairs Insulin Transcription and Secretion. Nat. Commun. 11, 1822. 10.1038/s41467-020-15529-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Mu J. H., Liu L. Y., Zhang H. Z. (2017). Upregulation of miR-802 Suppresses Gastric Cancer Oncogenicity via Targeting RAB23 Expression. Eur. Rev. Med. Pharmacol. Sci. 21, 4071–4078. [PubMed] [Google Scholar]
- Zhang Y. (2021). MiR-92d-3p Suppresses the Progression of Diabetic Nephropathy Renal Fibrosis by Inhibiting the C3/HMGB1/TGF-β1 Pathway. Biosci. Rep. 41 (9), BSR20203131. 10.1042/BSR20203131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Li T., Li J., Lu Q., Han C., Wang N., et al. (2016). miR-23b-3p Induces the Cellular Metabolic Memory of High Glucose in Diabetic Retinopathy through a SIRT1-Dependent Signalling Pathway. Diabetologia 59, 644–654. 10.1007/s00125-015-3832-0 [DOI] [PubMed] [Google Scholar]
- Zhao X. P., Chang S. Y., Liao M. C., Lo C. S., Chenier I., Luo H., et al. (2018). Hedgehog Interacting Protein Promotes Fibrosis and Apoptosis in Glomerular Endothelial Cells in Murine Diabetes. Sci. Rep. 8, 5958. 10.1038/S41598-018-24220-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Guan M., Jia Y., Wang D., Pang R., Lv F., et al. (2016). The Coordinated Roles of miR-26a and miR-30c in Regulating TGFβ1-Induced Epithelial-To-Mesenchymal Transition in Diabetic Nephropathy. Sci. Rep. 6, 37492. 10.1038/SREP37492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Chung A. C., Chen H. Y., Dong Y., Meng X. M., Li R., et al. (2013). miR-21 Is a Key Therapeutic Target for Renal Injury in a Mouse Model of Type 2 Diabetes. Diabetologia 56, 663–674. 10.1007/s00125-012-2804-x [DOI] [PubMed] [Google Scholar]
- Zhu E., Wang X., Zheng B., Wang Q., Hao J., Chen S., et al. (2014). miR-20b Suppresses Th17 Differentiation and the Pathogenesis of Experimental Autoimmune Encephalomyelitis by Targeting RORγt and STAT3. J. Immunol. 192, 5599–5609. 10.4049/jimmunol.1303488 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Xu J., Liang W., Li J., Feng L., Zheng P., et al. (2019). miR-98-5p Alleviated Epithelial-To-Mesenchymal Transition and Renal Fibrosis via Targeting Hmga2 in Diabetic Nephropathy. Int. J. Endocrinol. 2019, 4946181. 10.1155/2019/4946181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoja C., Corna D., Camozzi D., Cattaneo D., Rottoli D., Batani C., et al. (2002). How to Fully Protect the Kidney in a Severe Model of Progressive Nephropathy: A Multidrug Approach. J. Am. Soc. Nephrol. 13, 2898–2908. 10.1097/01.asn.0000034912.55186.ec [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the NCBI SRA archive with BioProject record PRJNA759746 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA759746/) and BioSample records SRX11993569, SRX11993570, SRX11993571, SRX11993572, SRX11993573, SRX11993574, SRX11993575, SRX11993576, SRX11993577, SRX11993578, SRX11993579, and SRX11993580.